Significance
Protein–protein interactions are important to all facets of life, but their experimental and computational characterization is arduous and frequently of uncertain outcome. The current study demonstrates both the power and limitation to study protein interactions by utilizing sophisticated statistical inference technology to derive protein contacts from available sequence databases, more precisely from the coevolution between residues, that are in contact across the interaction interface of two proteins. By studying homo-oligomeric protein interactions, the current study expands from anecdotal evidence of the performance of this technology to systematic evidence of its value across close to 2,000 interacting protein families.
Keywords: homo-oligomers, coevolution, direct coupling analysis, protein–protein interactions, big data analysis
Abstract
Proteins have evolved to perform diverse cellular functions, from serving as reaction catalysts to coordinating cellular propagation and development. Frequently, proteins do not exert their full potential as monomers but rather undergo concerted interactions as either homo-oligomers or with other proteins as hetero-oligomers. The experimental study of such protein complexes and interactions has been arduous. Theoretical structure prediction methods are an attractive alternative. Here, we investigate homo-oligomeric interfaces by tracing residue coevolution via the global statistical direct coupling analysis (DCA). DCA can accurately infer spatial adjacencies between residues. These adjacencies can be included as constraints in structure prediction techniques to predict high-resolution models. By taking advantage of the ongoing exponential growth of sequence databases, we go significantly beyond anecdotal cases of a few protein families and apply DCA to a systematic large-scale study of nearly 2,000 Pfam protein families with sufficient sequence information and structurally resolved homo-oligomeric interfaces. We find that large interfaces are commonly identified by DCA. We further demonstrate that DCA can differentiate between subfamilies with different binding modes within one large Pfam family. Sequence-derived contact information for the subfamilies proves sufficient to assemble accurate structural models of the diverse protein-oligomers. Thus, we provide an approach to investigate oligomerization for arbitrary protein families leading to structural models complementary to often-difficult experimental methods. Combined with ever more abundant sequential data, we anticipate that this study will be instrumental to allow the structural description of many heteroprotein complexes in the future.
Life on the molecular level is orchestrated by the interplay of many different biomolecules. A crucial component for the function is its structure, ranging from small monomers to complex homo- or heteromultimers. The full structural characterization of a biomolecule therefore typically precedes a detailed explanation of its functional mechanism. However, despite the incredible progress of structural characterization methods, many important biomolecules have not been structurally resolved. An intriguing alternative to often involved experimental measurements of 3D structures is taking advantage of the growing wealth of genetic sequential information via sophisticated statistical methods. Direct coupling analysis (DCA) (1, 2) and related tools (3, 4) develop a global model mimicking evolutionary fitness landscapes of protein families (5, 6) and quantify the coevolution of amino acid residue positions (7–9). In the context of protein structure prediction, these models allow extraction of residue–residue contacts from sequence information alone. Such information has proven useful in the prediction of tertiary protein structures (10–13); conformational transitions (14–16); and, similarly, RNA structures (17, 18).
To be successful, global statistical models must be trained on sufficiently large alignments of homologous sequences. It has been argued (2) that on the order of 1,000 properly aligned and sufficiently divergent sequences are adequate for accurate model learning. Scanning the latest Pfam database release (19), we observe a fundamental extension of the families amenable to statistical modeling (compare SI Appendix, Fig. S1). Whereas in the Pfam 5.0 release in the year 2000, only 33 families contained at least 1,000 sequences, this number has grown by a factor of more than 200: 6,783 in Pfam 28.0 (released in 2015) of a total of 16,230 included families. The median family size of 654 sequences is close to the required sequence number. Statistical sequence modeling will therefore strongly increase in importance over the next years.
Statistical modeling of single-protein families is the final aim: Proteins rarely work in isolation and monomeric form. Protein–protein interfaces coevolve to conserve the interaction and interaction specificity between proteins. To identify such coevolving protein–protein interfaces has been the original motivation for DCA development (1, 20), and this methodology has recently been extended beyond simple anecdotal cases to dozens of protein pairs (13, 21–25). Much more extensive datasets are needed for a solid statistical assessment of the quality of coevolution-based predictions, which tells us when DCA can reliably predict residue contacts between interacting proteins and how to overcome a possible failure. The main difficulty in going to the results of much larger datasets stems from the necessity to create joint multiple sequence alignments (MSAs) of interaction partners, where each line contains an interacting pair of proteins. Computationally, this problem becomes difficult by itself due to the existence of paralogs (26–28), and most work so far has concentrated on systems where interaction partners have colocalized genes (e.g., in common operons in the bacteria).
The present work addresses the challenge by focusing on homo-oligomerization rather than heteroprotein–heteroprotein interactions, thereby increasing the number of available datasets by more than one order of magnitude. The reason is simple: The creation of joint MSAs of interaction partners is simplified, because the identical sequence of both interaction partners allows one to work with MSAs for single proteins. On the contrary, the distinction between intraprotein and interprotein residue–residue contact predictions from coevolutionary couplings is nontrivial when studying homo-oligomers, and the existence of experimental monomer structures is needed to identify putative interprotein contacts as directly coevolving pairs with a long intramonomeric distance.
By coupling DCA with in silico molecular simulations, we propose an analysis approach that can be applied to modeling homo-oligomeric as well as hetero-oligomeric biological assemblies of protein complexes (Fig. 1). In this workflow, contact–residue pairings between proteins are extracted from sequence alignments of large protein families. Intraprotein contacts are eliminated by pruning those DCA-predicted contacts compatible with known monomeric structures. The remaining pairings are considered as potential interprotein contacts and are used to drive docking of monomeric structures into biological assemblies. Where individual proteins have not been structurally resolved, these proteins can typically be homology-modeled.
Fig. 1.
Analysis approach. Starting from a target amino acid sequence, we use Pfam to extract MSA of homologous sequences and exemplary monomer protein structures from the PDB. DCA is run on the MSA to predict a contact map. Interprotein residue contacts are obtained by pruning all those DCA-predicted residue pairs that are compatible with the known monomeric structure. To obtain a structural model for the biological assembly, we use two monomers (possible homology-modeled if not directly available in the PDB) and dock them guided by the DCA interprotein predictions. BA, biological assembly; CM, contact map.
The concentration on homo-oligomers allows us to go substantially beyond the current stage of coevolutionary analysis of interacting proteins by analyzing nearly 2,000 systematically selected and sufficiently abundant protein families (i.e., MSAs large enough to provide the statistical signal detected by DCA) with known structures (Fig. 2). This number of families makes the use of automated procedures essential. Together with the strict selection criteria of investigated protein families, this approach reduces possible artifacts coming from a subjective selection of systems and/or human data curation. The analysis of more than 750 families with large interfaces shows significantly enriched coevolutionary couplings across the interfaces, whereas small interfaces are rarely detected. However, even for large interfaces, the detection of strong coevolutionary couplings, and therefore the accuracy of the resulting contact predictions, is limited to roughly 50% of cases. To understand this observation better, and to open paths for overcoming this limited performance, we analyze in more detail the specific case of response regulator (RR) proteins involved in bacterial two-component signal transduction, where several subfamilies (characterized by different domain architectures) show different homodimerization modes. We show that due to the nonconservation of the dimerization mode in the entire Pfam (19) domain family of RRs, a straightforward application of DCA leads to a weakened signal as different dimerization modes are mixed. The restriction of the MSAs to proteins of presumably the same dimerization interface, however, strongly improves the strength and accuracy of the coevolutionary signal. In the case of RRs, we demonstrate that by subdividing the alignments based on protein domain architecture, sufficient contact information can be extracted from sequence to model the dimer structures in silico reliably and accurately, whereas the coevolutionary signal in the entire protein family is dominated by the common structural characteristics of all subfamilies (mostly tertiary contacts).
Fig. 2.
Overview of results for investigated protein families. To understand the effect of residue coevolution in homo-oligomerization, we investigate a total of 1,984 protein families (PF) from the Pfam database. They fulfill the requirements of sufficient sequence information and a known monomeric structure, which is a requirement to differentiate coevolving intraprotein contacts (i.e., tertiary contacts) from putative interprotein contacts (i.e., quaternary contacts). Larger assemblies have stronger signals, because only some residues forming the interface strongly coevolve. We thus subdivide the PF into two classes pending their known interface sizes. A total of 750 PF have a large known interface in their biological assembly. For 372 PF of these total PF, we can identify this interface through strongly coevolving residue pairs based purely on sequences. For the other 1,234 PF, no large biomolecular assemblies are known. A total of 1,088 PF, however, form a known small interface, and this interface can be identified for 189 PF. For the 146 PF possessing no known interface, the large majority do not show strongly coevolving residue pairs. For 24, however, we find strongly coevolving pairs indicating a possible biological homo-oligomeric assembly.
Finally, we analyze 142 of the nearly 2,000 large Pfam families without an annotated biological assembly. Interestingly, only a small number of these families show a strong nonmonomeric coevolutionary signal. Individual analysis of these cases does not provide consistent oligomerization signals, suggesting that, to date, many of the physiologically relevant homo-oligomerization structures with large and well-conserved interfaces in sufficiently populated protein families have already been experimentally described. In addition to this important finding, our procedure is applicable to the determination of heteroprotein complexes and the detection of alternative homo-oligomerization modes.
Results
Database Selection of Biological Oligomers.
This study aims to assess how readily interaction surfaces of proteins can be identified from large protein sequence alignments by analyzing coevolutionary signals between residue positions. To accomplish this goal systematically, we focused on homo-oligomer interactions. To this end, we scanned the Pfam 27.0 database (19) for domains with sufficient sequence diversity to apply DCA, as outlined in Methods. In addition, we required that at least one high-resolution protein structure be deposited in the Protein Data Bank (PDB) (29) to distinguish coevolving intraprotein from interprotein contacts as further discussed below. Our study thus provides an exhaustive analysis of coevolution across homo-oligomeric interfaces.
The above requirements resulted in a dataset of 1,984 protein domain families (Fig. 2). The dataset was further analyzed for the existence of an annotated biological homo-oligomeric assembly. Interestingly, only 146 protein families within the dataset had no annotated biological assembly, suggesting that the large majority of protein families are believed to form dimers or higher order structures of physiological relevance. Protein families with known biological assemblies were further subdivided based on size of the interface covered by the Pfam sequence models, because larger interaction surfaces can be expected to be more readily identified than those surfaces with few interaction contacts. The 1,838 protein families with annotated biological assemblies were thus further subdivided into 750 families with large biological assembly interfaces and 1,088 families with small biological assembly interfaces as detailed in Methods. The 750 families with large biological interfaces are our main focus of this study, but the other families were also analyzed for coevolutionary signals.
Intraprotein Versus Interprotein Contacts in Homo-Oligomers.
The coevolutionary detection of interprotein contacts in homo-oligomeric assemblies is, at the same time, easier and more complicated than the prediction of interprotein contacts in hetero-oligomers (compare Fig. 1). It is easier because we need a joined MSA of both interaction partners across the interface, which are identical in the case of homo-oligomers. It is therefore sufficient to have one MSA of a single protein. This fact, however, implies that the differentiation of intraprotein and interprotein contacts becomes nontrivial in the case of homo-oligomers. Without any exemplary monomeric structure, this problem remains currently unsolved. When an exemplary structure of the monomer exists, and such is the case treated in this work, we can interpret all those residue pairs as interprotein contact predictions, which are distant inside the monomer but show a strong coevolutionary signal. In other words, not realized false-positive (FP) intraprotein predictions are interpreted as possible interprotein predictions. As illustrated in SI Appendix, Fig. S1, this treatment may hide part of the protein–protein interface. Particularly in symmetrical assemblies, residue pairs may be in spatial vicinity both inside the monomer and across the interface of the complex. Actually, although only about 14.5% of all residue pairs are within 8 Å inside a single protein, we find about 32.3% of all interprotein contacts (8-Å cutoff) to also be intraprotein contacts (5.4% vs. 14.8% at a more stringent 5-Å distance cutoff). This phenomenon may possibly happen in alternative structures [e.g., by the mechanism of domain swapping (30), where internal contacts of the monomer become interface contacts in the dimer]. Disentangling the two is, again, a currently unsolved problem, and their physiological relevance is not always clear.
To shed light on the problem of hiding parts of the interface when concentrating on large internal distances, we typically use more than one distance cutoff for identifying intraprotein contacts. As a general tendency, we see that residue pairs with a strong coevolutionary signal, but at a large distance in the monomer (e.g., minimal atom distance above 12 Å), have a very large probability of being interprotein contacts (compare Fig. 3). Pairs at lower distances, but not directly in contact (e.g., between 8 Å and 12 Å) inside the monomer, might be accommodated as internal contacts by minor conformational changes or different residue choices, and thus have a lower probability of actually being interprotein contacts.
Fig. 3.
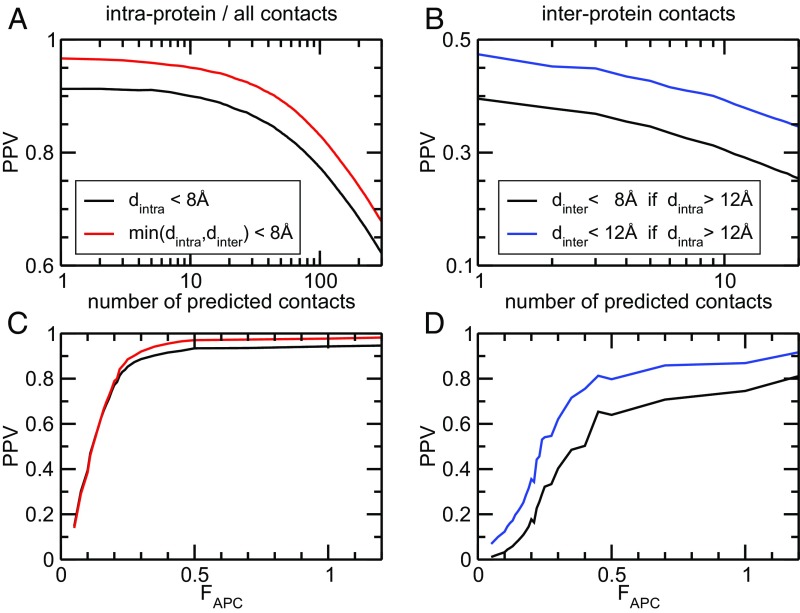
Average prediction accuracy for intraprotein and interprotein contacts. The figures report the performance of DCA for predicting intraprotein and interprotein contacts for the 750 selected families forming homo-oligomers. (A) Displayed is the positive predictive value PPV [PPV = TP/(TP + FP) = number of TP predictions/number of predictions] as a function of the number of predictions, and averaged over all 750 families. The black line takes into account contacts inside the monomer, whereas the red line also counts intraprotein contacts extracted from biological assemblies deposited in the PDB. Whereas the intraprotein contacts prediction is of high accuracy (average PPV = 90.2% for first 10 predictions per family, 77.5% for first 100 predictions), we find that a large fraction of FPs are actually interprotein contacts (combined PPV = 95.2% for 10 predictions, 83.2% for 100 predictions per family). (B) PPV is traced (same colors as in A) as a function of the DCA FAPC score; it shows that almost constantly high PPVs above 90% are obtained for FAPC > 0.3. (C and D) Analysis of predictions only, which are highly incompatible with the monomeric structure (minimum atom distance above 12 Å). We see that almost 40% are in contact between the proteins (8-Å distance cutoff) and more than 46% are in the same spatial vicinity (12-Å cutoff). We find again that high-scoring residue pairs with FAPC > 0.3 have a high probability of being in interprotein contact (average PPV above 80%).
Strongly Coevolving Residue Pairings Are Typically Internal or Interprotein Contacts.
To assess the accuracy of contact prediction in homo-oligomers, we selected the database of 750 protein domain families that (i) feature sufficient sequence space to enable statistical analysis, (ii) have at least one high-resolution structure deposited in the PDB annotated as a biological homo-oligomer, and (iii) have homo-oligomer interfaces of sufficient size (compare Methods).
When analyzing these 750 protein families, the first expected result is that most strongly coupled residue pairings represent intraprotein contacts defining the fold of the domain family (Fig. 3A). When only considering internal contacts, the positive predictive value (PPV), defined as true-positive (TP) contact predictions over all predictions [TP/(TP + FP)] for all 750 families, shows 90.2% accuracy for the top 10 highest scoring contact pairings. When also considering interprotein contacts, the PPV is further increased to 95.2%. In other words, on average, 95.2% of the top 10 highest scoring residue pairs across 750 protein families are either internal or interprotein contacts. When analyzing residue pairings according to their coevolutionary coupling, as measured by the FAPC score (31) (Methods), we find that pairings with a score over 0.3 have a PPV of about 90% (Fig. 3C). In other words, an FAPC score >0.3 provides an excellent prediction that two residues are in contact, thus providing a natural prediction cutoff across diverse protein families.
Enrichment of Coevolutionary Couplings Across Homo-Oligomeric Interfaces.
The accuracy of our interprotein contact prediction was further analyzed by two main measures. First, for each of the selected families, we extract the number of pairs of strongest coevolutionary signal that are not in contact inside the monomer (Methods) and determine the fraction of these predictions that are interprotein contacts in biological homo-oligomeric assemblies. Fig. 3B shows this quantity as a function of the number of interprotein contact predictions. We observe a clear enrichment in true interprotein contacts; more than 40% of the first interprotein predictions are in contact in at least one of the considered structures, compared with 2.9% of all residue pairs. However, despite this strong enrichment, we also observe that up to 60% of the first predictions are actually FP predictions (i.e., they are in contact neither in the monomer nor in the oligomer). To understand this finding better, we focused on the PPV according to the coevolutionary FAPC score. Sorting all internal noncontacts according to their scores, we find again that values above FAPC > 0.3 have a very high probability (>60%) of being in contact across the interaction interface, particularly if they are distant inside the monomer, (Fig. 3D). The PPV even grows to ca. 80% for FAPC > 0.5. Not all protein families show such strong coevolutionary coupling, however. Although a vast majority of families (714 of 750) show at least one residue pair with an internal distance above 8 Å and a coevolutionary FAPC score >0.2, this number drops rapidly when going to larger scores: 483 families have at least one coupling with FAPC > 0.3 and 190 families have at least one coupling with FAPC > 0.5. As stated before, not all of these pairs are necessarily quaternary contacts. Asking, for example, for at least five (respectively 10) internal noncontacts with FAPC > 0.3, we find only 236 (respectively 111) families; for FAPC > 0.5, this number is reduced to 55 (respectively 16). The full dependence of the cutoff of the DCA score is given in SI Appendix, Fig. S2.
The Size of the Homo-Oligomeric Interface Significantly Influences the Availability of Highly Correlated Interface Positions.
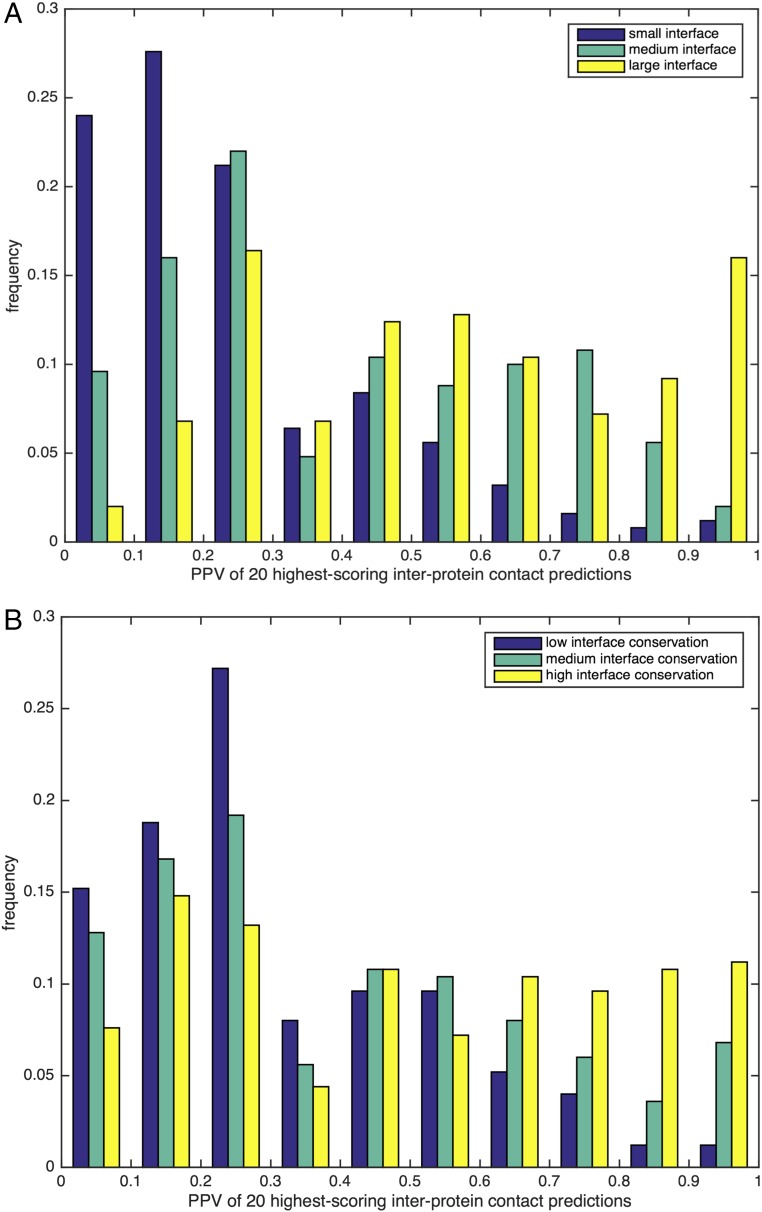
The high probability of being an interprotein contact if a large coevolutionary coupling was observed together with elevated intraprotein distance is very interesting. It underlines the observation that most direct couplings unveiled by DCA are actually based on physical contact and no other mechanisms (e.g., allosteric coupling), which, consequently, should be mediated by spatially connected networks of intermediate residues. However, a substantial number of protein families do not show any strong homo-oligomeric DCA signal. To understand this fact better, we have partitioned the dataset according to the size of the interface. In Fig. 4A, protein families are divided into three equally large groups containing the smallest, largest, and intermediate interface sizes, and histograms of the PPV for the first 20 homo-oligomeric contact predictions are displayed. We see that small interfaces have by far the largest probability of remaining undetected, whereas interfaces of high PPV are dominated by large interfaces. We conclude that the larger the interface is, the more readily it might exhibit strong coevolutionary signals, and that smaller interfaces do not necessarily have the same requirement for a significant coevolutionary signal.
Fig. 4.
Influence of interface size and conservation on the accuracy of interprotein contact prediction. (A) Histogram for the PPV [PPV = TP/(TP + FP)] of the first 20 contact predictions for the 750 selected oligomer-forming protein families. For the histogram, the dataset is divided into three classes according to their interface size (small, medium, and large), measured by the number of interprotein contacts. Whereas families with a large interface size dominate the highest accuracy cases, families with a small interface size typically lead to very bad PPVs. (B) Analogous histogram, now with the dataset of selected families with at least two alternative homo-oligomeric structures. More conserved interfaces (i.e., with less variability between the different representative structures of the concerned family) frequently show higher PPV values than variable interfaces. Because the coevolutionary signal for each oligomerization mode is specific to an appropriate selection of proteins, it becomes weak in the overall family alignment, which mixes different oligomerization modes.
The Conservation of the Homo-Oligomeric Interface Significantly Influences the Availability of Highly Correlated Interface Positions.
In Fig. 4B, we have analyzed the dataset according to the interface conservation (compare SI Appendix, section SI-3 for a precise description of how this conservation measure has been obtained). As a general idea, an interface is considered to be conserved if interfaces tend to strongly overlap over different available PDB structures. On the contrary, an interface is considered to be nonconserved if entirely different homo-oligomeric interfaces are found in the PDB. Dividing the dataset again into three classes, we see that more conserved interfaces have larger PPVs than variable ones. This result might be expected: A pair being in contact in one specific interface may coevolve in all proteins showing this interface, and may evolve independently in all proteins showing an alternative interface or no interface at all. Therefore, the coevolutionary signal for each individual interface becomes diluted if we consider the full Pfam MSA, and thus less easily detectable by DCA. We hypothesize that by focusing on a subalignment possessing a common oligomerization interface, the coevolutionary signal might become more evident, as long as the subalignment contains sufficient sequences. In fact, the issue of differing interaction modes might be a primary reason why DCA fails to predict interprotein contacts in many protein families that fulfill requirements of sequence availability and interface size because Pfam protein families commonly include proteins with similar folds but diverse function and activity.
RR Dimers: Nonconserved Dimerization Modes and Subfamily-Specific Coevolutionary Signals.
To test the hypothesis on differing interaction modes, we focused on one particular protein domain family with a known diverse set of protein–protein interfaces, the bacterial RRs defined by Pfam ID PF00072. It is an extremely abundant protein family used predominantly in bacterial signal transduction. RR domains are subject to phosphorylation by an associated histidine kinase. In response to phosphorylation, they alter their activity, most commonly by acting as transcription factors. Diverse DNA-binding domains are used by these RR proteins, and, accordingly, different subclasses oligomerize in different ways (compare Fig. 5 C, H, and I). Some subclasses of RRs are of single-domain architecture, and thus do not have additional output domains but rather act directly by forming heteroprotein interactions [e.g., those RRs that mediate bacterial chemotaxis as reviewed by Zschiedrich et al. (32)].
Fig. 5.
Intraprotein and interprotein contact predictions for different classes of RRs. RRs with DNA-binding domains dimerize when activated, but the dimerization mode differs between different DNA-binding domain families. Here, we analyze RRs having the three major DNA-binding domains: Trans_Reg_C, GerE, and LytTR. As C, F, and I show, they have substantially differing biological assemblies; the blue domain is positioned such that a visual comparison is facilitated. A, D, and G show the contact prediction using the full RR Pfam family (PF00072), and B, E, and H show the contact prediction using only the sub-MSA of RRs containing the corresponding DNA-binding domain. The triangles in the upper diagonals show the intraprotein contacts. Native contacts (d < 8 Å) are depicted by black dots, and residue pairs with native distances below 12 Å are shown by gray dots. The colored dots correspond to DCA predictions with FAPC > 0.3: Green dots are within a distance of 8 Å, orange dots are at a distance between 8 Å and 12 Å, and red dots are above a distance of 12 Å. The lower diagonals show the interprotein contact map. Black/gray dots correspond to distances below 8 Å/12 Å, which are not in contact inside the monomer, and thus potentially detectable by our method. Light-blue dots correspond to interprotein distances below 12 Å, which are not detectable by our method because they are also contained in the intraprotein contact map. The other colored dots report all DCA predictions with FAPC > 0.3 and dintra > 8 Å: green dots if interprotein distance (dinter) < 12 Å, orange dots if dinter > 12 Å but dintra < 12 Å, and red dots if minimum(dinter,dintra) [min(dinter,dintra)] > 12 Å. Although the full Pfam alignment results in only a very few strong coevolutionary signals with incompatibility to the monomeric structure (one residue pair with FAPC > 0.3 and dintra > 8 Å in all three structures), the subclass alignment results in strongly improved interprotein contact predictions. Green, orange, and red dots (FAPC > 0.3 and dintra > 8 Å) were used in the docking simulations.
The different DNA-binding domains found in transcription factor RRs belong to different Pfam families. Of the 151,337 RR sequences listed in Pfam, 45,431 have a Trans_Reg_C transcription factor domain, 20,829 have a GerE domain, and 6,269 have a LytTR domain. Other architectures (RR/Sigma54_activat/HTH_8,RR/HTH_18) are less frequent, besides the single-domain architecture, which covers 10,673 protein sequences. Whereas the activated dimer structure of the Trans_Reg_C-associated RR has long been known (33, 34), dimer structures of LytTR- and GerE-associated RRs have only been determined more recently (35, 36). All three classes of RRs show significantly different dimerization modes (Fig. 5, Right).
In Fig. 5, we show contact maps for all three classes, overlaid with DCA prediction, both for intraprotein (upper diagonals) and interprotein (lower diagonals) contact. The panels on the left (Fig. 5 A, D, and G) show the predictions from the full Pfam MSA. The interprotein prediction shows a mixture of contacts from different architectures, with a preference for the dominant Trans_Reg_C class. Consequently, for each individual architecture, many FP results are found. The picture changes substantially if we consider architecture-specific subalignments (Fig. 5 B, E, and H). The most evident change is the elimination of many FP predictions resulting from contacts in other architectures. The best TP signal is observed in the largest architectural subfamily, the Trans_Reg_C class (1, 2, 20). Substantial improvements are also obtained in the GerE-containing architecture. Note that for the third, smallest sub-MSA of the RR/LytTR architecture, a smaller improvement in terms of correctly predicted contacts is discovered. However, it has to be noted that the majority of native interprotein contacts in this case are also in close spatial vicinity inside each protein monomer (blue in Fig. 5), and thus hidden from our analysis as discussed before.
In summary, by analyzing the RR in detail, we find that by subdividing a specific family of proteins according to their domain architecture, we can strongly improve the predictive value of DCA. It can be expected that this idea is applicable to many protein families where DCA currently fails to detect an oligomerization signal. In addition, the data demonstrate for the RR protein family how one particular protein fold can evolve to accommodate various different interaction modes according to need, which, in this case, is connecting the homodimerization mode with the various different DNA-binding domains.
Coevolutionary Analysis on RR Subalignments Provides Sufficient Information for Accurate Structural Prediction of Protein-Complex Structures.
We next ask whether the contact information forthcoming from coevolutionary analysis of the three architecture-specific sub-MSAs is sufficient to dock monomeric structures and to determine the different biological assemblies. To this end, we use the above determined cutoff of FAPC > 0.3; all residue pairs with FAPC > 0.3 and intraprotein distance (dintra) > 8 Å are listed in Table 1. We consider them putative contacts (whether realized or not in the known dimer structures), and they are used in high-ambiguity–driven protein–protein docking (HADDOCK) (37) docking studies to determine whether accurate structural models of homodimers can be inferred from sequence-derived contact information (details are provided in Methods).
Table 1.
Investigated RRs, their predicted DCA contacts, and the contacts realized in the structure prediction of their homodimeric form
| Protein | Predicted contacts (FAPC > 0.3, dintra > 8 Å; ordered by DCA score) | Realized contacts in homodimer (dinter < 8 Å) |
| OmpR (PDB ID code 1nxs) | 84–104, 87–107, 33–57, 34–58, 35–57, 58–66, 84–106, 38–58, 88–106, 85–106, 92–113, 91–110, 91–111, 92–106, 87–104, 35–58, 92–110, 88–104, 65–89, 88–110, 89–106, 84–107, 34–57, 53–90, 53–93, 12–20 | 84–104, 87–107, 84–106, 88–106, 85–106, 92–113, 91–110, 91–111, 92–106, 87–104, 92–110, 88–104, 88–110, 84–107 |
| LyTR (PDB ID code 4cbv) | 101–120, 87–112, 66–74, 89–94, 99–104, 101–106, 83–87, 100–104, 99–107 | 101–120, 101–106, 99–107 |
| GerE (PDB ID code 4e7p) | 11–80, 42–72, 14–107, 43–72, 8–81, 46–72, 9–104 | 11–80, 14–107, 9–104 |
HADDOCK does not directly use the DCA-paired residues as contacts, but declares all residues in the pairings as potential interface residues. Therefore, docking results in several clusters of structures for each protein subfamily. DCA pairings can be used to rank clusters by the number of actually realized putative contacts, wherein the first-ranking dimer is considered as the likely physiologically relevant dimer. When comparing these dimer models with the experimental structures, we observe excellent agreement for all three subclasses of RR proteins (Fig. 6). The mean deviation for each structural dimer cluster with respect to the dimer structures was an impressive 1.1 Å for Transreg_c, 1.2 Å for LytTR, and 1.1 Å for GerE subfamilies, respectively. For each of these structures, several of the DCA-predicted residue pairs are realized as interprotein contacts. Others are not, and they all constitute residue pairings with intraprotein distances of 8 Å < dintra <12 Å, and thus likely represent intraprotein rather than interprotein contacts that might be realized in other members of the protein family or by conformational changes between inactive RR monomers and active RR dimers.
Fig. 6.
Prediction of homodimeric structures. The predicted interdomain contacts (compare Fig. 5) allow reliable predictions of homodimeric structures with up to crystal structure accuracy for the different RRs OmpR (PDB ID code 1nxs; A and D), LytTR (PDB ID code 4cbv; B and E), and GerE (PDB ID code 4e7p; C and F). (A–C) Displayed are the predicted homodimers (blue) overlaid with their known crystal structures; monomers are highlighted in green and purple. The backbone rmsds between the prediction and the crystal structures are 1.1 Å (OmpR), 1.1 Å (GerE), and 1.2 Å (LyTR). Interestingly, for LyTR, we predict the biological assembly 2. In the predictions, only part of the predicted interdomain contacts are realized (dinter < 8 Å; highlighted in green in D–F), whereas many predicted contacts are not formed (highlighted in orange in D–F). All these contacts, however, are close to being realized in the monomer as intra contacts within a threshold of 12 Å (dinter > 8 Å, dintra < 12 Å). None of the predicted contacts are >12 Å both inter and intra.
Surprisingly, we note that GerE has two annotated biological dimers in the PDB, one author-assigned and one PISA software-assigned. We reconstitute the latter, suggesting that this dimer is the physiologically relevant dimer for GerE type RR, whereas the author-assigned dimer is likely irrelevant.
We realize that the astonishing accuracy of the predicted dimer models with respect to experimental structures represents a best-case scenario, because the monomers used in the docking approach are actually derived from physiological dimers. In SI Appendix, we also explore what could be considered a worst-case scenario for the Trans_Reg_C subfamily, where the RR monomer structure was first homology-modeled based on the structure of a monomeric homolog and then used for docking studies. This approach resulted in slightly reduced, but still impressive, accuracy (compare SI Appendix, section SI-4, Figs. S4–S8, and Tables S1 and S2). As detailed in Methods and SI Appendix, we also compare the accuracy of HADDOCK docking with docking obtained by Magma (20, 38) using eSBMTools (39). With Magma, we realized the specific DCA pairings (compare SI Appendix, Fig. S6 and Tables S1 and S2). With HADDOCK, in a first step, all residues in the DCA pairings are considered as potential interface residues (“nonspecific pairings”), and the model with the highest number of DCA-pairings is then selected as the best prediction. Little difference in model accuracy was obtained with these substantially differing docking approaches, demonstrating that the accuracy of our approach is compatible with several different protein docking techniques.
In summary, these data demonstrate that sequence-derived contact information for subfamilies of protein folds can be used successfully to predict alternative physiological oligomerization modes for one large but diverse protein family. We anticipate that there are many other protein families where some but not all relevant oligomerization modes are currently known, and where this approach would be of significant utility. In addition, as evident for the example of GerE, a DCA approach represents a complement to PISA interaction software to determine the relevance of a specific oligomerization mode.
Coevolutionary Signal in Families Without Known Homo-Oligomeric Crystal Structures.
In our selection of homo-oligomeric structures, we have seen that almost all those residue pairs are actually homo-oligomeric interprotein contacts, which show a strong DCA signal while being distant in the protein’s tertiary structure. Strong DCA signals appear to be, with high probability, related to residue contacts.
At this point, an interesting question appears: How many strong signals are there in protein families without known homo-oligomeric structure? To this end, we have analyzed the 1,234 protein families that satisfy our selection criteria for sequence abundance and availability of high-resolution monomer crystal structure, without having a sufficiently large homo-oligomeric interface in the PDB. A total of 786 families have no residue pairings with FAPC > 0.3 at intraprotein distances above 12 Å, and 235 families have only one such large DCA signal. Of the remaining 213 families, 189 are actually annotated as homo-oligomeric biological units, but the size and quality of the interfaces resulted in exclusion by our selection criteria. Only 24 families have no known homo-oligomeric biological units. This observation suggests that over all sufficiently large Pfam families with available high-resolution structures, there are very few directly coevolving pairs that are not explainable via known intraprotein or interprotein contacts. In SI Appendix, we provide a list of all 448 families with at least one distant DCA high-scoring residue pair, together with these pairs, for further exploration.
Discussion
The rapidly expanding protein sequence databases provide us with the necessary raw material to explore the interplay between the evolutionary sequence variability in homologous protein families and the structural and functional aspects of proteins that are conserved across species. Coevolutionary modeling approaches, particularly global modeling approaches as represented by the DCA and related methods, have played an important role in detecting residue–residue contacts from directly statistically coupled residues. In the context of tertiary structure prediction of proteins, this approach has been tested across hundreds of protein families and, in turn, has led to a large number of in silico tertiary structure models (10–13) for a large variety of protein families, each one containing thousands of proteins but not having a single experimentally solved representative structure.
Success remains more limited in the case of protein–protein interactions, even if DCA was originally conceived for this case. So far, the largest scale studies remain limited to about 100 protein families (21).The reason is simple: Coevolutionary analysis requires large MSAs as inputs, with each line containing a pair of interacting proteins from the two interacting families under study. Due to the abundance of paralogs in a large fraction of protein families, the generation of such alignments is a hard task, and studies typically have been restricted to solvable cases where interacting proteins are colocalized along the genomes (e.g., in operons in the case of bacteria).
In this paper, we extend the analysis by about one order of magnitude, concentrating on homo-oligomeric interactions. Because both interaction partners have identical sequences, the analysis can be done on a single protein alignment. However, as a potential problem, we have identified that even in the case of the presence of monomeric protein structures, the distinction between intraprotein and interprotein contacts is frequently nontrivial. Many residue pairs are in contact both inside the protein and in-between the proteins. Large coevolutionary scores would be coherent with the monomeric structure, and no existing method might identify them as interprotein contacts. They are a priori hidden for our method; however, in principle, they might be used a posteriori as support for a docked oligomer model.
Despite this complication, we find that of the close to 2,000 protein families having sufficient sequence numbers and experimentally determined high-resolution crystal structures, only about 150 families have no assigned oligomeric biological unit and about 750 have a sufficiently large interface to be potential targets of coevolutionary analysis. This large dataset allows us to assess the strength and limitations carefully in applying methods like DCA to protein–protein interactions, ensuring reproducibility of results and avoiding bias.
First, we find that the majority of FP predictions of intraprotein contacts (i.e., residue pairs with a high DCA score but a large distance in the protein monomer) are actually interprotein contacts involved in homo-oligomerization. Remarkably, we find that almost all residue pairs with coevolutionary FAPC scores above 0.3 are either intraprotein or interprotein contacts (or both), demonstrating that the strongest couplings detected by DCA are contacts, and are not related to any other structural or functional aspects.
We also observe that not all families have such large coevolutionary scores corresponding to distant residue pairs in the monomer structure. Half of the considered 750 protein families with sufficiently large interfaces have few or weak oligomerization signals, and DCA fails to predict the oligomerization mode right away. However, we also find that the larger the interface is, the higher is also the probability of being detectable by DCA, whereas smaller interfaces are frequently missed. This finding immediately raises the question of whether methods like PconsC2 (40), which uses machine-learning ideas to combine DCA scores with contact patterns from existing protein structures (thereby discriminating locally coherent but potentially weak coevolutionary signals from strong but incoherent noise), can be adapted from intraprotein to interprotein contact prediction.
We were able to identify a second limiting factor for the capacity of DCA to detect interprotein contacts. In many protein families, the homo-oligomeric complex structure is not conserved even if the monomeric structure is almost unchanged. In these cases, the coevolutionary signal for each oligomeric structure is present only in part of the large MSA; it becomes weaker if the full MSA is analyzed. This general observation has motivated us to study in detail the case of bacterial RRs. A focus was on those bacterial RRs acting as dimeric transcription factors when activated. In this case, different DNA-binding domains correspond to different dimerization interfaces even in the common RR domain. When restricting the MSA to a domain architecture-specific sub-MSA, specific oligomerization signals emerged. For each of the three dominant domain architectures (Trans_Reg_C, GerE, and LytTR class), these signals were sufficiently precise to guide docking procedures to extremely high precision. When analyzing the full MSA, oligomeric signals faded out and mixtures of the different oligomeric interfaces were found. We see that understanding the origins of the limited accuracy of DCA in predicting interprotein contacts in the full Pfam MSA actually opens up a strategy for a finer scale analysis based on subfamilies. In future work, it will be interesting to explore similar cases in more detail, as well as the emergence of subfamily-specific signals when reducing the alignment depth.
Finally, we analyze the 142 families without an annotated biological assembly. Interestingly, only a small number of these families express a strong coevolutionary signal. Individual analyses of these cases do not provide consistent oligomerization signals. Therefore, not a single example of known tertiary but unknown quaternary structure is forthcoming from this study that would allow for in silico structure prediction. This finding suggests that, to date, many of the large and widely conserved homo-oligomerization interfaces (i.e., those homo-oligomerization interfaces where we would expect a clear coevolutionary signal) have already been experimentally described.
By their nature, experimental studies of protein structures are characterized by slow throughput and usually involve a single or, at best, a handful of examples of a specific family of proteins. The scientific literature is abundant with generalizations of protein interactions based on a single structure as representative of an entire protein family. One strength of DCA is that it collects all examples of a given protein family in one MSA, and forthcoming interaction data can thus be considered to be truly representative for most of the proteins of a given family. This fact presents another unique utility of DCA in aiding the researcher to determine whether broad generalizations are indeed possible based on an individual protein structure and its interactions or if appropriate subfamilies have to be extracted.
The case of GerE suggests the potential utility of DCA in identifying physiologically relevant interfaces in protein crystal structures. Of the two deposited alternative biological GerE assemblies, only one shows strong coevolutionary coupling across the interface, suggesting a physiological relevance resulting in selective pressure in evolution. A similar case was reported for Hsp70 chaperones, where supposed crystallographic artifacts were identified as coevolving contacts (41) and subsequently shown to be physiologically relevant (42). The global analysis presented here does not take this possibility into account systematically: A residue pair is considered a TP finding if it is found in contact in at least one assembly (i.e., in the union of all contact maps). However, we can refine the analysis to search systematically for distinct interfaces and for diversified DCA results for the different interfaces. We have done so for each PDB structure independently, and kept cases where (i) at least five pairs with high nontertiary coevolutionary signal (FAPC > 0.3, dintra > 12 Å) exist, (ii) one interface has more than 50% true contacts within these predictions, and (iii) a distinct interface has less than 10% true contacts. We find 27 cases, mostly with distinct interfaces inside a higher order oligomeric assembly. A simply understandable case is given by PDB ID code 1EA4 (43) (compare SI Appendix, Fig. S9; the structure consists of several DNA-bound dimers). The internal interface in each dimer is well predicted by DCA (six pairs with FAPC > 0.3, all in contact), and the interface between dimers shows no signal and is generated probably only by the DNA functioning as a joint scaffold. Although this last result would have a probability of 0.81 to appear in six randomly selected residue pairs, the six interprotein contacts are highly significant (P ∼ 10−5).
A full analysis of different homo-oligomer interfaces should also compare different PDB structures for the same protein family. However, caution is urged: As shown again in the case of RRs, only the major subfamily (Trans_Reg_C) has some small but detectable dimerization signal on the level of the full Pfam alignment, whereas interface signal corresponding to minor subfamilies becomes invisible in the full MSA. A careful study of individual cases would be needed to be able to distinguish cases where coevolutionary signals are completely absent for a crystallographic interface, or where they are present only in a limited subfamily.
Notably, by focusing on a large dataset, we defined a score cutoff, FAPC > 0.3, applicable across all protein families as an excellent determinant of whether residue pairs can be expected to make contact in the physiological structure. We expect this cutoff to be of significant value when expanding current efforts toward the identification of novel heteroprotein–heteroprotein interactions, and this application is one of our future goals. As mentioned earlier, due to the existence of amplified protein folds and paralogs, identifying the correct interaction partners for accurate generation of joint MSA is not trivial and needs to be solved to apply this technology to heterointeractions on a global level.
Methods
Selection of Protein Families and Oligomeric Structures.
To perform a large-scale analysis of coevolution in homo-oligomers, we have selected an exhaustive database of protein families from Pfam 27.0 according to the two following criteria: (i) Families are required to have, at 80% sequence identity, an effective sequence number of at least 500 [the definition of the effective sequence number follows the method of Morcos et al. (2)] so as to guarantee sufficient statistics to detect coevolutionary signals by DCA, and (ii) at least one high-resolution (<3 Å) structure with homo-oligomeric contacts in the biological unit must be present in the PDB structure.
This second step requires more precise description. Many protein families have more than one homo-oligomeric interface classified as a biological assembly in the PDB. These assemblies may be quite diversified, and a DCA prediction being in contact in at least one PDB structure can be considered a TP prediction. To this end, for each Pfam family:
-
i)
We have collected all PDB structures with a biological assembly that contains homo-oligomers between chains matching the Pfam family.
-
ii)
For each PDB structure, domain repeats in the same chain and different assemblies are taken into account. At this point, a homo-oligomeric assembly is uniquely characterized by the following list: (Pfam ID, PDB ID code, chain 1, chain 2, chain 1 domain number, chain 2 domain number, and biological assembly number).
-
iii)
We have created a mapping between the Pfam HMM (Hidden Markov Model) and each concerned chain, which allows us, for each pair of matched columns in the Pfam HMM, to calculate intraprotein and interprotein residue–residue distances. Distances are measured as minimal distances between heavy atoms.
-
iv)
PDB structures of low-sequence or interface coverage by the Pfam MSA [<30% of HMM positions matched, <15 interface residues (contact distance <8 Å)] are excluded from further analysis.
-
v)
Distances between two positions (columns) in the Pfam MSA are now defined as the minimum over all matched native distances in all retained PDB files for intraprotein and interprotein distances, respectively.
-
vi)
A last step of filtering removes remaining small interfaces of very low contact density [dinter < min(0.01, 0.1⋅dintra)], where the contact density is defined as the fraction of residue pairs of native distance below 8 Å, min is minimum, and dinter is the interprotein distance. At the final stage, the database includes 750 Pfam domain families matching 13,156 PDB structures, with a total number of 77,109 intrachain structure units and 54,065 interchain structure units.
As we see in Results, interfaces that are too small are not detectable by DCA. Finally, we have 750 Pfam families, which form our dataset.
Besides these homo-oligomeric families, we have also collected a database of all those families that fulfill our constraints on the sequence number and the existence of a high-resolution PDB structure but are not classified as a homo-oligomeric biological assembly or do not pass the tests on the interface size. There are a total of 1,234 of these families.
DCA.
DCA is based on the maximum-entropy modeling of protein families, with the aim of reproducing amino acid frequencies in single MSA columns, and frequencies of amino acid co-occurrences in pairs of MSA columns, via an otherwise unconstrained statistical model. For an aligned amino acid sequence of length L, these conditions lead to a generalized Potts model or Markov random field:
Parameters of this model, independently for each Pfam family, are calculated using the pseudo-likelihood maximization approach of Ekeberg et al. (31), with standard settings for the reweighting and the regularization parameters. Following the same reference, coupling strengths are characterized by the Frobenius norm, , with subsequent average-product correction . The dot(s) indicates an average over the concerned position(s). These FAPC scores are used to sort all residue pairs, and strong DCA couplings are interpreted as predictors for residue–residue contacts inside or between proteins.
Structure Prediction.
We used HADDOCK (37) to obtain the complex/dimers of RR_REC domains. HADDOCK is based on “sticky” surface regions between the provided chains.
Briefly, we isolated chain A from a representative PDB file for the different RRs Transreg_c (PDB ID code 1nxs), LytTR (PDB ID code 4cbv), and GerE (PDB ID code 4e7p). The resulting monomers were docked in HADDOCK by using all amino acids involved in predicted contacts (Table 1) as surface residues. This procedure results in several clusters of docked structures for the three RRs (Transreg_c, n = 2; GerE, n = 2; and LytTR, n = 3) that differ substantially. By counting the number of correctly realized DCA contacts for the different clusters, one can identify a unique conformation in best agreement with the DCA contact predictions in each case.
We stress that the used prediction technique differs from prior docking studies using coevolutionary direct information (20). Here, we use the DCA contact predictions not as pairwise constraints/energetic biases between residues (i,j) in the initial docking step but as one large surficial cluster formed by all involved residues in any predicted contact (i,j). In a second step, we discriminate between the predicted clusters by counting the number of specific contacts (i,j) as selection criteria. The main advantage of this two-step approach is to search a wider range of conformations in the initial step before deciding on the specific conformation in best agreement with the DCA contacts. We also performed docking studies, which used the DCA contacts as direct constraints with eSBMTools (39), which is more similar to prior work (compare SI Appendix; Dataset S1 lists inter-domain contacts for the considered PFAM families).
Supplementary Material
Acknowledgments
G.U. and M.W. acknowledge funding by the Agence Nationale de la Recherche via the project COEVSTAT (Grant ANR-13-BS04-0012-01). S.J.L. and A.S. received funding from the Impuls- und Vernetzungsfond of the Helmholtz association. A.S. received support from a Google Faculty Research Award. H.S. was funded by Grant GM106085 from the National Institute of General Medical Sciences, NIH.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1615068114/-/DCSupplemental.
References
- 1.Weigt M, White RA, Szurmant H, Hoch JA, Hwa T. Identification of direct residue contacts in protein-protein interaction by message passing. Proc Natl Acad Sci USA. 2009;106(1):67–72. doi: 10.1073/pnas.0805923106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morcos F, et al. Direct-coupling analysis of residue coevolution captures native contacts across many protein families. Proc Natl Acad Sci USA. 2011;108(49):E1293–E1301. doi: 10.1073/pnas.1111471108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones DT, Buchan DWA, Cozzetto D, Pontil M. PSICOV: Precise structural contact prediction using sparse inverse covariance estimation on large multiple sequence alignments. Bioinformatics. 2012;28(2):184–190. doi: 10.1093/bioinformatics/btr638. [DOI] [PubMed] [Google Scholar]
- 4.Kamisetty H, Ovchinnikov S, Baker D. Assessing the utility of coevolution-based residue-residue contact predictions in a sequence- and structure-rich era. Proc Natl Acad Sci USA. 2013;110(39):15674–15679. doi: 10.1073/pnas.1314045110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morcos F, Schafer NP, Cheng RR, Onuchic JN, Wolynes PG. Coevolutionary information, protein folding landscapes, and the thermodynamics of natural selection. Proc Natl Acad Sci USA. 2014;111(34):12408–12413. doi: 10.1073/pnas.1413575111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Figliuzzi M, Jacquier H, Schug A, Tenaillon O, Weigt M. Coevolutionary landscape inference and the context-dependence of mutations in beta-lactamase TEM-1. Mol Biol Evol. 2016;33(1):268–280. doi: 10.1093/molbev/msv211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schug A, Onuchic JN. From protein folding to protein function and biomolecular binding by energy landscape theory. Curr Opin Pharmacol. 2010;10(6):709–714. doi: 10.1016/j.coph.2010.09.012. [DOI] [PubMed] [Google Scholar]
- 8.de Juan D, Pazos F, Valencia A. Emerging methods in protein co-evolution. Nat Rev Genet. 2013;14(4):249–261. doi: 10.1038/nrg3414. [DOI] [PubMed] [Google Scholar]
- 9.Noel JK, Morcos F, Onuchic JN. Sequence co-evolutionary information is a natural partner to minimally-frustrated models of biomolecular dynamics. F1000 Res. 2016;5:5. doi: 10.12688/f1000research.7186.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marks DS, et al. Protein 3D structure computed from evolutionary sequence variation. PLoS One. 2011;6(12):e28766. doi: 10.1371/journal.pone.0028766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sułkowska JI, Morcos F, Weigt M, Hwa T, Onuchic JN. Genomics-aided structure prediction. Proc Natl Acad Sci USA. 2012;109(26):10340–10345. doi: 10.1073/pnas.1207864109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tian P, et al. Structure of a functional amyloid protein subunit computed using sequence variation. J Am Chem Soc. 2015;137(1):22–25. doi: 10.1021/ja5093634. [DOI] [PubMed] [Google Scholar]
- 13.Ovchinnikov S, et al. Large-scale determination of previously unsolved protein structures using evolutionary information. eLife. 2015;4:e09248. doi: 10.7554/eLife.09248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dago AE, et al. Structural basis of histidine kinase autophosphorylation deduced by integrating genomics, molecular dynamics, and mutagenesis. Proc Natl Acad Sci USA. 2012;109(26):E1733–E1742. doi: 10.1073/pnas.1201301109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morcos F, Jana B, Hwa T, Onuchic JN. Coevolutionary signals across protein lineages help capture multiple protein conformations. Proc Natl Acad Sci USA. 2013;110(51):20533–20538. doi: 10.1073/pnas.1315625110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sutto L, Marsili S, Valencia A, Gervasio FL. From residue coevolution to protein conformational ensembles and functional dynamics. Proc Natl Acad Sci USA. 2015;112(44):13567–13572. doi: 10.1073/pnas.1508584112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Leonardis E, et al. Direct-coupling analysis of nucleotide coevolution facilitates RNA secondary and tertiary structure prediction. Nucleic Acids Res. 2015;43(21):10444–10455. doi: 10.1093/nar/gkv932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weinreb C, et al. 3D RNA and functional interactions from evolutionary couplings. Cell. 2016;165(4):963–975. doi: 10.1016/j.cell.2016.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Finn RD, et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016;44(D1):D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schug A, Weigt M, Onuchic JN, Hwa T, Szurmant H. High-resolution protein complexes from integrating genomic information with molecular simulation. Proc Natl Acad Sci USA. 2009;106(52):22124–22129. doi: 10.1073/pnas.0912100106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ovchinnikov S, Kamisetty H, Baker D. Robust and accurate prediction of residue-residue interactions across protein interfaces using evolutionary information. eLife. 2014;3:e02030. doi: 10.7554/eLife.02030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hopf TA, et al. Sequence co-evolution gives 3D contacts and structures of protein complexes. eLife. 2014;3:3. doi: 10.7554/eLife.03430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feinauer C, Szurmant H, Weigt M, Pagnani A. Inter-protein sequence co-evolution predicts known physical interactions in bacterial ribosomes and the Trp operon. PLoS One. 2016;11(2):e0149166. doi: 10.1371/journal.pone.0149166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.dos Santos RN, Morcos F, Jana B, Andricopulo AD, Onuchic JN. Dimeric interactions and complex formation using direct coevolutionary couplings. Sci Rep. 2015;5:13652. doi: 10.1038/srep13652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodriguez-Rivas J, Marsili S, Juan D, Valencia A. Conservation of coevolving protein interfaces bridges prokaryote-eukaryote homologies in the twilight zone. Proc Natl Acad Sci USA. 2016;113(52):15018–15023. doi: 10.1073/pnas.1611861114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burger L, van Nimwegen E. Accurate prediction of protein-protein interactions from sequence alignments using a Bayesian method. Mol Syst Biol. 2008;4:165. doi: 10.1038/msb4100203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bitbol A-F, Dwyer RS, Colwell LJ, Wingreen NS. 2016. Inferring interaction partners from protein sequences. arXiv:1604.08354. [DOI] [PMC free article] [PubMed]
- 28.Gueudré T, Baldassi C, Zamparo M, Weigt M, Pagnani A. 2016. Simultaneous identification of specifically interacting paralogs and inter-protein contacts by direct-coupling analysis. arXiv:1605.03745. [DOI] [PMC free article] [PubMed]
- 29.Rose PW, et al. The RCSB Protein Data Bank: Views of structural biology for basic and applied research and education. Nucleic Acids Res. 2015;43(Database issue):D345–D356. doi: 10.1093/nar/gku1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y, Eisenberg D. 3D domain swapping: As domains continue to swap. Protein Sci. 2002;11(6):1285–1299. doi: 10.1110/ps.0201402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ekeberg M, Lövkvist C, Lan Y, Weigt M, Aurell E. Improved contact prediction in proteins: Using pseudolikelihoods to infer Potts models. Phys Rev E Stat Nonlin Soft Matter Phys. 2013;87(1):012707. doi: 10.1103/PhysRevE.87.012707. [DOI] [PubMed] [Google Scholar]
- 32.Zschiedrich CP, Keidel V, Szurmant H. Molecular mechanisms of two-component signal transduction. J Mol Biol. 2016;428(19):3752–3775. doi: 10.1016/j.jmb.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barbieri CM, Wu T, Stock AM. Comprehensive analysis of OmpR phosphorylation, dimerization, and DNA binding supports a canonical model for activation. J Mol Biol. 2013;425(10):1612–1626. doi: 10.1016/j.jmb.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Toro-Roman A, Mack TR, Stock AM. Structural analysis and solution studies of the activated regulatory domain of the response regulator ArcA: A symmetric dimer mediated by the alpha4-beta5-alpha5 face. J Mol Biol. 2005;349(1):11–26. doi: 10.1016/j.jmb.2005.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boudes M, et al. Structural insights into the dimerization of the response regulator ComE from Streptococcus pneumoniae. Nucleic Acids Res. 2014;42(8):5302–5313. doi: 10.1093/nar/gku110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park AK, Moon JH, Lee KS, Chi YM. Crystal structure of receiver domain of putative NarL family response regulator spr1814 from Streptococcus pneumoniae in the absence and presence of the phosphoryl analog beryllofluoride. Biochem Biophys Res Commun. 2012;421(2):403–407. doi: 10.1016/j.bbrc.2012.04.035. [DOI] [PubMed] [Google Scholar]
- 37.Dominguez C, Boelens R, Bonvin AM. HADDOCK: A protein-protein docking approach based on biochemical or biophysical information. J Am Chem Soc. 2003;125(7):1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 38.Schug A, et al. Computational modeling of phosphotransfer complexes in two-component signaling. Methods Enzymol. 2010;471:43–58. doi: 10.1016/S0076-6879(10)71003-X. [DOI] [PubMed] [Google Scholar]
- 39.Lutz B, Sinner C, Heuermann G, Verma A, Schug A. eSBMTools 1.0: Enhanced native structure-based modeling tools. Bioinformatics. 2013;29(21):2795–2796. doi: 10.1093/bioinformatics/btt478. [DOI] [PubMed] [Google Scholar]
- 40.Skwark MJ, Raimondi D, Michel M, Elofsson A. Improved contact predictions using the recognition of protein like contact patterns. PLOS Comput Biol. 2014;10(11):e1003889. doi: 10.1371/journal.pcbi.1003889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Malinverni D, et al. Large-scale conformational transitions and dimerization are encoded in the amino-acid sequences of Hsp70 chaperones. PLoS Comput Biol. 2015;11(6):e1004262. doi: 10.1371/journal.pcbi.1004262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sarbeng EB, et al. A functional DnaK dimer is essential for the efficient interaction with Hsp40 heat shock protein. J Biol Chem. 2015;290(14):8849–8862. doi: 10.1074/jbc.M114.596288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Costa M, et al. Plasmid transcriptional repressor CopG oligomerises to render helical superstructures unbound and in complexes with oligonucleotides. J Mol Biol. 2001;310(2):403–417. doi: 10.1006/jmbi.2001.4760. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.