Abstract
The high-throughput, high-resolution phenotyping enabled by metabolomics has been increasingly applied to a variety of questions in nephrology research. This article provides an overview of current metabolomics methodologies and nomenclature, citing specific considerations in sample preparation, metabolite measurement, and data analysis that investigators should understand when examining the literature or designing a study. Further, we review several notable findings that have emerged in the literature that both highlight some of the limitations of current profiling approaches, as well as outline specific strengths unique to metabolomics. More specifically, we review data on 1.) tryptophan metabolites and CKD onset, illustrating the interpretation of metabolite data in the context of established biochemical pathways; 2.) trimethylamine-N-oxide and cardiovascular disease in CKD, illustrating the integration of exogenous and endogenous inputs to the blood metabolome; and 3.) renal mitochondrial function in diabetic kidney disease and AKI, illustrating the potential for rapid translation of metabolite data for diagnostic or therapeutic aims. Finally, we review future directions, including the need to better characterize inter-person and intra-person variation in the metabolome, pool existing data sets to identify the most robust signals, and capitalize on the discovery potential of emerging non-targeted methods.
INTRODUCTION
The metabolome is the global collection of small molecules (typically <1500 daltons) – e.g. sugars, amino acids, organic acids, nucleotides, acylcarnitines, lipids, etc – in a cell or biologic specimen. Several online databases, such as the Human Metabolome Database (HMDB) and METLIN, catalog the growing compendium of metabolites that have been observed in biologic samples, ranging from ~103 to 104 analytes depending on the specimen of interest.1,2 Downstream of transcriptional and translational processes, metabolite levels reflect gene, transcript, and protein abundance and function. Thus, the systematic analysis of metabolites (or metabolomics) provides information that is both complementary and in some cases summative of these other molecular domains. Although our knowledge is incomplete, decades of research in biochemistry and metabolism have outlined the relationship between many metabolites and metabolic pathways, providing a framework for interpreting metabolomics data. This stands in contrast, for example, to our more nascent understanding of protein-protein interactions across the proteome. In addition, unlike its upstream counterparts, the metabolome also includes direct inputs from diet, the environment, and the microbiome. This ability to integrate endogenous and exogenous sources of variation represents an important strength for metabolomics in the study of complex human diseases.
The interactions between metabolism and kidney disease are particularly complex. First, from a clinical perspective, diabetes is the major cause of rising CKD prevalence worldwide. In turn, decreased kidney function directly impacts systemic metabolism as well, causing both peripheral insulin resistance and protein energy wasting.3,4 Second, the kidneys directly impact circulating metabolite levels, including both the net uptake of many metabolites through a variety of mechanisms—glomerular filtration, tubular secretion, and catabolism—and the net release (anabolism) of several amino acids and other metabolites.5–7 Recent studies reveal unanticipated hormone-like roles for select circulating metabolites,8–10 further increasing interest in how the kidney modulates the plasma metabolome. Third, the kidney is notable both for the degree and the complexity of its intrinsic metabolic activity. Second only to the heart in mitochondrial abundance, the kidney performs the energy-intensive task of solute and water reabsorption from >100 l of filtrate per day, with distinct cell populations performing different metabolic tasks in widely disparate oxygen tensions and osmotic environments across the nephron. How kidney metabolism is regulated and how alterations in kidney metabolism contribute to clinical disease requires further study. Taken together, the growing burden of CKD and the incomplete understanding of its metabolic underpinnings provides a strong rationale for metabolomics in nephrology research.
METHODOLOGIES
A full discussion of methodologies pertinent to metabolomics analyses, particularly emerging bioinformatic tools and flux analysis using isotopes, is beyond the scope of this review and the reader is directed to other recent review articles.11–15 Here, we outline several key technologies and concepts, highlighting important limitations that investigators should understand when examining the literature or designing a study. As a starting point, it is important to recognize that current metabolomics approaches are not as robust or comprehensive as some of the other ‘omic’ technologies, particularly genomics. Whereas nucleic acids are limited to a handful of chemical motifs of similar abundance, metabolites span a variety of compound classes with large differences in size and polarity, across a wide range of concentrations. As a result, no single technique is able to provide complete coverage of the metabolome, and for any given method there is likely to be heterogeneity in how well individual metabolites are measured. Although tools continue to improve, incomplete and non-overlapping coverage remains a central challenge in metabolite measurement, and also has relevance to sample preparation, data analysis, and synthesis of the literature.
Metabolite measurement
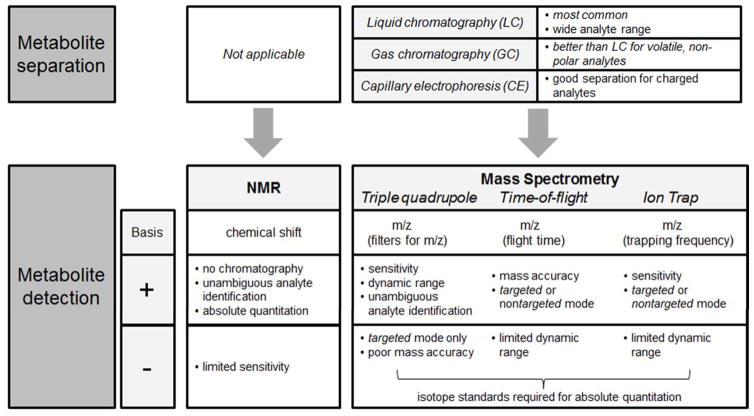
Nuclear magnetic resonance spectroscopy (NMR) and mass spectrometry are the two major technologies used in metabolomics (Figure 1). NMR utilizes the magnetic properties of select atomic nuclei (e.g. 1H, 13C, or 31P) to determine the structure and abundance of metabolites in a biological specimen. NMR requires relatively little sample preparation, with no upfront chromatography, and can provide absolute quantitation, but its sensitivity is limited to the detection of relatively abundant analytes (typically <100 in human biofluids). By contrast, mass spectrometer (MS)-based methods generally have superior sensitivity but rely on chromatographic separation to reduce sample complexity prior to mass analysis. Because different chromatographic conditions are better suited for different kinds of metabolites, many metabolomics laboratories will employ more than one chromatography-MS combination, necessitating the injection of different aliquots of the same experimental sample across methods. After chromatography, metabolites enter the MS and undergo ionization, and the MS then resolves metabolites on the basis of their mass-to-charge (m/z) ratios. Different kinds of MSs, including but not limited to triple quadrupole, time-of-flight, and ion trap instruments use distinct methods to resolve m/z, each with different strengths, weaknesses, and applications (Figure 1).
Figure 1.
Overview of metabolomics methodologies. Nuclear magnetic resonance (NMR) is robust, requiring relatively little sample preparation and no chromatographic separation. However, because of limited sensitivity and high data complexity, unambiguous identification is typically limited to less than a hundred metabolites. Mass spectrometry (MS)-based approaches have higher sensitivity and rely on a combination of chromatographic separation and mass-to-charge ratio (m/z) resolution for metabolite identification. In MS-based platforms, triple quadrupole instruments are generally used for targeted analyses, in which approximately hundreds of metabolites of known identity are measured, whereas time-of-flight and ion trap instruments are often used for nontargeted analyses of approximately thousands of metabolite peaks (only a subset of which have assigned identities). Relative advantages (+) and disadvantages (−) of the different approaches are detailed in the figure.
Although there is no consensus on nomenclature, some clarification on specific terms in the literature and how they relate to different methodologies warrant mention:
Targeted versus Non-targeted Analyses: in “targeted” profiling, the MS is configured only to ‘look for’ a pre-specified list of metabolites. This is often performed on a triple quadrupole MS, which can pass a given precursor ion through the first quadrupole, fragment this ion in the second quadrupole, and then pass a select product ion through the third quadrupole. For a given sample injection, the mass spectrometer can be set to monitor several dozen to >100 precursor ion/product ion combinations, i.e., a list of “targeted” metabolites. This approach has the advantage of high sensitivity for the metabolites of interest and a high degree of confidence in metabolite identities. A notable disadvantage of quadrupole-based MSs is relatively poor mass accuracy compared to time-of-flight and ion-trap MSs. These latter instruments are often used for “non-targeted” (or “un-targeted” or “global”) profiling, in which the instruments scan across a wide mass range and quantitate all detected ions. For a given sample injection, these instruments are able to distinguish several hundreds to >1000 analyte peaks based on accurate mass (i.e. the experimentally determined mass), with no need for ion fragmentation. However, these non-targeted methods are still subject to the limitations imposed by the given sample preparation and chromatography. Further, these methods raise additional analytical and computational hurdles, including handling and extraction of very large spectral data files, filtering out background noise, and data alignment to permit comparison across samples. At present, different software packages are used across different non-targeted metabolomics laboratories complicating attempts to compare and generalize results. One avenue to address this challenge includes the deposition of data, including raw NMR and mass spectrometry spectral data to a central repository, as endorsed by the Metabolomics Consortium Data Repository and Coordinating Center (http://www.metabolomicsworkbench.org/data/index.php) for NIH-sponsored studies.
Known versus Unknown Analytes: of the thousands of analyte peaks quantitated using non-targeted methods, only a subset will have an unambiguous metabolite identification (“known” metabolites), whereas the identity of many remain “unknown”, tagged on the basis of their retention time in chromatography and their m/z. The identities of unknown metabolites can sometimes be established after analysis of the fragmentation patterns of the parent ion, and when feasible the comparison of chromatographic and MS characteristics with commercial standards; these efforts, however are not always straight-forward or successful.
To date, many of the metabolomics studies in nephrology research, including the majority of studies discussed in this review, have applied NMR or targeted MS-based methods. However, increasing interest is being directed towards nontargeted MS-based approaches, in parallel with efforts across metabolomics laboratories to assign IDs to many of the resulting unknown analyte peaks.
Sample preparation
As noted, one of the advantages of NMR is the need for relatively little sample preparation prior to analysis. For liquid chromatography (LC)-MS based methods, samples usually undergo precipitation using an organic extraction solution, which precipitates out protein and halts metabolism while keeping the metabolites of interest in solution; extraction solutions that contain relatively greater aqueous content will be better suited for solubilizing polar compounds whereas solutions that are more organic will be better suited for lipids. Supernatants from these extractions are then either concentrated with a dry-down step or are directly injected into the LC-MS. For gas chromatography (GC)-MS, because samples are vaporized and undergo chromatography in the gas phase at high temperatures, sample preparation typically requires chemical derivatization prior to analysis. Because protocols for sample preparation often differ, they represent an important source of variation across metabolomics laboratories.
Typical specimens for metabolomics analysis include biofluids such as plasma, serum, or urine; tissue homogenates; and cell lysates or cultured media. “Matrix effects” preclude direct comparison of peak areas measure across different specimen types (unless absolute quantitation has been performed for the analytes of interest). A few small studies have directly compared results between serum and plasma, suggesting overall concordance in results, although serum appears to have higher levels of some metabolites including some reflective of the clotting process.16–19 One challenge with urinary analyses relates to the wide range of dilution, resulting in metabolite levels that may fall above or below the linear dynamic range of detection. For all biofluids, investigators should pay close attention to fasting status at the time of collection, as well as details on methods of collection, duration of storage, and freeze-thaw cycles. In cellular studies, it is important to note that there is no known ‘housekeeping’ metabolite, analogous to β-actin mRNA or protein levels, that permits straight-forward normalization of results. In these experiments, careful attention needs to be paid to analyzing similar cell numbers across conditions and/or use of parallel wells dedicated to counting cells or measuring surrogates for cell number such as protein levels.
Data analysis
As noted, a full discussion of data processing and reporting is beyond the scope of this review. Depending on the methodology, metabolite levels may be reported in either absolute concentrations or relative abundance in arbitrary units. Within a given data set, not all metabolite measures are equally robust, reflective of differences in metabolite abundance and stability, the availability of analyte standards, and issues specific to the given analytical methods— these differences can be quantified in part on the basis of analytic variation for each metabolite in blind replicate samples. Further, not all metabolites in biologic samples will have levels within the linear dynamic range of a given method, with some metabolites at the lower limit of detection and others at the saturation limit of the detector. Understanding this heterogeneity in quality within a data set is critical in interpreting both positive and negative findings. As with data processing and reporting, there is no agreement on optimal statistical approaches and significance thresholds, and these depend in part on the nature of the study, i.e. clinical biomarker studies versus mechanistic studies in cells. Given the inter-correlation between many metabolites, traditional statistical methods that examine each metabolite individually and apply Bonferroni-adjusted significance thresholds can be too conservative and increase the risk of false-negative associations. False discovery rate (FDR) is often used as an alternative, but has not necessarily gained wide acceptance among clinical reviewers. Other methods, e.g. principal component analysis (PCA), partial least squares discriminant analysis (PLS-DA), and cluster analysis can be helpful in reducing the complexity of high-dimensional data, but do not provide outputs like fold-change or odds ratios that are more intuitively grasped. Finally, as with other ‘omic’ analyses, there is interest in applying pathway analysis software to help visualize and extract biologic insights from metabolomics data sets. These tools can provide valuable information, but do not circumvent the need to understand data content and quality. For example, if pathway analysis is applied to a data set generated using a method optimized for nonpolar (lipid) compounds, it will likely highlight lipid biology as important for the given study question, when in fact this was a bias implicit in the method of metabolite measurement.
OVERVIEW OF THE LITERATURE
Given longstanding interest in metabolism in kidney disease, it is not surprising that metabolomic approaches have been rapidly incorporated into nephrology research. For example, a Pubmed search using the keywords “metabolomics” and “kidney or renal” yields 3, 8, 11, 29, 41, 50, 71, 68, 93, and 107 citations over the prior 10 years. These reports span human studies at the extremes of renal health (including normal kidney function and ESRD) and acuity of disease (AKI to CKD and after transplantation), as well as animal and in vitro experiments (Table 1). As noted, incomplete and non-overlapping coverage of metabolomics methods, as well as other issues common to all nephrology research, including limitations in sample size and power, confounding by baseline kidney function, imprecision of creatinine based GFR estimation, and lack of ideal animal models of disease, are important challenges in the field. Here, we review several notable findings that have emerged in the literature that highlight some of these challenges, but also outline strengths and themes unique to metabolomics. In each case, we review findings that have both clinical and basic underpinnings and that have been examined by more than one research team.
Table 1.
Representative metabolomics studies in nephrology research
| Area | Comment | References | |
|---|---|---|---|
| Cross-sectional clinical studies | Normal GFR | Association of serum metabolites with eGFR in >2800 individuals across two large cohorts | 51 |
| CKD | Plasma metabolites in 30 individuals with CKD stages 2–4 | 55 | |
| Paired plasma and urine metabolomics in 77 individuals with CKD stages 2–5 | 56 | ||
| ESRD | Plasma metabolites before and after hemodialysis in 44 individuals with ESRD | 57 | |
| Longitudinal clinical studies | Incident CKD | Baseline plasma and serum metabolite levels and the risk of future CKD (n >1000 in each study) | 7,20,21 |
| CKD Progression | Baseline plasma metabolites and CKD progression (n=400) | 26 | |
| CV outcomes in ESRD | Baseline plasma metabolites and 1-year cardiovascular death (n=500) | 39 | |
| Transplantation | Urine metabolites in serial samples from 57 kidney transplant patients with or without T cell mediated rejection | 58 | |
| AKI | Plasma metabolites and the risk of AKI following cardiac surgery (n=85) | 59 | |
| Plasma metabolites and the risk of AKI following transcatheter aortic valve replacement (n=44) | 60 | ||
| Metabolite profiles of plasma, kidney cortex, and medulla during AKI in mice | 61 | ||
| Mechanistic studies in tissues, animals, and cells | Mitochondria and AKI | 50 | |
| Diabetic kidney disease | 47,48,62 | ||
| Polycystic kidney disease | 63,64 | ||
| Tubulointerstitial fibrosis | 60,61 | ||
| Renal cell carcinoma | 65–67 |
Tryptophan Metabolites and CKD Prediction: established pathways as a framework for interpreting metabolomics data
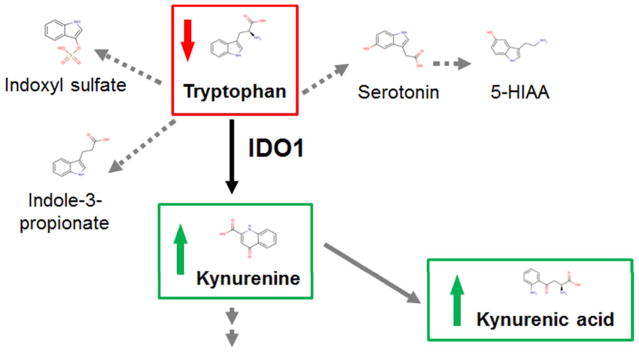
Plasma elevations of several tryptophan metabolites in individuals with advanced CKD have been recognized for many years, with particular interest in indoxyl sulfate as a potential uremic toxin. Whether alterations in tryptophan metabolism occur in early stages of renal disease had not been explicitly tested. With the increased throughput enabled by metabolomics, several groups have recently measured baseline plasma or serum metabolite levels in >1000 subjects each from the Cooperative Health Research in the Region of Augsburg S4/F4 (KORA) Study, the Framingham Heart Study (FHS), and the Atherosclerosis Risk in Communities (ARIC) Study and tested the association between baseline metabolite profiles and the subsequent development of CKD, defined as crossing below an eGFR threshold of 60 mL/min/1.73m2.7,20,21 In a smaller study, investigators profiled baseline plasma from 80 individuals seen at the Joslin Diabetes Center with diabetic nephropathy, 40 of whom subsequently progressed to ESRD.22 Although no unequivocal “winner” emerged from these studies, the strongest shared signal has been around tryptophan metabolites of the kynurenine pathway (Figure 2). For example, in KORA, the strongest association with incident CKD was with the baseline kynurenine-to-tryptophan ratio, with an odds ratio (OR) of 1.36 per SD (P = 0.003) after adjusting for eGFR and other CKD risk factors. The FHS study, which did not examine metabolite ratios, also highlighted kynurenine (OR per SD 1.49, P < 0.001) as well as its downstream metabolite kynurenic acid (OR per SD 1.53, P < 0.001) as markers of future CKD risk. Notably, there was no association between 5-HIAA or indoxyl sulfate, which result from alternative pathways of tryptophan breakdown, and incident CKD. In the Joslin study, kynurenine and kynurenic acid were not elevated in cases versus controls, but tryptophan levels were lower in cases. These findings across three prospective studies, highlighting overlapping but different components of a common pathway highlight one of the strengths of metabolomics: the ability to interpret data in the context of established biochemical pathways.
Figure 2.
Tryptophan metabolism and incident CKD. IDO1 catalyzes the first and rate-limiting step of the tryptophan catabolism via the kynurenine pathway. Alternative pathways of tryptophan metabolism result in the production of indoxyl sulfate and indole-3-propionate (via gut microbial tryptophan metabolism) or serotonin and 5-hydroxyindoleacetic acid (5-HIAA).
More specifically, the associations of tryptophan depletion and downstream metabolite excess with incident disease raise the possibility that tryptophan metabolism through IDO1, which encodes the first and rate-limiting step of the kynurenine pathway, is a marker and/or effector of renal injury. Notably, ischemia reperfusion injury has been shown to induce renal tubular Ido1 expression in mice, and Ido1−/− mice are protected from ischemic AKI.23 In addition, prior studies have shown that kynurenic acid can serve as a ligand for GPR35 expressed on leukocytes and promote inflammation and that kynurenine can promote vascular endothelial relaxation, suggesting potential deleterious roles for these molecules in renal disease.24,25
Although the studies cited above motivate interest in tryptophan metabolites as potential markers and even mediators of renal injury, several qualifications warrant mention. First, lack of overlap across metabolomics methods raises important challenges. For example, kynurenine and kynurenic acid were not reported in the ARIC Study. Second, whereas studies of incident CKD have compared individuals who do or do not cross an eGFR threshold of 60 ml/min/1.73 m2, identifying markers of progression among individuals with established disease is arguably of greater interest. To address this question, a recent analysis in the Chronic Renal Insufficiency Cohort (CRIC) Study profiled baseline plasma from 200 individuals each with eGFR slope < −3 ml/min/1.73m2/yr (cases) or between −1 and +1 ml/min/1.73m2/yr (controls), matched on baseline eGFR and proteinuria.26 Individuals in this study started with a mean eGFR of 43.3 mL/min/1.73 m2, compared to eGFRs of 81.0 (KORA), 93.6 (FHS), 105.3 (ARIC), and 81.0 mL/min/1.73 m2 (Joslin) in the studies of incident disease. In cross-sectional analysis, tryptophan was strongly positively correlated and kynurenine and kynurenic acid were strongly negatively correlated with eGFR. However, in longitudinal analysis none of these metabolites were associated with CKD progression and in general, no signals were as strong as the associations found in individuals with normal eGFR at baseline. As kidney function declines, it is possible that nonspecific accumulation dilutes the risk signal attributable to select metabolite alterations or that the effects of CKD on protein binding skew total and free metabolite levels, adding complexity to the interpretation of associations. Finally, although the studies cited above outline potential deleterious roles for kynurenine and kynurenic acid, a more recent study suggests a beneficial role for circulating kynurenic acid in cardiac ischemia. After finding that the cardioprotective effect of remote ischemic preconditioning could extend across mice with shared circulations (parabiosis), the authors performed metabolomics to identify circulating alpha-ketoglutarate as the key molecule that then drives hepatic production and secretion of kynurenic acid, which is necessary and sufficient to mediate cardiac protection.27 Whether or not this ultimately has relevance to renal injury, these data expand the compendium of studies that assign unanticipated, hormone-like roles to circulating metabolites.
TMAO and Cardiovascular Risk: examining exogenous inputs to the blood metabolome
Using nontargeted LC-MS, investigators identified three metabolites that were elevated and intercorrelated in the plasma of 75 patients who subsequently experienced a myocardial infarction, stroke or death compared to plasma from 75 age- and gender-matched controls who did not.28 Using a combination of analytical approaches, the investigators then identified these three molecules as betaine, choline, and trimethylamine-N-oxide (TMAO), and used targeted LC-MS assays to confirm the association between these metabolically related metabolites and cross-sectional cardiovascular phenotypes in ~1900 individuals. In a subsequent study, the same investigators found that increased plasma levels of TMAO are associated with increased risk of heart attack, stroke, or death (HR for highest vs. lowest quartile 2.54, P < 0.001) in the ensuing 3 years among 4007 patients undergoing elective coronary angiography.29 Plasma TMAO levels were also associated with 5-year mortality (HR per SD 1.75, P < 0.001) among 720 individuals with congestive heart failure.30 Analogous to the tryptophan metabolites outlined above, betaine, choline, and TMAO are biochemically interrelated (i.e. in an established pathway), with the additional twist that blood levels of TMAO are determined by a combination of gut microbial conversion of dietary cholines and L-carnitine into trimethylamine and hepatic oxidation of trimethylamine to TMAO. The critical dependence of blood TMAO appearance on gut flora has been demonstrated in elegant feeding studies of isotope labeled phosphatidylcholine, before and after gut flora suppression using antibiotics, in both humans and mice.28,29 Further, a functional role for TMAO in atherogenesis has been hypothesized, as TMAO has been shown to inhibit reverse cholesterol transport, increase platelet hyperresponsiveness, and promote macrophage foam cell formation in mice. 31,32 Taken together, this body of work highlights another strength of metabolomics-based discovery: the integration of endogenous and exogenous inputs, with the latter including diet and the microbiome, as well as potentially medications and other environmental exposures.
Because TMAO is primarily excreted via the kidney33 and is elevated with diminished GFR, including several fold in dialysis patients, a number of groups have examined the association between TMAO and cardiovascular (CV) disease among individuals with CKD.34–37 In a cross-sectional analysis, serum TMAO concentrations were found to correlate with atherosclerotic burden among 220 individuals with CKD who had undergone coronary angiography, and in exploratory analysis appeared to be associated with long-term mortality. 35 Similarly, in an independent cohort of 179 individuals with CKD, higher TMAO was associated with all-cause mortality. 36 In the largest CKD study to date, investigators used LC-MS to measure plasma TMAO, choline, and L-carnitine in 2529 individuals with stage 3b/4 CKD and found that TMAO was independently associated with CV events (HR per SD 1.23, P < 0.006) during 3 years follow-up.37 The association between TMAO and CV outcomes in ESRD has been less consistent. For example, in one study of 235 incident hemodialysis patients, baseline TMAO levels were not associated with time to death, cardiovascular hospitalization, or cardiovascular death.38 Similarly, in a metabolomics study of 500 incident dialysis patients, there was no association between baseline plasma TMAO and 1 year cardiovascular death.39 By contrast, a study of 1232 prevalent dialysis patients enrolled in the HEMO Study did identify an association between TMAO levels and CV outcomes, with the signal stronger in whites compared to blacks.40 In addition to its size, an important strength of this positive study was that it was able to account for residual renal function (likely a key contributor to TMAO clearance,41 and a significant predictor of mortality in dialysis patients).42
In addition to TMAO, metabolomics has been used to demonstrate the microbiome’s broad impact on the uremic solute profile,43 and targeted analyses have identified an association between some of these other metabolites and CV outcomes. For example, in a study of 400 incident dialysis patients, p-cresol sulfate was associated with a 62% higher risk of cardiovascular mortality and a 60% higher risk of first cardiovascular event and plasma phenylacetylglutamine was associated with a 37% higher risk of first cardiovascular event, per 1 SD higher solute concentration.44 Importantly, this study examined free metabolite levels, applying an ultrafiltration step during sample preparation that excludes protein-bound solutes. Most metabolomics studies do not utilize plasma ultrafiltrates, and thus measure total metabolite levels instead.
Oxidative Metabolism and Renal Injury: identifying small molecule mediators of renal injury and protection
The high density of mitochondria and significant energetic requirements for solute transport in renal tubular cells, particularly proximally and in the thick ascending limb, have long motivated interest in renal oxidative metabolism – clearly, this interest far predates the advent of metabolomics. For detailed reviews, for example on mitochondria in diabetic kidney disease and AKI, the reader is directed elsewhere.45,46 Here, we highlight recent studies that illustrate a specific strength of metabolomics: the ability to identify potential biomarkers that may also be causal and/or therapeutically modifiable mediators of disease.
Using GC-MS, investigators screened urine obtained from 158 individuals, including subjects with diabetes and CKD, diabetes without CKD, and healthy controls. Thirteen metabolites were decreased in the urine of individuals with diabetic CKD, and based on these findings, the authors pursued a variety of approaches including cytochrome C oxidase immunostaining and PPARγ-coactivator-1α (PGC1α) mRNA profiling in tissue and exosome mtDNA quantitation in urine to demonstrate decreased mitochondrial activity in human diabetic kidney disease samples.47 In addition to serving as a springboard for these more invasive studies, the urine metabolites highlighted in this study are attractive potential biomarkers of decreased renal mitochondrial function in diabetic kidney disease. In a separate study, the same group focused on fumarate, a TCA cycle intermediate that was found to be increased in the urine of mice with type 1 diabetic kidney disease compared to controls.48 Podocyte-specific overexpression of NADPH oxidase 4 (NOX4), a major source of basal ROS production in the kidney, both recapitulated several pathologic features of diabetic kidney disease in mice and specifically decreased cortical fumarate hydratase (FH) expression (FH catalyzes the hydration of fumarate to malate). Conversely, NOX4 inhibition increased FH expression, resulting in lower fumarate levels. Notably, fumarate has been purported to competitively inhibit HIF-1α proline hydroxylation, thus preventing its recognition and targeting for proteasomal degradation. Alternatively, fumarate is able to covalently modify cysteine residues, a reaction known as “succination”, a process shown to impact the cellular antioxidant response.49 Taken together, these observations outline a potential functional role for fumarate, an intermediate in the mitochondrial TCA cycle, in mediating the effects of oxidative stress in diabetic kidney disease.
Interest in renal oxidative metabolism extends to acute renal injury as well, with disruption of renal tubular mitochondria an early and prominent feature of different forms of experimental AKI. PGC1α, the master regulator of mitochondrial biogenesis, is highly expressed in the kidney and global knockout mice are more susceptible to AKI. Further, transgenic renal tubular expression of PGC1α is renoprotective and prevents the cellular lipid accumulation that occurs following ischemia reperfusion injury (IRI) in wild type mice.50 In order to further explore these observations, LC-MS based metabolomics was performed on kidney homogenates from PGC1α−/− and wild type mice as well as from mice subjected to IRI-AKI or sham surgery. Niacinamide emerged from this screen as a molecule of particular interest, because it was lower in kidneys from PGC1α−/− mice as well as following IRI, and because niacinamide is a major precursor in the salvage pathway of NAD, a critical coenzyme for energy metabolism. Notably, treating mice with intraperitoneal niacinamide led to increases in kidney NAD levels, reduced renal fat accumulation, and conferred renoprotection from both IRI and cisplatin mediated AKI. Renoprotection was evident even if niacinamide was administered after the ischemic or toxic insult had occurred. More work is needed to test the potential therapeutic value of niacinamide, or NAD fortification in general, in human AKI.
FUTURE DIRECTIONS
Despite substantial progress, there are numerous opportunities to enhance and expand the impact of metabolomics in nephrology research. As more metabolites are measured in more people, it is increasingly clear that renal function, even within the normal range, has a large impact on circulating metabolite levels. For example, 112 of 488 measured metabolites in 1735 participants of the KORA F4 study were associated with eGFR at a conservative Bonferroni adjusted significance threshold.51 Some of these metabolites have the potential to add prognostic information beyond serum creatinine, perhaps because they are more accurate markers of filtration, because they report on other renal functions such as tubular secretion,52 or because they may play a functional role in disease pathogenesis. Although investigators have begun to explore all of these possibilities for select metabolites, there has been a relative paucity of work characterizing short and long term variation and the sources of variation of the metabolome in health and in CKD. For a given metabolite, potential sources of variation will include exogenous sources as discussed above, as well as factors such as age, gender, genetic variation, comorbidities including diabetes, and even circadian oscillation.53 Addressing these gaps will require both physiologic studies with repeated sampling over time and in response to various stimuli, and pooling of data across large data sets in order to permit meta-analysis, examination of rare renal phenotypes, and integration of metabolomics with genomics and other functional genomics outputs. These latter attempts to synthesize increasingly large data sets of course will not circumvent the need for independent replication and careful clinical validation. Further, although this review has emphasized the value of interpreting metabolomics data in the context of established pathways, our knowledge of small molecule metabolism is certainly incomplete. For both clinical and basic studies, increasing utilization of nontargeted metabolomics methods will clearly expand the scope of discovery, and perhaps reveal novel metabolites or metabolite modifications of functional significance in disease. As an example, a recent nontargeted analysis of adipose tissue from mice overexpressing the Glut4 glucose transporter identified a completely novel class of lipids (fatty acid-hydroxy fatty acids) that is highly correlated with insulin sensitivity and that can signal through a G-protein coupled receptor (GPR-120) to enhance insulin-stimulated glucose uptake.54 Hopefully, analogous insights await in the study of kidney disease and its complications.
In conclusion, ongoing methodological improvements and a strong scientific rationale motivate significant interest in the application of metabolomics to renal research. At present, incomplete and non-overlap of metabolite coverage across methods, as well as variation introduced during sample preparation or even data analysis represent important limitations. Nevertheless, metabolomics has begun to make an impact across a wide range of topics in nephrology. In lieu of providing a comprehensive catalog of these studies, we focused on select findings that have been corroborated across independent research groups, are supported by both clinical and basic science observations, and highlight specific features of the metabolome, namely the organization of metabolites in established pathways, the integration of exogenous and endogenous inputs, and the potential for rapid translation for diagnostic or therapeutic purposes.
Acknowledgments
Funding: SK is supported by NIH grant K23 DK106479. EPR received support from NIH grants U01DK060990, K08-DK-090142, and the Extramural Grant Program of Satellite Healthcare, a not-for-profit renal care provider.
Footnotes
Financial Conflicts: None
References
- 1.Wishart DS, Jewison T, Guo AC, et al. HMDB 3.0--The Human Metabolome Database in 2013. Nucleic Acids Res. 2013;41:D801–7. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tautenhahn R, Cho K, Uritboonthai W, et al. An accelerated workflow for untargeted metabolomics using the METLIN database. Nat Biotechnol. 2012;30:826–8. doi: 10.1038/nbt.2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu H, Huang X, Arnlov J, et al. Clinical Correlates of Insulin Sensitivity and Its Association with Mortality among Men with CKD Stages 3 and 4. Clin J Am Soc Nephrol. 2014;9:690–7. doi: 10.2215/CJN.05230513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeFronzo RA, Alvestrand A, Smith D, et al. Insulin resistance in uremia. J Clin Invest. 1981;67:563–8. doi: 10.1172/JCI110067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Owen EE, Robinson RR. Amino acid extraction and ammonia metabolism by the human kidney during the prolonged administration of ammonium chloride. J Clin Invest. 1963;42:263–76. doi: 10.1172/JCI104713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tizianello A, De Ferrari G, Garibotto G, et al. Renal metabolism of amino acids and ammonia in subjects with normal renal function and in patients with chronic renal insufficiency. J Clin Invest. 1980;65:1162–73. doi: 10.1172/JCI109771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rhee EP, Clish CB, Ghorbani A, et al. A combined epidemiologic and metabolomic approach improves CKD prediction. J Am Soc Nephrol. 2013;24:1330–8. doi: 10.1681/ASN.2012101006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He W, Miao FJ, Lin DC, et al. Citric acid cycle intermediates as ligands for orphan G-protein-coupled receptors. Nature. 2004;429:188–93. doi: 10.1038/nature02488. [DOI] [PubMed] [Google Scholar]
- 9.Watanabe M, Houten SM, Mataki C, et al. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature. 2006;439:484–9. doi: 10.1038/nature04330. [DOI] [PubMed] [Google Scholar]
- 10.Ryan KK, Tremaroli V, Clemmensen C, et al. FXR is a molecular target for the effects of vertical sleeve gastrectomy. Nature. 2014 doi: 10.1038/nature13135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rhee EP, Gerszten RE. Metabolomics and cardiovascular biomarker discovery. Clin Chem. 2012;58:139–47. doi: 10.1373/clinchem.2011.169573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patti GJ, Yanes O, Siuzdak G. Innovation: Metabolomics: the apogee of the omics trilogy. Nat Rev Mol Cell Biol. 2012;13:263–9. doi: 10.1038/nrm3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.German JB, Hammock BD, Watkins SM. Metabolomics: building on a century of biochemistry to guide human health. Metabolomics. 2005;1:3–9. doi: 10.1007/s11306-005-1102-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sas KM, Karnovsky A, Michailidis G, et al. Metabolomics and diabetes: analytical and computational approaches. Diabetes. 2015;64:718–32. doi: 10.2337/db14-0509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zamboni N, Saghatelian A, Patti GJ. Defining the metabolome: size, flux, and regulation. Mol Cell. 2015;58:699–706. doi: 10.1016/j.molcel.2015.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Breier M, Wahl S, Prehn C, et al. Targeted metabolomics identifies reliable and stable metabolites in human serum and plasma samples. PLoS One. 2014;9:e89728. doi: 10.1371/journal.pone.0089728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wedge DC, Allwood JW, Dunn W, et al. Is serum or plasma more appropriate for intersubject comparisons in metabolomic studies? An assessment in patients with small-cell lung cancer. Anal Chem. 2011;83:6689–97. doi: 10.1021/ac2012224. [DOI] [PubMed] [Google Scholar]
- 18.Lin Z, Zhang Z, Lu H, et al. Joint MS-based platforms for comprehensive comparison of rat plasma and serum metabolic profiling. Biomed Chromatogr. 2014;28:1235–45. doi: 10.1002/bmc.3152. [DOI] [PubMed] [Google Scholar]
- 19.Ishikawa M, Tajima Y, Murayama M, et al. Plasma and serum from nonfasting men and women differ in their lipidomic profiles. Biol Pharm Bull. 2013;36:682–5. doi: 10.1248/bpb.b12-00799. [DOI] [PubMed] [Google Scholar]
- 20.Yu B, Zheng Y, Nettleton JA, et al. Serum metabolomic profiling and incident CKD among African Americans. Clin J Am Soc Nephrol. 2014;9:1410–7. doi: 10.2215/CJN.11971113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goek ON, Prehn C, Sekula P, et al. Metabolites associate with kidney function decline and incident chronic kidney disease in the general population. Nephrol Dial Transplant. 2013;28:2131–8. doi: 10.1093/ndt/gft217. [DOI] [PubMed] [Google Scholar]
- 22.Niewczas MA, Sirich TL, Mathew AV, et al. Uremic solutes and risk of end-stage renal disease in type 2 diabetes: metabolomic study. Kidney Int. 2014;85:1214–24. doi: 10.1038/ki.2013.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mohib K, Wang S, Guan Q, et al. Indoleamine 2,3-dioxygenase expression promotes renal ischemia-reperfusion injury. Am J Physiol Renal Physiol. 2008;295:F226–34. doi: 10.1152/ajprenal.00567.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang J, Simonavicius N, Wu X, et al. Kynurenic acid as a ligand for orphan G protein-coupled receptor GPR35. J Biol Chem. 2006;281:22021–8. doi: 10.1074/jbc.M603503200. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y, Liu H, McKenzie G, et al. Kynurenine is an endothelium-derived relaxing factor produced during inflammation. Nat Med. 2010;16:279–85. doi: 10.1038/nm.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rhee EP, Clish CB, Wenger J, et al. Metabolomics of Chronic Kidney Disease Progression: A Case-Control Analysis in the Chronic Renal Insufficiency Cohort Study. Am J Nephrol. 2016;43:366–74. doi: 10.1159/000446484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olenchock BA, Moslehi J, Baik AH, et al. EGLN1 Inhibition and Rerouting of alpha-Ketoglutarate Suffice for Remote Ischemic Protection. Cell. 2016;164:884–95. doi: 10.1016/j.cell.2016.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Z, Klipfell E, Bennett BJ, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang WH, Wang Z, Levison BS, et al. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med. 2013;368:1575–84. doi: 10.1056/NEJMoa1109400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang WH, Wang Z, Fan Y, et al. Prognostic value of elevated levels of intestinal microbe-generated metabolite trimethylamine-N-oxide in patients with heart failure: refining the gut hypothesis. J Am Coll Cardiol. 2014;64:1908–14. doi: 10.1016/j.jacc.2014.02.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koeth RA, Wang Z, Levison BS, et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med. 2013;19:576–85. doi: 10.1038/nm.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu W, Gregory JC, Org E, et al. Gut Microbial Metabolite TMAO Enhances Platelet Hyperreactivity and Thrombosis Risk. Cell. 2016;165:111–24. doi: 10.1016/j.cell.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Al-Waiz M, Mitchell SC, Idle JR, et al. The metabolism of 14C-labelled trimethylamine and its N-oxide in man. Xenobiotica. 1987;17:551–8. doi: 10.3109/00498258709043962. [DOI] [PubMed] [Google Scholar]
- 34.Tang WH, Wang Z, Kennedy DJ, et al. Gut microbiota-dependent trimethylamine N-oxide (TMAO) pathway contributes to both development of renal insufficiency and mortality risk in chronic kidney disease. Circ Res. 2015;116:448–55. doi: 10.1161/CIRCRESAHA.116.305360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stubbs JR, House JA, Ocque AJ, et al. Serum Trimethylamine-N-Oxide is Elevated in CKD and Correlates with Coronary Atherosclerosis Burden. J Am Soc Nephrol. 2016;27:305–13. doi: 10.1681/ASN.2014111063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Missailidis C, Hallqvist J, Qureshi AR, et al. Serum Trimethylamine-N-Oxide Is Strongly Related to Renal Function and Predicts Outcome in Chronic Kidney Disease. PLoS One. 2016;11:e0141738. doi: 10.1371/journal.pone.0141738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim RB, Morse BL, Djurdjev O, et al. Advanced chronic kidney disease populations have elevated trimethylamine N-oxide levels associated with increased cardiovascular events. Kidney Int. 2016;89:1144–52. doi: 10.1016/j.kint.2016.01.014. [DOI] [PubMed] [Google Scholar]
- 38.Kaysen GA, Johansen KL, Chertow GM, et al. Associations of Trimethylamine N-Oxide With Nutritional and Inflammatory Biomarkers and Cardiovascular Outcomes in Patients New to Dialysis. J Ren Nutr. 2015;25:351–6. doi: 10.1053/j.jrn.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kalim S, Clish CB, Wenger J, et al. A plasma long-chain acylcarnitine predicts cardiovascular mortality in incident dialysis patients. J Am Heart Assoc. 2013;2:e000542. doi: 10.1161/JAHA.113.000542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shafi T, Powe NR, Meyer TW, et al. Trimethylamine N-Oxide and Cardiovascular Events in Hemodialysis Patients. J Am Soc Nephrol. 2016 doi: 10.1681/ASN.2016030374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hai X, Landeras V, Dobre MA, et al. Mechanism of Prominent Trimethylamine Oxide (TMAO) Accumulation in Hemodialysis Patients. PLoS One. 2015;10:e0143731. doi: 10.1371/journal.pone.0143731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shafi T, Jaar BG, Plantinga LC, et al. Association of residual urine output with mortality, quality of life, and inflammation in incident hemodialysis patients: the Choices for Healthy Outcomes in Caring for End-Stage Renal Disease (CHOICE) Study. Am J Kidney Dis. 2010;56:348–58. doi: 10.1053/j.ajkd.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aronov PA, Luo FJ, Plummer NS, et al. Colonic contribution to uremic solutes. J Am Soc Nephrol. 2011;22:1769–76. doi: 10.1681/ASN.2010121220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shafi T, Meyer TW, Hostetter TH, et al. Free Levels of Selected Organic Solutes and Cardiovascular Morbidity and Mortality in Hemodialysis Patients: Results from the Retained Organic Solutes and Clinical Outcomes (ROSCO) Investigators. PLoS One. 2015;10:e0126048. doi: 10.1371/journal.pone.0126048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hallan S, Sharma K. The Role of Mitochondria in Diabetic Kidney Disease. Curr Diab Rep. 2016;16:61. doi: 10.1007/s11892-016-0748-0. [DOI] [PubMed] [Google Scholar]
- 46.Ralto KM, Parikh SM. Mitochondria in Acute Kidney Injury. Semin Nephrol. 2016;36:8–16. doi: 10.1016/j.semnephrol.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sharma K, Karl B, Mathew AV, et al. Metabolomics reveals signature of mitochondrial dysfunction in diabetic kidney disease. J Am Soc Nephrol. 2013;24:1901–12. doi: 10.1681/ASN.2013020126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.You YH, Quach T, Saito R, et al. Metabolomics Reveals a Key Role for Fumarate in Mediating the Effects of NADPH Oxidase 4 in Diabetic Kidney Disease. J Am Soc Nephrol. 2016;27:466–81. doi: 10.1681/ASN.2015030302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng L, Cardaci S, Jerby L, et al. Fumarate induces redox-dependent senescence by modifying glutathione metabolism. Nat Commun. 2015;6:6001. doi: 10.1038/ncomms7001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tran MT, Zsengeller ZK, Berg AH, et al. PGC1alpha drives NAD biosynthesis linking oxidative metabolism to renal protection. Nature. 2016;531:528–32. doi: 10.1038/nature17184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sekula P, Goek ON, Quaye L, et al. A Metabolome-Wide Association Study of Kidney Function and Disease in the General Population. J Am Soc Nephrol. 2016;27:1175–88. doi: 10.1681/ASN.2014111099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suchy-Dicey AM, Laha T, Hoofnagle A, et al. Tubular Secretion in CKD. J Am Soc Nephrol. 2015 doi: 10.1681/ASN.2014121193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nikolaeva S, Ansermet C, Centeno G, et al. Nephron-Specific Deletion of Circadian Clock Gene Bmal1 Alters the Plasma and Renal Metabolome and Impairs Drug Disposition. J Am Soc Nephrol. 2016 doi: 10.1681/ASN.2015091055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yore MM, Syed I, Moraes-Vieira PM, et al. Discovery of a class of endogenous mammalian lipids with anti-diabetic and anti-inflammatory effects. Cell. 2014;159:318–32. doi: 10.1016/j.cell.2014.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shah VO, Townsend RR, Feldman HI, et al. Plasma metabolomic profiles in different stages of CKD. Clin J Am Soc Nephrol. 2013;8:363–70. doi: 10.2215/CJN.05540512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Duranton F, Lundin U, Gayrard N, et al. Plasma and urinary amino acid metabolomic profiling in patients with different levels of kidney function. Clin J Am Soc Nephrol. 2014;9:37–45. doi: 10.2215/CJN.06000613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rhee EP, Souza A, Farrell L, et al. Metabolite profiling identifies markers of uremia. J Am Soc Nephrol. 2010;21:1041–51. doi: 10.1681/ASN.2009111132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Blydt-Hansen TD, Sharma A, Gibson IW, et al. Urinary metabolomics for noninvasive detection of borderline and acute T cell-mediated rejection in children after kidney transplantation. Am J Transplant. 2014;14:2339–49. doi: 10.1111/ajt.12837. [DOI] [PubMed] [Google Scholar]
- 59.Zacharias HU, Hochrein J, Vogl FC, et al. Identification of Plasma Metabolites Prognostic of Acute Kidney Injury after Cardiac Surgery with Cardiopulmonary Bypass. J Proteome Res. 2015;14:2897–905. doi: 10.1021/acs.jproteome.5b00219. [DOI] [PubMed] [Google Scholar]
- 60.Elmariah S, Farrell LA, Daher M, et al. Metabolite Profiles Predict Acute Kidney Injury and Mortality in Patients Undergoing Transcatheter Aortic Valve Replacement. J Am Heart Assoc. 2016;4:e002712. doi: 10.1161/JAHA.115.002712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wei Q, Xiao X, Fogle P, et al. Changes in metabolic profiles during acute kidney injury and recovery following ischemia/reperfusion. PLoS One. 2014;9:e106647. doi: 10.1371/journal.pone.0106647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sas KM, Nair V, Byun J, et al. Targeted Lipidomic and Transcriptomic Analysis Identifies Dysregulated Renal Ceramide Metabolism in a Mouse Model of Diabetic Kidney Disease. J Proteomics Bioinform. 2015;(Suppl 14) doi: 10.4172/jpb.S14-002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Priolo C, Henske EP. Metabolic reprogramming in polycystic kidney disease. Nat Med. 2013;19:407–9. doi: 10.1038/nm.3140. [DOI] [PubMed] [Google Scholar]
- 64.Hwang VJ, Kim J, Rand A, et al. The cpk model of recessive PKD shows glutamine dependence associated with the production of the oncometabolite 2-hydroxyglutarate. Am J Physiol Renal Physiol. 2015;309:F492–8. doi: 10.1152/ajprenal.00238.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Abu Aboud O, Donohoe D, Bultman S, et al. PPARalpha inhibition modulates multiple reprogrammed metabolic pathways in kidney cancer and attenuates tumor growth. Am J Physiol Cell Physiol. 2015;308:C890–8. doi: 10.1152/ajpcell.00322.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wettersten HI, Hakimi AA, Morin D, et al. Grade-Dependent Metabolic Reprogramming in Kidney Cancer Revealed by Combined Proteomics and Metabolomics Analysis. Cancer Res. 2015;75:2541–52. doi: 10.1158/0008-5472.CAN-14-1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lucarelli G, Galleggiante V, Rutigliano M, et al. Metabolomic profile of glycolysis and the pentose phosphate pathway identifies the central role of glucose-6-phosphate dehydrogenase in clear cell-renal cell carcinoma. Oncotarget. 2015;6:13371–86. doi: 10.18632/oncotarget.3823. [DOI] [PMC free article] [PubMed] [Google Scholar]