Abstract
Some natural product leads of drugs (NPLDs) have been found to congregate in the chemical space. The extent, detailed patterns, and mechanisms of this congregation phenomenon have not been fully investigated and their usefulness for NPLD discovery needs to be more extensively tested. In this work, we generated and evaluated the distribution patterns of 442 NPLDs of 749 pre-2013 approved and 263 clinical trial small molecule drugs in the chemical space represented by the molecular scaffold and fingerprint trees of 137,836 non-redundant natural products. In the molecular scaffold trees, 62.7% approved and 37.4% clinical trial NPLDs congregate in 62 drug-productive scaffolds/scaffold-branches. In the molecular fingerprint tree, 82.5% approved and 63.0% clinical trial NPLDs are clustered in 60 drug-productive clusters (DCs) partly due to their preferential binding to 45 privileged target-site classes. The distribution patterns of the NPLDs are distinguished from those of the bioactive natural products. 11.7% of the NPLDs in these DCs have remote-similarity relationship with the nearest NPLD in their own DC. The majority of the new NPLDs emerge from preexisting DCs. The usefulness of the derived knowledge for NPLD discovery was demonstrated by the recognition of the new NPLDs of 2013–2014 approved drugs.
The focus of drug discovery has moved from natural products (NPs) to technology-derived synthetic molecules for about 20 years1 without the anticipated drug productivity improvement2. Although being largely sidelined, NPs are still relevant3,4 with NP-related small molecule drugs representing 29.5% of the 132 FDA approved drugs in 2008–2012 (Supplementary Table S1), NP-related drugs include NPs and NP semi-synthetic derivatives, mimetics, and pharmacophore-guided synthetic molecules3. The NPs from which the NP-related drugs have been derived are named as the NP leads of drugs (NPLDs). There is a renewed interest in discovering drugs5 from NP privileged structures6 and derivative libraries7. The knowledge of the distribution of the NPLDs in the chemical space provides useful clues for prioritizing the relevant efforts.
Although NPs are in well-defined subspaces of the chemical space8, because of their enormous number9, structural diversity9,10 and molecular complexity11, only a fraction of NPs can be practically explored in the foreseeable future. Drug discovery efforts need to be prioritized towards the NPs with higher discovery potentials. The key questions are which NPs to explore and where to find them. Evidences suggest that some NPLDs may congregate in specific drug-productive regions of the chemical space. Certain NP chemical classes (e.g. steroids and nucleosides) are drug prolific12. Half of the drugs are made of dozens of molecular frameworks13. For many drugs, their individual molecular, physicochemical and topological pharmacophore properties14,15,16 and the corresponding principal components17,18,19,20 are constrained in specific ranges. Moreover, the GPCR, kinase and protease targeting agents have been reported to each cluster together in the chemical space1. These studies have consistently shown that a substantial percentage of the NPLDs congregate in the chemical space. However, the extent and the detailed distribution patterns of the congregation of NPLDs in the chemical space and the mechanisms leading to such patterns have not been fully investigated.
There is a need to study these questions from different structural and molecular binding perspectives to gain a deeper understanding of the structural characteristics of NPLDs and to find clues for guiding the search of new NPLDs. In this work, we determined the distribution patterns of the 348 and 94 NPLDs of 749 pre-2013 approved and 263 clinical trial small molecule drugs (Supplementary Tables S2, S3) in the chemical space represented by the molecular scaffold trees21 and the molecular fingerprint based hierarchical clustering tree22,23 of 137,836 non-redundant NPs3,24,25. The number of NPs profiled here is comparable to those of the earlier large-scale NP studies9,21,26. Molecular fingerprints were used for representing NPs in the hierarchical clustering tree because of its demonstrated effectiveness in structural similarity searching, and its extensive applications in drug lead discovery12,23,27,28,29,30.
The derived distribution patterns were studied from the perspective of preferential binding of NPLDs to the privileged target-sites for determining whether it contributes to the formation of these patterns. We also evaluated whether these patterns are distinguished from those of the bioactive NPs and how they evolve with time. We further tested whether the derived knowledge can be explored for NPLD discovery by applying it to retrospectively judge the development potential of the new NPLDs of 2013–2014 approved drugs uninvolved in the derivation of the NPLD distribution patterns. New technologies are expected to significantly expand the currently accessible NP chemical space31,32 and their potential impact is not reflected in this study.
Drug scaffolds have been well analyzed13 and drug distribution in the chemical space have been extensively studied from the perspectives of specific molecular and physicochemical properties1,14,15,16,17,18,19,20. To the best of our knowledge, our work is the first large-scale and systematic study of the detailed distribution patterns of the largest set of NPLDs in the chemical space from the perspectives of their molecular scaffolds and structures. The molecular scaffold analysis was intended for determining whether there is a significant change in the congregation patterns of the NPLDs in comparison to the previous studies13,33. The molecular structural analysis was intended for further probing the complex structural features of the NPLD congregation phenomenon and the underlying molecular mechanisms that might contribute to the clustering of NPLDs with particular focus on the possible influence of the binding of NPLDs and their derivatives to the privileged target sites.
Methods
We collected 442 NPLDs1,3,27,28,34,35 and the information about their NP origin1,28 from the literature. We also collected 169,037 NPs from the ZINC24, TCM-ID29, TCM@Taiwan25, and other literatures3. For database entries with multiple non-linked components, only the largest component was selected. Hydrogens were added and small fragments (counter ions, solvent molecules, etc.) were removed by using Corina, The number of NPs were reduced to 137,836 after removing the duplicate entries, small NPs with molecular weight <50 Daltons (drug leads are >100 Daltons30) and the NPs whose molecular fingerprints could not be computed by using available software tools such as PaDEL36. Duplicates were identified and removed by structural comparison based on a set of 98 molecular descriptors we have used for classifying bioactive molecules37 and implemented in the online server MODEL38, open-source software PaDEL36, and our own software, which can distinguish different molecules non-distinguishable by the 881-bit Pubchem molecular fingerprints.
In deriving the molecular scaffold trees of the 442 NPLDs and 137,836 NPs, Scaffold Hunter v2.3.021 was used to select the NPLDs and NPs with ring structures and to subsequently cluster them into molecular scaffold trees by using default rule set in the Scaffold Tree Generation window. The molecular fingerprint based hierarchical clustering tree of the 442 NPLDs and 137,836 NPs was generated by using the Matlab statistics toolbox with the structures of the NPs represented by 2D molecular fingerprints23 (specifically, the 881-bit PubChem substructure fingerprints computed by using PaDEL36) and with their similarity levels measured by the Tanimoto coefficient Tc22,23 and the complete linkage. Tc was used because it is the most popular similarity metric for molecular fingerprint based measurement of compound similarity23. Complete linkage was used because of its relatively good performance in clustering bioactive compounds in a recent comparative study39. The hierarchical tree graphs were generated by using EMBL automatic tree generator in iTOL version-1.8.140 with the distance of the NPs measured by the Tanimoto distance Td = 1-Tc. In analyzing the physicochemical landscapes of the NPs in specific regions of the chemical space, we used MODEL38 and Discovery studio 3.1.1 software to compute eight molecular descriptors frequently used for analyzing drug-like14,41,42,43 and lead-like43,44 features. These are molecular weight (MW), lipophilicity AlogP and logD, polarizability (PZ), and the number of O+N (ON), hydrogen bond donor (HD), hydrogen bond acceptor (HA), rotatable bond (RB), and rings (RI).
To determine whether the clustering of the NPLDs in specific sub-regions of the chemical space are statistically more significant than chance, our derived distribution patterns of the NPLDs with respect to those of the randomly shuffled NP communities were analyzed by the method used for determining the statistical significance of the phylogenetic clustering of traditional medicinal plants45. In this method, the mean Tanimoto distance MTd of the NPLDs in every NPLD-clustered sub-region was compared to the MTd values of these NPLDs in 60,000 randomly generated NP sub-regions. By using the algorithms implemented in the Phylocom: software46, a one-tailed P-value and a net relatedness index (NRI) were calculated for each sub-region. The P-value is the number of randomly selected NPs that are more clustered than the NPLDs in each sub-region divided by the number of runs (60,000 in this study). The desired significance level α of the P-value was further adjusted by Bonferroni correction to α′ = α/N (N is the number of independent statistical significance tests, which is 60 in this study)47. The NRI is a standardized effect size measure of the community structure in each sub-regions, which is the difference in average Td between the NPLDs and the 60,000 randomly generated NP sub-regions, and standardized by the standard deviation of the Td values in 60,000 randomly shuffled sets of NP communities. The sign of NRI informs whether the NPLDs are more clustered (NRI > 0) or more dispersed (NRI < 0) than the NPs in each sub-region. These quantities were calculated by using Phylocom v4.146 with the Td values of the NPLDs and NPs as input data.
Results and Discussion
Distribution profiles of NPLDs in the chemical space from the perspectives of molecular scaffolds and molecular structures
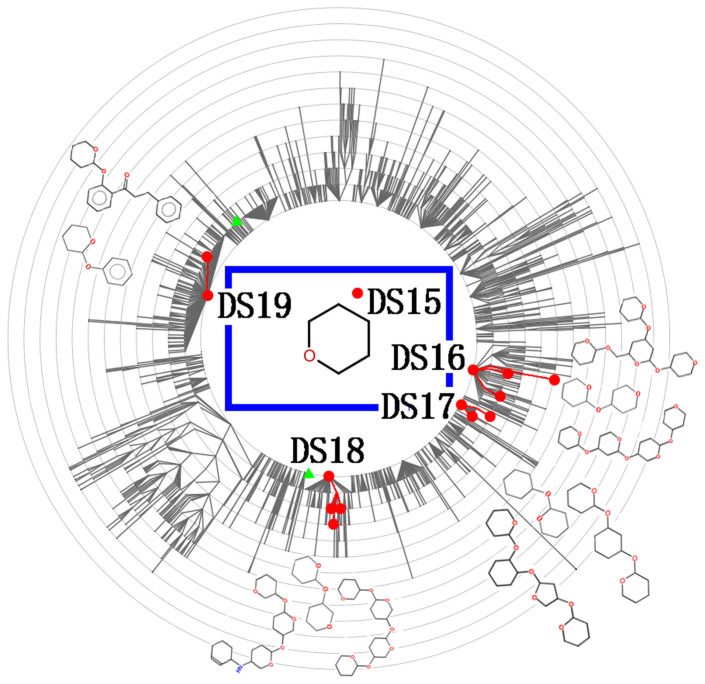
There are 411 NPLDs and 134,097 NPs with ring structures. These were grouped by Scaffold hunter21 into molecular scaffold trees of 39,051 scaffolds (114 are drug-productive). The distribution of the NPLD scaffolds in this large-scale analysis is similar to the previous findings13,33, the majority (62.7%) of the NPLDs of the approved drugs and a substantial percentage (37.4%) of the NPLDs of the clinical trial drugs congregate in 62 drug-productive scaffolds or scaffold parent-child sub-branches (DSs) labeled as DS1 to DS62 (Figure 1, Supplementary Table S4 and Figures S1–S5). A DS is defined as a scaffold with ≥2 NPLDs that have yielded ≥1 approved drug or a scaffold parent-child sub-branch with ≥2 NPLD-producing scaffolds that have yielded ≥1 approved drug. These DSs have collectively yielded 69.6% approved and 44.4% clinical trial drugs. The congregation of NPLDs in the DSs coupled with the earlier finding that the GPCR, kinase and protease targeting agents each are clustered together in the chemical space1 indicates that NPLDs of the same and different scaffolds against the same classes of targets may on a broader scale be clustered together in the chemical space. To facilitate the visualization of our generated scaffold trees by using Scaffold Hunter, the resulting scaffold database was exported as a SQL file that can be downloaded at http://bidd.nus.edu.sg/group/NPLD_Distribution/NP_ScaffoldHunter.zip.
Figure 1. Distribution of the natural product leads of approved and clinical trial drugs in branch 5 of the Scaffold-Hunter derived molecular scaffold trees of the 134,097 natural products and 411 natural product leads.
The drug-productive scaffolds or scaffold parent-child sub-branches (DSs) are indicated by red dots or red dots connected by red lines, which marked by the respective label DS15-DS19. The green triangles indicate the natural product leads outside the DSs. Some of the representative scaffolds in these DSs are shown in the Figure. The more complete sets of the representative scaffolds are shown in the Supplementary Figure S2.
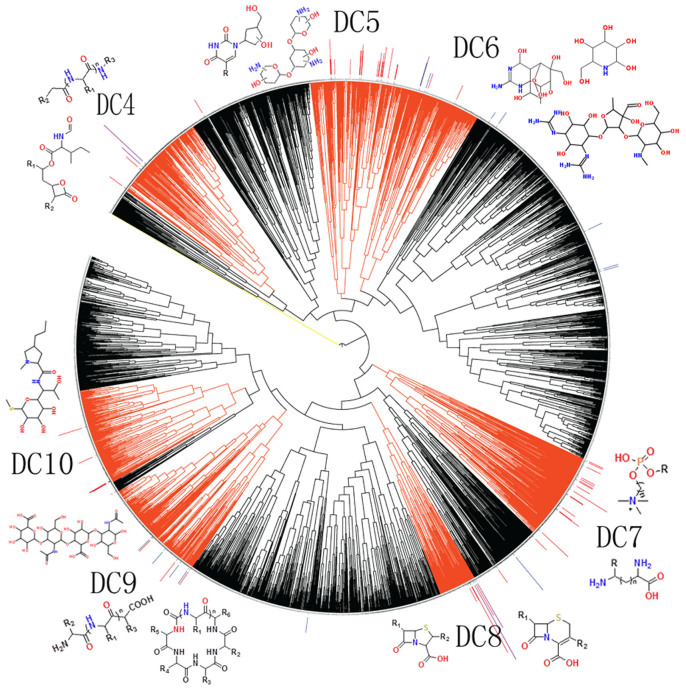
To probe the larger-scale distribution patterns of NPLDs in the chemical space from the perspective of molecular structures, we generated a molecular fingerprint based hierarchical clustering tree of the 442 NPLDs and 137,836 NPs. The derived tree is composed of 33 main branches (Supplementary Figure S6 and Table S5). Most (87.9%) branches are drug-productive, reflecting the fact that NPs primarily co-evolve and interact with proteins6 and a variety of chemical classes3,4 and target families48,49 have been therapeutically explored. Nonetheless, NPLDs within each branch are mostly clustered together, with 341 (77.2%) NPLDs (82.5% approved, 3.0% clinical trial) clustered in 60 drug-lead productive clusters (DCs) labeled as DC1 to DC60 (Figure 2, Supplementary S7–S10 and Table S6). A DC is defined as a relatively small region of the molecular fingerprint characterized chemical space with moderate to high concentration of NPLDs yielding ≥1 approved drug.
Figure 2. Distribution of the natural product leads of approved and clinical trial drugs in branch 3 of the substructure-fingerprint clustering tree of the 137,836 natural products and 442 natural product leads.
The drug-lead productive clusters are red-orange colored and marked by the respective cluster label DC4-DC10. The red, purple and blue lines on top of the clustering tree indicate the locations of the approved, approved + clinical trial, and clinical trial drug-leads with the height correlating with the number of approved + clinical trial drugs.
To facilitate the analysis of the clustered distribution of the NPLDs, we generated the heat map of the proximity matrix of 442 NPLDs against 137,836 non-redundant NPs. The proximity matrix was calculated by using molecular fingerprint Tanimoto distance Td between NPLDs and NPs with the row and column positions representing the NPLDs and NPs in the same order as their respective positions in the hierarchical clustering tree of the NPLDs and NPs. The heat map was created by using the heatmap.2 function of the gplots package in R with the red to yellow colors indicating the stronger to weaker structural similarity between the NPLDs and NPs. The heat map for branch 4 and 9 are shown in Supplementary Figures S11–S12 and those of the other branches can be downloaded from http://bidd.nus.edu.sg/group/NPLD_Distribution/NP_heatmaps.zip.
We found that 11.7% of the NPLDs in the DCs have remote-similarity relationships (0.57 ≤ Tc < 0.7) with the nearest NPLD in their own DC, and another 24.9% of the NPLDs in these DCs have intermediate-similarity relationship (0.7 ≤ Tc < 0.85) with their nearest NPLD in their own DC. Remote-similarity relationships have been reported in compounds with cross-pharmacology relationships50 and between a bioactive compound and its scaffold hopping parent bioactive compound51. Therefore, the DCs broadly cover the high-similarity to remote-similarity relationships for capturing similar activities, and cross-pharmacology and scaffold hopping types of relationships.
These DCs have collectively yielded 87.9% approved and 68.8% clinical trial drugs. In particular, 56.0% approved and 67.4% clinical trial NPLDs are clustered in 22 NPLD-prolific DCs (Table 1) that have collectively yielded 68.4% approved and 39.2% clinical trial drugs, which is consistent with the report that half of the drugs are made of dozens of molecular frameworks13. The NPLD-prolific DCs were ranked based on the ratio of the approved NPLDs to the NPs in each DC. Partly because of the inadequate exploration and partly because of the limited availability of the relevant information, these ratios may not fully reflect the reality but nonetheless provide useful indications. We found that 60% of the top-10 NPLD-prolific DCs with >100 searchable NPs in Table 1 are among top-ranked DCs with higher approved NPLD to NP ratios. Thus, drug productivity of these top-ranked DCs seems to arise from higher NPLD yields instead of the higher number of NPs explored. The top-ranked DC38 and DC8 in Table 1 were excluded because they have <100 searchable NPs. If counted, they are among the DCs with highest approved NPLD to NP ratios.
Table 1. Top-ranked drug lead productive clusters with higher number of approved leads in the natural product chemical space represented by 137,836 natural products and 442 natural product leads. The corresponding target site class(es) and superclass(es) are also listed.
| Drug Lead Cluster (Branch) | Drug Lead Molecular Scaffold Groups | No of NPs | No of Leads, Drugs (Approved/Trial) | Target Site Super Class | Target Site Class |
|---|---|---|---|---|---|
| DC19 (14) | Steroids & derivatives | 546 | 32/5, 85/8 | steroid sites | Nuclear receptor ligand sites |
| DC5 (3) | Pyrimidine nucleosides, Aminoglycosides | 225 | 19/0, 32/0 | nucleoside phosphate sites, aminoacyl-tRNA sites | DNA metab enzymes nucleoside phosphate, ribosome 30 s aminoacyl-tRNA sites |
| DC13 (4) | Purine nucleosides, Imidazole analogs & their oligopeptide hybrids | 264 | 17/2, 39/22 | Nucleoside sites, nucleoside phosphate sites | nucleoside receptor ligand & metabolism enzyme substrate sites, Nucleoside phosphate receptor ligand, DNA metab enzymes nucleoside phosphate sites |
| DC7 (3) | Amino acids with acyclic hydroxyl side chain & derivatives | 217 | 15/0, 23/0 | amino acid sites | amino acid receptor ligand & metab enzyme substrate sites |
| DC17 (14) | Macrolides, Polyenes, Spinosyns, Acarviosins | 117 | 15/4, 29/14 | amino acid phosphate sites, oligopeptide sites, lipopolysaccharide sites | phosphatase substrate, ribosome 23S peptidyl transferase, outer membrane lipopolysaccharide sites |
| DC21 (14) | Fatty acids & derivatives, Prostanoids | 423 | 8/1, 26/1 | fatty acid, cannabinoid, eicosanoid, retinoid, coenzyme A sites | retinoid receptor ligand, CoA metab enzyme substrate sites |
| DC28 (17) | Cardiac glycosides | 176 | 8/1, 12/1 | nucleoside phosphate sites | nucleoside phosphate metab enzyme substrate sites |
| DC38 (21) | Intermediate-sized linear and cyclic peptides | 33 | 7/1, 32/2 | Oligopeptide sites, lipopolysaccharide sites | exopeptidase substrate, Neuropeptide receptor ligand, outer membrane lipopolysaccharide sites |
| DC8 (3) | Beta-lactams | 92 | 6/2, 90/6 | peptidoglycan sites | ß-lactam binding protein peptidoglycan sites |
| DC29 (20) | Saponins, Triterpenoid glycosides, Macrocyclic lactones | 1696 | 6/1, 7/2 | nucleoside phosphate sites | steroid metab enzyme nucleoside phosphate, calcium channel DHP, chloride channel CBS sites |
| DC45 (30) | Tetracyclines, Capsaicinoids, Disulfide bromotyrosine derivatives | 447 | 6/3, 13/7 | aminoacyl-tRNA sites | ribosome 30 s aminoacyl-tRNA sites |
| DC49 (32) | Opiate alkaloids, Phenanthrene alkaloids | 457 | 6/1, 18/1 | amine sites, opiate sites | amine receptor ligand, opiate receptor ligand sites |
| DC10 (3) | Glycosaminoglycans, glucosamines, Lincosamides & derivatives | 237 | 5/0, 17/0 | oligopeptide sites | serine endopeptidase substrate sites |
| DC12 (4) | Purine base analogs, modified purine base analogs | 180 | 5/0, 9/0 | nucleoside sites, nucleoside phosphate sites | nucleoside receptor ligand, DNA metab enzymes nucleoside phosphate sites |
| DC14 (7) | Larger indole alkaloids | 4104 | 5/1, 15/16 | amine sites, oligopeptide sites | amine receptor ligand & transporter substrate, exopeptidase substrate sites |
| DC53 (10) | Cannabinoids, Diarylheptanoids, Dihydrostilbenoids, Small phenolic molecules with a long tail | 2531 | 5/4, 9/8 | fatty acid, cannabinoid, eicosanoid, retinoid sites | fatty acid metab enzyme substrate, retinoid receptor ligand, cannabinoid receptor ligand sites |
| DC24 (16) | Oligo-, Poly-, Cyclic- saccharides | 259 | 5/0, 5/0 | cyclic oligosaccharide drug delivery systems | cyclodextrin drug delivery systems |
| DC36 (20) | Large cyclic peptides | 265 | 5/6, 6/7 | nucleoside phosphate sites, sites within peptidoglycans, saccharide sites, lipopolysaccharide sites | calcium channel DHP, cell wall peptidoglycan, polysaccharide metab enzyme substrates, outer membrane lipopolysaccharide sites |
| DC40 (24) | Porphyrins, Prodiginines, Ergoline-, Ellipticine-, Epibatidine- alkaloids | 519 | 5/3, 8/4 | amine sites, nucleobase sites | amine receptors ligand, DNA intercalation sites |
| DC42 (26) | Indole-containing amino acid tryptophan analogs, Monoterpenoid indole alkaloids, Yohimbine alkaloids | 512 | 5/2, 6/4 | amine sites | amine receptor ligand sites |
| DC43 (28) | Tropane alkaloids | 29 | 5/0, 10/0 | amine sites | amine receptor ligand & transporter substrate sites |
| DC44 (28) | Catecholamines, Small alkaloids with an amine group | 358 | 5/0, 21/0 | amine, opiate sites | amine receptor, opiate receptor ligand sites |
Statistical significance of the clustering of NPLDs in the DCs
The statistical significance of the clustering of the NPLDs in every DC was evaluated by using the Phylocom software46 to calculate the P-value and NRI of the NPLDs against the chance clustering of the NPs in the DC from 60,000 sets of randomly selected NPs, as outlined in the Method section. We found that there are statistically more NPLDs in most of the DCs than expected by chance, with 78.3% of the DCs having P ≤ 0.0095 and additional 10% of the DCs having 0.011 ≤ P ≤ 0.0362 respectively (Table 2), which correspond to very strong (P ≤ 0.01) and strong (0.01 < P ≤ 0.05) presumption against null hypothesis respectively52. The P-value of the remaining 4 (6.7%) and 3 (5%) DCs are in the range of 0.0525 ≤ P ≤ 0.0736 and 0.1233 ≤ P ≤ 0.1857 respectively, which correspond to low (0.05 < P ≤ 0.1) and no (P > 0.1) presumption against null hypothesis respectively52. It is noted that each of these seven DCs has only 2 NPLDs and there is a possibility that the low statistical significance of these DCs are partly due to the few discovered NPLDs in these DCs.
Table 2. The statistical significance of the clustering of the NPLDs in every DC. MTd is the mean Tanimoto distance of the NPLDs in each DC, MTd.rnd is the mean Tanimoto distance in randomization, NRI is a standardized effect size measure of the community structure, and P-value is the number of randomly selected NPs that are more clustered than the NPLDs in each DC divided by the number of runs (60,000 in this study). P values in bold are the ones which remain significant after Bonferroni correction with conservative α′ = 0.05/60 = 0.000833.
| DC | Branch | No of NPLD | MTD | MTD.rnd | NRI | P-value |
|---|---|---|---|---|---|---|
| DC1 | 1 | 2 | 0.2642 | 1.8368 | 6.2835 | 0.00238 |
| DC2 | 1 | 2 | 0.1224 | 1.8373 | 6.9228 | 0.00117 |
| DC3 | 1 | 2 | 0.6296 | 1.8331 | 4.7303 | 0.00847 |
| DC4 | 3 | 4 | 1.0322 | 1.6705 | 6.1005 | 0.00047 |
| DC5 | 3 | 19 | 1.2111 | 1.6704 | 16.3408 | 0.00000 |
| DC6 | 3 | 5 | 1.2328 | 1.6704 | 5.2077 | 0.00142 |
| DC7 | 3 | 15 | 1.4909 | 1.6706 | 5.2466 | 0.00043 |
| DC8 | 3 | 6 | 1.0778 | 1.6706 | 8.2775 | 0.00003 |
| DC9 | 3 | 7 | 1.1073 | 1.6711 | 9.1502 | 0.00002 |
| DC10 | 3 | 5 | 0.8436 | 1.6704 | 9.8633 | 0.00000 |
| DC11 | 4 | 4 | 0.8826 | 1.5121 | 4.7221 | 0.00277 |
| DC12 | 4 | 5 | 0.6238 | 1.5122 | 8.4309 | 0.00000 |
| DC13 | 4 | 18 | 1.0751 | 1.5107 | 12.7659 | 0.00000 |
| DC14 | 7 | 5 | 0.8233 | 1.2803 | 4.4320 | 0.00177 |
| DC15 | 8 | 3 | 0.8090 | 1.5069 | 4.7523 | 0.00175 |
| DC16 | 9 | 3 | 0.4985 | 1.4882 | 5.2888 | 0.00145 |
| DC17 | 9 | 16 | 0.8576 | 1.4892 | 10.2529 | 0.00000 |
| DC18 | 9 | 2 | 1.0548 | 1.4913 | 1.5675 | 0.06920 |
| DC19 | 9 | 37 | 1.0487 | 1.4896 | 11.3267 | 0.00000 |
| DC20 | 10 | 2 | 0.1740 | 1.4029 | 5.8662 | 0.00135 |
| DC21 | 10 | 9 | 0.9145 | 1.4017 | 6.8067 | 0.00000 |
| DC22 | 10 | 3 | 0.7339 | 1.4007 | 4.5322 | 0.00078 |
| DC23 | 10 | 2 | 1.2542 | 1.3999 | 0.6864 | 0.15480 |
| DC24 | 11 | 5 | 0.8121 | 1.3910 | 5.5223 | 0.00100 |
| DC25 | 12 | 2 | 0.2934 | 1.2207 | 3.7910 | 0.00947 |
| DC26 | 12 | 2 | 0 | 1.2202 | 4.9798 | 0.00078 |
| DC27 | 12 | 8 | 0.9926 | 1.2208 | 2.9058 | 0.01428 |
| DC28 | 12 | 8 | 0.2007 | 1.2200 | 12.9182 | 0.00000 |
| DC29 | 12 | 7 | 0.7426 | 1.2203 | 5.5655 | 0.00043 |
| DC30 | 13 | 2 | 0.8474 | 1.3667 | 1.9145 | 0.06510 |
| DC31 | 14 | 5 | 1.0297 | 1.5449 | 4.2627 | 0.00100 |
| DC32 | 14 | 5 | 0.9406 | 1.5439 | 4.9565 | 0.00007 |
| DC33 | 14 | 4 | 1.1776 | 1.5454 | 2.5820 | 0.02413 |
| DC34 | 14 | 3 | 0.8209 | 1.5460 | 4.0057 | 0.00120 |
| DC35 | 14 | 6 | 0.6607 | 1.5452 | 8.3548 | 0.00000 |
| DC36 | 15 | 10 | 0.6860 | 1.3351 | 7.9798 | 0.00000 |
| DC37 | 16 | 3 | 0.6935 | 1.1435 | 3.4132 | 0.01103 |
| DC38 | 16 | 7 | 0.5400 | 1.1431 | 10.9642 | 0.00000 |
| DC39 | 17 | 3 | 0.4787 | 1.1665 | 5.0540 | 0.00093 |
| DC40 | 17 | 7 | 0.8464 | 1.1671 | 4.7313 | 0.00235 |
| DC41 | 17 | 2 | 0.6630 | 1.1649 | 2.3529 | 0.03623 |
| DC42 | 17 | 7 | 0.7352 | 1.1663 | 6.3612 | 0.00025 |
| DC43 | 20 | 5 | 0.4329 | 1.3241 | 17.2567 | 0.00000 |
| DC44 | 20 | 5 | 0.6912 | 1.3241 | 12.1159 | 0.00003 |
| DC45 | 20 | 7 | 0.9507 | 1.3238 | 10.0919 | 0.00002 |
| DC46 | 21 | 3 | 0.1372 | 0.7713 | 5.0125 | 0.00010 |
| DC47 | 21 | 4 | 0.5792 | 0.7708 | 1.8932 | 0.02985 |
| DC48 | 24 | 4 | 0.7416 | 1.0739 | 5.7931 | 0.00110 |
| DC49 | 24 | 6 | 0.5427 | 1.0740 | 14.5890 | 0.00000 |
| DC50 | 26 | 5 | 0.4218 | 0.7916 | 6.3884 | 0.00012 |
| DC51 | 26 | 2 | 0.6052 | 0.7909 | 1.1859 | 0.07355 |
| DC52 | 28 | 3 | 0.9049 | 1.1883 | 2.4123 | 0.03025 |
| DC53 | 28 | 9 | 0.9577 | 1.1880 | 5.9241 | 0.00070 |
| DC54 | 30 | 2 | 0.7262 | 0.8764 | 0.7311 | 0.18568 |
| DC55 | 32 | 3 | 0.3393 | 0.8538 | 4.4925 | 0.00152 |
| DC56 | 32 | 2 | 0.6884 | 0.8548 | 0.9532 | 0.12325 |
| DC57 | 32 | 2 | 0.2012 | 0.8550 | 3.7418 | 0.00427 |
| DC58 | 32 | 2 | 0.4916 | 0.8533 | 2.0560 | 0.05248 |
| DC59 | 32 | 3 | 0.2995 | 0.8535 | 4.8258 | 0.00077 |
| DC60 | 33 | 2 | 0 | 0.9059 | 5.0937 | 0.00040 |
To further provide a more conservative evaluation of the statistical significance of the clustering of NPLDs in the DCs, Bonferroni correction for study-wide hypothesis testing was performed. We found that, under the Bonferroni correction with α′ = 0.05/60, 48.3% DCs still have statistically strong or very strong significance against null hypothesis (Table 2). On the other hand, 16.7% DCs showed weak and 35% DCs showed no significance. It is noted that the majority (67.7%) of these weak or no significance DCs have 2–3 NPLDs in their respective DCs. The low number of NPLDs in each of these DCs likely leads to a higher tendency of forming a distribution pattern with weaker statistical significance that can be exposed by stricter statistic tests.
Molecular mechanisms that contribute to the clustering of NPLDs
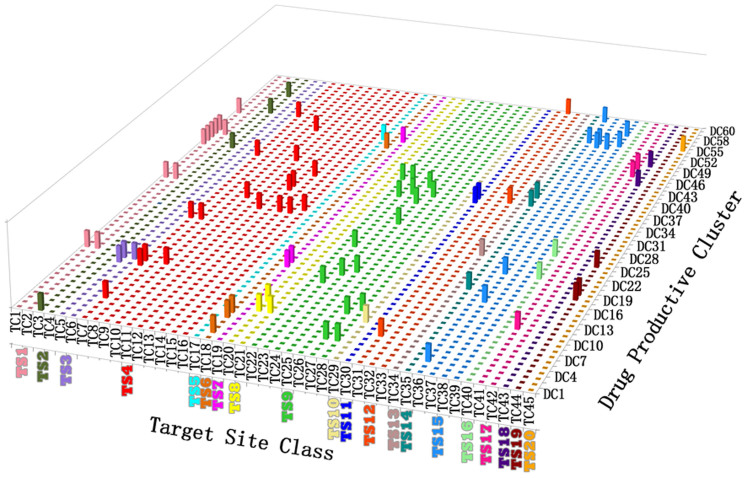
To determine what molecular mechanisms might contribute to the clustering of NPLDs within individual DCs with particular focus on the possible influence of the targets of their derived drugs, we evaluated the 203 targets of the 822 approved and clinical trial drugs of the 331 NPLDs in the 55 DCs with their target information available in the therapeutic target database53. We found that the targets of each individual DC are primarily from one to a few target classes (e.g. amine receptors) with their substrates/ligands from one to a few chemical classes (e.g. amines). This finding is based on the limited target information for 74.9% NPLDs and without considering the additional targets of the non-NPLDs in each DC. While the limited target information may not enable a full investigation of the influence of drug target-sites, it nonetheless provides useful hints about the key factors that promote the clustering of NPLDs. The 203 targets can be classified based on their target-sites into 45 target-site classes (TCs) labeled as TC1 to TC45, which collectively belong to 20 target-site super-classes (TSs) labeled as TS1 to TS20 (Supplementary Table S7). A TS is defined as a group of target-sites bound by substrates/ligands of a specific chemical class (e.g. amine binding sites) irrespective of their targets. A TC represents a sub-group of target-sites of a specific target class (e.g. amine transporters) bound by substrates/ligands of a specific chemical class.
The targets in 53 (96.4%) DCs are from 1–3 TCs (27, 19, 7 DCs from 1, 2, 3 TCs) with the majority (65.5%) from either 1 TC (27 DCs) or 2–3 TCs of 1 TS (9 DCs), and the remaining 2 DCs are from 4 TCs (Figure 3). This indicates that the similar target-site structural constraints are likely the key factors in promoting the clustering of NPLDs in individual DCs. The targets of approved and clinical trial drugs are highly selective in their numbers, druggability features, and systems profiles48,53,54,55, and the druggability features have been characterized by the affiliation of the family members of the studied target to the known drug targets56 and by the existence of a privileged binding site with unique physicochemical properties57 for enabling favorable binding by drug-like molecules58.
Figure 3. Distribution of the approved NP-related drugs, grouped into 45 target-site classes (TCs) of 20 target-site super-classes (TSs), in the drug-productive clusters DC1 to DC60.
TSs are colored as: TC1, TC2 of TS1 amine sites (LightCoral), TC3, TC4 of TS2 nucleobase sites (OliverGreen), TC5, TC6 of TS3 nucleoside sites (PalePurple), TC7-TC16 of TS4 nucleoside phosphate sites (Red), TC17 of TS5 cyclic nucleotide sites (Cyan), TC18 of TS6 aminoacyl-tRNA sites (Chocolate), TC19 of TS7 amino acid phosphate sites (Magenta), TC20, TC21 of TS8 amino acid sites (Yellow), TC22-TC28 of TS9 oligopeptide sites (Green), TC29 of TS10 peptidoglycan sites (PaleYellow), TC30 of TS11 peptidoglycan sites (Blue), TC31-TC33 of TS12 saccharide sites (OrangeRed), TC34 of TS13 cyclic oligosaccharide drug delivery systems (PaleBrown), TC35 of TS14 lipopolysaccharide sites (DarkCyan), TC36-TC39 of TS15 fatty acid, cannabinoid, eicosanoid, retinoid sites (PaleBlue), TC40 of TS16 coenzyme A & analog sites (PaleGreen), TC41, TC42 of TS17 microtubule sites (DeepPink), TC43 of TS18 opiate sites (Purple), TC44 of TS19 steroid sites (Brown), and TC45 of TS20 naphthoquinone sites (Orange).
Our revealed links between the clustering of NPLDs in individual DCs and the grouping of their targets in selected TCs are consistent with these findings. NPLDs in these DCs possess structural, physicochemical and/or pharmacophore features complementary to a privileged target-site, are at or near activity peaks against the target, and have good or amendable safety and pharmacokinetic properties. They either have or may be further optimized to gain such additional features as adequate metabolic stability59, metabolite safety60, absorption61 and physical forms62 to reach the drug sweet spots63 in the chemical space. Therefore, our revealed clustered patterns of NPLDs and their links to the selected TCs provide useful information and enable further study of the distribution profiles of the NPLDs in the chemical space particularly with respect to the relevant target-site classes.
Consistent with the reported clustering of GPCR, kinase and protease targeting agents in the chemical space1, the GPCR, kinase and protease TCs are primarily targeted by the selected chemical classes of NPLDs in specific DCs. For GPCRs, amine receptors (TC1) are primarily targeted by amines (DC31, DC44), ergoline alkaloids (DC40, DC41), and indole (DC14, DC42) and tropane (DC43) alkaloids, amino acid receptors (TC20) by amino acids (DC7) and oligopeptides (DC9), cannabinoid receptors (TC39) by cannabinoids (DC56) and cannabidiols (DC53), purine nucleoside receptors (TC5) by purines (DC12, DC13), opiate receptors (TC43) by opiate alkaloids (DC49), and monosaccharide receptors (TC32) by phenylpropanoids (DC60). Kinases (TC11) are primarily targeted by staurosporines (DC39). For the proteases, serine endopeptidases (TC28) are primarily targeted by glycosaminoglycans (DC10) and linear amino acid derivatives (DC4), proteasome (TC27) by oligopeptides (DC9), and exopeptidases (TC23) by phenethylamines (DC34), sesquiterpenes (DC22), larger indole alkaloids (DC14), and linear and cyclic peptides (DC38).
The other drug-prolific DCs are also closely linked to specific TCs (Table 1), with DC19 (steroids) linked to the nuclear receptor ligand binding sites (TC44), DC5 (aminoglycosides) to the DNA metabolism enzyme nucleoside phosphate (TC7) and ribosome 30 s subunit aminoacyl-tRNA (TC18) binding sites, DC17 (acarviosins) to the phosphatase substrate (TC19), ribosome 23S rRNA peptidyl transferase (TC25) and outer membrane lipopolysaccharide (TC35) sites, DC21 (fatty acids, prostanoids) to the retinoid receptor ligand (TC37) and coenzyme A analog metabolism enzyme substrate (TC40) binding sites, DC28 (cardiac glycosides) to the nucleoside phosphate metabolism enzymes substrate binding sites (TC10), and DC8 (β-lactams) to the β-lactam binding protein peptidoglycan binding sites (TC29).
Detailed analysis of the physicochemical landscape of the NPLD distribution profile reveals clues for searching the sweet spots in the DCs
While the knowledge of the clustered distribution patterns of the NPLDs in the DCs and the correlation to the TCs is useful for revealing the NPLD-like structural frameworks for targeting specific target classes, more detailed analysis is needed for identifying the NPLDs within each DC. On the other hand, drug-like14,41,42,43 and lead-like43,44 rules have been derived and extensively used for identifying drug leads on the basis of whether their specific physicochemical properties fall into certain drug-likeness or lead-likeness ranges. Optimal hydrophobic and hydrogen bond interactions, and thus the AlogP/logD and ON/HD/HA values, are important for optimizing NPLDs into drugs64. Therefore, additional clues for searching the new NPLDs may be obtained by studying the physicochemical landscapes of the known NPLDs and the NPs in the DCs.
We evaluated the physicochemical landscape of the NPLDs and NPs in branch 9 characterized by the eight physicochemical properties MW, AlogP, logD, ON, HD, HA, RB, RI, and PZ frequently used for analyzing drug-like14,41,42,43 and lead-like43,44 features (Supplementary Figure S13). This branch contains four DCs (DC16, DC17, DC18, and DC19). While, the NPs inside and outside these DCs have mixed MW values, there is a significantly higher concentration of NPs with either higher AlogP/logD values or higher ON/HD/HA values. In particular, regardless of their MW values, the NPLDs tend to be located at the peak of either AlogP/logD or ON/HD/HA. For instance, the NPLDs in DC17 (which include macrolides, polyenes, spinosyns and acarviosins) have peak MW values likely due to the added or enlarged hydrophobic groups (peak AlogP/logD values) to optimally interact with, e.g., the outer membrane lipopolysaccharide sites of TC35, or the added hydrogen bonding components (peak ON/HD/HA values) to optimally interact with, e.g., the phosphatase substrate sites of TC19. The NPLDs in DC19 (composed of steroids and derivatives) tend to have peak AlogP or logD values without significantly enlarged MW values over other NPs in the DC, possibly due to enhanced hydrophobic components within the steroid structural framework for achieving optimal hydrophobic interactions with the nuclear receptor ligand sites of TC44. Therefore, the tendency of the NPLDs to be located at either the AlogP/logD or the ON/HD/HA peaks in the DCs may be potentially used as an indicator for searching new NPLDs.
The distribution profiles of NPLDs with respect to bioactive NPs
The more clustered distribution of NPLDs (Figure 2 and Supplementary S7–10) are in contrast to the much less clustered distribution of the 48,216 bioactive NPs from the TCM@Taiwan database25 and the literatures3 (Supplementary Figures S14–17). Although the number and diversity of our collected bioactive NPs are limited in representing bioactive NPs, useful indications may be revealed. These bioactive NPs are more diversely distributed in 32 of the 33 branches with 78.8% of the bioactive NPs located outside the DCs. Therefore, NPLDs are distinguished from bioactive NPs in their tendency to more closely cluster together in the chemical space, which is consistent with the distribution pattern of drug-productive species families in the phylogenetic tree (drug-productive species families are more closely clustered than the species families of bioactive NPs)3. To investigate whether the more clustered distribution of NPLDs in the DCs is due to the more extensive exploration efforts towards these DCs, the exploration times of the 442 NPLDs, crudely estimated by the time since the first literature report, were compared to those of the 11,816 bioactive NPs inside and outside the DCs, which are largely comparable to each other (Supplementary Figures S18–21). Hence, there is no clear indication to link drug-productivity of the DCs to the biased exploration efforts.
The distribution profiles of NPLDs with respect to time and disease classes
Since 1988, the number of DCs has been gradually increased at an average rate of 3.2 new DCs per 5 years, and the majority (60.0%–69.0%) of the 15–32 new NPLDs approved in every five-year period from 1560 to 2012 are from preexisting DCs (Table 3). Drug discovery focus has been shifting in terms of targets, chemotypes, diseases and therapeutic strategies48,65. To study if novel drugs derived from shifted focuses are outside pre-existing DCs, we analyzed 27 new NPLDs approved in 1990–2012 each targeting a novel target previously unaddressed by an approved drug (Supplementary Table S8) and thus are novel NPLDs of the time. At the time of their first drug approval, 18 (66.7%) of these novel NPLDs were from preexisting DCs, suggesting that existing DCs remain good sources of novel NPLDs and drugs.
Table 3. chronological data of the natural product leads with the first approved drugs and the drug lead productive clusters during every five-year period from 1963 to 2012. The six drug lead clusters with only one approved drug plus one or more clinical trial drugs were not included here.
| Number of natural product leads with the first approved drug in period | Number of drug lead clusters in period | |||
|---|---|---|---|---|
| Period | Inside preexisting DCs | Outside preexisting DCs | Number of preexisting DCs | Number of new DCs |
| Pre–1963 | 56 | NA | 8 | NA |
| 1963–1967 | 7 | 20 | 8 | 7 |
| 1968–1972 | 4 | 9 | 15 | 3 |
| 1973–1977 | 9 | 8 | 18 | 2 |
| 1978–1982 | 17 | 56 | 20 | 13 |
| 1983–1987 | 20 | 20 | 33 | 5 |
| 1988–1992 | 20 | 12 | 38 | 6 |
| 1993–1997 | 20 | 9 | 44 | 2 |
| 1998–2002 | 19 | 10 | 46 | 3 |
| 2003–2007 | 9 | 6 | 49 | 1 |
| 2008–2012 | 11 | 6 | 50 | 4 |
The approved drugs from individual DCs largely target one to a few disease classes (Supplementary Figure S22). Specifically, 61.7% of DCs target one (DC1, DC2, DC11, DC18, DC23, DC26, DC39, DC46, DC51 and DC60), two (DC3, DC6, DC16, DC20, DC22, DC25, DC30, DC34, DC35, DC37, DC50, DC54, DC55, DC56, DC57 and DC59) or three (DC4, DC9, DC15, DC28, DC31, DC32, DC41, DC47, DC48, DC52 and DC58) disease classes. While the remaining DCs target multiple disease classes, most drugs from these DCs target a few disease classes. Anti-infectious and anti-parasite drugs are mostly from DC8 (87 drugs), DC5 (22 drugs), DC17 (19 drugs), DC45 (10 drugs), DC4 (9 drugs) and DC13 (8 drugs). Anticancer drugs are primarily from DC19 (31 drugs), DC13 (13 drugs), DC50 (9 drugs), DC38 (8 drugs), DC5 (7 drugs), DC14 (5 drugs) and DC46 (5 drugs). Circulatory system drugs are largely from DC38 (17 drugs), DC10 (12 drugs), DC28 (11 drugs), DC13 (9 drugs), DC44 (9 drugs) and DC21 (6 drugs). Nervous system drugs are mostly from DC49 (13 drugs), DC44 (7 drugs) and DC7 (5 drugs). Drugs for endocrine, nutritional and metabolic diseases are primarily from DC19 (13 drugs), DC26 (7 drugs), DC38 (6 drugs) and DC25 (5 drugs). Genitourinary system drugs are mostly from DC19 (15 drugs) and DC8 (5 drugs).
The usefulness of the knowledge of NPLD distribution profile for facilitating new NPLD discovery
The tendencies of NPLDs to cluster together in the chemical space and to preferentially bind to the privileged target-sites in the target-space may be explored for assessing the development potential of new NP leads. Based on the insights derived from our analysis, one can postulate that, apart from the ability of an NPLD or its derivatives to modulate a validated target, an NPLD may have a higher probability to be developed into a drug if it is inside a DC, near a DC (to form an expanded DC) or near an NPLD outside existing DCs (to form a new DC) in the chemical space, and if its target belongs to an existing TC or a new TC in an existing TS. This postulation was tested by the retrospective analysis of the new NPLDs of FDA approved drugs in 2013–June 201466,67 that were not used in the derivation of the NPLD distribution patterns and the target-site linkages. Our literature search led to the finding of 4 new small molecule NPLDs, 3 of which were retrospectively recognized as developable based on our postulation (Table 4). Specifically, the NPLD uridine monophosphate of sofosbuvir is inside DC5 and target TC7, the NPLD phlorizin of canagliflozin is near DC57 (Tc = 0.91 to the nearest NPLD) and target monosacharide transporter substrate sites as a new TC in TS12 (saccharide binding sites), and the imidazole-based NPLD (e.g. mizoribine) of luliconazole is inside DC13 and target a steroid metabolism enzyme substrate site in TS19 (steroid binding sites). Therefore, the insights derived from this and other studies of NPLD distribution profiles may be explored for facilitating the assessment of the development potential of NP leads.
Table 4. The list of natural product derived drugs approved by FDA from 2013 to 2014 June. The natural product leads, targets and affiliations to drug productive clusters and target-site classes are provided.
| Drug | Drug Type | NP Lead | NP Lead Type | Affiliation to drug lead productive cluster | target | Affiliation to target-site class |
|---|---|---|---|---|---|---|
| Canagliflozin | Small Molecule | Phlorizin | N | Near DC57 (tanimoto similarity coefficient 0.91 to the nearest NPLD) | SGLT2 | Monosacharide transporter substrate sites as a new TC in TS12 (saccharide binding sites) |
| Luliconazole | Small Molecule | Imidazole-based NP such as mizoribine | N | DC13 | Lanosterol demethylase | Steroid metabolism enzyme substrate sites in TS19 (steroid binding sites) |
| Sofosbuvir | Small Molecule | Uridine monophosphate | N | DC5 | HCV NS5B polymerase | TC7 |
| Vorapaxar | Small molecule | Himbacine | N | - | Protease-activated receptor-1 | TC22 |
| Simeprevir | Oligopeptide | HCV NS3/4A protease product oligopeptide | B (oligopeptide) | DC9 | HCV NS3/4A protease | TC28 |
| Mipomerse | Antisense | Section of mRNA of apolipoprotein B-100 | B (oligonucleotide) | - | Apolipoprotein B-100 | Lipid-binding sites in TS15 |
| Dalvance | Semisynthetic lipoglycopeptide | Lipoglycopeptide | B (lipoglycopeptide) | - | Cell wall | TC30 |
| Tanzeum | Peptide | Glucagon-like peptide-1 | B (peptide) | - | Glucagon-like peptide-1 receptor | TC22 |
Concluding Remarks
This study systematically exposed the clustered distribution profiles of NPLDs and revealed useful insights into the mechanisms that partly contribute to the formation of these profiles, i.e., the tendency of NPLDs to preferentially bind to the privileged target-sites. The insights from this and other studies of NPLD distribution profiles provide useful clues to and enable further studies of the lead sweet spots in the chemical space with respect to the corresponding target-sites. The distribution of NPLDs and the lead sweet spots in the chemical space is collectively influenced by potent binding to the target-sites and such additional factors as the optimization potential to reach the drug sweet spots in the chemical space63 with more adequate metabolic stability59, metabolite safety60, absorption61 and physical forms62). Further studies are needed for a deeper understanding of the collective influence of these multiple factors on the distribution of NPLDs in the chemical space. These advances coupled with expanded knowledge of lead-like and drug-like structures and physicochemical properties13 may enable more prioritized and rational exploration of the NP-subspaces for drug discovery.
Supplementary Material
Supplementary Information
Acknowledgments
We acknowledge the support from Ministry of Science and Technology, 863 Hi-Tech Program (2007AA02Z160), Key Special Project Grant 2009ZX09501-004, China and Singapore Academic Research Fund R-148-000-181-112.
Footnotes
The authors declare no competing financial interests.
Author Contributions Y.Z.C. and Y.Y.J. conceived and designed the project. Y.Z.C. and L.T. wrote the manuscript. L.T., F.Z. and C.Q. collected the data. L.T., F.Z., C.Q., C.Z., S.Y.C., P.Z., C.L.Z., C.Y.T., C.M.G. and Z.C. analyzed the data. All authors reviewed the manuscript.
References
- Lipinski C. & Hopkins A. Navigating chemical space for biology and medicine. Nature 432, 855–61 (2004). [DOI] [PubMed] [Google Scholar]
- Paul S. M. et al. How to improve R&D productivity: the pharmaceutical industry's grand challenge. Nat Rev Drug Discov 9, 203–14 (2010). [DOI] [PubMed] [Google Scholar]
- Zhu F. et al. Clustered patterns of species origins of nature-derived drugs and clues for future bioprospecting. Proc Natl Acad Sci U S A 108, 12943–8 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman D. J. & Cragg G. M. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J Nat Prod 75, 311–35 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. W. & Vederas J. C. Drug discovery and natural products: end of an era or an endless frontier? Science 325, 161–5 (2009). [DOI] [PubMed] [Google Scholar]
- Hert J., Irwin J. J., Laggner C., Keiser M. J. & Shoichet B. K. Quantifying biogenic bias in screening libraries. Nat Chem Biol 5, 479–83 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aquino C. et al. A biomimetic polyketide-inspired approach to small-molecule ligand discovery. Nat Chem 4, 99–104 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ertl P. & Schuffenhauer A. Cheminformatics analysis of natural products: lessons from nature inspiring the design of new drugs. Prog Drug Res 66, 217, 219–35 (2008). [DOI] [PubMed] [Google Scholar]
- Koch M. A. et al. Charting biologically relevant chemical space: a structural classification of natural products (SCONP). Proc Natl Acad Sci U S A 102, 17272–7 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetzel S., Bon R. S., Kumar K. & Waldmann H. Biology-oriented synthesis. Angew Chem Int Ed Engl 50, 10800–26 (2011). [DOI] [PubMed] [Google Scholar]
- Clardy J. & Walsh C. Lessons from natural molecules. Nature 432, 829–37 (2004). [DOI] [PubMed] [Google Scholar]
- Newman D. J. & Cragg G. M. Natural product scaffolds as leads to drugs. Future Med Chem 1, 1415–27 (2009). [DOI] [PubMed] [Google Scholar]
- Bemis G. W. & Murcko M. A. The properties of known drugs. 1. Molecular frameworks. J Med Chem 39, 2887–93 (1996). [DOI] [PubMed] [Google Scholar]
- Lipinski C. A., Lombardo F., Dominy B. W. & Feeney P. J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 46, 3–26 (2001). [DOI] [PubMed] [Google Scholar]
- Awale M. & Reymond J. L. Cluster analysis of the DrugBank chemical space using molecular quantum numbers. Bioorg Med Chem 20, 5372–8 (2012). [DOI] [PubMed] [Google Scholar]
- Lee M. L. & Schneider G. Scaffold architecture and pharmacophoric properties of natural products and trade drugs: application in the design of natural product-based combinatorial libraries. J Comb Chem 3, 284–9 (2001). [DOI] [PubMed] [Google Scholar]
- Paolini G. V., Shapland R. H., van Hoorn W. P., Mason J. S. & Hopkins A. L. Global mapping of pharmacological space. Nat Biotechnol 24, 805–15 (2006). [DOI] [PubMed] [Google Scholar]
- Singh N. et al. Chemoinformatic analysis of combinatorial libraries, drugs, natural products, and molecular libraries small molecule repository. J Chem Inf Model 49, 1010–24 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickerton G. R., Paolini G. V., Besnard J., Muresan S. & Hopkins A. L. Quantifying the chemical beauty of drugs. Nat Chem 4, 90–8 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awale M., van Deursen R. & Reymond J. L. MQN-mapplet: visualization of chemical space with interactive maps of DrugBank, ChEMBL, PubChem, GDB-11, and GDB-13. J Chem Inf Model 53, 509–18 (2013). [DOI] [PubMed] [Google Scholar]
- Wetzel S. et al. Interactive exploration of chemical space with Scaffold Hunter. Nat Chem Biol 5, 581–3 (2009). [DOI] [PubMed] [Google Scholar]
- Bocker A., Derksen S., Schmidt E., Teckentrup A. & Schneider G. A hierarchical clustering approach for large compound libraries. J Chem Inf Model 45, 807–15 (2005). [DOI] [PubMed] [Google Scholar]
- Willett P. Similarity-based virtual screening using 2D fingerprints. Drug Discov Today 11, 1046–53 (2006). [DOI] [PubMed] [Google Scholar]
- Irwin J. J., Sterling T., Mysinger M. M., Bolstad E. S. & Coleman R. G. ZINC: a free tool to discover chemistry for biology. J Chem Inf Model 52, 1757–68 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. Y. TCM Database@Taiwan: the world's largest traditional Chinese medicine database for drug screening in silico. PLoS One 6, e15939 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson J., Gottfries J., Muresan S. & Backlund A. ChemGPS-NP: tuned for navigation in biologically relevant chemical space. J Nat Prod 70, 789–94 (2007). [DOI] [PubMed] [Google Scholar]
- Saklani A. & Kutty S. K. Plant-derived compounds in clinical trials. Drug Discov Today 13, 161–71 (2008). [DOI] [PubMed] [Google Scholar]
- Zhu F. et al. Drug discovery prospect from untapped species: indications from approved natural product drugs. PLoS One 7, e39782 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. et al. Database of traditional Chinese medicine and its application to studies of mechanism and to prescription validation. Br J Pharmacol 149, 1092–103 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perola E. An analysis of the binding efficiencies of drugs and their leads in successful drug discovery programs. J Med Chem 53, 2986–97 (2010). [DOI] [PubMed] [Google Scholar]
- Wilkinson B. & Micklefield J. Mining and engineering natural-product biosynthetic pathways. Nat Chem Biol 3, 379–86 (2007). [DOI] [PubMed] [Google Scholar]
- Chiang Y. M., Chang S. L., Oakley B. R. & Wang C. C. Recent advances in awakening silent biosynthetic gene clusters and linking orphan clusters to natural products in microorganisms. Curr Opin Chem Biol 15, 137–43 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yongye A. B., Waddell J. & Medina-Franco J. L. Molecular scaffold analysis of natural products databases in the public domain. Chem Biol Drug Des 80, 717–24 (2012). [DOI] [PubMed] [Google Scholar]
- Newman D. J. & Cragg G. M. Natural products as sources of new drugs over the last 25 years. J Nat Prod 70, 461–77 (2007). [DOI] [PubMed] [Google Scholar]
- Butler M. S. Natural products to drugs: natural product-derived compounds in clinical trials. Nat Prod Rep 25, 475–516 (2008). [DOI] [PubMed] [Google Scholar]
- Yap C. W. PaDEL-descriptor: an open source software to calculate molecular descriptors and fingerprints. J Comput Chem 32, 1466–74 (2011). [DOI] [PubMed] [Google Scholar]
- Ma X. H. et al. Virtual screening of selective multitarget kinase inhibitors by combinatorial support vector machines. Mol Pharm 7, 1545–60 (2010). [DOI] [PubMed] [Google Scholar]
- Li Z. R. et al. MODEL-molecular descriptor lab: a web-based server for computing structural and physicochemical features of compounds. Biotechnol Bioeng 97, 389–96 (2007). [DOI] [PubMed] [Google Scholar]
- Smieja M., Warszycki D., Tabor J. & Bojarski A. J. Asymmetric clustering index in a case study of 5-HT1A receptor ligands. PLoS One 9, e102069 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccarelli F. D. et al. Toward automatic reconstruction of a highly resolved tree of life. Science 311, 1283–7 (2006). [DOI] [PubMed] [Google Scholar]
- Bhal S. K., Kassam K., Peirson I. G. & Pearl G. M. The Rule of Five revisited: applying log D in place of log P in drug-likeness filters. Mol Pharm 4, 556–60 (2007). [DOI] [PubMed] [Google Scholar]
- Vistoli G., Pedretti A. & Testa B. Assessing drug-likeness–what are we missing? Drug Discov Today 13, 285–94 (2008). [DOI] [PubMed] [Google Scholar]
- Lipinski C. A. Lead- and drug-like compounds: the rule-of-five revolution. Drug Discov Today Technol 1, 337–41 (2004). [DOI] [PubMed] [Google Scholar]
- Ritchie T. J. & Macdonald S. J. The impact of aromatic ring count on compound developability–are too many aromatic rings a liability in drug design? Drug Discov Today 14, 1011–20 (2009). [DOI] [PubMed] [Google Scholar]
- Saslis-Lagoudakis C. H. et al. Phylogenies reveal predictive power of traditional medicine in bioprospecting. Proc Natl Acad Sci U S A 109, 15835–40 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb C. O., Ackerly D. D. & Kembel S. W. Phylocom: software for the analysis of phylogenetic community structure and trait evolution. Bioinformatics 24, 2098–100 (2008). [DOI] [PubMed] [Google Scholar]
- Dunn O. J. Estimation of the Medians for Dependent-Variables. Annals of Mathematical Statistics 30, 192–197 (1959). [Google Scholar]
- Zheng C. J. et al. Therapeutic targets: progress of their exploration and investigation of their characteristics. Pharmacol Rev 58, 259–79 (2006). [DOI] [PubMed] [Google Scholar]
- Rask-Andersen M., Almen M. S. & Schioth H. B. Trends in the exploitation of novel drug targets. Nat Rev Drug Discov 10, 579–90 (2011). [DOI] [PubMed] [Google Scholar]
- Brianso F., Carrascosa M. C., Oprea T. I. & Mestres J. Cross-pharmacology analysis of G protein-coupled receptors. Curr Top Med Chem 11, 1956–63 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godden J. W., Stahura F. L. & Bajorath J. Anatomy of fingerprint search calculations on structurally diverse sets of active compounds. J Chem Inf Model 45, 1812–9 (2005). [DOI] [PubMed] [Google Scholar]
- Nuzzo R. Scientific method: statistical errors. Nature 506, 150–2 (2014). [DOI] [PubMed] [Google Scholar]
- Qin C. et al. Therapeutic target database update 2014: a resource for targeted therapeutics. Nucleic Acids Res 42, D1118–23 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overington J. P., Al-Lazikani B. & Hopkins A. L. How many drug targets are there? Nat Rev Drug Discov 5, 993–6 (2006). [DOI] [PubMed] [Google Scholar]
- Imming P., Sinning C. & Meyer A. Drugs, their targets and the nature and number of drug targets. Nat Rev Drug Discov 5, 821–34 (2006). [DOI] [PubMed] [Google Scholar]
- Hopkins A. L. & Groom C. R. The druggable genome. Nat Rev Drug Discov 1, 727–30 (2002). [DOI] [PubMed] [Google Scholar]
- Mason J. S., Bortolato A., Congreve M. & Marshall F. H. New insights from structural biology into the druggability of G protein-coupled receptors. Trends Pharmacol Sci 33, 249–60 (2012). [DOI] [PubMed] [Google Scholar]
- Cheng A. C. et al. Structure-based maximal affinity model predicts small-molecule druggability. Nat Biotechnol 25, 71–5 (2007). [DOI] [PubMed] [Google Scholar]
- Thompson T. N. Optimization of metabolic stability as a goal of modern drug design. Med Res Rev 21, 412–49 (2001). [DOI] [PubMed] [Google Scholar]
- Liebler D. C. & Guengerich F. P. Elucidating mechanisms of drug-induced toxicity. Nat Rev Drug Discov 4, 410–20 (2005). [DOI] [PubMed] [Google Scholar]
- Stenberg P., Bergstrom C. A., Luthman K. & Artursson P. Theoretical predictions of drug absorption in drug discovery and development. Clin Pharmacokinet 41, 877–99 (2002). [DOI] [PubMed] [Google Scholar]
- Gardner C. R., Walsh C. T. & Almarsson O. Drugs as materials: valuing physical form in drug discovery. Nat Rev Drug Discov 3, 926–34 (2004). [DOI] [PubMed] [Google Scholar]
- Hann M. M. & Keseru G. M. Finding the sweet spot: the role of nature and nurture in medicinal chemistry. Nat Rev Drug Discov 11, 355–65 (2012). [DOI] [PubMed] [Google Scholar]
- Tao L. et al. Nature's contribution to today's pharmacopeia. Nat Biotechnol 32, 979–80 (2014). [DOI] [PubMed] [Google Scholar]
- Bilanges B., Torbett N. & Vanhaesebroeck B. Killing two kinase families with one stone. Nat Chem Biol 4, 648–9 (2008). [DOI] [PubMed] [Google Scholar]
- Mullard A. 2013 FDA drug approvals. Nat Rev Drug Discov 13, 85–9 (2014). [DOI] [PubMed] [Google Scholar]
- DeFrancesco L. Drug pipeline: 1Q14. Nature Biotechnology 32, 409 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information