Abstract
Introduction
Friedreich's ataxia (FRDA) is an autosomal recessive neurodegenerative disease caused by expansion of a GAA·TTC triplet in the first intron of the FXN gene, encoding the essential mitochondrial protein frataxin. Repeat expansion results in transcriptional silencing through an epigenetic mechanism, resulting in significant decreases in frataxin protein in affected individuals. Since the FXN protein coding sequence is unchanged in FRDA, an attractive therapeutic approach for this disease would be to increase transcription of pathogenic alleles with small molecules that target the silencing mechanism.
Areas covered
We review the evidence that histone postsynthetic modifications and heterochromatin formation are responsible for FXN gene silencing in FRDA, along with efforts to reverse silencing with drugs that target histone modifying enzymes. Chemical and pharmacological properties of histone deacetylase (HDAC) inhibitors, which reverse silencing, together with enzyme target profiles and kinetics of inhibition, are discussed. Two HDAC inhibitors have been studied in human clinical trials and the properties of these compounds are compared and contrasted. Efforts to improve on bioavailability, metabolic stability, and target activity are reviewed.
Expert opinion
2-aminobenzamide class I HDAC inhibitors are attractive therapeutic small molecules for FRDA. These molecules increase FXN gene expression in human neuronal cells derived from patient induced pluripotent stem cells, and in two mouse models for the disease, as well as in circulating lymphocytes in patients treated in a phase Ib clinical trial. Medicinal chemistry efforts have identified compounds with improved brain penetration, metabolic stability and efficacy in the human neuronal cell model. A clinical candidate will soon be identified for further human testing.
Keywords: Friedreich ataxia, histone deacetylase inhibitor, heterochromatin, epigenetics, neurodegenerative disease
1. Introduction
Friedreich ataxia (FRDA, OMIM 229300) is an autosomal recessive neurodegenerative disorder caused by reduced levels of the essential mitochondrial protein frataxin [1]. Most affected individuals harbor a GAA•TTC triplet repeat expansion in the first intron of the nuclear FXN gene encoding frataxin [1]. These repeats induce heterochromatin formation [2–6], resulting in transcriptional silencing and decreased levels of frataxin protein. The available data are consistent with a negative correlation between repeat length, frataxin levels, age-of-onset and disease severity in patients [7]. A small proportion of FRDA patients (~4 to 5%) are compound heterozygous for the repeat expansion on one allele and loss-of-function mutations (missense, nonsense, indel) in the other FXN allele [8]. While largely a neurodegenerative disease, the major cause of death in FRDA is cardiomyopathy [9]. For those individuals with homozygous repeat expansions, the amounts of residual frataxin protein (ranging from 5 to 35% of the levels in unaffected individuals) correlate with the number of repeats and severity of clinical symptoms. Interestingly, heterozygous carriers of the repeat allele have approximately 50 – 60% of the frataxin levels found in unaffected individuals and are themselves healthy. Thus, increasing frataxin levels to those found in carriers is predicted to be therapeutically useful.
Frataxin is involved in the assembly of iron-sulfur clusters, and their transfer to mitochondrial enzymes and components of the electron transport chain (reviewed in [10, 11]). Given its role in mitochondrial protein homeostasis, frataxin deficiency results in impaired activities of Fe-S enzymes, altered cellular iron metabolism with iron accumulation in mitochondria, decreased mitochondrial energy production, and increased oxidative stress [12]. To address deficits caused by the loss of activity of Fe-S enzymes, antioxidants (e.g., idebenone and co-enzyme Q10 [13]) and iron chelators (e.g., deferiprone, [14]) have been proposed as therapeutics. However, no clear clinical evidence supporting a benefit of these approaches have so far been obtained in randomized controlled trials (reviewed in [15, 16]). While recent success in combination therapy has been reported [17], the alternative approach of replacing or increasing frataxin expression in patients is an appealing therapeutic strategy to slow or prevent disease progression. Gene therapy [18] or protein replacement therapy [19] can in principle be used for this purpose. Studies in animal models support these approaches, at least for the cardiac manifestations of the disease; however, clinical development depends on the resolution of many general problems in the field of gene and protein replacement therapy, in particular, targeted delivery, controlled expression, and, for gene therapy, potential genotoxicity. An alternative approach we have focused on is increasing output of FXN mRNA from the endogenous alleles by small molecule gene activating compounds. We expect that increases in FXN mRNA will be paralleled by similar increases in frataxin protein. The rationale for this approach is that the coding region of the FXN gene is not mutated in the majority of FRDA patients, hence frataxin protein structure is unchanged from unaffected individuals. Therefore, increasing levels of FXN mRNA and frataxin protein from the endogenous gene is an attractive therapeutic approach, and has been considered by many investigators in the field. A number of laboratories have reported identification of small molecule activators of FXN mRNA synthesis or frataxin protein in patient cells. These include histone deacetylase inhibitors [3, 20], erythropoietin alpha [21], dyclonine [22], gamma-interferon [23], SRC inhibitors [24], short oligonucleotides [25], and inhibitors of frataxin degradation [26]. With the exception of HDAC inhibitors and frataxin stabilizers, the mechanism of action of these compounds has yet to be elucidated. Here, we review the progress in the development of drugs targeting FXN gene silencing caused by the GAA•TTC repeat expansion. First, we summarize the evidence for the epigenetic basis of FXN gene silencing in Friedreich ataxia and how such silencing might be overcome with small molecule therapeutics.
2. Loss of frataxin in FRDA is due to heterochromatin-mediated transcriptional silencing
2.1 Evidence for transcriptional defects in FRDA cells
Within a relatively short time after the initial publication of the genetic basis for FRDA [1], two publications reported that expanded GAA•TTC repeats cause transcriptional silencing of the FXN gene [27, 28]. Wells and colleagues first showed length- and orientation-dependent inhibition of reporter gene expression with DNA constructs containing variable numbers of repeats [27]. Subsequently, the activity of the endogenous FXN gene was analyzed along with in vitro studies documenting inhibition of transcription by eukaryotic RNA polymerase II [28]. This report also showed that the repeats did not affect RNA processing of the primary transcript to the mature mRNA [28]. While one study has claimed that expanded GAA repeats affected RNA splicing in a reporter construct [29], there is no available evidence to suggest that GAA repeats affect RNA splicing [28]. There is also no evidence that long GAA-repeat intron 1 RNA is stable and could lead to an RNA-toxicity disease, such as found in myotonic dystrophy or fragile X-associated tremor/ataxia (reviewed in [30]). Indeed, efforts to find RNA foci containing GAA repeat RNA have been unsuccessful [31].
2.2 How do the repeats induce gene silencing?
Research into the mechanism whereby the GAA•TTC repeats cause transcriptional silencing has led to insights for therapeutic development. Within a few years after the discovery of the GAA•TTC repeats as the cause of FRDA, several groups reported that the repeats form unusual DNA structures in vitro, such as triplexes and an intramolecular structure known as “sticky” DNA [32–35]. These non-B type DNA structures were hypothesized to impede the progress of RNA polymerase II through the repeats and lead to stalled or aborted transcription [27]. We hypothesized that small molecules that exclusively bind B-type DNA might be able to drive the equilibrium between non-B structures and B-type DNA, thereby alleviating transcriptional repression. We showed that a pyrrole-imidazole polyamide that was designed to bind GAA•TTC repeat DNA did indeed reverse the “sticky” DNA conformation and resulted in increases in FXN mRNA in patient lymphoid cells [36]. However, this compound has limited brain penetration, limiting its utility as a therapeutic for FRDA [37]. Besides non-B DNA structures, a DNA-RNA triplex formed at the repeats could be responsible for blocking transcription elongation [38, 39]. In this regard, recent work suggests that R-loops are formed at expanded repeats and that these RNA-DNA hybrid structures are involved in the initiation of silencing or may impede passage of RNA polymerase through the repeat region [40]. However, further studies are necessary to fully elucidate the relevance of R-loops and other non-B DNA structures in FXN gene regulation.
2.3 Repeat induced heterochromatin formation at pathogenic FXN alleles
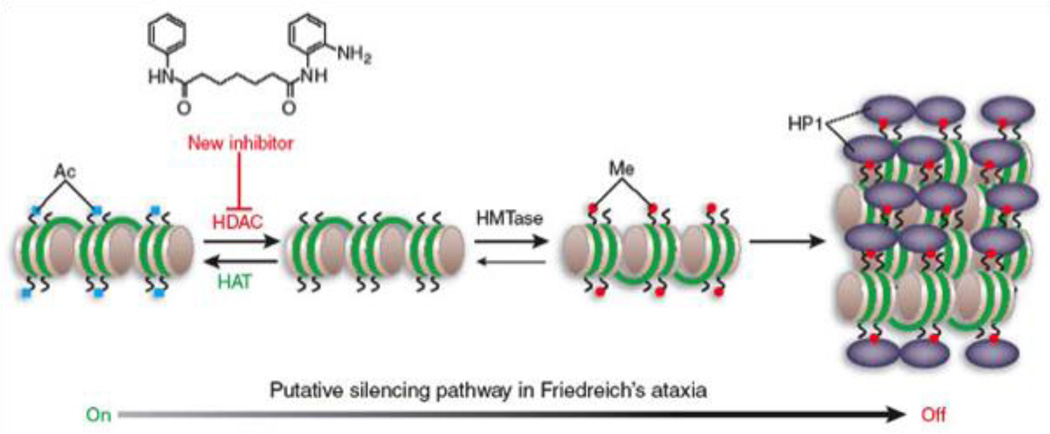
An alternative, but not mutually exclusive, mechanism for silencing pathogenic FXN alleles is epigenetic gene silencing through heterochromatin. Heterochromatin is characterized by histone hypoacetylation, histone H3 lysine 9 and lysine 27 methylation, and the association of histone deacetylase enzymes, specific histone methyltransferases and heterochromatin proteins, such as members of the HP1 family and polycomb group proteins (Figure 1). The first report in support of an epigenetic silencing mechanism in FRDA came from Festenstein and colleagues [2], who showed that a transgene containing GAA•TTC repeats was silenced in vivo, in a manner reminiscent of position effect variegated gene silencing. In this study, repeat-induced silencing was augmented by over-expression of the heterochromatin protein HP1, and the silenced transgene was packaged into condensed chromatin, as evidenced by resistance to nuclease digestion [2]. Interestingly, the repeats could be located outside of the transcribed region of the transgene, suggesting that models for a block in transcription elongation due to non-B DNA or RNA-DNA hybrids do not fully account for FXN gene silencing.
Figure 1. Model for epigenetic gene silencing in FRDA and activity of HDAC inhibitors.
Gene activity is governed by the balance of activities of the chromatin modifying enzymes, the histone acetyltransferases (HATs), histone deacetylases (HDACs), and histone methyltransferases (HMTases). Histone methylation (me at H3 lysine 9) provides a binding site for heterochromatin protein 1 (HP1), which has been shown to be involved in GAA repeat mediated gene silencing [2]. The HDAC inhibitor 4b was shown to reverse silencing [3]. Taken from Festenstein [54], with permission.
Several laboratories, including our own, have used chromatin immunoprecipitation (ChIP) methods to monitor the histone modifications on the endogenous FXN alleles in cell lines derived from Friedreich ataxia patients and in patient primary cells (peripheral lymphocytes) [3, 5, 41–43]. Yandim and colleagues have recently provided a detailed review of the literature on chromatin modifications associated with FXN gene silencing [44]. The general consensus from these studies is that the first intron of active FXN alleles in cells from unaffected individuals is enriched in acetylated histones H3 and H4, compared with the inactive alleles in Friedreich ataxia cells. Additionally, lysines 9 and 27 of histone H3 (H3K9 and H3K27, respectively) are highly methylated in Friedreich ataxia cells compared with the normal cells. Along with hypoacetylation, trimethylation of H3K9 is a hallmark of heterochromatin, and provides the binding site for heterochromatin protein HP1 [2]. H3K27 methylation is a mark of polycomb group mediated gene silencing [45]. Although the initial studies on FXN chromatin were conducted in Friedreich ataxia lymphoid cells, the same epigenetic differences between active and inactive FXN alleles have also been found in the affected tissues (brain and heart) from mouse models for the disease [4, 46] and in Friedreich ataxia autopsy brain, cerebellum, and heart [4]. Other studies have documented similar heterochromatin marks on pathogenic FXN alleles in neurons derived from patient induced pluripotent stem cells [31]. Several reports have suggested that the chromatin changes associated with pathogenic FXN alleles prevent transcript elongation by RNA polymerase II through expanded GAA•TTC repeats [42, 43, 47, 48] and one report suggested that both transcription initiation and elongation may be affected [42]. More recently, Bidichandani and colleagues have provided strong evidence that initiation of transcription at pathogenic FXN alleles is impaired by the presence of a nucleosome over the start-site both in patient cells [49] and in a mouse model [50]. Interestingly, the degree of promoter silencing and chromatin compaction is directly related to the length of the GAA•TTC repeats [51].
3. Histone deacetylase inhibitors correct frataxin deficiency in Friedreich ataxia
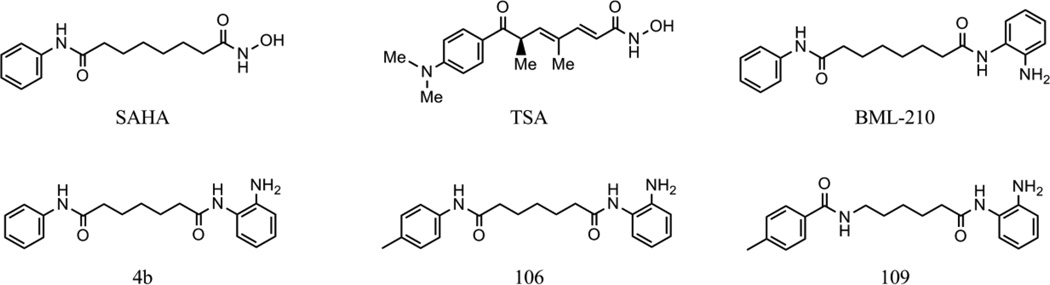
Having established that FXN gene silencing is largely due to the epigenetic chromatin modification state at the repeats and at the promoter element, we and other labs asked whether small molecule inhibitors of chromatin modifying enzymes might be identified that reactivate the gene in patient cells. Our laboratory and Festenstein [20] have focused on histone deacetylase (HDAC) inhibitors as potential therapeutics. Numerous studies have shown that small molecule HDAC inhibitors revert silent heterochromatin to an active chromatin conformation, and restore mRNA synthesis at genes that are silenced in various human diseases, including neurodegenerative and neuromotor diseases [52, 53]. A model for HDAC inhibitor reactivation of the FXN gene is shown in Figure 1 [54]. Eighteen HDAC enzymes have been identified in the human genome, including the zinc-dependent (class I, class II, and class IV) and the NAD+-dependent enzymes (class III or sirtuins). HDACs 1, 2, 3, and 8 belong to class I. Class II is further divided into class IIa (HDACs 4, 5, 7, and 9) and IIb (HDACs 6 and 10), according to their sequence homology and domain organization. HDAC 11 is the lone member of class IV. HDAC inhibitors fall into several chemical classes [55, 56], including small carboxylates (such as sodium butyrate, valproic acid, and sodium phenylbutyrate), hydroxamic acids (such as trichostatin A and suberoylanilide hydroxamic acid), benzamides (such as MS-275), epoxyketones (trapoxins), and cyclic peptides (including apicidin and depsipeptide). The structures of some of these compounds are shown in Figure 2. While the development of HDAC inhibitors as anti-cancer therapeutics is quite advanced [57], interest in the development of HDAC inhibitors for neurodegenerative and neuromotor diseases has increased in the past few years [53, 58].
Figure 2. Structures of the histone deacetylase inhibitors.
Based on our finding of epigenetic silencing marks associated with pathogenic FXN alleles, we asked whether HDAC inhibitors could reverse silencing in FRDA lymphoblasts and primary lymphocytes [3]. We screened a small collection of the HDAC inhibitors discussed above, and found that only a 2-aminobenzamide called BML-210 (Figure 2) was active in restoring FXN gene expression in FRDA cells. Increases in mature FXN mRNA were accompanied by increases in frataxin protein. Our qRT-PCR assays utilized primers that span separate exons, providing additional evidence that RNA processing is not affected by the repeat expansion [28]. We synthesized a number of derivatives of BML-210, including 4b and 106 (Figure 2), and found that these molecules as well were active. Although several compounds, such as suberoylanilide hydroxamic acid (SAHA) and trichostatin A (TSA), showed global cellular HDAC activity (monitored as increases in total histone acetylation), only 2-aminobenzamides increased acetylation at FXN gene chromatin and FXN gene expression. Chromatin immunoprecipitation experiments also showed that the 2-aminobenzamide HDAC inhibitor 4b increases acetylation at particular lysine residues of histones H3 and H4 within intron 1 of FXN (H3-K14, H4-K5, and H4-K12 [3]), while SAHA and TSA did not. Chromatin signatures indicate that histone H3K9 is a key residue for gene silencing through methylation and reactivation through acetylation, mediated by the HDAC inhibitor [31]. Taken together, our results suggest that a specific HDAC enzyme, or HDAC-protein complex, is involved in FXN silencing and 2-aminobenzamides selectively target this protein in cells.
Our initial results with benzamide HDAC inhibitors were obtained with lymphoblast cell lines and primary lymphocytes from FRDA patients [3, 41], but have been extended to two mouse FRDA models [41, 46, 59]. Importantly, the animal experiments document brain penetration of the 2-aminobenzamides and activity in the nervous system and report increases in FXN mRNA and frataxin protein. Bidichandani and colleagues have shown that HDACi 109 (Figure 2) is able to reverse nucleosome occupancy at the FXN promoter and relieve the transcription initiation defect in FRDA patient cells [60]. We recently found increases in FXN gene expression in human FRDA neuronal cells derived from patient induced pluripotent stem cells with 2-aminobenzamide HDAC inhibitors [31]. Additionally, we find that the 2-aminobenzamide HDAC inhibitors increase FXN expression selectively in patient cells, with minor effects on the normal alleles [61, 62], indicating that these compounds are likely acting on the mechanism of silencing induced by the repeats. Significant increase in expression occurs in >80% of patients tested through in vitro cell culture with the majority showing a greater than 2 fold increase [60, 61]. Since heterozygous individuals do not exhibit symptoms of Friedreich ataxia, the epigenetic effects of these compounds may provide a therapeutically useful increase in FXN messenger RNA in patient cells.
Along with studies documenting the activity of 2-aminobenzamide HDAC inhibitors, Festenstein and colleagues have reported that the class III HDAC inhibitor nicotinamide (vitamin B3) has similar properties [20]. These authors reported increases in FXN mRNA levels in patient lymphocytes, lymphoblast cell lines and in the YG8R mouse model. They reported opening of FXN chromatin to nuclease sensitivity, along with small increases in histone acetylation [20]. In contrast to the 2-aminobenzamides that are only active on pathogenic FXN alleles, nicotinamide was reported to increase FXN gene expression in cells from both patients and healthy individuals, suggesting a different mechanism of action for these two compounds. Additionally, millimolar concentrations of nicotinamide are required for activity while the 2-aminobenzamides are active in the low µM range. Another difference between these compounds is that nicotinamide reverses histone H3 methylation (H3K9 and H3K27) whereas benzamides had no effect on histone methylation levels [31]. This observation may suggest that nicotinamide is working through regulation of sirtuin activity on the histone methyltransferases, as reported for Sirt1 [63].
4. Which HDAC isoform is the target of 2-aminobenzamide inhibitors?
We felt that it was essential to know the cellular HDAC enzyme target of our inhibitors for future drug development. Previous studies with other benzamide-type HDAC inhibitors, such as MS-275, indicated that these compounds target the class I HDAC family [56, 64]. We find that the pimelic 2-amimobenzamides, exemplified by compound 106, are also class I histone deacetylase inhibitors, with a moderate preference for HDAC3 over HDACs 1 and 2, with little activity against the other class I HDAC, HDAC8, and little or no activity against class II HDACs [65]. Using an activity profiling approach [66, 67], we were able to identify class I HDACs as the cellular targets of the 2-aminobenzamides [68]. To understand why the 2-aminobenzamides but not other potent HDAC inhibitors, such as the hydoxamates SAHA and TSA, fail to activate FXN gene expression, we determined the kinetic parameters for these compounds with recombinant class I HDACs [65]. While the hydroxamates are rapid-on/rapid-off, classical competitive inhibitors, the 2-aminobenzamides inhibit HDACs 1 and 3 through a slow-on/slow-off mechanism [65]. This property can be documented both with recombinant enzymes and in cellular assays [65, 69]. Small interfering RNA approaches also point to a role for class I HDAC in FXN gene silencing in FRDA cell models [31].
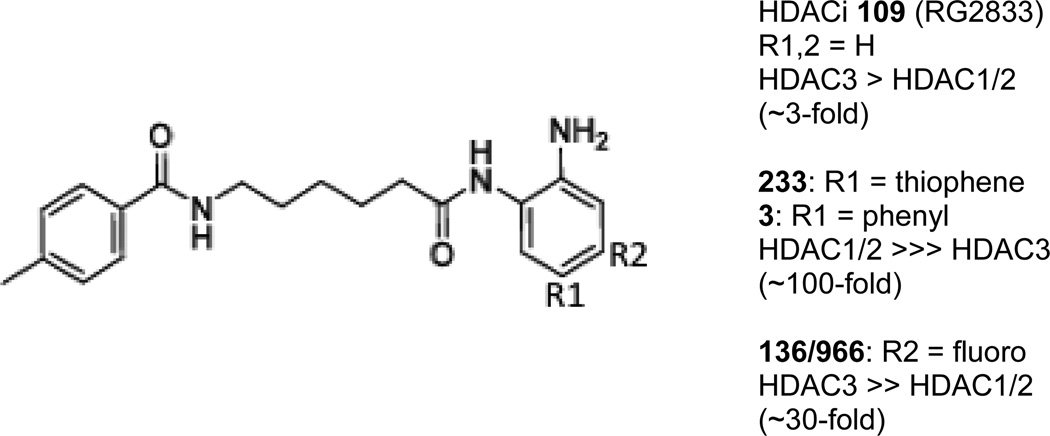
While we have established that 2-aminobenzamides are potent activators of FXN gene expression, given results with nicotinamide [20], the question remained as to whether compounds with other HDAC isoform selectivities would also activate FXN gene expression. To address this issue, we tested compounds with different selectivity profiles in the FRDA lymphoblast cell line [68] and in neuronal cells derived from patient iPSCs [31]. Appending phenyl or thiophene groups at the 5 position of the 2-aminobenzamide ring results in compounds that are ~100 to 300-fold selective for HDACs 1 and 2 over HDAC3 [68, 70, 71]. While HDAC inhibitors 3 (N1-(4-aminobiphenyl-3-yl)-N7-phenylheptanediamide [68], Figure 3) and 233 (N-(2-amino-5- (2-thienyl)phenyl)-7-nicotinoylamino-heptanamide [71], Figure 3) are potent inhibitors of recombinant HDAC1 and in cells, these compounds are without effect on FXN gene expression either in lymphoblasts [68] or iPSC-derived neurons [31]. Also an HDAC3-selective inhibitor (with a fluorine at the 4- position of the benzamide ring; 966; (E)-N-(2-amino-4-fluorophenyl)-3-(1-cinnamyl-1H-pyrazol-4-yl)acrylamide; ~30-fold selectivity for HDAC3 over HDAC1/2 [72], Figure 3) is without effect on FXN mRNA levels in lymphoblasts [51] and in neuronal cells [31]. Potent inhibitors of class II and class III (sirtuin) HDACs also fail to activate FXN expression in the FRDA cells, although each of these compounds is active against known substrates [68]. This latter finding with a potent class III inhibitor (6-chloro-2,3,4,9-tetrahydro-1H-carbazole-1-carboxamide [73]) is striking in light of the claim that nicotinamide is acting as a class III HDAC inhibitor [20]. Recent work has suggested that only compounds that exhibit slow-on/slow-off kinetic properties against the class I HDACs are active in restoring FXN gene expression in FRDA cell models [74].
Figure 3. Modifications to the benzamide scaffold create molecules with isoform selectivity.
The structure of HDACi 109 is shown, indicating the positions of the benzamide ring that can be modified to afford isotype selectivity (R1, R2) and the fold differences in HDAC inhibitory activity for selective compounds are shown at the right.
5. Potential off-target effects of HDACi treatment
As HDAC inhibitors have been shown to induce widespread changes in gene expression in mammalian cells, it was important to address the issue of off-target effects of benzamide HDACi treatment in both unaffected and FRDA cells and animal models. We used high-density microarrays to monitor the effects of HDACi 106 on gene expression in the brain, cerebellum and heart of KIKI and WT mice [46]. 655 genes were affected, showing small (less than two-fold) changes in expression. The relatively limited overall effect of HDACi 106 on expression profiles and the absence of significant dysregulation of apoptosis or tumorigenesis-related genes suggest that severe adverse effects from this compound class are unlikely, at least at the dosages used in this study. A second study utilized lymphocytes from unaffected individuals, carriers and FRDA patients and found that 822 genes had a fold change of 2 or greater in at least one of these cell sources [75]. The majority of the differentially expressed genes induced by HDACi 4b or 106 were shared in lymphocytes from controls, carriers and patients, as expected from the structural similarity and mechanism of action of these compounds. A control compound, lacking the essential 2-amino group of the benzamide ring that was inactive in FXN gene activation, had minimal effects on global gene expression. Overall, these two studies failed to reveal concerns for the use of 2-aminobenzamides as therapeutics for FRDA.
In a second approach to determine the off-target effects of 2-aminobenzamides, we synthesized an activity-based profiling probe (ABPP) of HDACi 106 [68]. In our first study, we used western blotting to monitor capture of class I HDACs by the probe, and indeed identified HDAC3 as a major target [68]. Subsequently, we used a quantitative proteomic method coupled with multidimensional protein identification technology (MudPIT) to identify the proteins captured by the ABPP 106 probe [76]. Nuclear proteins were extracted from FRDA patient iPSC-derived neural stem cells, and then reacted with a control and ABPP 106 probe. The proteins selectively bound by the active probe were determined by mass spectrometry. Beyond the class I HDACs, pathway analysis indicated that the targets of HDACi 106 are not only involved in transcriptional regulation, but also in posttranscriptional processing of mRNA and translational regulation. We did not find targets that would be highly indicative of adverse effects of the compounds. A similar study employing the related 2-aminobenzamide BML-210 found that the major target was the HDAC3-NCoR complex [77]. This study as well failed to uncover targets that would preclude the use of the 2-aminobenzamides as therapeutic for a chronic disease.
Another potential concern for the use of HDAC inhibitors in repeat expansion diseases comes from the finding that activating transcription of a GAA-repeat reporter construct in mammalian cells induces repeat instability, with expansions predominating [78]. To address the effect of HDAC inhibitors and repeat instability, we treated FRDA hiPSC-derived neurons with HDACi 109 for up to 16 days in culture and monitored GAA repeat numbers. While potent increases in FXN mRNA were observed over this incubation period, no effect on GAA repeat length was found [31]. In another study, HDAC complexes were found to induce repeat expansions and inhibition with HDACi 4b was found to inhibit such expansions [79]. Taken together, these results suggest that the 2-aminobenzamides do not induce repeat expansion, at least in cell culture experiments. Lastly, a study of HDACi 109 in the YG8R FRDA mouse model found no significant animal toxicity on long-term (5-month) exposure to the drug [59]. Taken together, these observations suggest that while benzamide HDAC inhibitors have multiple cellular targets, and have significant effects on gene expression, these off-target effects do not appear to raise significant concern for further development of this compound class for FRDA therapy.
6. Human clinical trials
It was found that reversing the orientation of the “left” amide in HDACi 106, resulting in compound 109 (Figure 2), improved brain penetration along with small improvements in HDAC enzyme inhibitory activity [41]. This molecule was found to be active in two mouse models for FRDA and in patient lymphocytes [41, 59]. In the YG8R mouse model, HDACi 109 increased frataxin protein expression in the brain and ameliorated motor deficits. HDACi 109 was also found to be non-toxic on prolonged 5-month chronic treatment [59]. Based on these encouraging results, HDACi 109/RG2833 (the drug formulation product of 109) was subjected to full preclinical, Investigational New Drug (IND) assessment by Repligen Corporation (Waltham, MA) and tested in a phase Ib safety trial in FRDA patients at San Luigi Gonzaga Hospital of the University of Turin, Italy [31]. RG2833 was well tolerated with no drug related adverse effects in this small 20 patient trial. Importantly, the drug was able to increase FXN gene expression in circulating lymphocytes in all but one patient (whose peripheral blood lymphocytes failed to respond to drug treatment ex vivo), and increases in FXN mRNA were observed at drug levels that were predicted to have an observable effect based on in vitro studies. The duration of compound treatment was insufficient to observe statistically significant increases in frataxin protein expression. Along with increases in FXN mRNA, ChIP assays documented increased acetylation at histone H3K9 at the FXN gene in circulating lymphocytes from drug-treated individuals. These findings provided a proof-of-principle for taking the 2-aminobenzamides forward as therapeutics for FRDA. However, assessment of drug metabolic byproducts in serum suggested that further medicinal chemistry efforts were warranted (see below).
Similar to the class I HDAC inhibitor, nicotinamide (vitamin B3) has also been assessed in a human clinical trial in FRDA patients [80]. In this study, FRDA patients were given escalating doses of nicotinamide, ranging from 2 to 8 grams, in both single and multiple doses, over periods of up to 8 weeks. Similar to the RG2833 study, FXN mRNA and protein levels were monitored, along with chromatin signatures. High doses (3.5 to 6 g) resulted in statistically significant increases in frataxin mRNA and protein and a reduction in repressive (methylation) histone modifications along FXN gene chromatin. In terms of safety, high doses of nicotinamide were accompanied by adverse reactions such as nausea (100% of patients in the high dose phase of the study) and vomiting (50%). These side effects were resolved after drug withdrawal or reduction, or with the use of anti-nausea drugs. As expected, this short-term study did not show significant changes in clinical measures of ataxia.
7. Improved pharmacological properties required for human therapy
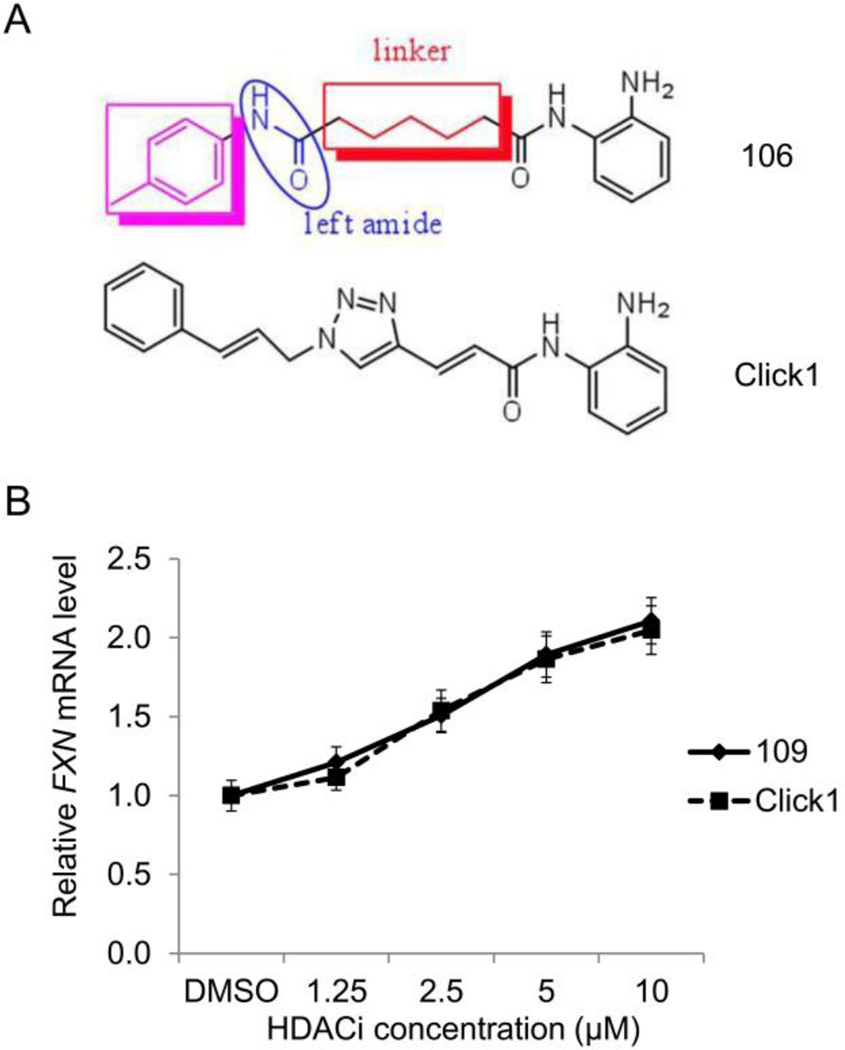
As mentioned above, we tested one Sirt1 selective inhibitor in FRDA lymphoid cells, and found no effect on FXN gene expression. However, to our knowledge, no other study testing other class III HDAC inhibitors in FRDA cells or animal models has been reported. Given the low activity and lack of specificity of nicotinamide, this would seem to be a natural next step in FRDA drug development. Although the 2-aminobenzamides are potent inducers of FXN gene expression in both cellular and animal models, and have potencies greater than 100-fold that reported for nicotinamide (both in vitro and in vivo), this compound class suffers from liabilities that need to be addressed for their use as human therapeutics for a chronic condition such as FRDA. The 2-aminobenzamides have relatively poor distribution to CNS tissue (0.15 brain to serum for HDACi 109) and rearrangements or scission of the amide bond adjacent to the phenylamine ring produce problematic metabolites, such as an inactive benzamidazole and o-phenylenediamine, respectively, the latter being a suspected carcinogen at very high doses (although far exceeding the levels produced by mg quantities of HDACi). Through a medicinal chemistry effort, two structural features that individually improve brain distribution and metabolic stability have been identified [81]. These are replacement of the “left” amide with groups less likely to form intermolecular hydrogen bonds such as ether, olefin, or ketone, each of which can improve brain penetration [81]. Additionally, introduction of an unsaturated linkage adjacent to the “right” amide substantially reduces formation of a benzimidazole metabolic byproduct (Figure 4a). On the basis of these results, we synthesized the new lead compound click-1, using Cu(I)-catalyzed click chemistry (Figure 4a) [81]. This synthetic route allows for the generation of compounds containing both modifications mentioned above, but introduces a triazole into the aliphatic linker region of the pimelic 2-aminobenzamide scaffold. We recently showed that this compound is highly active in the FRDA iPSC-derived neuronal cell model as an inducer of FXN gene expression, with comparable activity to HDACi 109/RG2833 (Figure 4b) [74]. Importantly, click-1 and related compounds demonstrated improved brain penetration (> 0.7 brain/plasma ratio) and stability to benzimidazole formation. Based on these findings, new generations of molecules have been synthesized and are currently in efficacy and preclinical testing. Among the derivatives synthesized, modifications of the cap group (Figure 4a) and in the linker region (introducing new heterocycles, phenyl groups and other modifications) are being assessed.
Figure 4. Compounds with improved pharmacological properties.
(A) Structure of 106, highlighting the cap group (purple), left amide, and linker region. Brain penetration can be improved by elimination of the left amide, and replacement with an ether, olefin, or ketone. Metabolic stability can be improved by introducing a non-saturated linkage adjacent to the right amide, which prevents formation of a benzimidazole. Cu(I)-catalyzed click chemistry was used to generate compound click-1, where a triazole is generated in the linker region of the histone deacetylase inhibitor [81]. (B) Efficacy of click-1 in neuronal cells derived from FRDA iPSCs, as measured by qRT-PCR, compared to HDACi 109.
8. Expert opinion
In the course of developing small molecule therapeutics for FRDA, the field has experienced both success in early human clinical trials [31, 80] and other results suggesting that further medicinal chemistry efforts are necessary to find compounds that will be effective, well tolerated and non-toxic in patients. We will consider both nicotinamide and the 2-aminobenzamides in the following section. Given the large amounts of nicotinamide needed for observed increases in frataxin and the observed side effects [80], the doses required for efficacy might be limiting for chronic treatment of a life-long condition such as Friedreich ataxia. Therefore, it seems essential to know the precise molecular targets involved in FXN gene activation by vitamin B3. Since nicotinamide is relatively non-specific and could well have multiple cellular targets, we argue that such knowledge is a prerequisite for further clinical development. With such data in hand, it might be reasonable to pursue new chemical entities as modulators of the relevant target of nicotinamide. While Festenstein and colleagues have argued that nicotinamide is acting as an HDAC inhibitor with selectivity for class III HDACs [20, 80], no studies have documented that down regulation of any class III HDAC (sirtuin) by small interfering RNA methods can reproduce the effects of nicotinamide in FRDA cells, as shown for the class I HDACs [74]. Likewise, no ChIP experiments have been published to document the association of a sirtuin with silent FXN alleles in FRDA cells. We believe that such data are essential for future development of a class III inhibitor for FRDA.
For both nicotinamide and the 2-aminobenzamides, the observed changes in chromatin postsynthetic modifications on FXN gene chromatin could well be a consequence of gene activation through a different mechanism of action rather than direct activation of FXN by HDAC inhibition. For example, we recently showed that other genes might be the direct target of the 2-aminobenzamides, leading to FXN gene activation [74]. By interrogating microarray data from neuronal cells treated with inhibitors of different specificity (109, 966 and 233; Figure 2), we identified two genes, H2AFY2 and PCGF2, that were specifically down regulated by the inhibitors targeting HDACs1–3 versus the more selective inhibitors. These results were validated by qRT-PCR methods using the pan-class I inhibitors 109 and click 1 (Figure 3) along with the HDAC3 (966) and HDAC1/2 (233) selective compounds. Both of these genes are known to be involved in transcriptional repression and hence repression of these genes could in turn have a positive impact on FXN gene chromatin structure and transcriptional activity. H2AFY2 encodes a macroH2A protein that has been implicated in X chromosome inactivation, gene repression and genomic imprinting (reviewed in [82]). PCGF2 is a negative regulator of developmentally important genes, and specifically involved in polycomb H3K27 methylation-mediated gene repression (reviewed in [45]). While these data provide a correlation between HDACi 109 treatment, up-regulation of FXN and down-regulation of these two gene products, future studies, such as siRNA silencing and ChIP assays, are needed to address whether these two proteins, macroH2A and Polycomb group ring finger 2 are indeed directly involved in FXN gene expression in FRDA neuronal cells.
While our recent findings [31] provide a proof of concept that patient derived neuronal cells can be a quantitative screening tool for the development of an epigenetic therapy for FRDA, the compound used in our clinical study, 109/RG2833, suffers from liabilities for chronic use as FRDA therapeutics; namely, less than optimal brain penetration, and conversion of the active molecule into inactive, potentially toxic metabolic products (benzimidazole and products of amidolysis) that are poorly eliminated in vivo [81, 83]. Other preclinical studies placed RG2833 and the benzimidazole in the high-risk category for inducing QTc prolongation [31]. Given the cardiac involvement in FRDA, molecules that produce this benzimidazole metabolite are unlikely to be useful for chronic treatment of FRDA. We therefore searched for chemical modifications to the pimelic 2-aminobenzamide scaffold that would circumvent these liabilities. As described earlier [81], replacement at the “left” amide in the original scaffold were found to improve brain penetration, and modifications adjacent to the “right” amide prevented cyclization to the benzimidazole, and also reduced amidolysis. Compounds with these features exhibit no loss of activity compared to HDACi 109 (Figure 3b). Thus, new derivatives are candidates for future clinical studies in FRDA. If oral delivery of an HDAC inhibitor can be effective at drug exposures that are well tolerated, it will provide impetus to further advance the 2-aminobenzamide class of compounds to target histone deacetylase inhibition as a viable therapeutic strategy for this devastating disease.
Article highlights.
Friedreich ataxia is an autosomal recessive neurodegenerative disease caused by a GAA·TCC triplet expansion in the first intron of the FXN gene, encoding the essential mitochondrial protein frataxin.
Frataxin is involved in the synthesis of iron-sulfur clusters, and transfer of Fe-S clusters to mitochondrial enzymes and components of the electron transport chain.
Patients suffer from both neurological symptoms and cardiomyopathy due to reduced levels of frataxin protein.
GAA·TCC repeats cause epigenetic gene silencing, reducing levels of FXN mRNA and protein, which can be reversed with histone deacetylase (HDAC) inhibitors.
Two HDAC inhibitors have been evaluated in human clinical studies, but both molecules require improvements for use in chronic diseases.
Medicinal chemistry efforts have revealed modifications that improve pharmacological properties of 2-aminobenzamide HDAC inhibitors, which may lead to compounds for future clinical investigation.
Acknowledgments
Funding
Studies in the Gottesfeld laboratory have been supported by grants from the National Institute of Neurological Disorders and Stroke, National Institutes of Health (R01 NS063856), the Friedreich’a Ataxia Research Alliance, GoFAR (Italy), Ataxia UK, and Friedreich’s Ataxia Society Ireland, and by a sponsored research agreement with BioMarin Pharmaceutical.
JM Gottesfeld is an inventor on US patent applications 20070219244, 20110021562, 20130210918 and US Patent 8,835,502 B2 (licensees, Repligen Corporation/BioMarin Pharmaceuticals). JM Gottesfeld serves as a consultant to BioMarin Pharmaceuticals. The terms of this arrangement are managed by the Scripps Research Institute. Intellectual property has been licensed by The Scripps Research Institute to Repligen Corporation and BioMarin Pharmaceuticals, and The Scripps Research Institute and JMG have a financial interest in this technology. We wish to thank Dr. James Rusche and his associates at Repligen Corporation for their invaluable contributions to this work by taking the HDAC inhibitors into a phase Ib human clinical trial. We also thank Dr. Shripad Bhagwat and his colleagues at BioMarin for taking these compounds further toward clinical stage development.
Footnotes
Declaration of interest
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
References
Reference annotations
* Of interest
** Of considerable interest
- 1.Campuzano V, Montermini L, Molto MD, Pianese L, Cossee M, Cavalcanti F, Monros E, Rodius F, Duclos F, Monticelli A, et al. Friedreich's ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science. 1996;271:1423–1427. doi: 10.1126/science.271.5254.1423. [DOI] [PubMed] [Google Scholar]
- 2.Saveliev A, Everett C, Sharpe T, Webster Z, Festenstein R. DNA triplet repeats mediate heterochromatin-protein-1-sensitive variegated gene silencing. Nature. 2003;422:909–913. doi: 10.1038/nature01596. [DOI] [PubMed] [Google Scholar]
- 3.Herman D, Jenssen K, Burnett R, Soragni E, Perlman SL, Gottesfeld JM. Histone deacetylase inhibitors reverse gene silencing in Friedreich's ataxia. Nat Chem Biol. 2006;2:551–558. doi: 10.1038/nchembio815. [DOI] [PubMed] [Google Scholar]
- 4.Al-Mahdawi S, Pinto RM, Ismail O, Varshney D, Lymperi S, Sandi C, Trabzuni D, Pook M. The Friedreich ataxia GAA repeat expansion mutation induces comparable epigenetic changes in human and transgenic mouse brain and heart tissues. Hum Mol Genet. 2008;17:735–746. doi: 10.1093/hmg/ddm346. [DOI] [PubMed] [Google Scholar]
- 5.Greene E, Mahishi L, Entezam A, Kumari D, Usdin K. Repeat-induced epigenetic changes in intron 1 of the frataxin gene and its consequences in Friedreich ataxia. Nucl. Acids Res. 2007;35:3383–3390. doi: 10.1093/nar/gkm271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Biase I, Chutake YK, Rindler PM, Bidichandani SI. Epigenetic Silencing in Friedreich Ataxia Is Associated with Depletion of CTCF (CCCTC-Binding Factor) and Antisense Transcription. PLoS One. 2009;4:e7914. doi: 10.1371/journal.pone.0007914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Metz G, Coppard N, Cooper JM, Delatycki MB, Durr A, Di Prospero NA, Giunti P, Lynch DR, Schulz JB, Rummey C, Meier T. Rating disease progression of Friedreich's ataxia by the International Cooperative Ataxia Rating Scale: analysis of a 603-patient database. Brain. 2013;136:259–268. doi: 10.1093/brain/aws309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cossee M, Durr A, Schmitt M, Dahl N, Trouillas P, Allinson P, Kostrzewa M, Nivelon-Chevallier A, Gustavson KH, Kohlschutter A, Muller U, Mandel JL, Brice A, Koenig M, Cavalcanti F, Tammaro A, De Michele G, Filla A, Cocozza S, Labuda M, Montermini L, Poirier J, Pandolfo M. Friedreich's ataxia: point mutations and clinical presentation of compound heterozygotes. Ann Neurol. 1999;45:200–206. doi: 10.1002/1531-8249(199902)45:2<200::aid-ana10>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 9.Tsou AY, Paulsen EK, Lagedrost SJ, Perlman SL, Mathews KD, Wilmot GR, Ravina B, Koeppen AH, Lynch DR. Mortality in Friedreich ataxia. J Neurol Sci. 2011;307:46–49. doi: 10.1016/j.jns.2011.05.023. [DOI] [PubMed] [Google Scholar]
- 10.Pastore A, Puccio H. Frataxin: a protein in search for a function. J Neurochem. 2013;126(Suppl 1):43–52. doi: 10.1111/jnc.12220. [DOI] [PubMed] [Google Scholar]
- 11.Maio N, Rouault TA. Iron-sulfur cluster biogenesis in mammalian cells: New insights into the molecular mechanisms of cluster delivery. Biochim Biophys Acta. 2015;1853:1493–1512. doi: 10.1016/j.bbamcr.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martelli A, Puccio H. Dysregulation of cellular iron metabolism in Friedreich ataxia: from primary iron-sulfur cluster deficit to mitochondrial iron accumulation. Front Pharmacol. 2014;5:130. doi: 10.3389/fphar.2014.00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parkinson MH, Schulz JB, Giunti P. Co-enzyme Q10 and idebenone use in Friedreich's ataxia. J Neurochem. 2013;126(Suppl 1):125–141. doi: 10.1111/jnc.12322. [DOI] [PubMed] [Google Scholar]
- 14.Pandolfo M, Hausmann L. Deferiprone for the treatment of Friedreich's ataxia. J Neurochem. 2013;126(Suppl 1):142–146. doi: 10.1111/jnc.12300. [DOI] [PubMed] [Google Scholar]
- 15.Puccio H, Anheim M, Tranchant C. Pathophysiogical and therapeutic progress in Friedreich ataxia. Rev Neurol (Paris) 2014;170:355–365. doi: 10.1016/j.neurol.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 16.Strawser CJ, Schadt KA, Lynch DR. Therapeutic approaches for the treatment of Friedreich's ataxia. Expert Rev Neurother. 2014;14:949–957. doi: 10.1586/14737175.2014.939173. [DOI] [PubMed] [Google Scholar]
- 17.Elincx-Benizri S, Glik A, Merkel D, Arad M, Freimark D, Kozlova E, Cabantchik I, Hassin-Baer S. Clinical Experience With Deferiprone Treatment for Friedreich Ataxia. J Child Neurol. 2016 doi: 10.1177/0883073816636087. [DOI] [PubMed] [Google Scholar]
- 18.Perdomini M, Belbellaa B, Monassier L, Reutenauer L, Messaddeq N, Cartier N, Crystal RG, Aubourg P, Puccio H. Prevention and reversal of severe mitochondrial cardiomyopathy by gene therapy in a mouse model of Friedreich's ataxia. Nat Med. 2014;20:542–547. doi: 10.1038/nm.3510. [DOI] [PubMed] [Google Scholar]
- 19.Vyas PM, Tomamichel WJ, Pride PM, Babbey CM, Wang Q, Mercier J, Martin EM, Payne RM. A TAT-Frataxin fusion protein increases lifespan and cardiac function in a conditional Friedreich's Ataxia mouse model. Hum Mol Genet. 2011 doi: 10.1093/hmg/ddr554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan PK, Torres R, Yandim C, Law PP, Khadayate S, Mauri M, Grosan C, Chapman-Rothe N, Giunti P, Pook M, Festenstein R. Heterochromatinization induced by GAA-repeat hyperexpansion in Friedreich's ataxia can be reduced upon HDAC inhibition by vitamin B3. Hum Mol Genet. 2013;22:2662–2675. doi: 10.1093/hmg/ddt115. [DOI] [PubMed] [Google Scholar]
- 21.Acquaviva F, Castaldo I, Filla A, Giacchetti M, Marmolino D, Monticelli A, Pinelli M, Sacca F, Cocozza S. Recombinant human erythropoietin increases frataxin protein expression without increasing mRNA expression. Cerebellum. 2008;7:360–365. doi: 10.1007/s12311-008-0036-x. [DOI] [PubMed] [Google Scholar]
- 22.Sahdeo S, Scott BD, McMackin MZ, Jasoliya M, Brown B, Wulff H, Perlman SL, Pook MA, Cortopassi GA. Dyclonine rescues frataxin deficiency in animal models and buccal cells of patients with Friedreich's ataxia. Hum Mol Genet. 2014;23:6848–6862. doi: 10.1093/hmg/ddu408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tomassini B, Arcuri G, Fortuni S, Sandi C, Ezzatizadeh V, Casali C, Condo I, Malisan F, Al-Mahdawi S, Pook M, Testi R. Interferon gamma upregulates frataxin and corrects the functional deficits in a Friedreich ataxia model. Hum Mol Genet. 2012;21:2855–2861. doi: 10.1093/hmg/dds110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cherubini F, Serio D, Guccini I, Fortuni S, Arcuri G, Condo I, Rufini A, Moiz S, Camerini S, Crescenzi M, Testi R, Malisan F. Src inhibitors modulate frataxin protein levels. Hum Mol Genet. 2015;24:4296–4305. doi: 10.1093/hmg/ddv162. [DOI] [PubMed] [Google Scholar]
- 25.Li L, Matsui M, Corey DR. Activating frataxin expression by repeat-targeted nucleic acids. Nat Commun. 2016;7:10606. doi: 10.1038/ncomms10606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rufini A, Fortuni S, Arcuri G, Condo I, Serio D, Incani O, Malisan F, Ventura N, Testi R. Preventing the ubiquitin-proteasome-dependent degradation of frataxin, the protein defective in Friedreich's ataxia. Hum Mol Genet. 2011;20:1253–1261. doi: 10.1093/hmg/ddq566. [DOI] [PubMed] [Google Scholar]
- 27.Ohshima K, Montermini L, Wells RD, Pandolfo M. Inhibitory effects of expanded GAA.TTC triplet repeats from intron I of the Friedreich ataxia gene on transcription and replication in vivo. J Biol Chem. 1998;273:14588–14595. doi: 10.1074/jbc.273.23.14588. [DOI] [PubMed] [Google Scholar]
- 28.Bidichandani SI, Ashizawa T, Patel PI. The GAA triplet-repeat expansion in Friedreich ataxia interferes with transcription and may be associated with an unusual DNA structure. Am J Hum Genet. 1998;62:111–121. doi: 10.1086/301680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baralle M, Pastor T, Bussani E, Pagani F. Influence of Friedreich ataxia GAA noncoding repeat expansions on pre-mRNA processing. Am J Hum Genet. 2008;83:77–88. doi: 10.1016/j.ajhg.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zoghbi HY, Orr HT. Trinucleotide repeat disorders. Ann Rev. Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 31.Soragni E, Miao W, Iudicello M, Jacoby D, Demercanti S, Clerico M, Longo F, Piga A, Ku S, Campau E, Du J, Penalver P, Rai M, Madara JC, Nazor K, O'Connor M, Maximov A, Loring JF, Pandolfo M, Durelli L, Gottesfeld JM, Rusche JR. Epigenetic therapy for Friedreich's ataxia. Ann Neurol. 2014;76:489–508. doi: 10.1002/ana.24260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wells RD. DNA triplexes and Friedreich ataxia. FASEB J. 2008;22:1625–1634. doi: 10.1096/fj.07-097857. [DOI] [PubMed] [Google Scholar]
- 33.Sakamoto N, Ohshima K, Montermini L, Pandolfo M, Wells RD. Sticky DNA, a self-associated complex formed at long GAA*TTC repeats in intron 1 of the frataxin gene, inhibits transcription. J Biol Chem. 2001;276:27171–27177. doi: 10.1074/jbc.M101879200. [DOI] [PubMed] [Google Scholar]
- 34.Gacy AM, Goellner GM, Spiro C, Chen X, Gupta G, Bradbury EM, Dyer RB, Mikesell MJ, Yao JZ, Johnson AJ, Richter A, Melancon SB, McMurray CT. GAA instability in Friedreich's Ataxia shares a common, DNA-directed and intraallelic mechanism with other trinucleotide diseases. Mol Cell. 1998;1:583–593. doi: 10.1016/s1097-2765(00)80058-1. [DOI] [PubMed] [Google Scholar]
- 35.Mariappan SV, Catasti P, Silks LA, 3rd, Bradbury EM, Gupta G. The high-resolution structure of the triplex formed by the GAA/TTC triplet repeat associated with Friedreich's ataxia. J Mol Biol. 1999;285:2035–2052. doi: 10.1006/jmbi.1998.2435. [DOI] [PubMed] [Google Scholar]
- 36.Burnett R, Melander C, Puckett JW, Son LS, Wells RD, Dervan PB, Gottesfeld JM. DNA sequence-specific polyamides alleviate transcription inhibition associated with long GAA.TTC repeats in Friedreich's ataxia. Proc Natl Acad Sci U S A. 2006;103:11497–11502. doi: 10.1073/pnas.0604939103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harki DA, Satyamurthy N, Stout DB, Phelps ME, Dervan PB. In vivo imaging of pyrrole-imidazole polyamides with positron emission tomography. Proc. Natl. Acad. Sci. U.S.A. 2008;105:13039–13044. doi: 10.1073/pnas.0806308105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grabczyk E, Usdin K. Alleviating transcript insufficiency caused by Friedreich's ataxia triplet repeats. Nucleic Acids Res. 2000;28:4930–4937. doi: 10.1093/nar/28.24.4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grabczyk E, Usdin K. The GAA*TTC triplet repeat expanded in Friedreich's ataxia impedes transcription elongation by T7 RNA polymerase in a length and supercoil dependent manner. Nucleic Acids Res. 2000;28:2815–2822. doi: 10.1093/nar/28.14.2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Groh M, Lufino MM, Wade-Martins R, Gromak N. R-loops associated with triplet repeat expansions promote gene silencing in Friedreich ataxia and fragile X syndrome. PLoS Genet. 2014;10:e1004318. doi: 10.1371/journal.pgen.1004318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rai M, Soragni E, Chou CJ, Barnes G, Jones S, Rusche JR, Gottesfeld JM, Pandolfo M. Two new pimelic diphenylamide HDAC inhibitors induce sustained frataxin upregulation in cells from Friedreich's ataxia patients and in a mouse model. PLoS One. 2010;5:e8825. doi: 10.1371/journal.pone.0008825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kumari D, Biacsi RE, Usdin K. Repeat expansion affects both transcription initiation and elongation in friedreich ataxia cells. J Biol Chem. 2011;286:4209–4215. doi: 10.1074/jbc.M110.194035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim E, Napierala M, Dent SY. Hyperexpansion of GAA repeats affects post-initiation steps of FXN transcription in Friedreich's ataxia. Nucleic Acids Res. 2011;39:8366–8377. doi: 10.1093/nar/gkr542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yandim C, Natisvili T, Festenstein R. Gene regulation and epigenetics in Friedreich's ataxia. J Neurochem. 2013;126(Suppl 1):21–42. doi: 10.1111/jnc.12254. [DOI] [PubMed] [Google Scholar]
- 45.Di Croce L, Helin K. Transcriptional regulation by Polycomb group proteins. Nat Struct Mol Biol. 2013;20:1147–1155. doi: 10.1038/nsmb.2669. [DOI] [PubMed] [Google Scholar]
- 46.Rai M, Soragni E, Jenssen K, Burnett R, Herman D, Gottesfeld JM, Pandolfo M. HDAC inhibitors correct frataxin deficiency in a Friedreich ataxia mouse model. PLoS ONE. 2008;3:e1958. doi: 10.1371/journal.pone.0001958. doi 1910.1371/journal.pone.0001958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Punga T, Buhler M. Long intronic GAA repeats causing Friedreich ataxia impede transcription elongation. EMBO Mol Med. 2010;2:120–129. doi: 10.1002/emmm.201000064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li Y, Lu Y, Polak U, Lin K, Shen J, Farmer J, Seyer L, Bhalla AD, Rozwadowska N, Lynch DR, Butler JS, Napierala M. Expanded GAA repeats impede transcription elongation through the FXN gene and induce transcriptional silencing that is restricted to the FXN locus. Hum Mol Genet. 2015;24:6932–6943. doi: 10.1093/hmg/ddv397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chutake YK, Costello WN, Lam C, Bidichandani SI. Altered nucleosome positioning at the transcription start site and deficient transcriptional initiation in Friedreich ataxia. J Biol Chem. 2014;289:15194–15202. doi: 10.1074/jbc.M114.566414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chutake YK, Costello WN, Lam CC, Parikh AC, Hughes TT, Michalopulos MG, Pook MA, Bidichandani SI. FXN Promoter Silencing in the Humanized Mouse Model of Friedreich Ataxia. PLoS One. 2015;10:e0138437. doi: 10.1371/journal.pone.0138437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chutake YK, Lam C, Costello WN, Anderson M, Bidichandani SI. Epigenetic promoter silencing in Friedreich ataxia is dependent on repeat length. Ann Neurol. 2014;76:522–528. doi: 10.1002/ana.24249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Di Prospero NA, Fischbeck KH. Therapeutics development for triplet repeat expansion diseases. Nat Rev Genet. 2005;6:756–765. doi: 10.1038/nrg1690. [DOI] [PubMed] [Google Scholar]
- 53.Kazantsev AG, Thompson LM. Therapeutic application of histone deacetylase inhibitors for central nervous system disorders. Nat. Rev. Drug Discov. 2008;7:854–868. doi: 10.1038/nrd2681. [DOI] [PubMed] [Google Scholar]
- 54.Festenstein R. Breaking the silence in Friedreich's ataxia. Nat Chem Biol. 2006;2:512–513. doi: 10.1038/nchembio1006-512. [DOI] [PubMed] [Google Scholar]
- 55.Cole PA. Chemical probes for histone-modifying enzymes. Nat Chem Biol. 2008;4:590–597. doi: 10.1038/nchembio.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bradner JE, West N, Grachan ML, Greenberg EF, Haggarty SJ, Warnow T, Mazitschek R. Chemical phylogenetics of histone deacetylases. Nat. Chem. Biol. 2010;6:238–243. doi: 10.1038/nchembio.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ahuja N, Sharma AR, Baylin SB. Epigenetic Therapeutics: A New Weapon in the War Against Cancer. Annu Rev Med. 2016;67:73–89. doi: 10.1146/annurev-med-111314-035900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Irwin MH, Moos WH, Faller DV, Steliou K, Pinkert CA. Epigenetic Treatment of Neurodegenerative Disorders: Alzheimer and Parkinson Diseases. Drug Dev Res. 2016;77:109–123. doi: 10.1002/ddr.21294. [DOI] [PubMed] [Google Scholar]
- 59.Sandi C, Pinto RM, Al-Mahdawi S, Ezzatizadeh V, Barnes G, Jones S, Rusche JR, Gottesfeld JM, Pook MA. Prolonged treatment with pimelic o-aminobenzamide HDAC inhibitors ameliorates the disease phenotype of a Friedreich ataxia mouse model. Neurobiol Dis. 2011;42:496–505. doi: 10.1016/j.nbd.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chutake YK, Lam CC, Costello WN, Anderson MP, Bidichandani SI. Reversal of epigenetic promoter silencing in Friedreich ataxia by a class I histone deacetylase inhibitor. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Plasterer HL, Deutsch EC, Belmonte M, Egan E, Lynch DR, Rusche JR. Development of frataxin gene expression measures for the evaluation of experimental treatments in Friedreich's ataxia. PLoS One. 2013;8:e63958. doi: 10.1371/journal.pone.0063958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Soragni E, Xu C, Plasterer HL, Jacques V, Rusche JR, Gottesfeld JM. Rationale for the development of 2-aminobenzamide histone deacetylase inhibitors as therapeutics for Friedreich ataxia. J Child Neurol. 2012;27:1164–1173. doi: 10.1177/0883073812448533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vaquero A, Scher M, Erdjument-Bromage H, Tempst P, Serrano L, Reinberg D. SIRT1 regulates the histone methyl-transferase SUV39H1 during heterochromatin formation. Nature. 2007;450:440–444. doi: 10.1038/nature06268. [DOI] [PubMed] [Google Scholar]
- 64.Beckers T, et al. Distinct pharmacological properties of second generation HDAC inhibitors with the benzamide or hydroxamate head group. Int. J. Cancer. 2007;121:1138. doi: 10.1002/ijc.22751. [DOI] [PubMed] [Google Scholar]
- 65.Chou CJ, Herman D, Gottesfeld JM. Pimelic diphenylamide 106 is a slow, tight-binding inhibitor of class I histone deacetylases. J. Biol. Chem. 2008;283:35402–35409. doi: 10.1074/jbc.M807045200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Evans MJ, Cravatt BF. Mechanism-based profiling of enzyme families. Chem Rev. 2006;106:3279–3301. doi: 10.1021/cr050288g. [DOI] [PubMed] [Google Scholar]
- 67.Salisbury CM, Cravatt BF. Activity-based probes for proteomic profiling of histone deacetylase complexes. Proc Natl Acad Sci U S A. 2007;104:1171–1176. doi: 10.1073/pnas.0608659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu C, Soragni E, Chou CJ, Herman D, Plasterer HL, Rusche JR, Gottesfeld JM. Chemical probes identify a role for histone deacetylase 3 in Friedreich's ataxia gene silencing. Chem. Biol. 2009;16:980–989. doi: 10.1016/j.chembiol.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lauffer BE, Mintzer R, Fong R, Mukund S, Tam C, Zilberleyb I, Flicke B, Ritscher A, Fedorowicz G, Vallero R, Ortwine DF, Gunzner J, Modrusan Z, Neumann L, Koth CM, Lupardus PJ, Kaminker JS, Heise CE, Steiner P. Histone deacetylase (HDAC) inhibitor kinetic rate constants correlate with cellular histone acetylation but not transcription and cell viability. J Biol Chem. 2013;288:26926–26943. doi: 10.1074/jbc.M113.490706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Methot JL, Chakravarty PK, Chenard M, Close J, Cruz JC, Dahlberg WK, Fleming J, Hamblett CL, Hamill JE, Harrington P, Harsch A, Heidebrecht R, Hughes B, Jung J, Kenific CM, Kral AM, Meinke PT, Middleton RE, Ozerova N, Sloman DL, Stanton MG, Szewczak AA, Tyagarajan S, Witter DJ, Secrist JP, Miller TA. Exploration of the internal cavity of histone deacetylase (HDAC) with selective HDAC1/HDAC2 inhibitors (SHI-1-2) Bioorg Med Chem Lett. 2008;18:973–978. doi: 10.1016/j.bmcl.2007.12.031. [DOI] [PubMed] [Google Scholar]
- 71.Jia H, Pallos J, Jacques V, Lau A, Tang B, Cooper A, Syed A, Purcell J, Chen Y, Sharma S, Sangrey GR, Darnell SB, Plasterer H, Sadri-Vakili G, Gottesfeld JM, Thompson LM, Rusche JR, Marsh JL, Thomas EA. Histone deacetylase (HDAC) inhibitors targeting HDAC3 and HDAC1 ameliorate polyglutamine-elicited phenotypes in model systems of Huntington's disease. Neurobiol Dis. 2012;46:351–361. doi: 10.1016/j.nbd.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Malvaez M, McQuown SC, Rogge GA, Astarabadi M, Jacques V, Carreiro S, Rusche JR, Wood MA. HDAC3-selective inhibitor enhances extinction of cocaine-seeking behavior in a persistent manner. Proc Natl Acad Sci U S A. 2013;110:2647–2652. doi: 10.1073/pnas.1213364110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Napper AD, Hixon J, McDonagh T, Keavey K, Pons JF, Barker J, Yau WT, Amouzegh P, Flegg A, Hamelin E, Thomas RJ, Kates M, Jones S, Navia MA, Saunders JO, DiStefano PS, Curtis R. Discovery of indoles as potent and selective inhibitors of the deacetylase SIRT1. J Med Chem. 2005;48:8045–8054. doi: 10.1021/jm050522v. [DOI] [PubMed] [Google Scholar]
- 74.Soragni E, Chou CJ, Rusche JR, Gottesfeld JM. Mechanism of Action of 2-Aminobenzamide HDAC Inhibitors in Reversing Gene Silencing in Friedreich's Ataxia. Front Neurol. 2015;6:44. doi: 10.3389/fneur.2015.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Coppola G, Burnett R, Perlman S, Versano R, Gao F, Plasterer H, Rai M, Sacca F, Filla A, Lynch DR, Rusche JR, Gottesfeld JM, Pandolfo M, Geschwind DH. A gene expression phenotype in lymphocytes from Friedreich ataxia patients. Ann Neurol. 2011;70:790–804. doi: 10.1002/ana.22526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shan B, Xu C, Zhang Y, Xu T, Gottesfeld JM, Yates JR., 3rd Quantitative proteomic analysis identifies targets and pathways of a 2-aminobenzamide HDAC inhibitor in Friedreich's ataxia patient iPSC-derived neural stem cells. J Proteome Res. 2014;13:4558–4566. doi: 10.1021/pr500514r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bantscheff M, Hopf C, Savitski MM, Dittmann A, Grandi P, Michon AM, Schlegl J, Abraham Y, Becher I, Bergamini G, Boesche M, Delling M, Dumpelfeld B, Eberhard D, Huthmacher C, Mathieson T, Poeckel D, Reader V, Strunk K, Sweetman G, Kruse U, Neubauer G, Ramsden NG, Drewes G. Chemoproteomics profiling of HDAC inhibitors reveals selective targeting of HDAC complexes. Nat Biotechnol. 2010;29:255–265. doi: 10.1038/nbt.1759. [DOI] [PubMed] [Google Scholar]
- 78.Ditch S, Sammarco MC, Banerjee A, Grabczyk E. Progressive GAA·TTC Repeat Expansion in Human Cell Lines. PLoS Genet. 2009;5:e1000704. doi: 10.1371/journal.pgen.1000704. doi:1000710.1001371/journal.pgen.1000704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Debacker K, Frizzell A, Gleeson O, Kirkham-McCarthy L, Mertz T, Lahue RS. Histone deacetylase complexes promote trinucleotide repeat expansions. PLoS Biol. 2012;10:e1001257. doi: 10.1371/journal.pbio.1001257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Libri V, Yandim C, Athanasopoulos S, Loyse N, Natisvili T, Law PP, Chan PK, Mohammad T, Mauri M, Tam KT, Leiper J, Piper S, Ramesh A, Parkinson MH, Huson L, Giunti P, Festenstein R. Epigenetic and neurological effects and safety of high-dose nicotinamide in patients with Friedreich's ataxia: an exploratory, open-label, dose-escalation study. Lancet. 2014 doi: 10.1016/S0140-6736(14)60382-2. [DOI] [PubMed] [Google Scholar]
- 81.Xu C, Soragni E, Jacques V, Rusche JR, Gottesfeld JM. Improved Histone Deacetylase Inhibitors as Therapeutics for the Neurodegenerative Disease Friedreich's Ataxia: A New Synthetic Route. Pharmaceuticals. 2011;4:1578–1590. doi: 10.3390/ph4121578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Biterge B, Schneider R. Histone variants: key players of chromatin. Cell Tissue Res. 2014;356:457–466. doi: 10.1007/s00441-014-1862-4. [DOI] [PubMed] [Google Scholar]
- 83.Beconi M, Aziz O, Matthews K, Moumne L, O'Connell C, Yates D, Clifton S, Pett H, Vann J, Crowley L, Haughan AF, Smith DL, Woodman B, Bates GP, Brookfield F, Burli RW, McAllister G, Dominguez C, Munoz-Sanjuan I, Beaumont V. Oral Administration of the Pimelic Diphenylamide HDAC Inhibitor HDACi 4b Is Unsuitable for Chronic Inhibition of HDAC Activity in the CNS In Vivo. PLoS One. 2012;7:e44498. doi: 10.1371/journal.pone.0044498. [DOI] [PMC free article] [PubMed] [Google Scholar]