 A series of emissive ribonucleoside purine mimics, all comprised of an isothiazolo[4,3-d]pyrimidine core, was prepared using a divergent pathway involving a key Thorpe–Ziegler cyclization.
A series of emissive ribonucleoside purine mimics, all comprised of an isothiazolo[4,3-d]pyrimidine core, was prepared using a divergent pathway involving a key Thorpe–Ziegler cyclization.
Abstract
A series of emissive ribonucleoside purine mimics, all comprised of an isothiazolo[4,3-d]pyrimidine core, was prepared using a divergent pathway involving a key Thorpe–Ziegler cyclization. In addition to an adenosine and a guanosine mimic, analogues of the noncanonical xanthosine, isoguanosine, and 2-aminoadenosine were also synthesized and found to be emissive. Isothiazolo 2-aminoadenosine, an adenosine surrogate, was found to be particularly emissive and effectively deaminated by adenosine deaminase. Competitive studies with adenosine deaminase with each analogue in combination with native adenosine showed preference for the native substrate while still deaminating the isothiazolo analogues.
Introduction
With nucleosides so vital to life processes, a wide range of analogues has been made over the years, both for medicinal chemistry purposes, as well as biochemical and biological tools.1 Among the latter, fluorescent nucleosides are particularly attractive as both means for biophysical analyses and for fabricating effective discovery assays. Given that the canonical nucleobases found in DNA and RNA are practically nonemissive,2 and a limited number of naturally occuring emissive nucleosides exist,3 the design and synthesis of isomorphic fluorescent nucleosides is of great importance and yet, rather challenging.4 From a synthetic perspective, the construction of purine-like isomorphic nucleoside mimics, particularly C-nucleoside analogues, presents formidable difficulties in linking the heterocycle to the ribose moiety.5 From a photophysical viewpoint, the unpredictable nature of fluorescent molecules, particularly within the context of nucleic acids, propagates a trial and error approach.6 Taken together, the compounded challenges have resulted in very few analogues which are truly isomorphic and isofunctional.
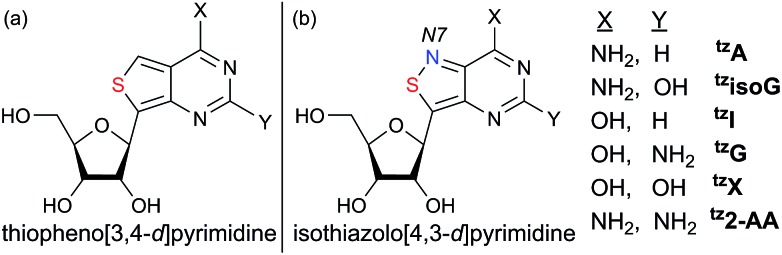
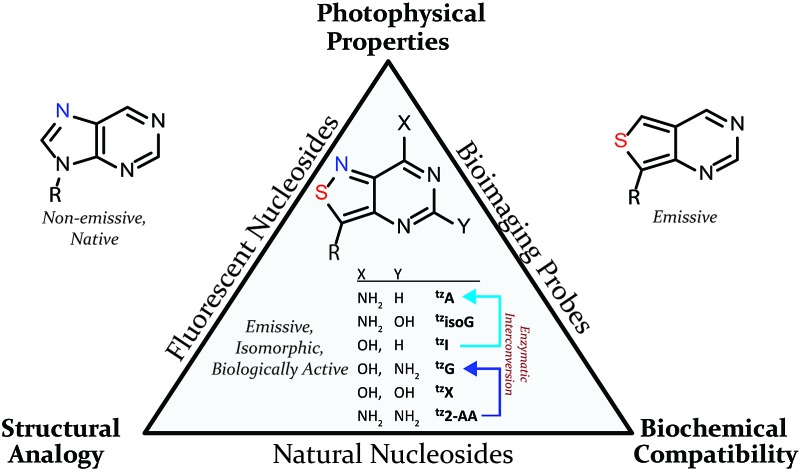
In 2011 our group designed and synthesized a highly emissive family of fluorescent RNA analogues based on a thiopheno[3,4-d]pyrimidine core (Fig. 1a).7 These molecules were utilized in a wide variety of applications and facilitated real-time monitoring of biologically relevant processes.8 One caveat of the purine-derived RNA analogues, thieno-adenosine (thA, Fig. 1a, X = NH2, Y = H) and thieno-guanosine (thG, Fig. 1a, X = OH, Y = NH2), was the lack of a distinct hydrogen bond acceptor and basic site, equivalent to the N7 of native purine systems (Fig. 1b). This caveat was elegantly exemplified in both the design of an assay based on an N7-dependent enzyme, adenosine deaminase (ADA), and a related enzyme without an N7 dependency, ADAR2, through the deamination of thieno-adenosine to thieno-inosine (thI Fig. 1a, X = OH, Y = H).8b,d
Fig. 1. Thiopheno vs. isothiazolo family.
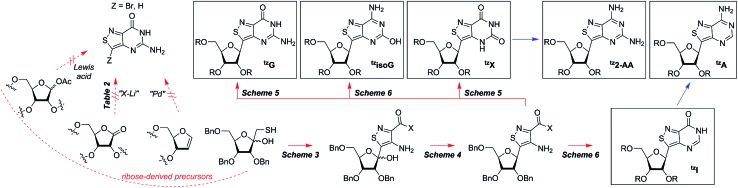
We hypothesized that if this hydrogen bond acceptor was to be reinstated through the replacement of a thiophene with an isothiazole ring (Fig. 1), the fluorescent properties of the molecule, albeit distinct, would likely be preserved while enhancing isofunctionality by restoring the ability to engage in hydrogen bonding. The isothiazolo[4,3-d]pyrimidine-based RNA alphabet, reported in 2015, indeed addressed the deficiency associated with the thiopheno family of emissive nucleosides.9 Subjected to the same conditions as thA, the isothiazolo adenosine analogue tzA was indeed shown to be deaminated ten-fold faster than the thiopheno analogue by ADA. Given that these newly synthesized molecules were both emissive and isofunctional, we have sought to complete this family of RNA nucleoside analogues by expanding it to non-canonical members, to both fully assess the unique electronic properties of this series and increase the fluorescent molecular toolkit for monitoring RNA-based processes.
Noncanonical nucleosides are naturally occurring and have been found in a wide variety of biological contexts.10 Isoguanine, for example, is a reported product of oxidative damage to DNA.11 2-Aminoadenosine is a known analogue of adenosine that engages in three hydrogen bonds of the W-C type, adding considerable stability to the double helix.12 Xanthine is a known product of deamination of nucleobases, a sign of miscoding and mutagenesis in DNA and RNA.13 Xanthosine is methylated during the early stages of caffeine biosynthesis while xanthosine monophosphate (XMP) is an intermediate for the biosynthesis of guanosine monophosphate from inosine monophosphate.14 Emissive analogs of such noncanonical nucleosides can enable the fabrication of biophysical assays that target biochemical pathways, involving such purines. Furthermore, possessing a systematically modified series of emissive nucleosides, which is based on a single heterocyclic core, also enables greater insight into electronic trends of such molecules, an aspect of fluorescent molecule design that is often difficult to accurately predict.15
Given the unpredictable features of fluorescent nucleosides, particularly characteristics related to their excited state dynamics, an inherit risk exists in devoting time and resources to extensively exploring synthetic pathways.16 Considering the highly emissive nature of the thiophene core, we found the corresponding isothiazolo counterparts to be nevertheless worth pursuing. Herein we report the synthetic design as well as the photophysical properties of new purine-inspired fluorescent nucleoside analogues. Perhaps unexpectedly, the syntheses presented diverse challenges, likely reflecting the scarce knowledge regarding the isothiazole nucleus and its reactivity. We thus elaborate not only on the successful pathways, but rather also on failed attempts, providing further insight into the chemistry of this largely unexplored heterocycle.17
Results and discussion
Isothiazolo nucleobase construction
In traditional nucleoside synthesis, the heterocyclic core is commonly furnished before being linked to a sugar moiety via established glycosylation methods.18 For the development of fluorescent nucleosides, constructing the heterocyclic framework first allows one to preliminarily evaluate the photophysical properties of a new nucleobase. This, however, does not address the synthetic hurdles and whether or not the heterocycle can ultimately be subjected to common methods of C-glycoside bond formation.5b The isothiazole core brings a unique challenge to the formation of the key carbon–carbon glycoside bond. In particular, the alkylation of C5 on functionalized isothiazoles, while previously observed, has only been reported for a limited substrate scope.19
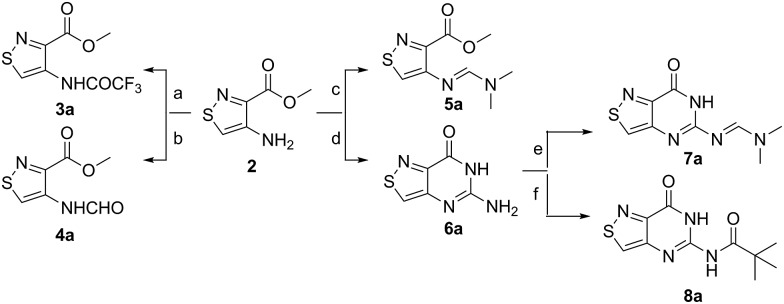
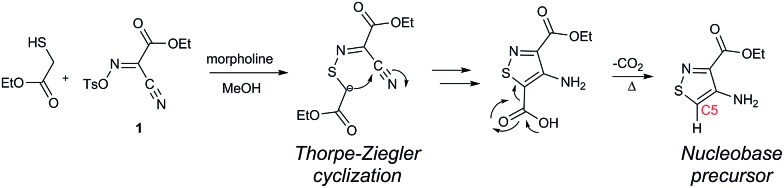
A priori, several approaches to forming our desired nucleosides may be employed (Fig. 2). While the Friedel–Crafts type glycosylation is classically the most common reaction used for the construction of C–C glycoside bonds, in some instances palladium may be used to catalyze a Heck-type coupling between nucleobases and cyclic enol ethers in good yield and high diastereomeric ratios.20 Lithiated heterocycles may also be used as nucleophiles with lactone- or aldehyde-derived carbohydrate scaffolds as electrophiles.5b,21 To extensively test the reactivity of the isothiazole's C5 in such reactions, a number of derivatives were first prepared (Scheme 1). The functionalized isothiazole core was synthesized using known methodology taking advantage of a modified Thorpe–Ziegler cyclization (Fig. 3).22
Fig. 2. The synthetic family tree of an isothiazolo alphabet and potential synthetic routes.
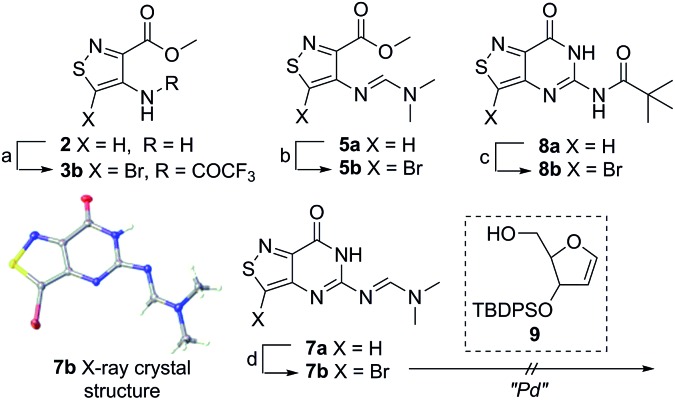
Scheme 1. Isothiazolo functional library. Conditions: (a) trifluoroacetic anhydride, Py, 0 °C to rt, 90 min, 72% yield; (b) KOCN, H2O, acetic acid, rt, 12 h, 70% yield; (c) dimethylformamide dimethyl acetal, DMF, rt, 12 h, 45% yield; (d) chloroformamidine hydrochloride, dimethylsulfone, 125 °C, 1 h, 74% yield; (e) dimethylformamide dimethyl acetal, DMF, rt, 12 h, 68% yield; (f) piv-Cl, Py, rt to 70 °C, 1 h, 61% yield.
Fig. 3. Thorpe–Ziegler cyclization.
Glycosylation attempts
An N-tosyl derivative of ethyl 2-cyano-2-(hydroxyimino)acetate (1, commonly known as Oxyma) was cyclized with ethyl thioacetate, yielding the basic isothiazole scaffold (Fig. 3). This pathway provided a derivative furnished with α-amino ester functionality (2), which can be used to construct the fused, functionalized pyrimidine ring. After decarboxylation, the heterocycle was furnished with varying functionalities, including a formamide, dimethyl imine, trifluoroacetamide, and guanidinylated species to provide diverse candidates for exploring glycosylation reactions. Starting with the Friedel–Crafts approach, the starting materials 3a–8a were treated with SnCl4 or BF3·OEt2 and an O-acetylated ribofuranose as a coupling partner and found to be unreactive.24 Given these results and the absence of any detectable glycosylation products, other viable routes were explored.
We attempted to make use of palladium catalysis as an alternate approach to form the desired bond. Heck coupling of heterocycles to form C-glycosides, while frequently useful, is reported to be inconsistent and highly varied depending on the specific heterocycle.20 To prepare suitable substrates, a variety of halogenated precursors were synthesized (Scheme 2). In an attempt to make the isothiazole C5-halogenated derivative more susceptible to cross-coupling reactions, the exocyclic amine was protected as a triflouroacetamide. The silyl-protected furanose glycal (9) was used as a coupling partner (see Table S1† for conditions).25 Unreacted starting materials were predominately recovered when subjecting substrates 3b, 7b and 8b to various palladium catalysts, including Pd(OAc)2, Pd(PPh3)4, Pd(Ph3P)2Cl2, and (Pd)2(dba)3.
Scheme 2. Bromination of derivatives. Reagents and conditions: (a) Br2, AcOH, 55 °C, 1 h, 73% yield; (b) Br2, AcOH, 50 °C, 2 h, 41% yield; (c) (i) Br2, AcOH, 50 °C, 1 h, (ii) TFAA, Py, 0 °C to rt, 9 h, 60% overall; (d) Br2, AcOH, 50 °C, 2 h, 63% yield.23 .
In an effort to test a reversal of the substrate roles in our palladium coupling substrates, the non-halogenated nucleobase 7a was tested and found to successfully couple with iodobenzene (see Scheme S1†). The success of this latter coupling with the failures of previous coupling reactions suggests that, perhaps, the primary challenge of the reaction of 3b, 7b and 8b lies in the oxidative addition step. While the “polarity-reversed” coupling of 7a was promising, we found that syntheses of a vinylogous halogenated ribose derivative capable of cross-coupling with our substrate would not be viable. With palladium catalysis not feasible for developing a scalable route to our nucleosides, chemically harsher methods were considered for activating the isothiazole C-5 for alkylation.
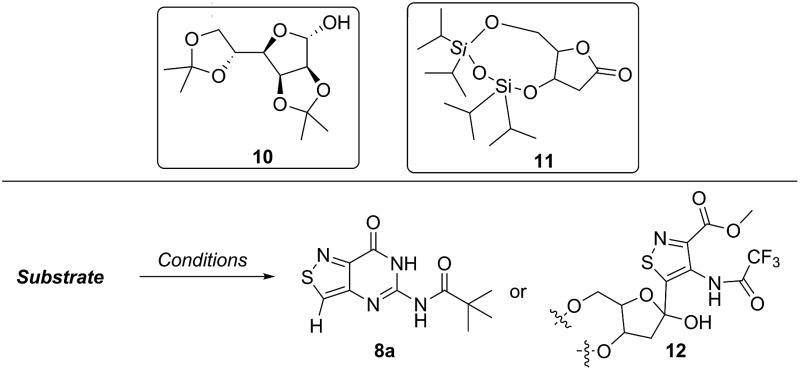
Lithiated heterocycles may be used to alkylate ribose or glucose derivatives, commonly functionalized as a lactone but occasionally as a lactol or an aldehyde.5b Using 3b and 8b, lithium halogen exchange reactions were tested (Table 1). Upon treatment of a pivaloyl-protected substrate (8b) with n-butyllithium, lithium halogen exchange was evident from the debromination of the starting material after quenching with water. Reaction of the piv-protected material with n-, sec-, and tert-butyllithium also returned dehalogenated material upon attempted alkylation of ketal-protected ribofuranose (10) or silylated deoxy-ribonolactone (11). Attempts to enhance the reactivity of the lithiated substrate with TMEDA failed. Treatment of the trifluoroacetylated compound (3b) lead to a trace amount of 12 that was not cleanly isolable with primarily decomposition of the starting material. Attempts to subject 7b to the same conditions showed reactivity of the substrate but no desired compound. The failure of these common methods thus necessitated taking a less traditional bottom-up approach to the construction of the desired nucleosides.
Table 1. Lithiated heterocycle alkylation attempts.
| ||
| Substrate | Conditions a | Product |
| 8b | a, b, c, d, e | 8a |
| 3b | b, e | 12 (trace) |
aConditions: (a) n-BuLi, THF, –78 °C to rt; (b) 10 or 11, n-BuLi, THF, –78 °C to rt; (c) 10 or 11, n-BuLi, TMEDA, THF, –78 °C to rt; (d) 10 or 11, s-BuLi, THF, –78 °C to 0 °C; (e) 10 or 11, t-BuLi, THF, –78 °C to –40 °C.
Thorpe–Ziegler cyclization approach
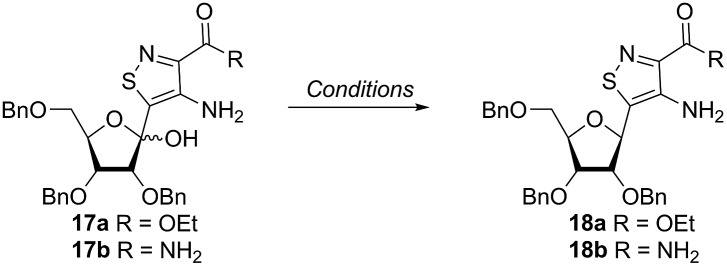
Inspired by previous syntheses of modified C-nucleosides, a different strategy was ultimately pursued to construct the isothiazolo-based glycoside.26 Returning to the Thorpe–Ziegler cyclization used to construct the isothiazole nucleobase precursor 2, we found that the originally published method for preparing isothiazoles could also apply to an α-thio ketone (as opposed to the thioglycolate, Fig. 3).22 We surmised that using a ribofuranose-derived lactol prefunctionalized with a primary thiol would likely suffice as a substrate for the proposed cyclization. Thiol 15 would therefore be necessary to act as the functional handle for installing the isothiazole ring (Scheme 3). Starting from the known benzyl-protected ribonolactone (13), the lactone was subjected to alkylation via lithium–halogen exchange with diiodomethane and furnished with a thiol via substitution and reduction of a thioester.27,28 The cyclization was then accomplished in good yield to provide derivatives with amide (17a) and ester (17b) functionalities, thus providing access to the desired scaffolds necessary for constructing the fused pyrimidine systems of the desired nucleosides.9
Scheme 3. Thiol construction and cyclization. Conditions: (a) CH2I2, MeLi, toluene, –78 °C, 1 h, 67%; (b) potassium thioacetate, DMF, rt, 6 h, 74%; (c) Et2O, LiAlH4, 0 °C to rt, 1 h, >90%; (d) 16a or 16b, morpholine, MeOH, 0 °C to rt, 12 h, 67% (17a), 79% (17b).
From the cyclized substrates 17a and 17b, stereoselective reduction of the furanosyl lactol was required to provide β nucleosides. While furanosyl oxocarbenium-based reductions reportedly show a higher preference for the formation of the β isomer in the case of ribose, several exceptions have been reported to provide primarily the α configuration.27,29 The cyclized substrates (17a, 17b) were subjected to Lewis-acid mediated reduction with triethylsilane (Scheme 4). All conditions cleanly gave the desired diastereomer, confirmed via crystal structure analyses of the final products. Note that both borohydride reductions followed by Mitsunobu conditions gave similar results (Scheme 4 conditions (c) and (d)).
Scheme 4. Reduction of lactol. Conditions: (a) 17a, BF3·OEt2, triethylsilane, DCM, –78 °C to rt, 4 h, 67% yield; (b) 17b, BF3·OEt2, triethylsilane, DCM, –78 °C to rt, 4 h, 79% yield. (c) 17a, (i) NaBH4, MeOH, 0 °C, 4 h, (ii) DIAD, PPh3, DCM, 0 °C to rt, 12 h, 30% over 2 steps; (d) (i) 17a, l-selectride, MeOH, 0 °C, 4 h, (ii) DIAD, PPh3, DCM, 0 °C to rt, 12 h, 40% over 2 steps.
Isothiazolo-pyrimidine functional diversification
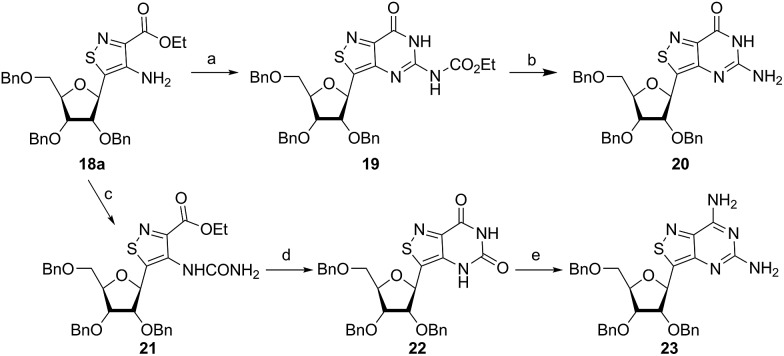
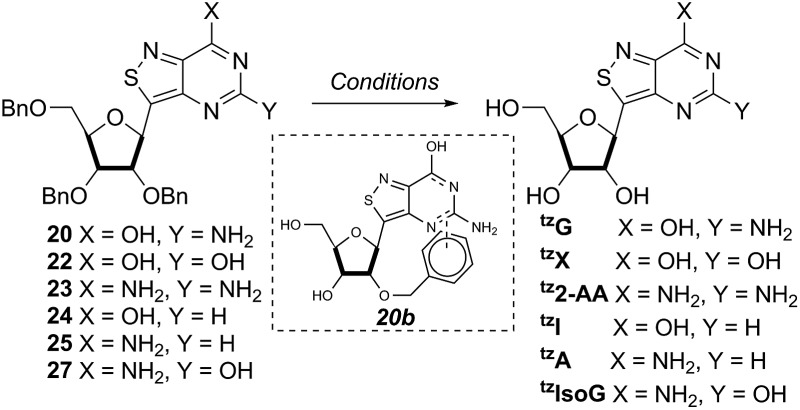
With the key substrate 18a in hand, construction of the fused pyrimidine was implemented via guanidinylation to furnish the tzG analog as well as the non-canonical tzisoG, tz2-AA and tzX. Typical methods used for guanidinylation of heterocycles to form fused pyrimidine rings often require multistep reactions involving harsh conditions including strong bases or excessive heat. Initially, 1,2,4-triazole-1-carboxamidine and chloroformamidine were used in an attempt to guanidinylate 18a and fabricate the guanosine analogue 20.30 Both reagents led to decomposed or deglycosylated products. A mild method for guanidinylation of our isothiazole species was therefore sought. Employing ethoxycarbonyl isothiocyanate and EDC in a mild 1-pot, 2 step synthesis, as reported by Fabis et al., gave the desired product, starting from the ester-substituted substrate 18a (Scheme 5).31 The same method, used with the amide 18b, yielded the isoguanosine analogue 26. The diamino purine analogue (23) was also found to be formed in low (<5%) yield but could not be cleanly isolated. The xanthosine derivative (22) was synthesized through the urea intermediate (21) upon treatment with sodium isocyanate and subsequent ring closure with sodium methoxide. To synthesize the 2-aminoadenosine derivative in greater yield than the previously discussed reaction, a more straightforward approach was used. The protected tzX (22) was treated with POCl3 followed by ammonia in methanol to give the protected tz2-AA (23) in moderate yield (Scheme 5).
Scheme 5. Protected tzG (20), tzX (22), tz2-AA (23) synthesis. Conditions: (a) (i) ethoxycarbonyl isothiocyanate, MeCN, rt, 6 h; (ii) EDCl, HMDS, rt, 24 h, 63% over 2 steps; (b) 1 M NaOH, MeOH, 80 °C, 1 h; (c) KOCN, H2O, acetic acid, rt, 12 h, 79%; (d) NaOMe, MeOH, rt, 12 h, 89%; (e) (i) POCl3, pyridine, 100 °C, 2 h; (ii) NH3, MeOH, 80 °C, 24 h, 40% over 2 steps.
Using the amide precursor 18b, the synthesis of the adenosine analogue tzA was completed through the inosine analogue (24) using triethylorthoformate and acetic anhydride. Conversion of the amide oxygen to an amine proceeded relatively smoothly, albeit with low yields, through the use of P2S5 and ammonia (Scheme 6).
Scheme 6. Synthesis of protected tzA (25) and tzisoG (27). Conditions: (a) triethyl orthoformate, acetic anhydride, 100 °C, 69% yield; (b) (i) P2S5, Py, 120 °C, (ii) NH3, MeOH, 80 °C, 80% crude yield; (c) ethoxycarbonyl isothiocyanate, EDC, HMDS, MeCN, rt, 72 h, 40% yield; (d) 1 M NaOH, MeOH, 80 °C, 1 h, 90% crude yield.
Benzyl ether removal
Debenzylation of the substrates was initially attempted with the protected tzG derivative 20 via palladium-catalyzed hydrogenolysis with either H2/Pd or transfer hydrogenation using ammonium formate (Table 2). With palladium hydroxide, only double deprotection (monitored by ESI-MS) was observed with the third benzyl group, presumably on the 2′ oxygen, left intact. This could perhaps be due to a combination of catalyst poisoning and reduced electron density of the last benzyl group arising from stacking with the adenosine ring as has previously been proposed (and depicted in 20b, Table 2).32 In a reported synthesis of fused thiophene-containing nucleoside analogues, Seley et al. used BF3-etherate and ethanethiol to remove benzyl ethers.33 When tried with tzG, we found that, interestingly, ethanethiol and BF3-etherate yielded very little deprotected substrate. Upon replacing ethanethiol with 1,2-ethanedithiol complete debenzylation of most substrates was found to occur within 48 hours.34
Table 2. Nucleoside deprotection conditions.
| |||
| Substrate | Cond. a | Time (h) | Product |
| 20 | a | 72 | n.r. |
| 20 | b | 72 | Monobenzyl |
| 20 | c | 72 | Dibenzyl |
| 20 | d | 24 | n.r. |
| a 20, b 25, c 24, d 27, e 23, f 22 | e | a48, b40, c48, d72, e96, f72 | tzG (59%), tzA (45%), tzI (30%), tzIsoG (51%), tz2-AA (54%), tzX (68%) |
aConditions: (a) Pd/C, H2, MeOH, rt, 90 psi; (b) Pd/C, ammonium formate, 60 °C, MeOH, sealed container; (c) Pd(OH)2, H2, rt to 40 °C; (d) ethanethiol, BF3·OEt2, DCM, rt; (e) 1,2-ethanedithiol, BF3·OEt2, DCM, rt.
To summarize, each isothiazole purine analogue was successfully synthesized from the commonly derived benzyl-protected thiol-substituted ribofuranose derivative 15. While the most common methods for C-glycosylation of heterocycles were unsuccessful with our isothiazole-based nucleobases, the Thorpe–Ziegler cyclization approach proved to be effective in completing the preparation of the isothiazolo[3,4-d]pyrimidine family.
Crystal structures
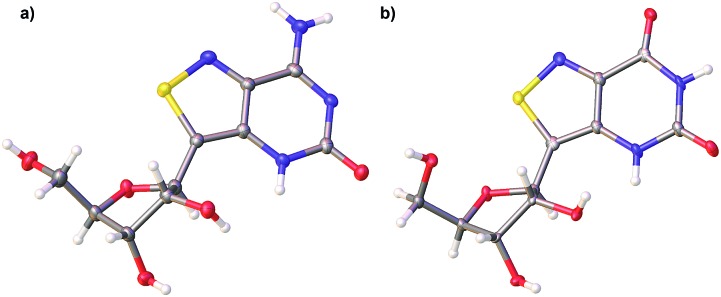
With all nucleosides in hand, crystal structure determination was necessary for final confirmation of connectivity and absolute configuration (Fig. 4). Originally, crystals of the adenosine and guanosine analogues were grown and found to all have the correct stereochemistry and functionality.9 The isothiazolo xanthosine crystals were found to exhibit two different sugar conformations in a unit cell. One conformer was found to closely resemble the native xanthosine when overlaid with the published crystal structure (see ESI Fig. S1†).35 Note that while the crystal structure of the isothiazolo 2-aminoadenosine was not determined (due to the poor quality of the crystals obtained), we are confident in the formation of the correct product as reinforced by the transformation to isothiazolo guanosine during deamination with adenosine deaminase discussed below.
Fig. 4. X-ray crystal structures of isothiazolo[3,4-d]pyrimidine analogs: (a) tzisoG (b) tzX.
Photophysical properties
The photophysical features of the newly synthesized noncanonical purine analogues were thoroughly analyzed and compared to tzA, tzG, and tzI. In investigating emission and absorption changes relative to pH and sensitivity towards changes in polarity, we have identified certain trends, which depend on the nucleosides' heteroatomic substitution.
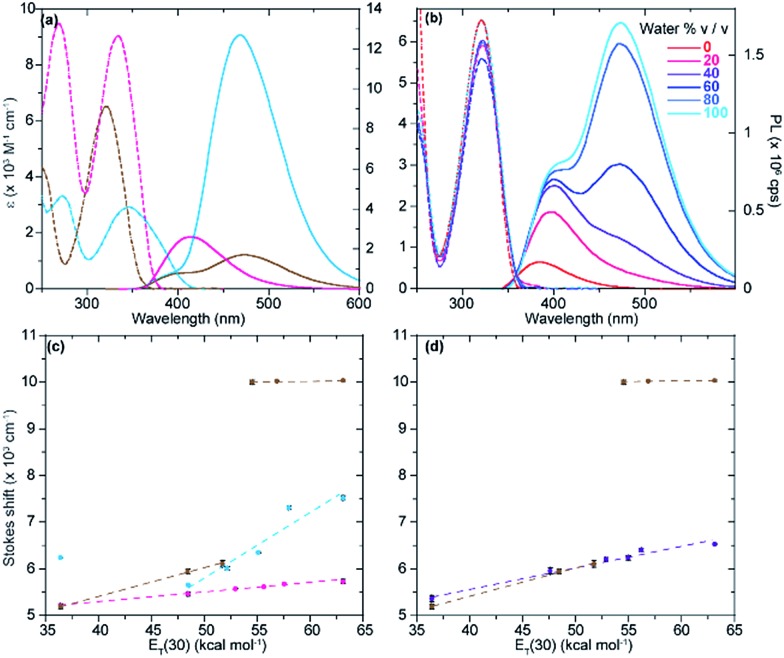
The absorption spectra of tzisoG, tzDAP, and tzX in aqueous solution have shown bathochromic-shifted maxima in comparison to the corresponding native nucleosides ranging from 321 nm for tzX to 346 nm for tz2-AA, the most red-shifted ground-state absorption spectra in the isothiazolo[4,3-d] pyrimidine-based nucleoside family (Fig. 5).
Fig. 5. (a) Absorption (dashed lines) and emission (solid lines) spectra of tzisoG (magenta), tz2-AA (cyan) and tzX (brown) in water. (b) Absorption (dashed lines) and emission (solid lines) traces in water, dioxane and mixture thereof for tzX. (c) Stokes shift correlation versus solvent polarity (E T(30)) of water/dioxane mixtures for tzisoG (magenta), tz2-AA (cyan) and tzX (brown). (d) Stokes shift correlation versus solvent polarity (E T(30)) of water/dioxane mixtures for tzX (brown) in comparison to the structural isomer tzU (purple).
Sensitivity towards environmental polarity was determined by recording the absorption and the emission spectra of tzisoG, tzX and tz2-AA in water, dioxane and binary mixtures of the two (Table 3, Fig. 5 and S6a–c†).36 It is reported in cm–1 (kcal mol–1)–1 and is equal to the slope of the linear fit shown in Fig. 5c, correlating the Stokes shifts and E T(30),37 a microscopic solvent polarity parameter. The value obtained for tzisoG (20 ± 1) is very similar to that of tzA (28 ± 2) and five times lower than tzG (102 ± 7) (Fig. S6d†). The importance of the N2 exocyclic amine in conferring high sensitivity to environmental changes is evident by the remarkable polarity sensitivity of tz2-AA (140 ± 2), where the exocyclic amine in position 2 is added to the tzA skeleton, enhancing the sensitivity to environmental polarity by a factor of five (Fig. S6f†).
Table 3. Photophysical properties of isothiazolo[4,3-d]pyrimidine nucleoside analogs.
| Solvent | λ abs (ε) a | λ em (Φ) a | Φε | Stokes shift a | Polarity sensitivity b | pK a(abs) c | pK a(em) c | |
| tzisoG | Water | 334 (9.14) | 413 (0.05) | 365 | 5.73 | 20 | 3.4, 11.5 | 3.90, 11.20 |
| Dioxane | 342 (8.6) | 416 (0.03) | 345 | 5.21 | ||||
| tzX | Water | 321 (6.61) | 472 (0.04) | 264 | 10.03 | 61, 3.0 | 9.03 | 1.98, 8.5 |
| Dioxane | 320 (6.60) | 384 (<0.01) | 13 | 5.10 | ||||
| tz2-AA | Water | 346 (2.85) | 447 (0.27) | 712 | 7.52 | 140 | 6.91 | 2.55, 5.93 |
| Dioxane | 349 (3.08) | 468 (0.22) | 616 | 6.24 | ||||
| tzA d | Water | 338 (7.79) | 410 (0.05) | 413 | 5.23 | 28 | 4.25 | 3.29 |
| Dioxane | 342 (7.42) | 409 (0.03) | 193 | 4.76 | ||||
| tzC d | Water | 325 (5.4) | 411 (0.05) | 289 | 6.42 | 10 | 2.9 | 2.46, 10.38 |
| Dioxane | 333 (5.03) | 419 (0.04) | 181 | 6.14 | ||||
| tzG d | Water | 333 (4.87) | 459 (0.25) | 1203 | 8.27 | 102 | 3.55, 8.51 | 9.88 |
| Dioxane | 339 (4.65) | 425 (0.17) | 539 | 6.01 | ||||
| tzU d | Water | 312 (5.17) | 392 (0.01) | 41 | 6.53 | 45 | 2.2, 8.88 | 8.9 |
| Dioxane | 314 (5.2) | 377 (<0.01) | 21 | 5.36 | ||||
| tzI d | Water | 316 (7.6) | 377 (0.01) | 46 | 5.13 | 12.7 | 9.26 | 7.83 |
| Dioxane | 315 (6.6) | 372 (<0.01) | 26 | 4.79 |
a λ abs, ε, λ em and Stokes shift are reported in nm, 103 M–1 cm–1, nm and 103 cm–1 respectively. All photophysical values reflect the average of at least three independent measurements.
bSensitivity to solvent polarity reported in cm–1 (kcal mol–1)–1 is equal to the slope of the linear fit in Fig. 6b.
cpK a values reflect the average over three independent measurements and are equal to the inflection point determined by the fitting curves in Fig. 6c and d.
dFrom previous work.9 See ESI Table S4 for expanded data including experimental errors.
The polarity sensitivity of tzX (61 ± 2) was similar to that of the previously reported N-glycosylated isomer, tzU (45 ± 4), in low water-content binary mixtures (up to 40% v/v water in dioxane).9 We noted the presence of a double emission band, perhaps a result by water-mediated tautomerization in the excited state.38 While the ground state absorption spectra did not show any change in the different binary solvent mixtures, the intensity ratio of the two emission maxima (PL472/PL384) changed from 0.07 in dioxane to 2.84 in pure water due to the lower-energy band intensity enhancement in water-enriched mixtures (Fig. S6b†).
Previously, the quantum yields of the purine analogues tzA and tzG were found to be 0.05 and 0.25, respectively. We found a striking reversal in quantum yield values, when comparing tzG to tzisoG (0.047 ± 0.004) and tzA to tz2-AA (0.27 ± 0.03). Again, the changes seemingly occur due to the presence of the amino group at C2. A five-fold enhancement of quantum yield of tz2-AA compared to tzA is noteworthy and comparable to the reported quantum yield of the native purine-based 2-aminoadenosine riboside, in comparison to that of native adenosine.39 Literature reports suggest that, in the case of the dramatic enhancement of fluorescence quantum yield seen for 2-aminopurine compared to adenine, the exocyclic N2 may shape the excited state manifold such that a barrier exists to avoid conical intersections, thus preventing ultrafast nonradiative decay.40 We can only speculate that in the isothiazole series, tz2-AA exhibits similar electronic trends arising from the presence of the amino group at position 2. In contrast, the xanthosine analogue tzX is quenched in dioxane while the quantum yield in water is significantly higher (0.043 ± 0.003), especially in comparison to the N-glycosylated tzU (0.008 ± 0.001).
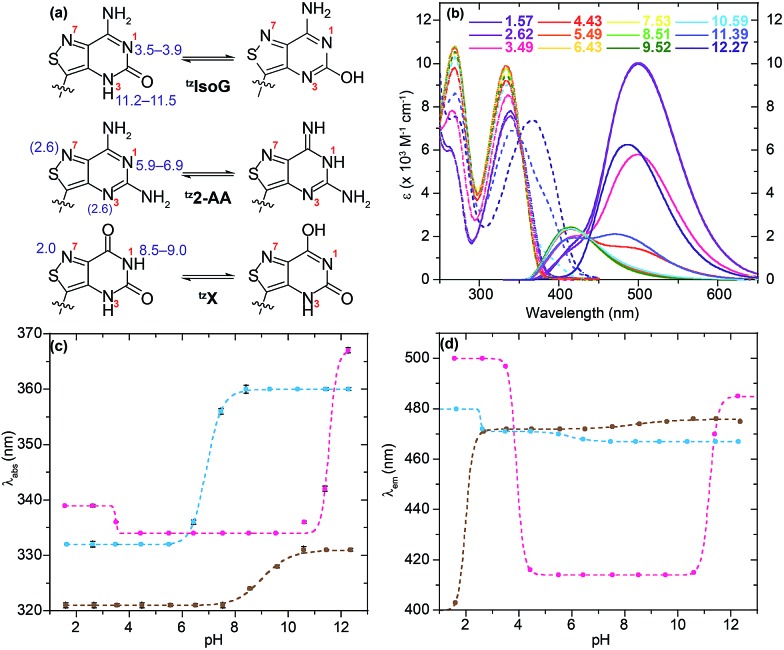
All isothiazolo-based nucleoside analogues reveal a remarkable sensitivity to pH, facilitating the extraction of pK a values (Fig. 6c and d). The absorption spectra of tzisoG displayed small hypso- and hyperchromic effects when the pH was elevated from 1.5 to 6.4, while the emission spectra showed an isosbestic point at 432 nm, suggesting deprotonation of the N1 [pK a 3.4 ± 0.1 (abs) to 3.90 ± 0.01 (em)] (Fig. 6c and d, magenta line). Another deprotonation event, assigned to the heterocyclic nitrogen at position 3, was depicted by 30 nm red-shifted ground state spectra and isosbestic points at 350 nm in absorption and 438 nm in the emission spectra within the pH range of 9.5 to 12.2. This yields a pK a value range of 11.20 ± 0.01 (em) to 11.5 ± 0.1 (abs), similar to the values of the native isoguanine nucleobase and ribonucleoside (pK a 3.8–4.0 and 11.0–11.1).38,41
Fig. 6. (a) Schematic representation of possible isothiazolo-pyrimidine nucleoside tautomers with labelled nitrogen atoms (red) and suggested pK a values (blue) corresponding to multiple possible protonation/deprotonation sites (parenthesis). (b) Absorption (dashed lines) and emission (solid lines) spectra of tzX as a function of water solution pH. (c) Absorption maxima and (d) emission maxima variation versus pH for tzisoG (magenta), tz2-AA (cyan) and tzX (brown).
Absorption spectra of tz2-AA showed a bathochromic shift from 332 to 360 nm with a well-defined isosbestic point at 346 nm upon deprotonation of N1 (pK a 6.91 ± 0.01) (Fig. 6c, cyan line) moving from mildly acidic to basic pH conditions (pH 4.4–8.4). The same pH dependent equilibrium was detected by a shift of the emission spectra to 467 nm and the presence of an isosbestic point at 410 nm, providing a pK a of 5.93 ± 0.02. This value corresponds well to the pK a value reported for 2-aminoadenosine (pK a 5.4–5.9).39 In addition, the emission spectra showed a remarkable bathochromic shift at lower pHs, which might be associated with either an additional protonation event involving N3 or N7 (pK a 2.55) or with other processes (e.g., tautomerization) in the excited state.
With regard to the pH sensitivity of tzX, the presence of isosbestic points at 334 and 437 in the absorption and emission-based pH titration, respectively, were suggestive of N1 deprotonation (pK a 9.03 ± 0.05, 8.5 ± 0.2) (Fig. 6c and d, brown line). These values compared well to those of the native xanthine (pK a 9.6).38 The emission spectra of tzX depicted a remarkable 70 nm red-shifted fluorescence maxima at lower pHs, which might indicate a protonation/deprotonation of N7 (pK a 1.98 ± 0.01).
For each of the noncanonical nucleoside analogues, the emission spectra displayed shoulder peaks at certain pHs. We speculate that pH-dependent excited state processes may yield distinct emissive states. To shed light on the origin of these peaks, excitation and emission spectra were recorded at varying wavelengths.
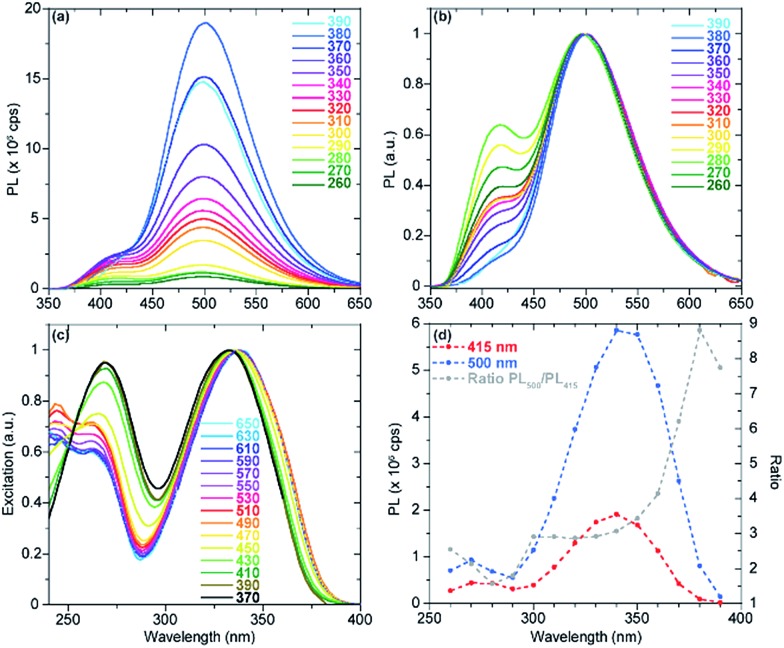
In the case of tzisoG, the acid–base equilibria were further investigated by recording emission spectra at pH 3.49, 4.43 and 11.39, upon excitation at different wavelengths. This analysis confirmed that the overall fluorescence signals (Fig. 7 and S3a–f†) were a linear combination of the emission spectra of two distinct species (Fig. 7d and S3g–l†). We speculate that the second observed species in the excited state may be a tautomer of tzIsoG (Fig. 6a), proposed to be a favorable one based on previous studies of isoguanosine.38,41
Fig. 7. (a) tzisoG emission spectra at pH 3.49, recorded upon excitation at different wavelengths and normalized for the corresponding absorbance intensity. (b) tzisoG emission spectra at pH 3.49, recorded upon excitation at different wavelengths and normalized to unit in intensity. (c) Excitation spectra normalized to unit recorded at selected emission spectra wavelengths, covering the whole emission band of aqueous solutions of tzisoG at pH 3.49. (d) Reconstructed excitation spectra of tzisoG at pH 3.49, plotting the emission intensities at the relative maxima 415 (red) and 500 (blue) nm upon excitation at different wavelengths. The grey line represents the relative ratio of the emission intensities upon excitation at different wavelengths.
With regard to tz2-AA, the emission spectra recorded at pH 1.63 upon excitation at different wavelengths (Fig. S4a and d†), were characterized by a shoulder and maxima at 395 and 480 nm, respectively. The excitation spectra did not display significant variations (Fig. S4g†), nevertheless suggesting the presence of two species with very close absorption maxima, relatively shifted to one another by 1–1.5 nm (the limit of our instrument). A similar investigation confirmed the presence of two different species at pH 6.42, near the pK a value (Fig. S4b, e, h and k†) and no further detectable deprotonation in basic media, pH 11.43 (Fig. S4c, f and i†). While the existence of a second species arising near the pK a may suggest a second tautomer existing in the excited state (Fig. 6a), we note the proposed structure is only speculated and inspired by previous computational studies of 2,6-diaminopurine.42
Lastly, comparing the emission spectra of tzX at pH 1.57 and 6.49 upon excitation at different wavelengths shows an inversion of the two emission bands over five pH units (Fig. S4a, b and d, e†). In acidic media, the higher energy band at 403 nm was followed by a shoulder at 472 nm, while at neutral pH the lower energy band was predominant. We speculate that this inversion in relative band intensity, without clear differences in the excitation spectra (Fig. S4g and h†), could likely be due to different populations of two excited state tautomeric forms of tzX, shown in Fig. 6a, at different pHs. The same analysis, carried out at pH 11.39 (Fig. S5c, f, i and l†) clearly showed the contribution of two different species to the absorption and emission spectra due to partial deprotonation of tzX around the pK a value.
In summary, each noncanonical analogue displays unique photophysical properties, which are sensitive to changes in pH and polarity. Absorption and emission spectra have allowed us to obtain pK a values, which are generally close to those of the native counterparts. The quantum yield values associated with each analogue granted further insight into structural determinants that affect the overall fluorescence features of the nucleosides. In particular, the presence of the exocyclic amine on the pyrimidine ring of tzG and tz2-AA seems to be a key determinant in maintaining a high quantum yield of these purine surrogates.
Enzymatic interconversion
While we have previously demonstrated the potential utility of isomorphic purine analogues in enzymatic interconversions,8b,9 we sought to extend it to tz2-AA, our newly-synthesized noncanonical adenosine analogue. With tz2-AA possessing a significantly higher quantum yield relative to tzA, the non-canonical nucleoside, if accepted as an adenosine deaminase (ADA) substrate, may possess an advantage for inhibitor discovery assays. Furthermore, we also sought to showcase the interconversion between two different modified nucleosides, beyond that of tzA to tzI, by directly converting tz2-AA to tzG. Lastly, we wished to investigate the substrate selectivity for our isothiazolo-based analogues compared to the native adenosine in an effort to showcase the elevated isomorphicity of our synthetic analogues.
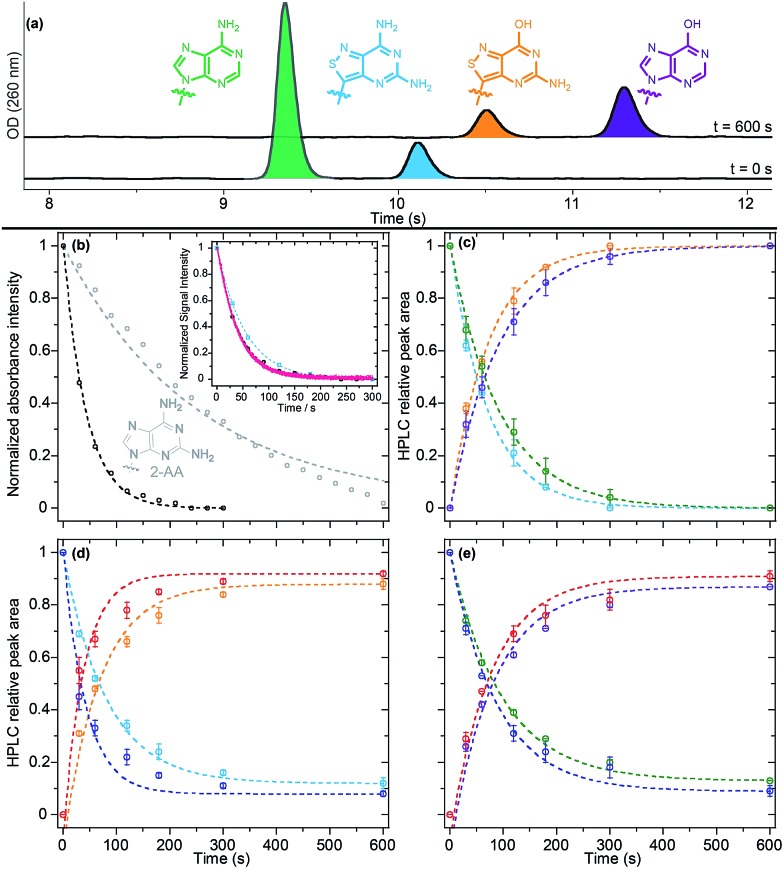
To evaluate the activity of tz2-AA with adenosine deaminase, we subjected our substrate to ADA under previously published conditions and followed the reaction via absorption, emission, and HPLC (Fig. 8a).9 The half-life of the deamination reaction was initially determined using steady state absorption and real-time emission spectroscopy for the conversion of tz2-AA into tzG (t 1/2 = 29 ± 1 and 28.4 ± 0.5 s for absorption and emission, respectively). Different from the native nucleosides (t 1/2 = 57 ± 4 and 207 ± 2 s for A and 2-aminoadenosine (2-AA) respectively, Table 4), the reaction half-life of tz2-AA was found to be similar to that of tzA.
Fig. 8. (a) Enzymatic competitive deamination of tz2-AA and native A to provide tzG and inosine as monitored by HPLC traces at T = 0 s and T = 600 s. (b) Enzymatic deamination of native 2-AA (grey) and tz2-AA (black) monitored by change in absorption spectra intensity at 290 and 368 nm respectively. Inset: ADA-mediated deamination of tz2-AA followed by absorption spectroscopy at 368 nm (black), real-time emission at 475 nm (pink) and HPLC relative peak area variation (cyan) at different time-points for tz2-AA. (c) Enzymatic competitive deamination of tz2-AA (cyan) and native A (green) to provide tzG (orange) and inosine (purple) monitored by HPLC relative peak area variation at different time-points. (d) Enzymatic competitive deamination of tz2-AA (cyan) and tzA (blue) to provide tzG (orange) and tzI (red) monitored by HPLC relative peak area variation at different time-points. (e) Enzymatic competitive deamination of tzA (blue) and native A (green) to provide tzI (red) and inosine (purple) monitored by HPLC relative peak area variation at different time-points.
Table 4. Enzyme deamination half-life.
| Substrate |
t
1/2 (s) |
HPLC-based conversion a (%) | ||
| UV-vis | PL | HPLC | ||
| A b | 57 | — | — | 100 |
| tzA b | 39 | 33 | — | 99 |
| 2-AA | 207 | — | — | 100 |
| tz2-AA | 29 | 28 | 33 | 100 |
| A/tzA | — | — | 66/54 | 87/94 |
| A/tz2-AA | — | — | 64/59 | 100/100 |
| tzA/tz2-AA | — | — | 30/56 | 92/88 |
aHPLC peak area quantification over a 600 s reaction time.
bFrom previous work.9
To gain insight into the selectivity of the enzyme, the deamination reaction was performed on binary combinations of A, tzA and tz2-AA (with a total substrate to enzyme ratio of 1.1 U μmol–1) under pseudo first order conditions. HPLC analyses at different time-points (Fig. 8b–d) evidenced that the enzymatic half-life of the adenosine conversion to inosine was slightly increased in the presence of tzA or tz2-AA (t 1/2 = 66 ± 1 and 64.3 ± 0.4 s, respectively, in the presence of each analogue) in comparison to the reaction in which adenosine was the only substrate (t 1/2 = 57 ± 4 s). On the other hand, both tzA and tz2-AA conversion kinetics were decreased two fold when the native adenosine was present (t 1/2 = 54.5 ± 0.7 and 58.8 ± 0.4 s, respectively, vs. t 1/2 = 33.2 ± 0.2 and 29 ± 1 s), suggesting a higher binding affinity for the native adenosine relative to the analogues. Interestingly, when mixing the two fluorescent analogs tzA and tz2-AA, we noticed that tzA had a similar effect as that of A on the reaction half-life of tz2-AA (t 1/2 = 56 ± 3 s) while maintaining a high rate of conversion to tzI (t 1/2 = 29.7 ± 0.9 s).
Finally, it is worth noting that tzI, as the deaminated product of tzA, seems to show a slight inhibitory effect, as is also reported for the corresponding native inosine.43 HPLC-based quantification of the enzymatic reaction after 600 seconds (Table 4 and Fig. 8) have shown that, different from the native nucleosides and tz2-AA, all competitive enzymatic deamination reactions in which tzA was present did not proceed to full conversion. As previously reported, the conversion to tzI was almost quantitative (99%) for tzA as the sole substrate for the enzyme. This potential slight inhibitory effect was more pronounced in the competitive assays of tzA in the presence of native A. In both cases, after 10 minutes, neither of the substrates were completely converted to the corresponding product.
Overall, when comparing the relative enzymatic deamination half-life of A, tzA, and tz2-AA and their corresponding binary isomolar mixtures, we surmise that, as single enzymatic substrates, tzA and tz2-AA are converted to their respective products faster than the native A. tzA was also converted faster than tz2-AA, likely due to the increased electronic perturbations caused by the added exocyclic amino group.
Enzyme-mediated deamination via ADA is suggested to proceed through an SNAr-like mechanism, via the addition of OH followed by NH3 elimination at C6. It has been suggested that modifications that decrease the aromatic character of the heterocycle and promote rehybridization may increase the reaction rate.8d,44 We propose that this indeed leads to the increase in deamination rates of tz2-AA and tzA (Fig. 8a), compared to A. We would also speculate that this reasoning applies for the difference in rates between tz2-AA and the native 2-AA riboside. Their diminished performance in competition reactions, however, likely reflects their lower affinity to the enzyme.
While ADA is a validated drug target, growing evidence suggest that misregulation of A to I editing in mRNA is associated with human disease, and that tRNA deamination and the enzymes involved may also impact human pathologies.45 The range of our emissive adenosine analogues (thA, tzA, tz2-AA) are all viable substrates for adenosine deaminases and thus can serve as substrates for the development of high throughput inhibitor discovery assays for such transformations. Beyond the advantages of fluorescence-based tools over HPLC, NH3 detection or absorption measurements, the range of emissive analogues with distinct deamination rates provides opportunities to fine tune such discovery assays.
Conclusions
In summary, we have completed an entire family of canonical and noncanonical isothiazolo[4,3-d]pyrimidine analogues, providing not only insight into synthetic intricacies of developing a unique structural scaffold, but also into its electronic features via the observed photophysical trends. Lastly, we have demonstrated the ability of our nucleosides to enzymatically interconvert to other emissive purine-based nucleoside analogues through deamination via adenosine deaminase with high rate and selectivity. We predict these nucleosides will behave as faithful fluorescent purine surrogates in other biologically relevant systems.
Acknowledgments
We thank the National Institutes of Health for generous support (GM 069773), the Chemistry & Biochemistry MS Facility, and the UCSD X-ray crystallography Facility.
Footnotes
References
- Jordheim L. P., Durantel D., Zoulim F., Dumontet C. Nat. Rev. Drug Discovery. 2013;12:447–464. doi: 10.1038/nrd4010. [DOI] [PubMed] [Google Scholar]
- Callis P. R. Annu. Rev. Phys. Chem. 1983;34:329–357. doi: 10.1146/annurev.physchem.48.1.271. [DOI] [PubMed] [Google Scholar]
- Favre A., Thomas G. Annu. Rev. Biophys. Bioeng. 1981;10:175–195. doi: 10.1146/annurev.bb.10.060181.001135. [DOI] [PubMed] [Google Scholar]
- (a) Sinkeldam R. W., Greco N. J., Tor Y. Chem. Rev. 2010;110:2579–2619. doi: 10.1021/cr900301e. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Wilhelmsson L. M. Q. Rev. Biophys. 2010;43:159–183. doi: 10.1017/S0033583510000090. [DOI] [PubMed] [Google Scholar]; (c) Wierzchowski J., Antosiewicz J. M., Shugar D. Mol. BioSyst. 2014;10:2756–2774. doi: 10.1039/c4mb00233d. [DOI] [PubMed] [Google Scholar]
- (a) Shaban M. A. E. Adv. Heterocycl. Chem. 1998;70:163–337. [Google Scholar]; (b) Stambasky J., Hocek M., Kocovsky P. Chem. Rev. 2009;109:6729–6764. doi: 10.1021/cr9002165. [DOI] [PubMed] [Google Scholar]
- Nir E., Kleinermanns K., Grace L., de Vries M. S. J. Phys. Chem. A. 2001;105:5106–5110. [Google Scholar]
- Shin D., Sinkeldam R. W., Tor Y. J. Am. Chem. Soc. 2011;133:14912–14915. doi: 10.1021/ja206095a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (a) Liu W., Shin D., Tor Y., Cooperman B. S. ACS Chem. Biol. 2013;8:2017–2023. doi: 10.1021/cb400256h. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Sinkeldam R. W., McCoy L. S., Shin D., Tor Y. Angew. Chem., Int. Ed. 2013;52:14026–14030. doi: 10.1002/anie.201307064. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) McCoy L. S., Shin D., Tor Y. J. Am. Chem. Soc. 2014;136:15176–15184. doi: 10.1021/ja5039227. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Mizrahi R. A., Shin D., Sinkeldam R. W., Phelps K. J., Fin A., Tantillo D. J., Tor Y., Beal P. A. Angew. Chem., Int. Ed. 2015;54:8713–8716. doi: 10.1002/anie.201502070. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Sholokh M., Sharma R., Shin D., Das R., Zaporozhets O. A., Tor Y., Mely Y. J. Am. Chem. Soc. 2015;137:3185–3188. doi: 10.1021/ja513107r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovira A. R., Fin A., Tor Y. J. Am. Chem. Soc. 2015;137:14602–14605. doi: 10.1021/jacs.5b10420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (a) Huang R. M., Chen Y. N., Zeng Z. Y., Gao C. H., Su X. D., Peng Y. Mar. Drugs. 2014;12:5817–5838. doi: 10.3390/md12125817. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Mccloskey J. A., Nishimura S. Acc. Chem. Res. 1977;10:403–410. [Google Scholar]; (c) Hagervall T. G., Ericson J. U., Esberg K. B., Ji-nong L., Björk G. R. Biochim. Biophys. Acta, Gene Struct. Expression. 1990;1050:263–266. doi: 10.1016/0167-4781(90)90178-5. [DOI] [PubMed] [Google Scholar]
- Cheng Q. Y., Gu J. D., Compaan K. R., Schaefer H. F. Chem.–Eur. J. 2012;18:4877–4886. doi: 10.1002/chem.201102415. [DOI] [PubMed] [Google Scholar]
- Chollet A., Kawashima E. Nucleic Acids Res. 1988;16:305–317. doi: 10.1093/nar/16.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang B., McFaline J. L., Burgis N. E., Dong M., Taghizadeh K., Sullivan M. R., Elmquist C. E., Cunningham R. P., Dedon P. C. Proc. Natl. Acad. Sci. U. S. A. 2012;109:2319–2324. doi: 10.1073/pnas.1118455109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulikowska E., Kierdaszuk B., Shugar D. Acta Biochim. Pol. 2004;51:493–531. [PubMed] [Google Scholar]
- (a) Sholokh M., Improta R., Mori M., Sharma R., Kenfack C., Shin D., Voltz K., Stote R. H., Zaporozhets O. A., Botta M., Tor Y., Mely Y. Angew. Chem., Int. Ed. 2016;55:7974–7978. doi: 10.1002/anie.201601688. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Chawla M., Poater A., Oliva R., Cavallo L. Phys. Chem. Chem. Phys. 2016;18:18045–18053. doi: 10.1039/c6cp03268k. [DOI] [PubMed] [Google Scholar]; (c) Samanta P. K., Pati S. K. Phys. Chem. Chem. Phys. 2015;17:10053–10058. doi: 10.1039/c5cp00134j. [DOI] [PubMed] [Google Scholar]; (d) Liu H. X., Song Q. X., Yang Y., Li Y., Wang H. J. J. Mol. Model. 2014:20. doi: 10.1007/s00894-014-2100-4. [DOI] [PubMed] [Google Scholar]; (e) Samanta P. K., Manna A. K., Pati S. K. J. Phys. Chem. B. 2012;116:7618–7626. doi: 10.1021/jp301752k. [DOI] [PubMed] [Google Scholar]
- (a) Jones A. C., Neely R. K. Q. Rev. Biophys. 2015;48:244–279. doi: 10.1017/S0033583514000158. [DOI] [PubMed] [Google Scholar]; (b) Crespo-Hernandez C. E., Martinez-Fernandez L., Rauer C., Reichardt C., Mai S., Pollum M., Marquetand P., Gonzalez L., Corral I. J. Am. Chem. Soc. 2015;137:4368–4381. doi: 10.1021/ja512536c. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Middleton C. T., de La Harpe K., Su C., Law Y. K., Crespo-Hernandez C. E., Kohler B. Annu. Rev. Phys. Chem. 2009;60:217–239. doi: 10.1146/annurev.physchem.59.032607.093719. [DOI] [PubMed] [Google Scholar]; (d) Wierzchowski J. Nucleosides, Nucleotides Nucleic Acids. 2014;33:626–644. doi: 10.1080/15257770.2014.913065. [DOI] [PubMed] [Google Scholar]; (e) De Camillis S., Miles J., Alexander G., Ghafur O., Williams I. D., Townsend D., Greenwood J. B. Phys. Chem. Chem. Phys. 2015;17:23643–23650. doi: 10.1039/c5cp03806e. [DOI] [PubMed] [Google Scholar]
- Scifinder structure search shows near 369 thousand hits for structures containing an isothiazole ring compared to near 6 million hits for structures containing thiazole rings
- Vorbruggen H. and Ruh-Pohlenz C., in Organic Reactions, John Wiley & Sons, Inc., N. Y., 2000, vol. 55, pp. 1–630. [Google Scholar]
- (a) Bunch L., Krogsgaard-Larsen P., Madsen U. J. Org. Chem. 2002;67:2375–2377. doi: 10.1021/jo0162134. [DOI] [PubMed] [Google Scholar]; (b) Kaae B. H., Krogsgaard-Larsen P., Johansen T. N. J. Org. Chem. 2004;69:1401–1404. doi: 10.1021/jo035578g. [DOI] [PubMed] [Google Scholar]; (c) Christoforou I. C., Koutentis P. A. Org. Biomol. Chem. 2006;4:3681–3693. doi: 10.1039/b607442a. [DOI] [PubMed] [Google Scholar]; (d) Ioannidou H. A., Koutentis P. A. Org. Lett. 2011;13:1510–1513. doi: 10.1021/ol200196m. [DOI] [PubMed] [Google Scholar]
- Wellington K. W., Benner S. A. Nucleosides, Nucleotides Nucleic Acids. 2006;25:1309–1333. doi: 10.1080/15257770600917013. [DOI] [PubMed] [Google Scholar]
- Grimmett M. R., Iddon B. Heterocycles. 1995;41:1525–1574. [Google Scholar]
- Gewald K., Bellmann P. Liebigs Ann. Chem. 1979:1534–1546. [Google Scholar]
- A crystal structure of 7b was used to initially structurally elucidate the synthesized nucleobase, due to the lack of any remaining characteristic 1H's for NMR analysis
- Typical procedure: isothiazolo substrate (1.0 eq.) was placed in flame-dried round bottom flask and dissolved in acetonitrile (0.1 M final conc.) followed by addition of 1-O-acetyl-tri-benzoyl-β-d-ribofuranose (1.1 eq.) and corresponding Lewis acid (2.0 eq.). The reaction is left to stir at 65 °C
- Silver salts (e.g., AgF) have been previously reported to play a critical role in enhancing palladium-catalyzed arylations and have proven crucial in the palladium coupling of isothiazole heterocycles. In an effort to avoid silyl-incompatible fluoride salts, Ag2CO3 was used as a base with a variety of common palladium catalysts and ligands tested
- Wamhoff H., Berressem R., Nieger M. J. Org. Chem. 1994;59:1912–1917. [Google Scholar]
- van Rijssel E. R., van Delft P., Lodder G., Overkleeft H. S., van der Marel G. A., Filippov D. V., Codee J. D. Angew. Chem., Int. Ed. 2014;53:10381–10385. doi: 10.1002/anie.201405477. [DOI] [PubMed] [Google Scholar]
- Bessieres B., Morin C. Synlett. 2000:1691–1693. [Google Scholar]
- (a) Matulic-Adamic J., Beigelman L. Tetrahedron Lett. 1997;38:203–206. [Google Scholar]; (b) Matulic-Adamic J., Beigelman L. Tetrahedron Lett. 1997;38:1669–1672. [Google Scholar]
- Typical procedure: 18a (1 eq., 0.2 mmol) is placed in round bottom with stir bar. Guanidinylation reagent (HCl salt) is added followed by dimethylsulfone (40 eq., 8 mmol) and triethylamine (3 eq., 0.6 mmol) and brought to stir at 125 °C
- Lecoutey C., Fossey C., Rault S., Fabis F. Eur. J. Org. Chem. 2011;2011:2785–2788. [Google Scholar]
- (a) Christensen L. F., Broom A. D. J. Org. Chem. 1972;37:3398–3401. doi: 10.1021/jo00795a003. [DOI] [PubMed] [Google Scholar]; (b) Hotoda H., Takahashi M., Tanzawa K., Takahashi S., Kaneko M. Tetrahedron Lett. 1995;36:5037–5040. [Google Scholar]
- Seley K. L., Zhang L., Hagos A., Quirk S. J. Org. Chem. 2002;67:3365–3373. doi: 10.1021/jo0255476. [DOI] [PubMed] [Google Scholar]
- Preliminary results suggest that deprotection of protected tzIsoG 26a leads to thioketalization of the amide oxygen with ethanedithiol
- Lesyng B., Marck C., Saenger W. Z. Naturforsch., C: Biochem., Biophys., Biol., Virol. 1984;39:720–724. [Google Scholar]
- The polarity sensitivity parameter for each nucleoside was extrapolated by plotting the calculated Stokes shift versus solvent polarity values E T(30) and applying a linear fit (Fig. 5c)
- (a) Reichardt C. Chem. Rev. 1994;94:2319–2358. [Google Scholar]; (b) Sinkeldam R. W., Tor Y. Org. Biomol. Chem. 2007;5:2523–2528. doi: 10.1039/b707719j. [DOI] [PubMed] [Google Scholar]
- Rogstad K. N., Jang Y. H., Sowers L. C., Goddard W. A. Chem. Res. Toxicol. 2003;16:1455–1462. doi: 10.1021/tx034068e. [DOI] [PubMed] [Google Scholar]
- Ward D. C., Reich E., Stryer L. J. Biol. Chem. 1969;244:1228–1237. [PubMed] [Google Scholar]
- Serrano-Andres L., Merchan M., Borin A. C. Proc. Natl. Acad. Sci. U. S. A. 2006;103:8691–8696. doi: 10.1073/pnas.0602991103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (a) Shugar D., Kierdaszuk B. J. Biosci. 1985;8:657–668. [Google Scholar]; (b) Seela F., Wei C., Kazimierczuk Z. Helv. Chim. Acta. 1995;78:1843–1854. [Google Scholar]
- Gengeliczki Z., Callahan M. P., Svadlenak N., Pongor C. I., Sztaray B., Meerts L., Nachtigallova D., Hobza P., Barbatti M., Lischka H., de Vries M. S. Phys. Chem. Chem. Phys. 2010;12:5375–5388. doi: 10.1039/b917852j. [DOI] [PubMed] [Google Scholar]
- Alunni S., Orru M., Ottavi L. J. Enzyme Inhib. Med. Chem. 2008;23:182–189. doi: 10.1080/14756360701475233. [DOI] [PubMed] [Google Scholar]
- (a) Véliz E. A., Easterwood L. M., Beal P. A. J. Am. Chem. Soc. 2003;125:10867–10876. doi: 10.1021/ja029742d. [DOI] [PubMed] [Google Scholar]; (b) Mizrahi R. A., Shin D. W., Sinkeldam R. W., Phelps K. J., Fin A., Tantillo D. J., Tor Y., Beal P. A. Angew. Chem., Int. Ed. 2015;54:8713–8716. doi: 10.1002/anie.201502070. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Pokharel S., Jayalath P., Maydanovych O., Goodman R. A., Wang S. C., Tantillo D. J., Beal P. A. J. Am. Chem. Soc. 2009;131:11882–11891. doi: 10.1021/ja9034076. [DOI] [PubMed] [Google Scholar]
- (a) Cortes A., Gracia E., Moreno E., Mallol J., Lluis C., Canela E. I., Casado V. Med. Res. Rev. 2015;35:85–125. doi: 10.1002/med.21324. [DOI] [PubMed] [Google Scholar]; (b) Morabito M. V., Ulbricht R. J., O'Neil R. T., Airey D. C., Lu P. C., Zhang B., Wang L., Emeson R. B. Mol. Pharmacol. 2010;77:895–902. doi: 10.1124/mol.109.061903. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Maas S., Kawahara Y., Tamburro K. M., Nishikura K. RNA Biol. 2006;3:1–9. doi: 10.4161/rna.3.1.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Torres A. G., Batlle E., de Pouplana L. R. Trends Mol. Med. 2014;20:306–314. doi: 10.1016/j.molmed.2014.01.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.