Abstract
Metabolism is a key health risk factor for exposures to pro-carcinogenic polycyclic aromatic hydrocarbons (PAHs) such as dibenzo[def,p]chrysene (DBC), an IARC classified 2A probable human carcinogen. Human exposure to PAHs occurs primarily from the diet in non-smokers. However, little data is available on the metabolism and pharmacokinetics in humans of high molecular weight PAHs (≥4 aromatic rings), including DBC. We previously determined the pharmacokinetics of DBC in human volunteers orally administered a micro-dose (29 ng; 5 nCi) of [14C]-DBC by accelerator mass spectrometry (AMS) analysis of total [14C] in plasma and urine. In the current study, we utilized a novel “moving wire” interface between ultra-performance liquid chromatography (UPLC) and the AMS to detect and quantify parent DBC and its major metabolites. The major [14C] product identified in plasma was unmetabolized [14C]-DBC itself, (Cmax= 18.5 ± 15.9 fg/mL, Tmax= 2.1 ± 1.0 h), whereas the major metabolite was identified as [14C]-(+/−)-DBC-11,12-diol (Cmax= 2.5 ± 1.3 fg/mL, Tmax= 1.8 h). Several minor species of [14C]-DBC metabolites were also detected for which no reference standards were available. Deconjugated and conjugated metabolites were detected in urine with [14C]-(+/−)-DBC-tetraol identified as the major metabolite, 88.7% of which was detected upon enzymatic deconjugation (Cmax= 35.8 ± 23.0 pg/pool, Tmax= 6–12 h pool). [14C]-DBC-11,12-diol, of which 94.4% was conjugated and identified in urine (Cmax= 29.4 ± 11.6 pg/pool, Tmax= 6–12 h pool). Parent [14C]-DBC was not detected in the urine. This is the first dataset to assess metabolite profiles and associated pharmacokinetics of a carcinogenic PAH in human volunteers at an environmentally relevant dose, providing the data necessary for translation of high dose laboratory animal models to human translation for environmental health risk assessment.
INTRODUCTION
PAHs
Polycyclic aromatic hydrocarbons (PAHs) are ubiquitous environmental pollutants produced by combustion of organic materials, often originating from industrial activity, automotive exhaust, and forest fires. The largest source of PAH exposure in non-smokers is through the diet, contributing to up to 95% of PAHs such as benzo[a]pyrene (BaP).1,2 Dietary exposures are most often associated with contamination of soils for food crops or produced through food preparation (i.e. grilling or smoking) or storing liquids in smoke charred containers.3,4
Of the 126 EPA priority pollutants, 16 are PAHs.5 Dibenzo[def,p]chrysene (DBC, also known as dibenzo[a,l]pyrene), is not included in this list; however it has been shown in pre-clinical cancer models to be a more potent and efficacious carcinogen than (BaP) with an accepted BaP equivalence (BaPeq) or Relative Potency Factor (RPF) of 30 and a range of 10–40 BaPeq.6,7 When metabolically activated, DBC is a potent DNA mutagen that causes a variety of cancers and morbidities in several animal models including mouse offspring following a single exposure to pregnant dams.8–16 The sensitivity of the offspring in developing T-cell lymphoblastic leukemia (T-ALL) is Cytochrome P450 (CYP)1b1-dependent.13 DBC is now an IARC class 2A probable human carcinogen.17 However, DBC is rarely included in the analyses of food products, in part because in the past it has been difficult to accurately quantify, and reliable estimates of human exposures are largely unknown. DBC has been detected in common human food sources.18
Metabolic Activation of PAHs
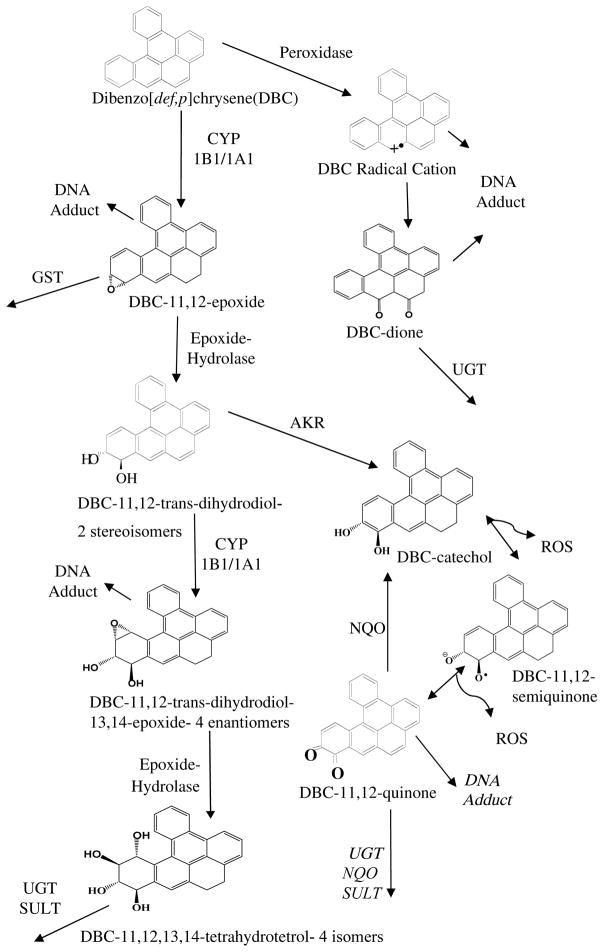
Metabolism is a key factor in potential health risks following exposure to PAHs, including DBC (Figure 1).12,19,20–25 The carcinogenicity of high molecular weight bay and fjord region containing PAHs, such as BaP and DBC, results from epoxygenation by CYP1A1 or CYP1B1, followed by epoxide hydrolase (EH) hydrolysis to the DHD.26–28 A second epoxygenation produces the ultimate carcinogen, DHD-epoxide (DHDE). In the case of DBC, this CYP-dependent activation produces 4 enatiomers of 11,12-DHD-13,14-E.29 DBC-DHDEs are electrophilic and readily form DNA adducts; the most potent is the (−)-anti-trans-11,12-diol-13,14-epoxide, primarily adducting to adenosine (dA).30 The DHDEs can be hydrolyzed to four tetraols, often the major urinary products following conjugation by UGT or SULT.31
Figure 1. Metabolic Activation of DBC.
Pro-carcinogenic DBC is activated to reactive intermediate metabolites by several enzymatic processes. Peroxidase activation can form a radical DBC species that adduct DNA. The CYP1B1 or CYP1A1 pathways can convert DBC to several metabolites that form DNA adducts, be conjugated for elimination or further metabolized to more reactive intermediates. The primary DBC carcinogenic metabolite is the (−)-anti-trans-11,12-diol-13,14-epoxide. The 11,12-trans-dihydrodiol is additionally a substrate for to aldo-keto reductase (AKR) formation of a catechol which can then undergo redox cycling. As information on the fate of DBC quinones was not available we relied on the prototypical PAH, benzo[a]pyrene (BaP), for which evidence has been published with respect to DNA adduction and further metabolism by UDP-glucuronosyl transferase (UGT), NADPH quinone oxidoreductase (NQO) and sulfatase (SULT)51–54. It is acknowledge that the reactivity and metabolism of DBC quinones may differ from BaP.
Pharmacokinetics of DBC
The complexity of high molecular weight PAH biological activation requires well-controlled pharmacokinetic studies in animals and humans to reduce uncertainties associated with human health risk predictions based upon high dose studies in animals. DBC produces tumors in offspring of mice following in utero exposures.14,15 More recently, pregnancy was shown to significantly impact PAH metabolism and pharmacokinetics in mice.32–34 Crowell et al., observed a 2- to 10-fold reduction in P450 activities during pregnancy in B6129F1 mice that significantly altered the pharmacokinetics of DBC and its major metabolites compared to non-pregnant mice.33
To extrapolate results from high-dose studies in mice to relevant human exposures, a physiologically based pharmacokinetic (PBPK) model was developed from in vitro studies in mouse and human tissues and in vivo pharmacokinetic studies in mice.19,33–35 To evaluate the accuracy of PBPK model predictions for humans, we performed the first pharmacokinetic study in human volunteers exposed to an ultra-low, but environmentally relevant, oral dose of DBC.36 Volunteers were orally administered a micro-dose (29 ng) of [14C]-DBC (5 nCi) with plasma and urine analyzed for total [14C] over 72 hours by accelerator mass spectrometry (AMS) at the Lawrence Livermore National Laboratory (LLNL) Center for Mass Spectrometry (CAMS). The 1 mV Bio-AMS provides attomole sensitivity for [14C] electrostatically detected by a particle detector.37 This allows reliable detection of potential carcinogens that are administered at environmentally relevant doses predicted to yield de minimus risk to volunteers. Human volunteers, administered a micro-dose of [14C]-DBC, were found to have a T1/2 of 5.8 hours for total [14C] in plasma, which agrees with PBPK model simulations scaled from rodent data assuming the majority of [14C] was associated with parent DBC.35,36
The purpose of the current study was to quantify [14C]-DBC and metabolites in human urine and plasma utilizing a recently developed novel liquid sample ultra performance liquid chromatography (UPLC)-AMS interface.38,39 This data provides the first environmentally relevant human metabolic data following exposure to a defined ultra-low dose of a carcinogenic PAH. The data will be critical for ongoing and future evaluations of PBPK model predictions and human health risk assessments.
MATERIALS AND METHODS
Human Volunteers
Volunteers were recruited from 2011 to 2012 to participate in a previously reported pharmacokinetic study of [14C]-DBC oral absorption and elimination.36 Additional plasma and urine samples were archived, per volunteer informed consent agreements, for metabolite pharmacokinetics as the technology to do so was developed. Volunteer demographics are included in Table 1. As previously described, a capsule dose (≥ 98% radiochemical purity), of [14C]-DBC, containing 29 ng and 5 nCi [14C], was administered overnight to fasted volunteers with water.36
Table 1.
Volunteer Demographics
| Volunteer | Sex | Ethnicity | Age | BMI |
|---|---|---|---|---|
| V5 | F | Caucasian | 65 | 34 |
| V8 | M | Caucasian | 26 | 24 |
| V9 | F | Caucasian | 56 | 32 |
| V10 | M | Caucasian | 20 | 23 |
| V11 | M | Caucasian | 43 | 35 |
| V13 | M | Caucasian | 36 | 34 |
The Oregon State University Institutional Review Board (IRB) approved the protocol and informed consent documents under approval #3853. The protocol described the method of recruitment (local papers, campus flyers, Craig’s list, etc.) and the informed consent, signed by each volunteer stated that their samples would be send to LLNL for analysis followed by transfer of the coded data to PNNL. With respect to recruitment and enrollment criteria, healthy adults between ages 20–65 were considered. Inclusion criteria included: age 20–65; healthy; non-smoking; not using medications that can affect gut motility; no history of gastrointestinal surgeries; kidney or liver disease; GI diseases such as Crohn’s, ulcerative colitis, or gastritis. Women volunteers must be post-menopausal or have had surgical sterilization to eliminate any possibility for fetal exposure. A recent routine medical examination (within 4 weeks) assuring good general health was required. This screening assessment and examination was conducted by a licensed physician. The screening assessment included menopausal history and a urine pregnancy test for all women. The most recent approval, pertinent to this manuscript, provided IRB approval to continue data analysis without further volunteer enrollment (“Microdosing to Determine the Pharmacokinetics of PAHs”, IRB #3853, April 12, 2016). The volunteer specimen transfer and analysis at LLNL was approved by their IRB (#00000285) and the data analysis at PNNL by their IRB (#52725).
Plasma
Archived aliquots of 1 mL plasma from 0, 0.25, 0.5, 0.75, 1.0, 1.5, 2.0, 2.5, 3, 4, 8, 12 and 24 hour time points were thawed from −80°C storage and prepared by the methods previously described.36 Samples were dried by speed evaporation at room temperature, reconstituted with 100 μL ethyl acetate, transferred to UPLC inserts, dried under vacuum and reconstituted with 50 μL acetonitrile. They were then stored at −80°C until UPLC injection. A analytical system recovery of 76% was determined based on the method previously described.36
Urine
All urine voided during the 72-hour cycle was collected. Urine was pooled by 0–6, 6–12, 12–24, 24–48 and 48–72 hour batches for homogenized sampling and volume recording by pool and stored at −80°C. Two aliquots (1 mL each) of pooled urine samples were adjusted to pH 7 with 0.063 M sulfuric acid and 0.12 M sodium hydroxide. One aliquot was treated with 2,000 Fleischman units of β-glucuronidase containing sulfatase activity (Sigma, St. Louis, MO) to generate total metabolites present in urine. All samples (β-glucuronidase treated and untreated) were vortexed and incubated at 37°C for 18 hours in a shaking water bath. Samples were extracted thrice with 1 mL ethyl acetate, the organic phase transferred to a separate culture tube for nitrogen evaporation at 35° C to dryness. Samples were stored at −80° C until reconstitution with 50 μL acetonitrile for UPLC.
Urine system recovery was determined by spike and recovery experiments. Recovery was 30.0% ± 3.5% S.D. of a 400 fg [14C]-BaP/100 μL spike solution from n=6 urine samples with 14.1% relative standard deviation (RSD) using the experimental preparation and AMS detection methods. Urine treated with β-glucuronidase had a 21.4 % ± 3.6 % S.D. spike recovery and 17.0% RSD from 6 samples. Applying these recovery constants from extracted samples analyzed by UPLC-AMS restored total [14C] values to matched samples that were previously analyzed as graphite converted un-extracted whole urine by solid sample AMS.
UPLC
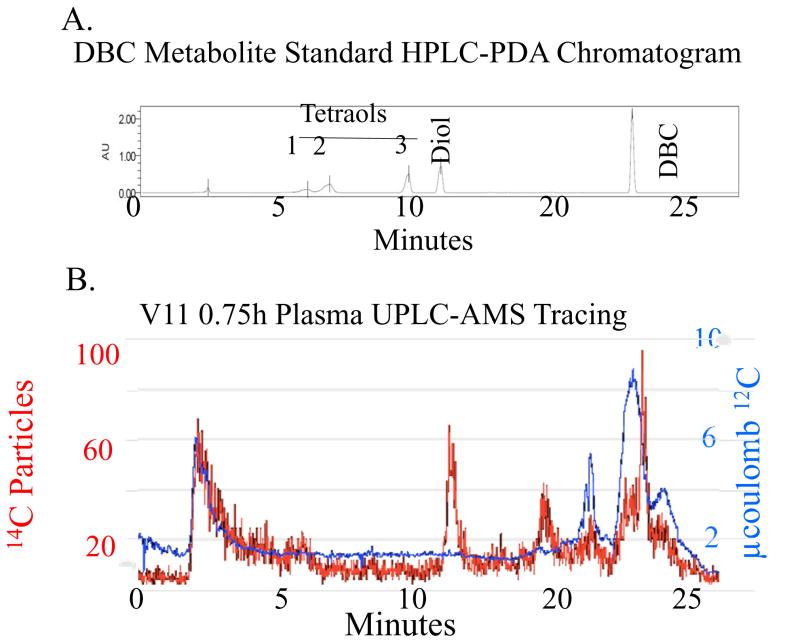
The syn-(+/−)-DBC-11,12,13,14-tetraol, anti-(+/−)-DBC-11,12,13,14-tetraol standard, and DBC-11,12-diol were obtained from the Oregon State University (OSU) PAH repository.40 The original custom synthesis was performed by Dr. Shantu Amin and colleagues at the Huck Institutes of the Life Sciences, Penn State University (Hershey, PA). A Waters Acquity H Series UPLC was utilized with a C18, 150 × 2.1 mm, 2.6 μm, 100 Å column (Phenomenex 00F-4462-AN) and 2.1 mm guard column (Phenomenex AJ0-8782) at 25°C. Detection was with a photo diode array (PDA). The gradient solvent program was acetonitrile:water, 0–3 minutes 45:55 isocratic, 3–18 minutes 45:55 to 100:0, 18–21 minutes at 100:0 and 21–23 minutes to return to starting conditions. The flow rate was 0.12 mL~min−1. [14C]-DBC and metabolite standards were found to elute as follows: [14C]-DBC, 20.5 minutes, [14C]-DBC-11,12-diol, 12.5 minutes and [14C]-DBC racemic tetraols, 6.5, 7.5, and 11 minutes, (Figure 2B). The tetraols were previously found to be comprised of a peak eluting at 6.5 minutes containing two tetraols, a (+/−) and a (+/+)-11,12,13,14-tetraol. A peak eluting at 7.5 minutes was identified as a (+/−)-11,12,13,14-tetraol followed by a (+/−)-11,12,13,14-tetraol eluting at 11.0 minutes.
Figure 2. [14C]-DBC Metabolite Profile in Plasma from a Representative Volunteer 0.75 hours after Dosing.
A. [14C] particles were detected from plasma extracts by AMS (left axis). The [14C] particles detected per peak were converted to fg DBC · mL−1 using the specific activity of [14C]-DBC and sample processing correction factors. [12C] measurements were used to determine and remove the endogenous biological [14C] (right axis). B. Non-labeled DBC and DBC metabolite standards provided retention time data utilizing PDA detection.
UPLC-AMS
UPLC-AMS analysis utilized an interface that converts UPLC eluent to CO2.39 Briefly, UPLC eluent was collected on a moving nickel wire, passed through a drying oven to remove volatile solvents, combusted to CO2, which was then transported to the gas-accepting ion source for subsequent ionization and quantification of [14C] via AMS.41 AMS detection retention time data was corrected to UPLC retention time by subtracting the in-system time difference between photo diode array and PDA, determined by an injection start signal from the PDA and the analysis of standards.
[12C] and [14C] particles were detected independently during the same UPLC-AMS analysis (Figure 2A, right and left axes). The contribution from endogenous carbon was estimated to be 103 zmol [14C]/μg total carbon, which is the approximate ratio in contemporary biological tissue, allowing endogenous [14C] to be independently quantified and removed from downstream [14C] particle counts in a single UPLC-AMS sample run (Figure 2A, right axis). The advantage over traditional UPLC methods is the AMS system allows distinction of matrix noise from analyte signal, reducing the need for inefficient or complex cleanup of low concentration samples, such as solid phase extraction or QuEChERS methods, many of which are inappropriate for PAH analysis. All reported data included corrections for endogenous [14C] contributions.42
Data Quality Assurance
AMS accuracy was determined prior to every UPLC injection by applying 5 μL of a reference [14C]-sucrose solution (IAEA-C6), containing 1 μg total carbon/μL and 148 zmol [14C]/μg C, to the liquid sample interface. Total error of analytic samples was determined by the non-linear uncertainty propagation method, including errors in precision, total carbon, and [14C] particle detection.43 Error tolerance was limited to 30% RSD of total error. All values containing error ≥ 30% RSD were reported as >%RSD (Supplemental Tables 1–3) and eliminated from data interpretation.
Analytic background noise, reported in [14C] counts·second−1, was determined from a 2·σ baseline mean of each sample from an area of [14C] chromatogram that did not contain a [14C] peak attributable to [14C]-DBC.44 Signal was defined as total [14C] particle counts less background and endogenous [14C] contributions. The lower limit of quantitation (LLOQ) was determined by the signal to noise (S/N) method, with a S/N ≤ 5 cut off.45,46 All data points below this value were reported as <S/N and were eliminated from data interpretation (Supplemental Tables 1–3).
Pharmacokinetic Analyses
Analysis of data utilized an Excel based pharmacokinetic (PK) add-on developed at Allergan, Inc. (Irvine, CA) as was performed previously with total [14C] analysis.36 Metabolite peaks were assessed individually or as the sums of peaks associated with [14C]-DBC-tetraol stereoisomers at a given time point. PK formulae relied upon non-compartmental analysis of six functions: peak concentrations in plasma (Cmax), time of peak plasma concentration (Tmax), plasma elimination half-life (T1/2), apparent elimination rate constant (kel) and area under the plasma concentration curve (AUC0-t) using standard regression techniques. Alpha phase classification covered primarily 0–8 hours, while the beta phase was considered to be 8–12 hours based upon visual inspection of the [14C] vs time plasma profiles. The α-AUC 0–8 hour analysis was the focus of this work as [14C]-DBC is rapidly metabolized from plasma, resulting in a slow β-phase elimination curve that remained flat and near background from 12–72 hours.
RESULTS
Plasma
All volunteer data is reported numerically in Supplemental Table 1 and all pharmacokinetic parameters are reported in Table 2. A representative graphical representation of plasma [14C]-DBCeq data from a representative volunteer by time is depicted in Figure 3. The major plasma component following [14C]-DBC dosing is parent [14C]-DBC, with a Cmax of 18.5 ± 15.9 fg·mL−1 (range 3.0–35.0 fg·mL−1), and α-AUC0–8 of 128.7 ± 73.5 fg·h·mL−1 (range 57.0–220.7 fg·h·mL−1). There is a large range of plasma [14C]-DBC concentrations, likely related to differing absorption and clearance rates in a heterogeneous and small population of volunteers. The [14C]-DBC Tmax was 2.1 ± 1.0 hours (range 1.0–3.0 h). The major plasma metabolite identified by available reference standards was (+/−)-[14C]-DBC-11,12-DHD, with a Cmax of 2.6 ± 1.3 fg·mL−1 (range 0.6–3.6 fg·mL−1), and an α-AUC0–8 of 11.9 ± 3.0 fg·h·mL−1 (range 8.1–15.0 fg·h·mL−1). The (+/−)-[14C]-DBC-11,12-DHD Tmax was 1.7 ± 0.9 hours (range 1.0–3.0 hours), which had a narrower range than the [14C]-DBC Tmax and appeared slightly earlier.
Table 2.
Plasma Pharmacokinetic Data by Volunteer and Metabolite*
| Volunteer | S. Front | M1 | Diol | M5 | M6 | M7 | M8 | DBC | Sum | |
|---|---|---|---|---|---|---|---|---|---|---|
| Tmax (h) | V5 | 3.0 | - | 3.0 | 3.0 | 3.0 | 1.0 | - | 3.0 | 3.0 |
| V8 | 2.5 | - | 1.0 | 1.0 | 1.0 | 1.0 | - | 1.0 | 1.0 | |
| V9 | 1.5 | 2.0 | 1.5 | 1.5 | 1.5 | 1.5 | 1.5 | 1.5 | 1.5 | |
| V13 | 3.0 | 1.0 | 1.5 | 1.5 | 1.5 | 1.5 | 3.0 | 3.0 | 3.0 | |
| Avg. | 2.5 | 1.5 | 1.7 | 1.7 | 1.7 | 1.2 | 2.2 | 2.1 | 2.1 | |
| S.D. | 0.7 | 0.7 | 0.9 | 0.9 | 0.9 | 0.3 | 1.1 | 1.0 | 1.0 | |
|
|
||||||||||
| Cmax (fg/mL) | V5 | 3.8 | ND | 3.6 | 2.7 | 2.0 | 1.6 | ND | 3.0 | 73.0 |
| V8 | 6.9 | ND | 2.4 | 1.1 | 1.2 | 0.7 | ND | 27.9 | 36.8 | |
| V9 | 3.3 | 0.8 | 3.1 | 1.3 | 2.7 | 2.0 | 4.0 | 35.0 | 52.9 | |
| V13 | 2.7 | 0.5 | 0.6 | 0.3 | 1.2 | 0.7 | 1.9 | 7.3 | 13.1 | |
| Avg. | 4.2 | 0.7 | 2. 6 | 1.3 | 1.8 | 1.2 | 3.0 | 18.5 | 44.0 | |
| S.D. | 1.9 | 0.2 | 1.3 | 1.0 | 0.7 | 0.7 | 1.4 | 15.9 | 25.3 | |
|
|
||||||||||
| α-AUC0–8 hr (fg/h/mL) | V5 | 17.9 | ND | 15.0 | 8.8 | 8.5 | 2.8 | ND | 220.7 | 277.0 |
| V8 | 22.3 | ND | 8.1 | 3.0 | 0.2 | 1.1 | ND | 57.0 | 99.6 | |
| V9 | 9.6 | 1.8 | 13.5 | 2.0 | 5.7 | 4.0 | 6.4 | 153.3 | 299.0 | |
| V13 | 38.0 | 1.2 | 11.0 | 1.3 | 8.6 | 2.0 | 14.0 | 83.9 | 169.3 | |
| Avg. | 22.0 | 1.5 | 11.9 | 4.0 | 5.8 | 2.5 | 10.2 | 128.7 | 211.3 | |
| S.D. | 11.0 | 0.4 | 3.0 | 3.3 | 3.9 | 1.2 | 5.4 | 73.5 | 93.5 | |
|
|
||||||||||
| α-Kel (fg/h) | V5 | 1.0 | ND | 0.2 | 0.3 | 0.3 | 0.7 | ND | 0.3 | 0.3 |
| V8 | 0.2 | ND | 0.2 | 0.2 | 0.7 | 0.9 | ND | 0.8 | 0.6 | |
| V9 | 0.4 | 0.9 | 0.1 | 0.3 | 0.9 | 0.6 | 1.1 | 0.3 | 0.1 | |
| V13 | 0.2 | 1.0 | 0.5 | 1.0 | 0.2 | 0.2 | 0.3 | 0.4 | 0.3 | |
| Avg. | 0.5 | 1.0 | 0.3 | 0.5 | 0.5 | 0.6 | 0.7 | 0.4 | 0.3 | |
| S.D. | 0.4 | 0.1 | 0.2 | 0.3 | 0.3 | 0.3 | 0.5 | 0.2 | 0.2 | |
|
|
||||||||||
| α-T½ (h) | V5 | 0.7 | ND | 3.4 | 2.0 | 2.2 | 0.9 | ND | 2.4 | 2.7 |
| V8 | 3.1 | ND | 3.3 | 3.7 | 1.0 | 0.7 | ND | 0.9 | 1.0 | |
| V9 | 1.7 | 0.7 | 5.0 | 2.0 | 0.7 | 1.2 | 0.6 | 2.0 | 8.6 | |
| V13 | 3.0 | 0.7 | 1.4 | 0.7 | 2.9 | 3.2 | 2.2 | 1.9 | 2.4 | |
| Avg. | 2.1 | 0.7 | 3.3 | 2.1 | 1.7 | 1.5 | 1.4 | 1.8 | 3.7 | |
| S.D. | 1.1 | 0.0 | 1.5 | 1.2 | 1.0 | 1.1 | 1.1 | 0.6 | 3.4 | |
The pharmacokinetic parameters of plasma [14C]-DBC UPLC-AMS data was determined using non-compartmental computational models. Where insufficient >LLOQ data was available, PK analysis was not determined (ND).
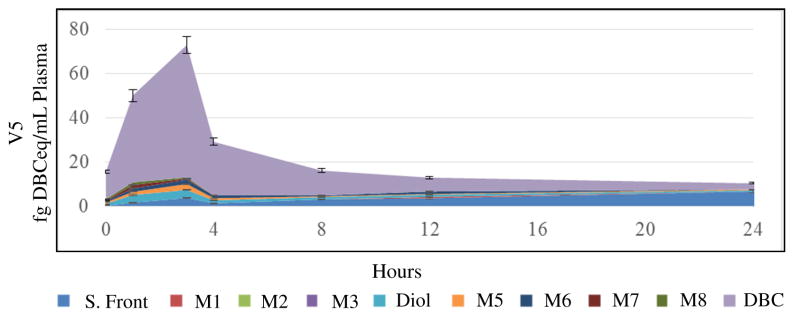
Figure 3. Profiles of Plasma [14C]-DBC and [14C]-DBC Metabolites over 24 Hours in a Representative Volunteer.
The plasma metabolite profile over the first 24 hours following dosing was compared to [14C]-DBC in plasma. The most abundant species in plasma was parent [14C]-DBC, with peaks eluting with the solvent front (more polar than tetrols), putatively identified as conjugates of [14C]-DBC metabolites.
Several [14C]-DBC metabolites were detected for which no reference standards were available, including a large [14C] peak eluting with the mobile phase solvent front, possibly [14C]-DBC-metabolite conjugates. Metabolites with a retention time between (+/−)-[14C]-DBC-11,12-DHD (12.5–13.5 minutes) and [14C]-DBC (20.6–22.0 minutes) were present and likely [14C]-DBC-quinones and/or hydroxylated [14C]-DBC (M5 (14.8–15.8 minutes), M6 (16.5–17.5 minutes), M7 (18.2–19.3 minutes), and M8 (19.6–20.5 minutes), Figures 2 and 3, Table 2, Supplemental Table 1)).47 Individually, these metabolites were minor relative to [14C]-DBC, (+/−)-[14C]-DBC-11,12-DHD, and the solvent front peaks. [14C]-DBC metabolite concentrations were more consistent across volunteers than parent [14C]-DBC concentrations (Tables 2 and 3). (+/−)-[14C]-DBC-11,12-DHD was the most consistent with a 25.2 % RSD, compared to [14C]-DBC with 57.1 % RSD (Tables 2 and 3).
Table 3.
Plasma AUC Normalization by Volunteer BMI and by [14C]-DBC Concentrations
| S. Front | M1 | Diol | M5 | M6 | M7 | M8 | DBC | |
|---|---|---|---|---|---|---|---|---|
| α-AUC (fg/h/mL) | 54.2 | 27.6 | 25.2 | 81.6 | 67.8 | 50.7 | 52.8 | 57.1 |
| α-AUC(BMI norm) | 54.0 | 31.5 | 16.1 | 68.3 | 65.6 | 61.0 | 49.1 | 47.2 |
| α-AUC(DBC norm) | 110.9 | 228.4 | 89.5 | 101.7 | 150.0 | 150.9 | 194.1 |
Metabolite plasma concentrations were compared over time as Area Under the Curve (AUC, fg·h·mL−1) for the alpha phase (peak to trough) (Table 2). To determine if tissue distribution contributed to the large range of volunteer plasma metabolite concentrations, plasma AUC values were normalized by volunteer BMI (α-AUC· (ng [14C]-DBC administered · volunteer BMI−1)), the % RSD was calculated across volunteers (α-AUC(BMI norm)) and compared to the % RSD of non-normalized plasma AUC (α-AUC) (Table 3). The % RSD was not consistently or greatly affected by volunteer BMI normalization, indicating that BMI and subsequent distribution was not a major contributor to the variation in plasma concentrations by volunteer at this environmentally relevant dose. To determine if [14C]-DBC absorption directly related to plasma metabolite concentrations, the individual plasma metabolite AUCs were normalized as a percentage of [14C]-DBC AUC by volunteer (metabolite AUC·[14C]-DBC AUC−1·100), the % RSD was determined across volunteers (α-AUC (DBC norm) ) and compared to non-normalized plasma AUC (α-AUC). The metabolite % RSDs increased several fold following normalization by the parent [14C]-DBC AUC, indicating that there is not a direct relationship between plasma [14C]-DBC concentrations and plasma [14C]-DBC metabolite concentrations (Table 3).
Urine
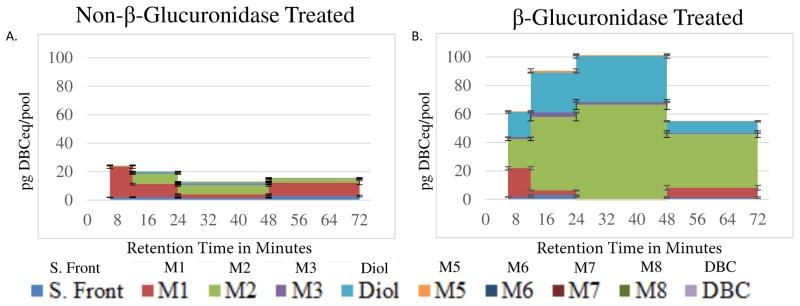
The untreated urine data set had low signal for most metabolites, decreasing the S/N and reducing the number of data points that met quality assurance limits (Supplemental Table 3). This is due to the very low level of unconjugated metabolites in urine available to be extracted with ethyl acetate (Figure 4A). The majority of urine metabolites exist as conjugates and are only available for ethyl acetate extraction after enzymatic hydrolysis (Figure 4B). Urine [14C]-DBC metabolites, analyzed with and without β-glucuronidase treatment, are reported as free (unconjugated, total (unconjugated plus enzyme hydrolyzed), and conjugated (total minus free) (Table 4). Only DBC tetraol and DBC diol were represented, with tetraol isomer M2 as the major peak. Combined, the total tetraol isomers were more heavily represented than the diols (Table 4). There was not a consistent trend in metabolite formation by concentration, over time or across volunteers.
Figure 4. β-Glucuronidase Cleavage of Urine DBC Metabolites.
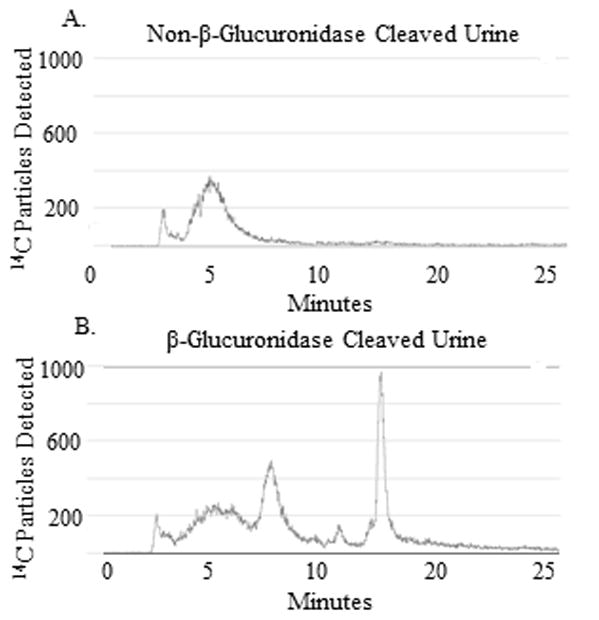
A. UPLC-AMS analysis of the 6–12 hour urine pool, not treated with β-glucuronidase, from a representative volunteer. Non-β-glucuronidase treated urine contains few retained metabolites and a low [14C] signal. B. UPLC-AMS analysis of urine from the same pool, treated with β-glucuronidase, contained several metabolites that were free to be extracted, separated, and detected.
Table 4.
Deconjugated Urine [14C]-DBC Metabolites (fg/mL) Tetraols
| Pool | S. Front | M1 | M2 | Sum (M1+2) | Diol | Sum Diol+Tetraols | |
|---|---|---|---|---|---|---|---|
| V5 | 0–6 | - | - | - | - | - | - |
| 6–12 | - | - | 19.8 | 19.8 | - | 19.8 | |
| 12–24 | 0.9 | - | 44.4 | 44.4 | 28.1 | 72.5 | |
| 24–48 | 0.2 | - | 61.4 | 61.4 | 31.1 | 92.5 | |
| 48–72 | - | - | 35.0 | 35.0 | - | 35.0 | |
|
|
|||||||
| V6 | 0–6 | 1.7 | - | - | - | - | - |
| 6–12 | - | - | 25.3 | 25.3 | - | 25.3 | |
| 12–24 | - | - | 20.8 | 20.8 | 38.3 | 59.1 | |
| 24–48 | - | 18.6 | - | 18.6 | - | 18.6 | |
| 48–72 | - | - | - | - | - | - | |
|
|
|||||||
| V8 | 0–6 | - | 18.9 | 22.5 | 41.4 | 20.5 | 61.9 |
| 6–12 | 11.4 | - | - | - | - | - | |
| 12–24 | 17.0 | 68.5 | 42.8 | 111.3 | 34.7 | 146.0 | |
| 24–48 | - | - | - | - | - | - | |
| 48–72 | - | - | - | - | - | - | |
|
|
|||||||
| V9 | 0–6 | 3.8 | 8.9 | 2.8 | 11.7 | 6.0 | 17.7 |
| 6–12 | 1.9 | 3.9 | - | 3.9 | 5.5 | 9.4 | |
| 12–24 | - | 8.3 | - | 8.3 | 9.4 | 17.7 | |
| 24–48 | - | - | - | - | - | - | |
| 48–72 | - | 1.1 | - | 1.1 | - | 1.1 | |
|
|
|||||||
| V10 | 0–6 | 10.4 | 55.7 | 21.8 | 77.5 | 16.3 | 93.8 |
| 6–12 | - | - | - | - | 28.0 | 28.0 | |
| 12–24 | - | - | - | - | 24.5 | 24.5 | |
| 24–48 | 1.0 | 3.9 | - | 3.9 | - | 3.9 | |
| 48–72 | - | 1.5 | - | 1.5 | - | 1.5 | |
The percentage of metabolites conjugated in urine was determined by subtracting the non-β-glucuronidase treated urine metabolite concentration from the metabolite concentration detected in β-glucuronidase treated urine. The excess metabolite remaining following enzymatic cleavage (relative to non-cleaved sample) is assumed to be added to the pool by enzymatic cleavage, freeing the metabolite for organic extraction. The reduction in the solvent front and M1 following enzymatic treatment (relative to non-cleaved samples) indicates that the un-identified peaks may be a conjugated species of [14C]-DBC, while the high percentage increase in M2 and [14C]-DBC-DHD indicates that the majority of these species exist in plasma as conjugates.
As a relative percent of [14C]-DBC-metabolites formed, calculated from Supplemental Tables 2 and 3, [14C]-DBC-tetraol (M2), across volunteers and time points was 88.7 % ± 6.5 % conjugated in urine while (+/−)-[14C]-DBC-11,12-DHD was 94.4% ± 1.7 % conjugated. These values were consistent across time points, indicating that there is not a temporal ratio of conjugated [14C]-DBC metabolites relative to unconjugated [14C]-DBC metabolites in urine.
Graphically, [14C]-DBC urine data from a representative volunteer is reported in Figure 5. Conjugated [14C]-DBC-tetraol M2 was the major urine metabolite with a Cmax of 35.8 ± 23.0 pg/pool (range 6.1–68.0 pg/pool), a Tmax of 6–12 hour (range 0–6 to 12–24 hours). Interestingly, M2 (a (+/+)-11,12,13,14-tetraol isomer) was only detected in the plasma of volunteer 10 at two time points (8 and 12 hours; Supplemental Table 1). [14C]-DBC-tetraols M1–M3 were not well represented in plasma, with only M1 consistently present and M3, a (+/−)11,12,13,14-tetraol never detected. Comparatively, [14C]-DBC-tetraols M1–M3 were predominant in β-glucuronidase treated urine samples (Table 4, Supplemental Table 2, and Figure 5). The conjugated (+/−)-[14C]-DBC-11,12-DHD was another major urine metabolite with a Cmax of 29.4 ± 11.6 pg/pool (range 9.9–39.8 pg/pool). The urinary (+/−)-[14C]-DBC-11,12-DHD Tmax of 6 – 12 hours (range 0–6 to 12–24 hours) appeared later than in plasma (Tmax of 1.7 ± 0.9 hours) (Figure 3). The unidentified metabolite M5, putatively a [14C]-DBC-quinone or hydroxy-[14C]-DBC (Supplemental Tables 2 and 3) was observed in the urine of two volunteers, V9 and V10, but was not consistently present (Table 4). In contrast, M5 was present in all volunteers’ plasma. In urine, unidentified metabolites M6–M8, as well as parent [14C]-DBC, were not detected.
Figure 5. Urine [14C]-DBC Metabolite Profile from a Representative Volunteer over 72 Hours.
A. The urine [14C]-DBC metabolite profile from extracts of urine not treated with β-glucuronidase, or B. from β-glucuronidase (with sulfatase activity)-treated urine extracts. The [14C]-DBC-DHD is only present following enzyme treatment, indicating that it is in a conjugated form in urine. The putative [14C]-DBC-dione is increased following enzyme treatment only slightly, while other metabolite fractions were mostly present following enzymatic cleavage, indicating that they exist in the conjugated form in urine.
The low yield of unconjugated [14C]-DBC-tetraol (M2) in plasma (Figure 3, Supplemental Table 1), whereas conjugated [14C]-DBC-tetraol (M2) is the major conjugated urinary metabolite (Figures 4 and 5, Table 4), indicates that [14C]-DBC-tetraol (M2) is rapidly conjugated after formation in GI or liver and eliminated in urine (or feces which were not collected in this study). In contrast, [14C]-DBC-tetraol (M1) is more readily excreted un-conjugated in urine. [14C-]-DBC was the major plasma component following [14C]-DBC ingestion, with an average Tmax of 2.1 hours (Table 2). There was not a delayed curve of metabolite appearance in plasma concurrent with a reduction in parent [14C]-DBC. However, there was an increase in conjugated metabolite appearance in the urine (Figures 4 and 5, Table 4) following the cumulative [14C]-DBCeq Tmax of 12–24 hour pool in plasma (Figure 3, Table 3). Both conjugated (+/−)-[14C]-DBC-11,12-DHD and conjugated [14C]-DBC-tetraol (M2) were found in urine at these timepoints (Figure 5, Table 4). Unfortunately, a full set of matched urine and plasma samples was not available from all volunteers due to a change in methods that prevented analysis of all intended samples. Incubations of non-labeled DBC with human expressed CYP1B1 or pooled human liver microsomes exhibited at almost identical metabolite profile with DBC-11,12-DHD as the major metabolite with a minor unidentified metabolite eluting later in the gradient (Figure S1). These incubations were performed at 10 μM DBC, approximately 8 orders of magnitude higher than the Cmax for [14C]-DBC in this study. The yield of metabolites (0.5–3% of DBC) is much lower than observed in plasma after microdosing. In addition, HPLC with the standard anti-(+/−)-r-11, t-12-dihydroxy-t-13,14-epoxy-11,12,13,14-tetrahydrodibenzo[def,p]chrysene (NCI Chemical Carcinogen Reference Standards Repository) elutes prior to DBC-11,12-dihydrodiol demonstrating that the DPBDE carcinogenic metabolite is not present in plasma or urine after micro-dosing (Figure S1).
DISCUSSION
Utilizing the specificity of a [14C] radiolabel, UPLC metabolite separation, and the enhanced sensitivity achieved with the new liquid sample interface for AMS, we were able to determine the environmentally relevant metabolite profile of a IARC class 2A carcinogenic PAH in human volunteers with de minimus risk.17 We were able to detect and quantify [14C]-DBC and (+/−)-[14C]-DBC-11,12-DHD and [14C]-DBC-tetraol metabolites, previously detected in plasma and urine in high dose rodent studies, in human volunteers following an oral environmentally relevant dose. Additionally, we detected several novel minor metabolite peaks (M5–M8, Figures 4 and 5, Tables 2 and 4, Supplemental Tables 1–3,) as well as polar metabolites, including conjugates eluting in the solvent front that collectively may be important in understanding human metabolism and potential PAH risk. As DBC is a pro-carcinogen, risk from exposure is determined by rates of detoxication versus formation of reactive metabolite species or redox products capable of covalent binding with DNA and other macromolecules (Figure 1).
[14C]-DBC metabolites appeared rapidly in plasma, with [14C]-DBC, (+/−)-[14C]-DBC-DHD and [14C]-DBC-tetraol (M1 isomer) all apparent by the 0.5 hour time point. The rapid [14C]-DBC metabolite appearance in plasma suggests possible GI and/or liver first-pass metabolism, presumably via CYP-dependent oxygenation. In mice, single, double and triple Cyp1a1, 1a2 and 1b1 gene knockout models have demonstrated the critical role of the Cyp1 family in mouse PAH metabolism.47 Mice that were orally exposed to BaP had a drastic variance in circulating plasma BaP concentrations by Cyp1a1 status, with a 25-fold plasma concentration increase in Cyp1a1(−/−) mice relative to Cyp1a1(+/+) mice and a 75-fold plasma concentration increase in Cyp1a1(−/−)/Cyp1b1(−/−) double knockout mice relative to wild type mice.48 Inter-individual variation in expression of isoforms of the CYP1 family provide a potential partial explanation for variability in plasma concentrations and urine metabolite profiles among volunteers. There are a number of human genetic polymorphisms in the CYP1 family, including several CYP1A1 and CYP1B1 alleles that could result in reduced Kcat with [14C]-DBC. For example, the allelic variant CYP1B1*3 (leu432val), results in reduced expression and altered metabolism of PAHs and has been positively correlated to susceptibility to lung cancer.49 The CYP1B1*3 allelic variant is found in 37% of Caucasians and 23% of Japanese individuals.50 Volunteers were required to be non-smokers with no gut motility issues, but there was no control for diets high in PAHs or other potential CYP inducing exposures. In addition to the CYP1 family, a number of additional phase 1 and phase 2 enzymes, active toward PAHs, (Figure 1) are induced by various environmental factors and also exist in humans as a number of allelic variants. When incubated with high (10 μM) concentrations of DBC, expressed human CYP1B1.1 yields predominantly the 11,12-DHD, as do human liver microsomes (Figure S1).
The extent and short period of time associated with appearance of [14C]-DBC metabolites in plasma may seem surprising. The average Cmax of [14C]-DBC corresponded to 60 fM (range, 10–115 fM). This illustrates the efficiency of DBC metabolism at ultra-low concentrations and the likelihood of a significant first pass metabolism in liver (and perhaps intestine). The yield of metabolites after a 30 minute incubation with human CYP1B1 or liver microsomes yielded a much lower percent of metabolites (predominately DBC-11,12-DHD). Incubations with human S9 gave essentially the same yield and metabolite profile as the microsomes (data not shown). The formation of DBC-11,12-diol (plasma) and DBC-11,12,13,14-tetraol (urine) could be of concern with respect to DNA adduction. However, we did not observe any peak corresponding to the 11,12-DHD-13,14-epoxide. This compound would be expected to hydrolyze in aqueous solutions but we know it is stable enough to observe in a reverse-phase system containing 30% water (Figure S1). We did not measure DNA adducts in this study but in an ongoing study with a BaP dose twice that of DBC (183 compared to 96 pmoles), the level of [14C]-BaP DNA adduction was below the limit of detection in PBMCs 72 hours after dosing.
DBC is known to be poorly absorbed in rodent models. In mice treated with unlabeled or [3H]-DBC, only about 3% of an orally administered dose was recovered in urine, with the great majority eliminated in feces.32,33 As we have previously demonstrated, only 1.2 ± 0.5% of the 29 ng oral dose of [14C]-DBC was eliminated in human urine, which could indicate similar poor oral bioavailability as seen in rodents. However, it is also possible that a significant portion of the [14C]-DBC absorbed from the GI is metabolized in liver and excreted through the biliary system and eliminated in feces (which was not collected in this study). The urinary profile of [14C]-DBC-metabolites was similar to that seen previously in rodent models.33 The major urinary products, in humans and rodents, were DBC-tetraols and to a lesser extent, DBC-diols and their conjugates.
Isotopic labeling and UPLC-AMS allows specific detection of metabolites (Figure 2). DBC standards require custom synthesis, as they are not commercially available. Relying upon isotopic detection, we were able to uniquely gate several consistent metabolites, for which we did not have reference standards, to compare across time points, volunteers, and biological matrices. Putatively, we assume that the longer retention time, less polar, metabolites (M5–M8) (Figures 2–4, Table 2) are likely [14C]-DBC-quinones and/or hydroxylated [14C]-DBC. We were not able to chemically identify these compounds with MS/MS techniques due to low sample extract concentrations.
Ongoing Bio-AMS PAH research includes [14C]-benzo[a]pyrene (BaP) administered to human volunteers in a 46 ng environmentally relevant dose and pharmacokinetic analysis by volunteers genotyped for CYP1B1 and GSTM1 allelic variants. A major advantage of UPLC-AMS over traditional solid graphite converted AMS samples, is the ability to include internal standards. Plasma and urine from [14C]-BaP administered volunteers will include an internal [14C]-DBC spike to correct for individual sample extraction efficiency, providing more precise analytical control relative to separate spike and recover extraction efficiency experiments.
Future applications of Bio-AMS include online UPLC-AMS coupled with simultaneous MS/MS for samples of sufficient chemical concentration. Compounds with less toxicity risk, such as drugs, can be administered at a dose with a lower specific activity and/or in doses for AMS retention time and isotopic label identification of novel peaks and precise quantification. Simultaneously, peaks can be chemically identified with an online MS/MS.
Supplementary Material
Supplemental Figure 1. DBC Metabolism by Human CYP1B1 and Liver Microsomes. A. HPLC of DBC, DBC-11,12-DHD and DBC-11,12-DHD-13,14-Epoxide. CRN (coronene) was used as an internal standard. B. Incubations were performed with 0.05 nmol of human CYP1B1 (Human CYP1B1 plus P450 reductase Supersomes or C. One mg/ml of pooled human liver microsomes (both from Gentest, BD Biosciences).
Supplemental Table 1. Plasma [14C]-DBC UPLC-AMS Metabolite Data. A. Individual volunteer plasma levels of [14C]-DBCeq are given in fg/mL for sampling from 0–48 hours. B. This table provides the propagated error for plasma values of each volunteer, again in fg [14C]-DBCeq/mL. The quality control of sample data included elimination of samples that were below the limit of detection (<LOD), defined as a signal to noise ratio of 5 or higher. Samples failing to meet the quality standard of a %RSD of ≥ 30 were also eliminated to maintain data quality (>%RSD). Samples that were not detected were reported as N.D. Samples that were not available for analysis were reported as N.A.
Supplemental Table 2. Deconjugated Cleaved Urine [14C]-DBC UPLC-AMS Metabolite Data. A. Individual volunteer levels of each metabolite in pools of urine collected at 6–12, 12–24, 24–48 and 48–72 hours post-dosing. The samples were treated with β-glucuronidase as described in materials and methods and reported as fg [14C]-DBC/pool. B. The values reported in A. are adjusted for propagated error as described for Supplemental Table 1B.
Supplemental Table 3. Native Urine [14C]-DBC UPLC-AMS Metabolite Data. A. Individual volunteer levels of each metabolite in pools of urine collected at 6–12, 12–24, 24–48 and 48–72 hours post-dosing. The samples were not treated with deconjugating enzymes and the results are reported as fg [14C]-DBC/pool. B. The values reported in A. are adjusted for propagated error as described for Supplemental Table 1B.
Acknowledgments
Funding. This study was funded by PHS grants P42ES016465, K.C. Donnelly Supplement P42ES016465, P41GM103483, P01CA90890 and T32ES07060 (E.P.M). AMS was performed at the Research Resource for Biomedical AMS which is operated at LLNL under the auspices of the U.S. Department of Energy under contract DE-AC52-07NA27344 and National Institute of General Medical Sciences, Biomedical Technology Research Resources (BTRR) under grant number P41GM103483.
ABREVIATIONS
- ACN
acetonitrile
- AKR
aldo-keto reductase
- AMS
accelerator mass spectrometry
- AUC
area under the concentration curve
- BaP
benzo(a)pyrene
- BaPeq
toxicity equivalence related to that of BaP
- CAMS
center for accelerator mass spectrometry
- Bio-AMS
CAMS at Lawrence Livermore detected to LC-AMS for identification of [14C]-labeled samples
- Cmax
maximum concentration
- CYP
cytochrome P450
- DBC
dibenzo(def,p)chrysene
- DHD
DBC (+/−) DBC-11,12-dihydrodiol
- DHDE
DBC (+/−)- 11,12-dihydrodiol-13,14-epoxide
- DBC
tetraol- 11,12,13,14-tetra-hydroxy-DBC (stereochemistry undefined)
- EH
epoxide hydrolase
- IARC
International Agency for Research on Cancer
- LLNL
Lawrence Livermore National Laboratory
- LLOQ
lower limit of quantitation
- Mx
DBC metabolite where x refers to the order of elution from the UPLC
- OSU
Oregon State University
- PAH
polycyclic aromatic hydrocarbon
- PDA
photo diode array
- PBMCs
peripheral blood mononuclear cells
- RPF
Relative Potency Factor
- %RSD
% relative standard deviation
- S/N
signal to noise ratio
- SULT
sulfotransferase
- Tmax
time of maximum concentration
- UGT
UDP-glucuronosyl transferase
- UPLC
ultra performance liquid chromatography.
Footnotes
The authors declare no competing financial interests.
References
- 1.IARC. Polynuclear aromatic compounds, Part 1: chemical, environmental, and experimental data. World Health Organization International Agency For Research On Cancer; Lyon, France: 1983. Monographs on the Evaluation of Carcinogenic Risks to Humans. [PubMed] [Google Scholar]
- 2.Menzie CA, Potocki BB, Santodonato J. Exposure to carcinogenic PAHs in the environment. Environmn Sci Technol. 1992;26:1278–1284. [Google Scholar]
- 3.Dennis MJ, Massey RC, Cripps G, Venn I, Howarth N, Lee G. Factors affecting the polycyclic aromatic hydrocarbon content of cereals, fats and other food-products. Food Addit Contam. 1991;8:517–539. doi: 10.1080/02652039109374004. [DOI] [PubMed] [Google Scholar]
- 4.Jakszyn P, Agudo A, Albanez R, Garcia-Closas R, Pera G, Amiano P, Gonzalez CA. Development of a food database of nitrosamines, heterocyclic amines, and polycyclic aromatic hydrocarbons. J Nutr. 2004;134:2011–2014. doi: 10.1093/jn/134.8.2011. [DOI] [PubMed] [Google Scholar]
- 5.EPA; Code of Federal Regulations PART 423 - Steam electric power generating point of source category. E.P. Agency, editor. Title 40 - Protection of Environment. Chapter 1 - Environmental protection agency subchapter N - Effluent guidelines and standards. 2007
- 6.Luch A. In: Molecular, Clinical and Environmental Toxicology: Volume 1: Molecular Toxicology. Luch A, editor. Vol. 1. Springer Science and Business Media; 2009. [Google Scholar]
- 7.EPA. U.S Development of a relative potency factor (RPF) approach for polycyclic aromatic hydrocarbon (PAH) mixtures. United States Environmental Protection Agency; 2010. [Google Scholar]
- 8.Guttenplan JB, Kosinska W, Zhao ZL, Chen KM, Aliaga C, Del Tondo J, Cooper T, Sun YW, Zhang SM, Jiang K, Bruggeman R, Sharma AK, Amin S, El-Bayoumy K. Mutagenesis and carcinogenesis induced by dibenzo[a,l]pyrene in the mouse oral cavity: a potential new model for oral cancer. Internatl J Cancer. 2012;130:2783–2790. doi: 10.1002/ijc.26344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang M, Liu X, Basu SS, Zhang L, Kushman ME, Harvey RG, Blair IA, Penning TM. Metabolism and distribution of benzo[a]pyrene-7,8-dione (B[a]P-7,8-dione) in human lung cells by liquid chromatography tandem mass spectrometry: detection of an adenine B[a]P-7,8-dione adduct. Chem Res Toxicol. 2012;25:993–1003. doi: 10.1021/tx200463s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeMarini DM, Hanley NM, Warren SH, Adams LD, King LC. Association between mutation spectra and stable and unstable DNA adduct profiles in Salmonella for benzo[a]pyrene and dibenzo[a,l]pyrene. Mutat Res. 2011;714:17–25. doi: 10.1016/j.mrfmmm.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 11.Mahadevan B, Dashwood WM, Luch A, Pecaj A, Doehmer J, Seidel A, Pereira C, Baird WM. Mutations induced by (−)-anti-11R, 12S-dihydrodiol-13S,14R-epoxide of dibenzo[a,l]pyrene in the coding region of the hypoxanthine phosphoribosyltransferase (Hprt) gene in Chinese hamster V79 cells. Environm Mol Mutag. 2003;41:131–139. doi: 10.1002/em.10136. [DOI] [PubMed] [Google Scholar]
- 12.Melendez-Colon VJ, Luch A, Seidel A, Baird WM. Comparison of cytochrome P450- and peroxidase-dependent metabolic activation of the potent carcinogen dibenzo[a,l]pyrene in human cell lines: Formation of stable DNA adducts and absence of a detectable increase in apurinic sites. Cancer Res. 1999;59:1412–1416. [PubMed] [Google Scholar]
- 13.Castro DJ, Baird WM, Pereira CB, Giovanini J, Löhr CV, Fisher KA, Yu Z, Gonzalez FJ, Krueger SK, Williams DE. Fetal mouse Cyp1b1 and transplacental carcinogenesis from maternal exposure to dibenzo(a,l)pyrene. Cancer Prev Res. 2008;1:128–134. doi: 10.1158/1940-6207.CAPR-07-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castro DJ, Löhr CV, Fisher KA, Pereira CB, Williams DE. Lymphoma and lung cancer in offspring born to pregnant mice dosed with dibenzo[a,l]pyrene: The importance of in utero vs. lactational exposure. Toxicol Appl Pharmacol. 2008;233:454–458. doi: 10.1016/j.taap.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu Z, Loehr C, Fischer KA, Louderback K, Krueger SK, Dashwood RH, Kerkvliet NI, Pereira CB, Jennings-Gee J, Dance ST, Miller MS, Bailey GS, Williams DE. In utero exposure of mice to dibenzo[a,l]pyrene produces lymphoma in the offspring: Role of the aryl hydrocarbon receptor. Cancer Res. 2006;66:755–762. doi: 10.1158/0008-5472.CAN-05-3390. [DOI] [PubMed] [Google Scholar]
- 16.Shorey LE, Castro DJ, Baird WM, Siddens LK, Löhr CV, Matzke MM, Waters KM, Corley RA, Williams DE. Transplacental carcinogenesis with dibenzo[def,p]chrysene (DBC): Timing of maternal exposures determines target tissue response in offspring. Cancer Lett. 2012;317:49–55. doi: 10.1016/j.canlet.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.IARC. Monographs on the Evaluation of Carcinogenic Risks to Humans, in Some non-heterocyclic polycyclic aromatic hydrocarbons and some exposures. World Health Organization International Agency for Research on Cancer; Lyon, France: 2010. [Google Scholar]
- 18.Veyrand B, Sirot V, Durand S, Pollono C, Marchand P, Dervilly-Pinel G, Tard A, Leblanc JC, Le Bizec B. Human dietary exposure to polycyclic aromatic hydrocarbons: results of the second French Total Diet Study. Environm Internat. 2013;54:11–17. doi: 10.1016/j.envint.2012.12.011. [DOI] [PubMed] [Google Scholar]
- 19.Cavalieri EL, Rogan EG. Central role of radical cations in metabolic-activation of polycyclic aromatic hydrocarbons. Xenobiotica. 1995;25:677–688. doi: 10.3109/00498259509061885. [DOI] [PubMed] [Google Scholar]
- 20.Shou MG, Krausz KW, Gonzalez FJ, Gelboin HV. Metabolic activation of the potent carcinogen dibenzo[a,l]pyrene by human recombinant cytochromes P450, lung and liver microsomes. Carcinogenesis. 1996;17:2429–2433. doi: 10.1093/carcin/17.11.2429. [DOI] [PubMed] [Google Scholar]
- 21.Conney AH, Chang RL, Jerina DM, Wei SJ. Studies on the metabolism of benzo[a]pyrene and dose-dependent differences in the mutagenic profile of its ultimate carcinogenic metabolite. Drug Metabol Rev. 1994;26:125–163. doi: 10.3109/03602539409029788. [DOI] [PubMed] [Google Scholar]
- 22.Shimada T, Fujii-Kuriyama Y. Metabolic activation of polycyclic aromatic hydrocarbons to carcinogens by cytochromes P450 1A1 and 1B1. Cancer Sci. 2004;95:1–6. doi: 10.1111/j.1349-7006.2004.tb03162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Penning TM, Ohnishi ST, Ohnishi T, Harvey RG. Generation of reactive oxygen species during the enzymatic oxidation of polycyclic aromatic hydrocarbon trans-dihydrodiols catalyzed by dihydrodiol dehydrogenase. Chem Res Toxicol. 1996;9:84–92. doi: 10.1021/tx950055s. [DOI] [PubMed] [Google Scholar]
- 24.Meinl W, Ebert B, Glatt H, Lampen A. Sulfotransferase forms expressed in human intestinal Caco-2 and TC7 cells at varying stages of differentiation and role in benzo[a]pyrene metabolism. Drug Metabol Dispos. 2008;36:276–283. doi: 10.1124/dmd.107.018036. [DOI] [PubMed] [Google Scholar]
- 25.Olson KC, Sun D, Chen G, Sharma AK, Amin S, Ropson IJ, Spratt TE, Lazarus P. Characterization of dibenzo[a,l]pyrene-trans-11,12-diol (dibenzo[def,p] chrysene) glucuronidation by UDP-glucuronosyltransferases. Chem Res Toxicol. 2011;24:1549–1559. doi: 10.1021/tx200178v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shimada T, Oda Y, Gillam EM, Guengerich FP, Inoue K. Metabolic activation of polycyclic aromatic hydrocarbons and other procarcinogens by cytochromes P450 1A1 and P4501B1 allelic variants and other human cytochromes P450 in Salmonella typhimurium NM 2009. Drug Metabol Dispos. 2001;29:1176–1182. [PubMed] [Google Scholar]
- 27.Shimada T, Gillam EM, Sutter TR, Strickland PT, Guengerich FP, Yamazaki H. Oxidation of xenobiotics by recombinant human cytochrome P450 1B1. Drug Metabol Dispos. 1997;25:617–622. [PubMed] [Google Scholar]
- 28.Buters JTM, Mahadevan B, Quintanilla-Martinez L, Gonzalez FJ, Greim H, Baird WM, Luch A. Cytochrome p450 1B1 determines susceptibility to dibenzo[a,l]pyrene-induced tumor formation. Chem Res Toxicol. 2002;15:1127–1135. doi: 10.1021/tx020017q. [DOI] [PubMed] [Google Scholar]
- 29.Ralston SL, Coffing SL, Seidel A, Luch A, Platt KL, Baird WM. Stereoselective activation of dibenzo[a,l]pyrene and its trans-11,12-dihydrodiol to fjord region 11,12-diol 13,14-epoxides in a human mammary carcinoma MCF-7 cell-mediated V79 cell mutation assay. Chem Res Toxicol. 1997;10:687–693. doi: 10.1021/tx9700275. [DOI] [PubMed] [Google Scholar]
- 30.Prahalad AK, Ross JA, Nelson GB, Roop BC, King LC, Nesnow S, Mass MJ. Dibenzo[a,l]pyrene-induced DNA adduction, tumorigenicity, and Ki-ras oncogene mutations in strain A/J mouse lung. Carcinogenesis. 1997;18:1955–1963. doi: 10.1093/carcin/18.10.1955. [DOI] [PubMed] [Google Scholar]
- 31.Hecht SS, Carmella SG, Villalta PW, Hochalter JB. Analysis of phenanthrene and benzo[a]pyrene tetraol enantiomers in human urine: relevance to the bay region diol epoxide hypothesis of benzo[a]pyrene carcinogenesis and to biomarker studies. Chem Res Toxicol. 2010;23:900–908. doi: 10.1021/tx9004538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun YW, El-Bayoumy K, Aliaga C, Awad AS, Gowda K, Amin S, Chen KM. Tissue distribution, excretion, and pharmacokinetics of the environmental pollutant dibenzo[def,p]pyrene in mouse. Chem Res Toxicol. 2015;28:1427–1433. doi: 10.1021/acs.chemrestox.5b00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crowell SR, Sharma AK, Amin SG, Soelberg JJ, Sadler NC, Wright AT, Baird WM, Williams DE, Corley RA. Impact of pregnancy on the pharmacokinetics of dibenzo[def,p]chrysene in mice. Toxicol Sci. 2013;135:48–62. doi: 10.1093/toxsci/kft124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crowell SR, Hanson-Drury S, Williams DE, Corley RA. In vitro metabolism of benzo[a]pyrene and dibenzo[def,p] chrysene in rodent and human hepatic microsomes. Toxicol Lett. 2014;228:48–55. doi: 10.1016/j.toxlet.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crowell SR, Amin SG, Anderson KA, Krishnegowda G, Sharma AK, Soelberg JJ, Williams DE, Corley RA. Preliminary physiologically based pharmacokinetic models for benzo[a]pyrene and dibenzo[def,p]chrysene in rodents. Toxicol Appl Pharmacol. 2011;257:365–376. doi: 10.1016/j.taap.2011.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Madeen E, Corley RA, Crowell S, Turteltaub K, Ognibene T, Malfatti M, McQuistan TJ, Garrard M, Sudakin D, Williams DE. Human in vivo pharmacokinetics of [14C]dibenzo[def,p]chrysene by accelerator mass spectrometry following oral microdosing. Chem Res Toxicol. 2015;28:126–134. doi: 10.1021/tx5003996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ognibene TJ, Bench G, Vogel JS, Peaslee GF, Murov S. A high-throughput method for the conversion of CO2 obtained from biochemical samples to graphite in septa-sealed vials for quantification of C-14 via accelerator mass spectrometry. Analy Chem. 2003;75:2192–2196. doi: 10.1021/ac026334j. [DOI] [PubMed] [Google Scholar]
- 38.Thomas AT, Stewart BJ, Ognibene TJ, Turteltaub KW, Bench G. Directly coupled high-performance liquid chromatography-accelerator mass spectrometry measurement of chemically modified protein and peptides. Analy Chem. 2013;85:3644–3650. doi: 10.1021/ac303609n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ognibene TJ, Thomas AT, Daley PF, Bench G, Turteltaub KW. An interface for the direct coupling of small liquid samples to AMS. Nucl Instr Methods Phys Res B. 2015;361:173–177. doi: 10.1016/j.nimb.2015.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hobbie KA, Peterson ES, Barton ML, Waters KM, Anderson KA. Integration of data systems and technology improves research and collaboration for a superfund research center. J Lab Autom. 2012;17:275–283. doi: 10.1177/2211068212448428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomas AT, Ognibene T, Daley P, Turteltaub K, Radousky H, Bench G. Ultrahigh efficiency moving wire combustion interface for online coupling of high-performance liquid chromatography (HPLC) Analy Chem. 2011;83:9413–9417. doi: 10.1021/ac202013s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ognibene TJ, Salazar GA. Installation of hybrid ion source on the 1-MV LLNL BioAMS spectrometer. Nucl Instr Methods Phys Res Sect B. 2013;294:311–314. doi: 10.1016/j.nimb.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lehotay SJ, de Kok A, Hiemstra M, Van Bodegraven P. Validation of a fast and easy method for the determination of residues from 229 pesticides in fruits and vegetables using gas and liquid chromatography and mass spectrometric detection. J AOAC Inter. 2005;88:595–614. [PubMed] [Google Scholar]
- 44.Tellinghuisen J. Statistical error propogation. J Phys Chem. 2001;105:3917–3921. [Google Scholar]
- 45.Dolan JW. The role of the signal-to-noise ratio in precision and accuracy. LC- GC. Europe. 2006;19:12. [Google Scholar]
- 46.Tiwari G, Tiwari R. Bioanalytical method validation: An updated review. Pharm Methods. 2010;1:25–38. doi: 10.4103/2229-4708.72226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shou M, Krausz KW, Gonzalez FJ, Gelboin HV. Metabolic activation of the potent carcinogen dibenzo[a,l]pyrene by human recombinant cytochromes P450, lung and liver microsomes. Carcinogenesis. 1996;17:2429–2433. doi: 10.1093/carcin/17.11.2429. [DOI] [PubMed] [Google Scholar]
- 48.Nebert DW, Shi Z, Gálvez-Peralta M, Uno S, Dragin N. Oral benzo[a]pyrene: understanding pharmacokinetics, detoxication, and consequences-Cyp1 knockout mouse lines as a paradigm. Mol Pharmacol. 2013;84:304–313. doi: 10.1124/mol.113.086637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu JY, Yang Y, Liu ZZ, Xie JJ, Du YP, Wang W. Association between the CYP1B1 polymorphisms and risk of cancer: a meta-analysis. Mol Genet Genom. 2015;290:739–765. doi: 10.1007/s00438-014-0946-x. [DOI] [PubMed] [Google Scholar]
- 50.Inoue K, Asao T, Shimada T. Ethnic-related differences in the frequency distribution of genetic polymorphisms in the CYP1A1 and CYP1B1 genes in Japanese and Caucasian populations. Xenobiotica. 2000;30:285–295. doi: 10.1080/004982500237677. [DOI] [PubMed] [Google Scholar]
- 51.Huang M, Blair IA, Penning TM. Identification of stable benzo[a]pyrene-7,8-dione-DNA adducts in human lung cells. Chem Res Toxicol. 2013;26:685–692. doi: 10.1021/tx300476m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang L, Huang M, Blair IA, Penning TM. Interception of benzo[a]pyrene-7,8-dione by UDP blucuronosyltransferases (UGTs) in human lung cells. Chem Res Toxicol. 2013;26:1570–1578. doi: 10.1021/tx400268q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang L, Huang M, Blair IA, Penning TM. Detoxication of benzo[a]pyrene-7,8-dione by sulfotransferase (SULTs) in human lung cells. J Biol Chem. 2012;287:29909–29920. doi: 10.1074/jbc.M112.386052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Joseph P, Jaiswal AK. NAD(P)H:quinone oxidoreductase (DT diaphorase) specifically prevents the formation of benzo[a]pyrene quinone-DNA adducts generated by cytochrome P4501A1 and P450 reductase. Proc Natl Acad Sci (USA) 1994;91:8413–8417. doi: 10.1073/pnas.91.18.8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. DBC Metabolism by Human CYP1B1 and Liver Microsomes. A. HPLC of DBC, DBC-11,12-DHD and DBC-11,12-DHD-13,14-Epoxide. CRN (coronene) was used as an internal standard. B. Incubations were performed with 0.05 nmol of human CYP1B1 (Human CYP1B1 plus P450 reductase Supersomes or C. One mg/ml of pooled human liver microsomes (both from Gentest, BD Biosciences).
Supplemental Table 1. Plasma [14C]-DBC UPLC-AMS Metabolite Data. A. Individual volunteer plasma levels of [14C]-DBCeq are given in fg/mL for sampling from 0–48 hours. B. This table provides the propagated error for plasma values of each volunteer, again in fg [14C]-DBCeq/mL. The quality control of sample data included elimination of samples that were below the limit of detection (<LOD), defined as a signal to noise ratio of 5 or higher. Samples failing to meet the quality standard of a %RSD of ≥ 30 were also eliminated to maintain data quality (>%RSD). Samples that were not detected were reported as N.D. Samples that were not available for analysis were reported as N.A.
Supplemental Table 2. Deconjugated Cleaved Urine [14C]-DBC UPLC-AMS Metabolite Data. A. Individual volunteer levels of each metabolite in pools of urine collected at 6–12, 12–24, 24–48 and 48–72 hours post-dosing. The samples were treated with β-glucuronidase as described in materials and methods and reported as fg [14C]-DBC/pool. B. The values reported in A. are adjusted for propagated error as described for Supplemental Table 1B.
Supplemental Table 3. Native Urine [14C]-DBC UPLC-AMS Metabolite Data. A. Individual volunteer levels of each metabolite in pools of urine collected at 6–12, 12–24, 24–48 and 48–72 hours post-dosing. The samples were not treated with deconjugating enzymes and the results are reported as fg [14C]-DBC/pool. B. The values reported in A. are adjusted for propagated error as described for Supplemental Table 1B.