Abstract
Newly synthesized proteins are triaged between biosynthesis and degradation to maintain cellular homeostasis, but the decision-making mechanisms are unclear. Here, we reconstitute the core reactions for membrane targeting and ubiquitination of nascent tail-anchored membrane proteins to understand how their fate is determined. The central six-component triage system is divided into an uncommitted client-SGTA complex, a self-sufficient targeting module, and an embedded but self-sufficient quality control module. Client-SGTA engagement of the targeting module induces rapid, private, and committed client transfer to TRC40 for successful biosynthesis. Commitment to ubiquitination is dictated primarily by comparatively slower client dissociation from SGTA and non-private capture by the Bag6 subunit of the quality control module. This provides a paradigm for how priority and time are encoded within a multi-chaperone triage system.
Protein biosynthesis and quality control pathways are precisely balanced to provide nascent proteins an initial opportunity to mature, while favoring degradation over time (1–3). Deviations from this balance lead to premature degradation of normal maturation intermediates or persistence of misfolded proteins, either of which can cause disease (2, 4, 5). Although accurate triage between biosynthesis and degradation is essential for maintaining protein homeostasis, a mechanistic understanding of protein triage for any pathway has been hampered by the lack of a fully reconstituted system that faithfully recapitulates both the biosynthetic and quality control outcomes of a nascent protein.
To achieve this goal, we chose the pathway of tail-anchored (TA) membrane protein insertion as a model. TA proteins engage a conserved and well-defined pathway for targeting and insertion at the ER (6–8), which in mammals is monitored by an embedded quality control (QC) pathway (9, 10) to degrade products that fail targeting (fig. S1). Studies on the yeast TA targeting pathway (7, 11–13) and the mammalian TA targeting (6, 14–16) and ubiquitination (9, 10) reactions suggest that six core factors constitute a minimal mammalian triage system amenable to complete reconstitution.
Three of the factors (SGTA, Bag6, and TRC40) can recognize and shield the transmembrane domain (TMD) of a TA protein client (6, 16–18). TRC40, the targeting factor, delivers TA proteins to the ER for insertion (6), while Bag6, a quality control factor, recruits the E3 ligase RNF126 for TA protein ubiquitination (10). However, the role of SGTA in either of these outcomes is less well established (19, 20). We found that depleting SGTA from an in vitro translation lysate impaired nascent TA protein capture by TRC40 and Bag6 (Fig. 1A and fig. S2A), with concomitant reductions in ER insertion and ubiquitination (fig. S2, B and C). Conversely, depleting Bag6 (with its tightly associated Ubl4A and TRC35 subunits) (16) or TRC40 caused retention of TA protein on SGTA (fig. S2D). Thus, SGTA can act upstream of, and facilitates, both targeting and ubiquitination, suggesting that TA proteins bound to SGTA are uncommitted to either fate.
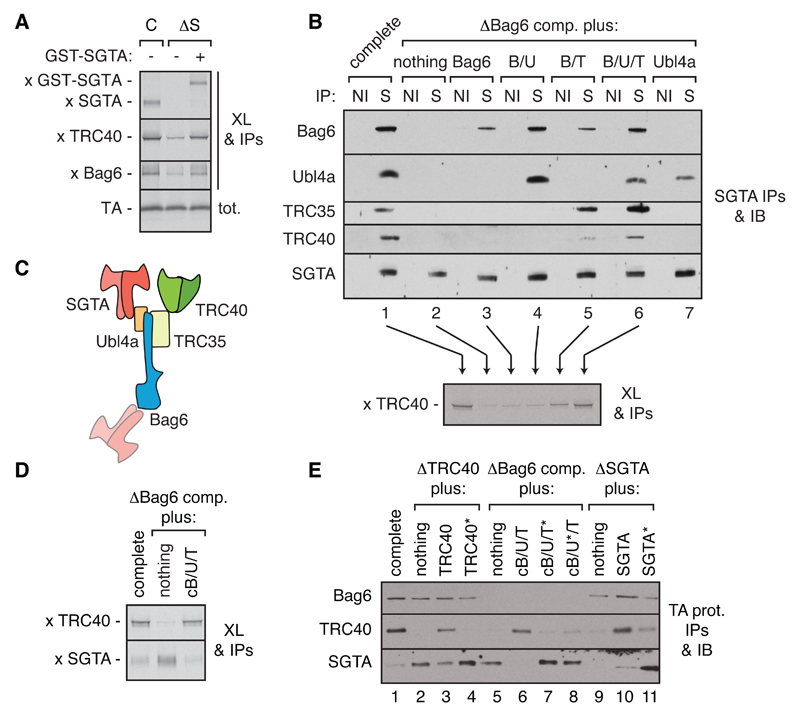
Fig. 1. Functional architecture of the TA protein triage system.
(A) Translation reactions of a 35S-labeled TA protein containing the Vamp2 TMD in complete (C) or SGTA-depleted (∆S) rabbit reticulocyte lysate (RRL) without or with recombinant GST-SGTA were subjected to chemical crosslinking (XL), immunoprecipitations (IPs) of the indicated factors, separation by SDS-PAGE, and visualization of TA-crosslinked products with autoradiography. (B) RRL was depleted of the Bag6 complex (∆Bag6 comp.) and replenished with recombinant Bag6 sub-complexes (fig. S3) as indicated. The lysates were analyzed by co-IP using non-immune (NI) or anti-SGTA (S) antibodies followed by immunoblot (top), or tested for interaction of a newly translated TA protein (Vamp2) with TRC40 by crosslinking and anti-TRC40 IPs (bottom). (C) Organization of the TA triage system deduced from the interactions in panel (B). (D) Interactions made by a newly translated TA protein (Vamp2) were assayed by crosslinking and IPs from control or Bag6-depleted RRL without or with the cBag6 complex (cB/U/T; see fig. S4A). (E) Interactions made by a newly translated FLAG-tagged TA substrate (Vamp2) in the indicated lysates (fig. S4B) were analyzed by anti-FLAG IP and immunoblotting for the indicated factors. ‘∆’ indicates immunodepletion and ‘*’ indicates a point mutant that, by homology to yeast proteins, disrupts intermolecular interactions.
Depletion-replenishment experiments replacing endogenous proteins with recombinant variants revealed the functional roles for all of the factors and interactions. The three-protein Bag6 complex was essential for optimal TA protein capture by TRC40 and bridges the SGTA-TRC40 interaction via Ubl4A and TRC35, respectively (14, 21) (Fig. 1, B and C, and fig. S3). Absence of either factor in Bag6 subcomplexes impaired SGTA-TRC40 bridging and TA protein capture by TRC40 (Fig. 1B). SGTA can also interact weakly with the N-terminal UBL domain of Bag6 (Fig. 1, B and C), but this was dispensable for TRC40 capture since only the C-terminal 110 residues of Bag6 (termed cBag6) could form a complex with Ubl4A and TRC35 (fig. S4A) (14) to fully replace the complete Bag6 complex in mediating TA protein capture by TRC40 (Fig. 1D). Point mutations that disrupt interactions between the cBag6 complex and either SGTA or TRC40 (22, 23) reduced TA protein transfer to TRC40 (Fig. 1E and fig. S4b). The complementary N-terminal part of Bag6 (nBag6, residues 1-1007), while dispensable for TRC40 capture, was sufficient to restore TA protein ubiquitination to a lysate depleted of the Bag6 complex (fig. S4, C to E). Thus, considered with earlier observations (9–11, 14, 16), we can assign each TA protein fate to different subsets of interacting factors: the targeting module consists of SGTA and TRC40 bridged by the cBag6 complex, while the QC module consists of SGTA, nBag6, and RNF126.
Analyses with purified factors rigorously established the sufficiency of these modules. In these experiments, radiolabeled TA protein is synthesized with purified E. coli translation factors supplemented with the desired TMD-binding chaperone (fig. S5). This orthogonal system produces homogeneous TA-chaperone complexes that can be used for downstream functional assays (24). Isolated TA-TRC40 complex was competent for ER targeting and insertion (Fig. 2A), but not ubiquitination by RNF126 (fig. S6). By contrast, the TA-Bag6 complex could not mediate insertion (data not shown), but allowed ubiquitination via RNF126 recruitment to Bag6’s N-terminal UBL domain (Fig. 2B).
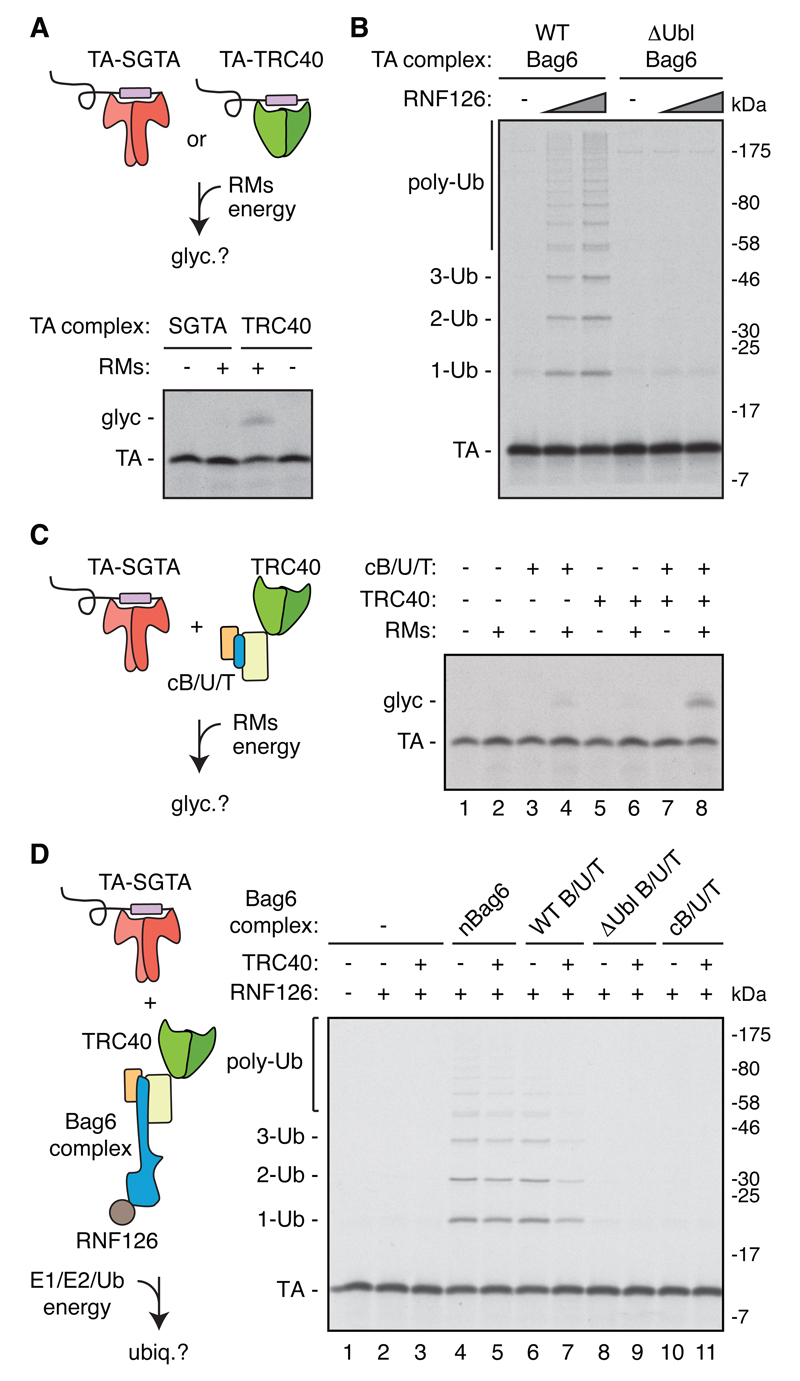
Fig. 2. Reconstitution of TA protein triage with purified factors.
(A) Recombinant TA-SGTA and TA-TRC40 complexes (fig. S5; TA protein contains the Vamp2 TMD) were assayed for insertion into ER-derived rough microsomes (RMs) by TA protein glycosylation (glyc). (B) TA protein (Vamp2) in complex with recombinant Bag6 or ∆Ubl-Bag6 was assayed for ubiquitination by RNF126. (C) TA-SGTA complex (Vamp2) was incubated with the indicated components and analyzed for insertion into RMs by glycosylation. The small amount of insertion seen in lane 4 is due to residual TRC40 that copurified with the RMs. (D) TA-SGTA complex (Vamp2) was incubated with the indicated components and assayed for TA protein ubiquitination. All reactions were conducted with 650 nM of each TRC component.
The TA-SGTA complex in isolation was not competent for insertion (Fig. 2C) or RNF126-mediated ubiquitination (Fig. 2D, lanes 1-3), but could lead to either fate if the minimal targeting (Fig. 2C) or QC module was provided (Fig. 2D, lanes 4-5, and fig. S6). The complete system containing the entire Bag6 complex and TRC40 (Fig. 2D, lane 7) produced a low level of ubiquitination, a ~70% reduction relative to the QC module (lanes 4-5) or the complete system lacking TRC40 (lane 6). Because TRC40 did not inhibit ubiquitination by the isolated QC module (lanes 4-5), reduced ubiquitination in the complete system is due to preferential TA protein capture by TRC40 (via the targeting module) at the expense of capture by Bag6. This mirrors observations made in total cytosol, where targeting is the primary outcome, while normally low levels of ubiquitination increase if targeting fails. Thus, a completely recombinant system can faithfully recapitulate TA protein triage.
To understand client flow within the triage system, we introduced a photo-crosslinking residue in the center of the TA protein TMD by site-specific amber codon suppression (fig. S7) and prepared this modified TA protein in complex with Ca2+-calmodulin (CaM) (fig. S8). TA protein chaperoned by CaM can be synchronously released by adding the Ca2+-chelator EGTA. The subsequent interactions made by the TA protein’s TMD can be monitored by UV-induced crosslinking of samples flash-frozen at defined times (Fig. 3A). This strategy revealed that TA protein released from CaM is captured rapidly (in 2 sec) and completely by SGTA, nBag6, and TRC40 when each is the only available chaperone (Fig. 3B, lanes 4-6). Combining all three chaperones at equimolar amounts (lane 7) revealed a rank order for the proportion of free TA captured by SGTA (55±4%) > nBag6 (36±4%) >> TRC40 (9±1%) (Fig. 3C). Competitive capture using the estimated ratio of endogenous chaperones (fig. S9) showed even stronger preference for SGTA capture (69±2%). Considered together with the possibility that general chaperones like Hsp70 might further funnel substrates to SGTA (11), and that a substantial proportion of TRC40 is membrane-bound and unavailable for capture (6), we conclude that the majority of TA substrates emerging into the cytosol initially engage SGTA. Consistent with TRC40 being a poor competitor for free TA protein, isolated TRC40 (but not SGTA or Bag6) fails to capture a highly hydrophobic client before it is likely to aggregate (fig. S10A). This explains why more hydrophobic TA proteins are also more dependent on SGTA for efficient engagement of TRC40 in total lysate (fig. S10B).
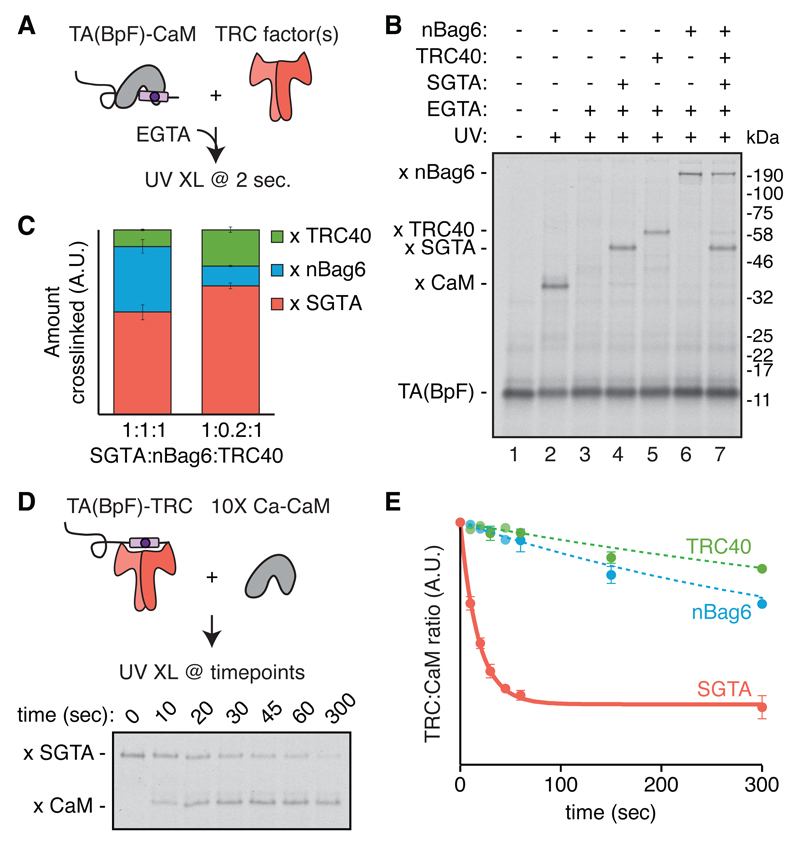
Fig. 3. TMD binding and release by individual chaperones.
(A) TA protein (Sec61β) containing a photocrosslinker (BpF) in the TMD (fig. S7) in complex with CaM (fig. S8) was synchronously released by addition of EGTA in the presence of the indicated components. The reactions were flash frozen after 2 sec and analyzed for TMD interactions by UV crosslinking on dry ice. (B) Autoradiography of the experiment described in (A) with 750 nM each of SGTA, TRC40, and nBag6. (C) Quantification of the competitive capture of free TA(BpF) as in (B), with equimolar or physiological ratios (1:0.2:1) of SGTA:nBag6:TRC40. SGTA is held constant at 750 nM (see fig. S9). Shown are mean ± SEM, n=3 for each condition. (D) TA(BpF) (Sec61β) in complex with 750 nM SGTA, TRC40, or nBag6 was mixed with 10-fold molar excess of CaM and monitored over time by photo-crosslinking. The autoradiography of the timecourse of SGTA complexes is shown. (E) Quantification of TA protein release from the indicated chaperone to CaM (n=3 for release from SGTA; n=2 for release from TRC40 and Bag6; see also fig. S11).
Photo-crosslinking assays of TA protein release from each chaperone using excess CaM to sequester released product showed that TA protein dissociates rapidly from SGTA, (t1/2 of ~12 sec; Fig. 3, D and E). Dissociation from Bag6 and TRC40 was at least 15- and 30-fold slower than from SGTA, respectively (Fig. 3E and fig. S11). Thus, the chaperone that is least committed to stable binding (SGTA) is also most favored for capturing free TA protein, while the most committed chaperone (TRC40) is least favored for initial capture. A nascent TA protein would therefore preferentially initiate triage on SGTA, consistent with its assignment as an upstream and uncommitted factor of the pathway.
TA protein transfer from SGTA to Bag6 complex (B/U/T) was two times slower than the rate of TA protein dissociation from SGTA (Fig. 4A). Transfer was also observed to the isolated QC module of Bag6, but was nearly absent to ΔUbl Bag6, which cannot interact with SGTA (fig. S12). This suggested that TA protein released from SGTA is primarily recaptured by SGTA, unless a high local concentration of Bag6 interacting with SGTA permits some capture by Bag6. Consistent with this interpretation, excess CaM in the SGTA to B/U/T transfer reaction led to TA protein loss from SGTA at a rate comparable to spontaneous dissociation, and appeared on CaM at the partial expense of Bag6 (Fig. 4A). Thus, the mechanism of TA protein transfer from SGTA to Bag6 involves spontaneous dissociation from SGTA and capture by nearby Bag6. At equal local concentrations of Bag6 and SGTA, competitive capture assays (Fig. 3B) suggest that ~40% of released TA protein would engage Bag6 and the remainder would be recaptured by SGTA.
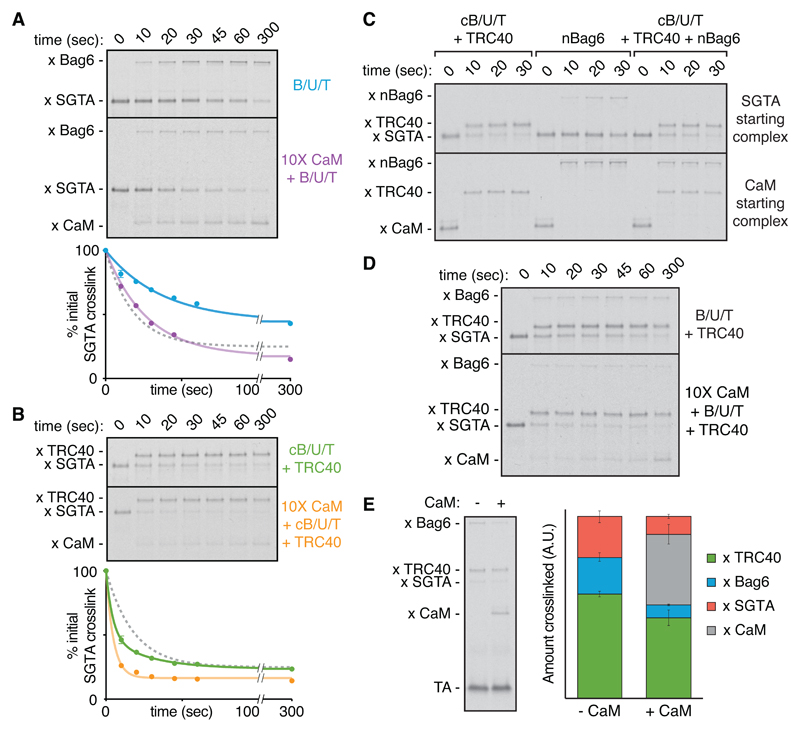
Fig. 4. Pathways of TA protein flux through the triage system.
(A) Photocrosslinking timecourse (top) and quantification (bottom) of TA(BpF) (Sec61β) transfer from SGTA to the Bag6 complex (B/U/T) without (n=3) or with (n=1) 10X CaM. Where applicable, mean ± SEM values are displayed with a first-order exponential decay fit. The gray dotted curve refers to the rate of spontaneous TA protein release from SGTA to 10X CaM from Fig. 3B. (B) Photocrosslinking timecourse (top) and quantification (bottom) of TA(BpF) (Sec61β) transfer from SGTA to TRC40 via cB/U/T without (n=3) or with (n=1) 10X CaM. (C) TA(BpF) (Sec61β) in complex with SGTA (top) or CaM (bottom) was incubated with the indicated factors and analyzed for TMD interactions at various times by photocrosslinking. (D) Photocrosslinking timecourse of TA(BpF) (Sec61β) transfer from SGTA in the presence of the full Bag6 complex and TRC40 without or with 10X CaM. (E) Direct comparison of TA (BpF) photocrosslinking interactions in the complete triage reaction after 5 min without (n=3 used for quantification) or with 10X CaM (n=2). The extent of TA protein interaction with Bag6 and TRC40 was quantified (right; mean ± SEM). All reactions contained 750 nM of each TRC factor.
Stable TA protein interaction with Bag6 (Fig. 3E and fig. S11) and prompt ubiquitination (fig. S13) would minimize excessive cycles of TA protein release and recapture by SGTA. Although the downstream steps remain to be studied, the ubiquitinated TA protein-Bag6 complex would presumably be a strong substrate for proteasome binding via the Bag6 Ubl domain and client ubiquitin(s). This would effectively commit most Bag6-bound TA proteins to degradation. Even if RNF126 or the proteasome system is temporarily unavailable, the relatively slow TA protein dissociation from Bag6 (Fig. 3E and fig. S11) and inefficient transfer back to SGTA (fig. S14) suggests that reversion to an uncommitted state is probably minimal under physiologic conditions. Thus, the rate of TA protein transfer to Bag6 imposes a time limit for how long clients remain in an uncommitted state on SGTA.
TA protein transfer from SGTA to TRC40 via the cBag6 complex displayed entirely different behavior. Transfer was two-fold faster than the rate of spontaneous TA protein dissociation from SGTA (Fig. 4B). Importantly, neither the cBag6 complex nor TRC40 alone triggers TA protein release from SGTA (fig. S15), arguing against an interaction-triggered transfer. Given that TRC40 competes poorly for free TA protein (Fig. 3, B and C), this transfer reaction appears to be a concerted handover. Indeed, TRC40 acquisition of TA protein from SGTA was comparably efficient in the presence of excess CaM, whose only effect was to minimize SGTA recapture (Fig. 4B). Similarly, the SGTA-to-TRC40 transfer was not competed by nBag6, which readily intercepted the otherwise efficient substrate transfer from CaM to TRC40 (Fig. 4C). This illustrates that the SGTA-to-TRC40 transfer occurs without a free TA protein intermediate, and is hence defined as ‘private.’ TA protein release from SGTA during the private transfer reaction required TRC40 to be competent for substrate binding (fig. S16). This suggests a mechanism of TA protein partitioning between closely juxtaposed SGTA and TRC40, explaining why it is private and how it can occur faster than the TA protein dissociation rate from SGTA.
Transfer from SGTA to TRC40 in the complete triage system, where Bag6 is present at high local concentration, is essentially indistinguishable from that seen with only the targeting module (Fig. 4D). The small amount of substrate capture by Bag6 is presumably due to the small proportion that dissociates from SGTA within the ~10 seconds needed to complete the private transfer to TRC40, and is probably the same population accessible to CaM in the reaction containing only the targeting module (Fig. 4B). Consistent with this interpretation, excess CaM in the complete reaction preferentially competes with the QC module for TA protein, with minimal effect on the targeting module (Fig. 4E). Thus, transfer of TA protein from SGTA to TRC40 is fast and private, while TA protein transfer from SGTA to Bag6 is slower and involves a transient chaperone-free intermediate.
Our results rationalize how three chaperones with similar client specificities can nevertheless be organized to encode both priority and time in a molecular triage reaction (fig. S17). The abilities of the three chaperones to compete for free TA protein are inversely related to their ability to retain the bound client (Fig. 3). Taking into account their relative abundances in the cytosol (fig. S9) and the ability of SGTA to sample housekeeping chaperones (11), nascent clients preferentially engage SGTA, the least committed chaperone that affords the most options. From this starting point, the distinct mechanisms of TA protein transfer from SGTA to Bag6 versus TRC40 (Fig. 4), combined with the embedding of Bag6 within the targeting module (Fig. 1), determine both priority and timing of triage.
Biosynthesis is the higher priority due to the rapid and private mechanism of transfer to TRC40. The time allowed for this prioritized fate is limited by the rate of spontaneous TA protein dissociation from SGTA, combined with the number of times SGTA recaptures TA protein for additional transfer attempts. Recapture is competed by Bag6, which is at a high local concentration due to its ability to interact with SGTA. Were Bag6 not embedded at the transfer site, released TA protein would be recaptured by SGTA repeatedly, increasing the risk of aggregation by prolonging the dwell time of a membrane protein in the cytosol. Based on ~40% capture by Bag6 relative to SGTA at equal concentrations, we estimate that ~80% of substrates would be committed to Bag6 within three cycles of SGTA recapture.
A time limit on private transfer to TRC40 means that after the initial unimpeded transfer attempt, further delays progressively favor ubiquitination. The rate of substrate transfer is likely influenced by the biophysical features of the TRC40 binding site (17, 24) and steric constraints imposed by the yet undefined architecture of SGTA relative to TRC40. Thus, membrane proteins that do not meet these criteria, which have presumably evolved to favor ER-destined TA proteins, are degraded by default. This implies that the rate of TA protein transfer from SGTA to TRC40, combined with the normal off rate for different types of clients from SGTA, increases the fidelity of client selection for ER targeting.
Because TA protein dissociation from SGTA follows first order kinetics with a t1/2 of 12 sec, ~25% would dissociate within 5 sec, almost half of which would be captured by Bag6. Although transfer to TRC40 is fast, some loss seems inevitable. Consistent with this, we see that even under optimal conditions in cytosolic extract or in pulse-labeled cells, ~10% of a TA substrate is lost to Bag6 (data not shown). The benefit of an embedded QC module in avoiding aggregation presumably offsets the cost of constitutively degrading a low level of otherwise productive maturation intermediates. Yeast, which do not have an embedded QC module, may favor overall efficiency to drive rapid growth, while mitigating failure by utilizing disaggregases (25) and asymmetric aggregate partitioning to mother cells (26).
QC modules embedded within a biosynthetic pathway, differential kinetics of client engagement and release, and a combination of private and non-private transfer reactions are principles that are probably generally applicable beyond the TA protein system. Both the cytosol and endoplasmic reticulum contain polypeptide-binding proteins that recognize similar features (typically hydrophobicity) and contain scaffolding proteins that bring chaperones and QC factors together (2, 3, 27, 28). Although analysis of client handling by chaperones (29) or ubiquitin ligases (30) in isolation have provided considerable mechanistic insights, an understanding of their roles in protein triage necessarily requires quantitative analyses of their functions as a collective. The triage system reconstituted in this study provides a roadmap for analogous cellular biosynthesis pathways.
Supplementary Material
One Sentence Summary.
Complete reconstitution of a protein triage pathway reveals how a quality control factor imposes a time limit on protein biosynthesis without directly competing for clients.
Acknowledgements
We thank N. Sinha and M. Mariappan for help during the early stages of this project, R. Keenan for reagents, L. Miller for comments on the manuscript, T. Rapoport and Rapoport lab members for equipment access, and Hegde lab members for discussions. This work was supported by the UK Medical Research Council (MC_UP_A022_1007 to R.S.H), a MRC career development fellowship, a St John’s College Title A fellowship, and Harvard Medical School startup funds (to S.S.), and a Life Sciences Research Foundation supported by the Ellison Medical Foundation/AFAR (to M.C.R-B.). S.S. and R.S.H designed the study. S.S. performed the experiments. M.C.R-B. provided reagents and advice for ubiquitination assays. M.H.K. generated most PURE translation system components. S.S. and R.S.H. wrote the manuscript with input from all authors.
References and Notes
- 1.Shao S, Hegde RS. Target Selection during Protein Quality Control. Trends Biochem Sci. 2016;41:124–137. doi: 10.1016/j.tibs.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 2.Guerriero CJ, Brodsky JL. The delicate balance between secreted protein folding and endoplasmic reticulum-associated degradation in human physiology. Physiol Rev. 2012;92:537–576. doi: 10.1152/physrev.00027.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Esser C, Alberti S, Hohfeld J. Cooperation of molecular chaperones with the ubiquitin/proteasome system. Biochim Biophys Acta. 2004;1695:171–188. doi: 10.1016/j.bbamcr.2004.09.020. [DOI] [PubMed] [Google Scholar]
- 4.Wolff S, Weissman JS, Dillin A. Differential Scales of Protein Quality Control. Cell. 2014;157:52–64. doi: 10.1016/j.cell.2014.03.007. [DOI] [PubMed] [Google Scholar]
- 5.Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 6.Stefanovic S, Hegde RS. Identification of a targeting factor for posttranslational membrane protein insertion into the ER. Cell. 2007;128:1147–1159. doi: 10.1016/j.cell.2007.01.036. [DOI] [PubMed] [Google Scholar]
- 7.Schuldiner M, et al. The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell. 2008;134:634–645. doi: 10.1016/j.cell.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hegde RS, Keenan RJ. Tail-anchored membrane protein insertion into the endoplasmic reticulum. Nat Rev Mol Cell Biol. 2011;12:787–798. doi: 10.1038/nrm3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hessa T, et al. Protein targeting and degradation are coupled for elimination of mislocalized proteins. Nature. 2011;475:394–397. doi: 10.1038/nature10181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodrigo-Brenni MC, Gutierrez E, Hegde RS. Cytosolic quality control of mislocalized proteins requires RNF126 recruitment to Bag6. Mol Cell. 2014;55:227–237. doi: 10.1016/j.molcel.2014.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang F, Brown EC, Mak G, Zhuang J, Denic V. A chaperone cascade sorts proteins for posttranslational membrane insertion into the endoplasmic reticulum. Mol Cell. 2010;40:159–171. doi: 10.1016/j.molcel.2010.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang F, Whynot A, Tung M, Denic V. The mechanism of tail-anchored protein insertion into the ER membrane. Mol Cell. 2011;43:738–750. doi: 10.1016/j.molcel.2011.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mariappan M, et al. The mechanism of membrane-associated steps in tail-anchored protein insertion. Nature. 2011;477:61–66. doi: 10.1038/nature10362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mock J-Y, et al. Bag6 complex contains a minimal tail-anchor-targeting module and a mock BAG domain. Proc Natl Acad Sci USA. 2015;112:106–111. doi: 10.1073/pnas.1402745112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vilardi F, Stephan M, Clancy A, Janshoff A, Schwappach B. WRB and CAML are necessary and sufficient to mediate tail-anchored protein targeting to the ER membrane. PLoS ONE. 2014;9:e85033. doi: 10.1371/journal.pone.0085033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mariappan M, et al. A ribosome-associating factor chaperones tail-anchored membrane proteins. Nature. 2010;466:1120–1124. doi: 10.1038/nature09296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leznicki P, Warwicker J, High S. A biochemical analysis of the constraints of tail-anchored protein biogenesis. Biochem J. 2011;436:719–727. doi: 10.1042/BJ20101737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leznicki P, Clancy A, Schwappach B, High S. Bat3 promotes the membrane integration of tail-anchored proteins. J Cell Sci. 2010;123:2170–2178. doi: 10.1242/jcs.066738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wunderley L, Leznicki P, Payapilly A, High S. SGTA regulates the cytosolic quality control of hydrophobic substrates. J Cell Sci. 2014;127:4728–4739. doi: 10.1242/jcs.155648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leznicki P, High S. SGTA antagonizes BAG6-mediated protein triage. Proc Natl Acad Sci USA. 2012;109:19214–19219. doi: 10.1073/pnas.1209997109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu Y, Cai M, Yang Y, Huang L, Ye Y. SGTA recognizes a noncanonical ubiquitin-like domain in the Bag6-Ubl4A-Trc35 complex to promote endoplasmic reticulum-associated degradation. Cell Rep. 2012;2:1633–1644. doi: 10.1016/j.celrep.2012.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chartron JW, Suloway CJM, Zaslaver M, Clemons WM. Structural characterization of the Get4/Get5 complex and its interaction with Get3. Proc Natl Acad Sci USA. 2010;107:12127–12132. doi: 10.1073/pnas.1006036107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chartron JW, VanderVelde DG, Clemons WM. Structures of the Sgt2/SGTA dimerization domain with the Get5/UBL4A UBL domain reveal an interaction that forms a conserved dynamic interface. Cell Rep. 2012;2:1620–1632. doi: 10.1016/j.celrep.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mateja A, et al. Protein targeting. Structure of the Get3 targeting factor in complex with its membrane protein cargo. Science. 2015;347:1152–1155. doi: 10.1126/science.1261671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doyle SM, Wickner S. Hsp104 and ClpB: protein disaggregating machines. Trends Biochem Sci. 2009;34:40–48. doi: 10.1016/j.tibs.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 26.Coelho M, Tolić IM. Asymmetric damage segregation at cell division via protein aggregate fusion and attachment to organelles. Bioessays. 2015;37:740–747. doi: 10.1002/bies.201400224. [DOI] [PubMed] [Google Scholar]
- 27.Assimon VA, Southworth DR, Gestwicki JE. Specific Binding of Tetratricopeptide Repeat Proteins to Heat Shock Protein 70 (Hsp70) and Heat Shock Protein 90 (Hsp90) Is Regulated by Affinity and Phosphorylation. Biochemistry. 2015;54:7120–7131. doi: 10.1021/acs.biochem.5b00801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Christianson JC, et al. Defining human ERAD networks through an integrative mapping strategy. Nat Cell Biol. 2012;14:93–105. doi: 10.1038/ncb2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Joachimiak LA, Walzthoeni T, Liu CW, Aebersold R, Frydman J. The structural basis of substrate recognition by the eukaryotic chaperonin TRiC/CCT. Cell. 2014;159:1042–1055. doi: 10.1016/j.cell.2014.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stein A, Ruggiano A, Carvalho P, Rapoport TA. Key steps in ERAD of luminal ER proteins reconstituted with purified components. Cell. 2014;158:1375–1388. doi: 10.1016/j.cell.2014.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shimizu Y, Ueda T. PURE technology. Methods Mol Biol. 2010;607:11–21. doi: 10.1007/978-1-60327-331-2_2. [DOI] [PubMed] [Google Scholar]
- 32.Shao S, Hegde RS. A calmodulin-dependent translocation pathway for small secretory proteins. Cell. 2011;147:1576–1588. doi: 10.1016/j.cell.2011.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mateja A, et al. The structural basis of tail-anchored membrane protein recognition by Get3. Nature. 2009;461:361–366. doi: 10.1038/nature08319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sharma A, Mariappan M, Appathurai S, Hegde RS. In vitro dissection of protein translocation into the mammalian endoplasmic reticulum. Methods Mol Biol. 2010;619:339–363. doi: 10.1007/978-1-60327-412-8_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ozawa K, et al. High-yield cell-free protein synthesis for site-specific incorporation of unnatural amino acids at two sites. Biochem Biophys Res Commun. 2012;418:652–656. doi: 10.1016/j.bbrc.2012.01.069. [DOI] [PubMed] [Google Scholar]
- 36.Stefer S, et al. Structural basis for tail-anchored membrane protein biogenesis by the Get3-receptor complex. Science. 2011;333:758–762. doi: 10.1126/science.1207125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jonikas MC, et al. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science. 2009;323:1693–1697. doi: 10.1126/science.1167983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gristick HB, et al. Mechanism of Assembly of a Substrate Transfer Complex during Tail-anchored Protein Targeting. Journal of Biological Chemistry. 2015;290:30006–30017. doi: 10.1074/jbc.M115.677328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rome ME, Rao M, Clemons WM, Shan S-O. Precise timing of ATPase activation drives targeting of tail-anchored proteins. Proc Natl Acad Sci USA. 2013;110:7666–7671. doi: 10.1073/pnas.1222054110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gristick HB, et al. Crystal structure of ATP-bound Get3–Get4–Get5 complex reveals regulation of Get3 by Get4. Nat Struct Mol Biol. 2014;21:437–442. doi: 10.1038/nsmb.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Favaloro V, Vilardi F, Schlecht R, Mayer MP, Dobberstein B. Asna1/TRC40-mediated membrane insertion of tail-anchored proteins. J Cell Sci. 2010;123:1522–1530. doi: 10.1242/jcs.055970. [DOI] [PubMed] [Google Scholar]
- 42.Krysztofinska EM, et al. Structural and functional insights into the E3 ligase, RNF126. Sci Rep. 2016;6:26433. doi: 10.1038/srep26433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leznicki P, et al. The Association of BAG6 with SGTA and Tail-Anchored Proteins. PLoS ONE. 2013;8:e59590. doi: 10.1371/journal.pone.0059590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leznicki P, et al. Binding of SGTA to Rpn13 selectively modulates protein quality control. J Cell Sci. 2015;128:3187–3196. doi: 10.1242/jcs.165209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kulak NA, Pichler G, Paron I, Nagaraj N, Mann M. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat Methods. 2014;11:319–324. doi: 10.1038/nmeth.2834. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.