Abstract
BRAF drives tumorigenesis by coordinating the activation of RAS/RAF/MEK/ERK oncogenic signaling cascade. However, upstream pathway(s) governing BRAF kinase activity and protein stability remains undefined. Here, we report that in primary cells with active APCFZR1, APCFZR1 earmarks BRAF for ubiquitination-mediated proteolysis, while in cancer cells with APC-free FZR1, FZR1 suppresses BRAF through disrupting BRAF dimerization. Moreover, we identified FZR1 as a direct target of ERK and CYCLIN D1/CDK4 kinases. Phosphorylation of FZR1 inhibits APCFZR1, leading to elevation of a cohort of oncogenic APCFZR1 substrates to facilitate melanomagenesis. Importantly, CDK4 and/or BRAF/MEK inhibitors restore APCFZR1 E3 ligase activity, which might be critical for their clinical effects. Furthermore, FZR1 depletion co-operates with AKT hyper-activation to transform primary melanocytes, while genetic ablation of Fzr1 synergizes with Pten loss, leading to aberrant co-activation of BRAF/ERK and AKT signaling in mice. Our findings therefore reveal a reciprocal suppression mechanism between FZR1 and BRAF in controlling tumorigenesis.
Keywords: BRAF, FZR1, ubiquitination, phosphorylation, melanoma, tumorigenesis
INTRODUCTION
The Anaphase-Promoting Complex/Cyclosome (APC/C, also named APC) ubiquitin E3 ligase is essential for cell cycle progression through the M/G1 phases largely by controlling the proteolytic degradation of key cell cycle regulators including mitotic cyclins and DNA replication factors (1). Notably, unlike CDC20, which has restricted function in M phase, its close homologue, Fizzy-related protein 1 (FZR1, also named Cdh1), associates with the APC core complex in late M phase and early G1 phase and determines G1 phase cell cycle fate decisions (2). During the remainder of the cell cycle, phosphorylation of FZR1 by CDK kinases abolishes the interaction between FZR1 and the APC core complex, therefore inhibiting APC-dependent functions of FZR1 (3–6). Although the APC-dependent functions of FZR1 have been well documented (7), how APC-free FZR1 participates in various cellular processes is just beginning to be uncovered (8). To this end, we have recently demonstrated an APC-independent function of FZR1 in positive regulation of the ubiquitin E3 ligase activity of SMURF1 to influence osteoblasts differentiation (9). However, APC-independent functions of FZR1 in controlling tumorigenesis, as well as in other cellular or tissue contexts, remain largely elusive.
There is mounting evidence indicating a tumor suppressive role for FZR1 (8). Consistent with this notion, most FZR1 substrates including mitotic and S phase cyclins (10), mitotic kinases (11) and DNA replication factors (12), are frequently overexpressed in a wide spectrum of human malignancies (8). Moreover, FZR1 deletions, reduced expression and mutations are found in various human tumor tissues (13, 14). Furthermore, while FZR1 homozygous deletion results in embryonic lethality, FZR1 heterozygous mice are more susceptible to developing epithelial tumors (15). In addition, our recent studies revealed a crucial role of APCFZR1 in controlling melanocytes differentiation and pigmentation (16). However, the molecular mechanisms underlying how loss of FZR1 induces tumorigenesis still remain largely unclear. Hence, it is important to define the major downstream oncogenic signaling pathway(s) that are negatively regulated by the FZR1 tumor suppressor, which will further define the critical role of FZR1 in tumorigenesis.
The RAF family of protein kinases consist of ARAF, BRAF and CRAF isoforms, which play a central role in driving tumorigenesis through activation of the MEK/ERK oncogenic signaling cascade (17). Notably, although cancer-associated BRAF mutants were found in over 60% melanoma and thyroid cancer patients (18), in other types of cancers, BRAF genetic status is largely wild type (WT) (19). Therefore, it is important to understand mechanistically how BRAF is aberrantly upregulated or hyper-activated in human cancers with WT-BRAF. This valuable information will provide further insights to guide novel targeted therapy strategies to efficiently treat cancer patients carrying WT-BRAF.
On the other hand, the BRAFV600E oncogenic mutation has become a major drug target in developing targeted therapeutics against BRAFV600E-driven cancers including melanoma (20, 21). Although anti-BRAFV600E inhibitors including vemurafenib (PLX4032) (22) and dabrafenib (23) were approved in treating melanoma patients harboring this mutation, drug resistance is frequently reported (24), suggesting a limitation of single agent treatment. Recent clinical trials have adopted combinational strategies by using BRAFV600E inhibitor together with another compound targeting either MEK, CDK4/6, PI3K or HSP90 to increase efficiency and to improve survival (25, 26). However, mechanistically how these combinational therapies suppress tumor growth remains poorly defined. Our findings reported here illustrate an inverse correlation between the ability of FZR1 to suppress BRAF activity and the aggressiveness of tumor developmental stages.
RESULTS
Depletion of FZR1 Leads to BRAF Accumulation and Subsequent Activation of ERK
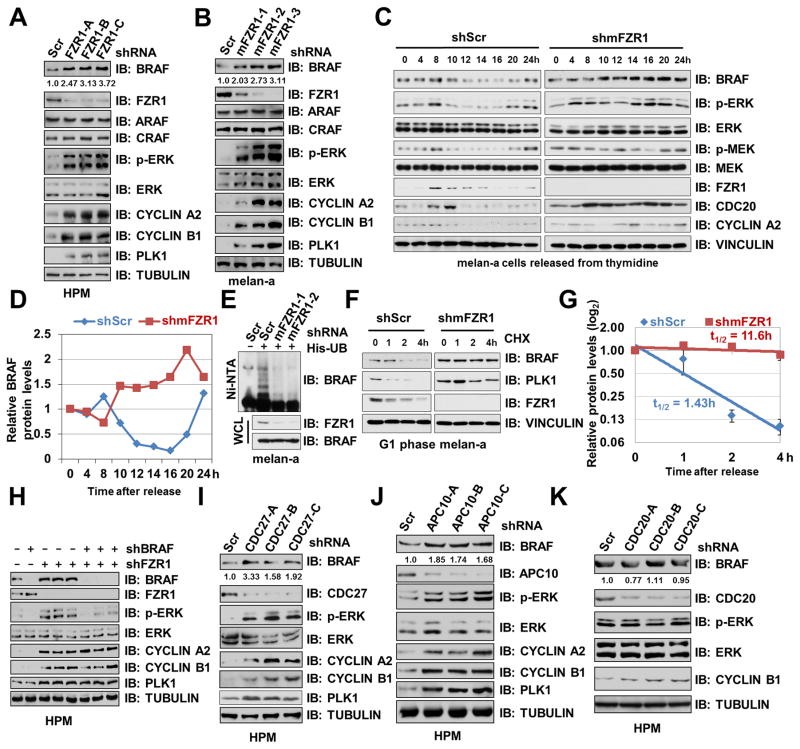
We and others have previously demonstrated that acute depletion of FZR1 in human primary fibroblasts leads to premature senescence (27, 28). Consistent with a critical role for BRAF/ERK signaling in triggering senescence in melanocytes (29), we found that BRAF and p-ERK, but not other RAF proteins, were significantly upregulated in FZR1-depleted human primary fibroblasts (Supplementary Fig. S1A), human primary melanocytes (HPM) (Fig. 1A) and mouse melanocytes melan-a (Fig. 1B). Inhibiting APCFZR1 using a specific APC inhibitor Apcin (30), which blocks the FZR1 substrate binding pocket, also resulted in an upregulation of BRAF and its downstream p-ERK levels (Supplementary Fig. S1B). In keeping with the oscillating nature of APCFZR1 E3 ligase activity during cell cycle progression (1), in primary melanocytes-derived melan-a cells, both endogenous BRAF and p-ERK levels decreased in the G1 phase where APCFZR1 is most active (Fig. 1C). Notably, depletion of endogenous FZR1 in melan-a cells resulted in stabilization of BRAF coupled with elevated p-ERK across the cell cycle, supporting the notion that the BRAF signaling pathway is negatively regulated by APCFZR1 in a cell cycle-dependent manner (Fig. 1C–D and Supplementary Fig. S1C). In further support of BRAF being a putative APCFZR1 downstream ubiquitin substrate, ubiquitination of endogenous BRAF was suppressed in FZR1-depleted melanocytes (Fig. 1E), resulting in an extended half-life of endogenous BRAF (Fig. 1F–G).
Figure 1. Depletion of FZR1 Leads to BRAF Accumulation and Subsequent Activation of ERK.
(A–B) Depletion of FZR1 led to an elevation of BRAF abundance and its downstream MEK/ERK activities in human primary melanocytes (HPM) and murine melanocytes melan-a. Immunoblot (IB) analysis of HPM (A) or melan-a (B) infected with control (shScr) or indicated shFZR1 lentiviral shRNA constructs. The infected cells were selected with 1 μg/ml puromycin for 72 hours before harvesting. BRAF band intensities were quantified using ImageJ, normalized to corresponding TUBULIN band intensities, and then normalized to shScr.
(C–D) In the absence of FZR1, BRAF protein levels failed to fluctuate across the cell cycle. (C) IB analysis of whole cell lysates (WCL) derived from primary melanocyte-derived melan-a cells synchronized at the G1/S boundary by double-thymidine block then released back into the cell cycle for the indicated periods of time. (D) Quantification of BRAF band intensities. BRAF bands were normalized to VINCULIN, then normalized to the t = 0 time point.
(E) Ubiquitination of endogenous BRAF was attenuated in FZR1-depleted melanocytes. IB analysis of WCL and Ni-NTA (Ni-nitrilotriacetic acid) affinity precipitates derived from melan-a cells infected with the indicated lentiviral shRNA and His-ubiquitin constructs. Cells were pretreated with 10 μM MG132 for 10 hours before harvesting.
(F–G) The half-life of BRAF was extended in FZR1-depleted melanocytes in early G1 phase. melan-a cells were infected with the indicated lentiviral shRNA constructs for 24 hours. Non-infected cells were eliminated by selection with 1 μg/ml puromycin for 48 hours. Cells were then synchronized by double thymidine block (12) and released back into the cell cycle for 14 hours (early G1 phase). 20 μg/ml cycloheximide (CHX) was added to the resulting cells for the indicated time periods before harvesting for IB analysis (F). (G) Quantification of BRAF band intensities was plotted as mean ± SD from three independent experiments, BRAF bands were normalized to VINCULIN, then normalized to the t = 0 time point.
(H) Further depletion of BRAF suppressed the activation of ERK upon FZR1 knockdown. IB analysis of HPMs infected with the indicated lentiviral shRNA constructs. The infected cells were selected with 1 μg/ml puromycin for 72 hours before harvesting.
(I–K) Depletion of APC core subunit CDC27 or APC10, but not CDC20, led to BRAF accumulation and ERK activation. IB analysis of HPMs infected with control (shScr) or the indicated shCDC27 (I), shAPC10 (J) or shCDC20 (K) lentiviral shRNA constructs. The infected cells were selected with 1 μg/ml puromycin for 72 hours before harvesting. BRAF band intensities were quantified using ImageJ, normalized to corresponding TUBULIN band intensities, and then normalized to shScr.
Additional depletion of endogenous BRAF in FZR1-knockdown HPM (Fig. 1H) and melan-a cells (Supplementary Fig. S1D) largely suppressed FZR1-depletion triggered p-ERK elevation, suggesting that FZR1 inhibits ERK activation primarily through BRAF. Furthermore, depletion of other APC complex subunits, such as CDC27 or APC10, also resulted in BRAF accumulation and ERK activation (Fig. 1I–J). In contrast, depletion of endogenous CDC20 failed to elevate either BRAF or p-ERK levels (Fig. 1K). Together, these data suggest that in primary cells, FZR1, but not CDC20, negatively regulates BRAF abundance and subsequent ERK activation largely in an APC-dependent manner.
Depletion of FZR1 Triggers Senescence in Primary Melanocytes
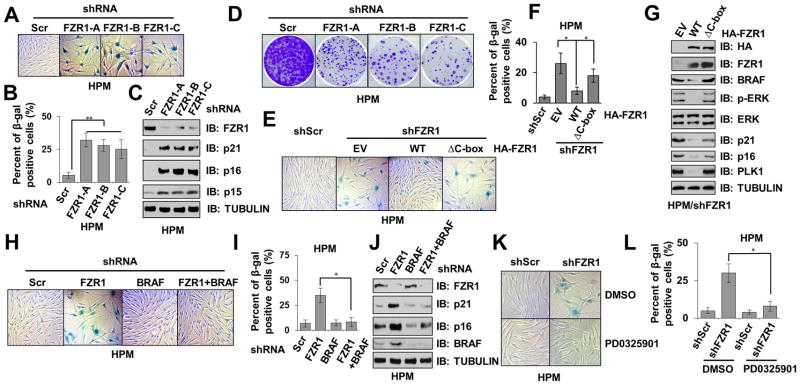
As hyper-activation of the RAF/MEK/ERK signaling cascade has been closely linked to nevi formation composed of senescent melanocytes (29), we sought to investigate whether acute depletion of FZR1 could also result in similar senescent phenotype in primary melanocytes as it does in human primary fibroblasts (27). Notably, compared with control cells, a marked increase of Senescence-Associated β-galactosidase (SA-β-gal) positive cells was observed in FZR1-depleted HPM (Fig. 2A–B) and melan-a cells (Supplementary Fig. S2A–B), indicating an accumulation of senescent cells 14 days after FZR1 depletion. Depletion of FZR1 also led to elevated expression of CDK inhibitors, p16INK4A, p15INK4B and p21WAF1 in HPM (Fig. 2C) and p21WAF1 in melan-a cells (Supplementary Fig. S2C), driving cell cycle arrest upon FZR1 depletion (Fig. 2D and Supplementary Fig. S2D). As hyper-activation of the RAS/RAF/ERK pathway has been reported to cause premature senescence (31, 32), our results suggest that loss of a negative regulatory mechanism of BRAF/ERK signaling pathway by FZR1 may lead to the onset of premature senescence. In support of this notion, we found that compared to normal human skin, human nevi, which is composed of largely senescent melanocytes (33), exhibited relatively lower FZR1 expression (34) (Supplementary Fig. S2E).
Figure 2. Depletion of Endogenous FZR1 Triggers Premature Senescence in Primary Melanocytes.
(A–B) Depletion of FZR1 in melanocytes triggered premature senescence. Control (shScr) or shFZR1 infected human primary melanocytes (HPM) were subjected to SA-β-gal staining assays 14 days after viral infection. The pictures showed one representative experiment (A) out of three independent experiments (B). Data are represented as mean ± SD, n=3. ** p < 0.01, Student’s t test.
(C) FZR1 knockdown resulted in the accumulation of CDK inhibitors including p21 and p16 in human primary melanocytes. Immunoblot (IB) analysis of whole cell lysates (WCL) derived from various HPMs generated in (A).
(D) Depletion of FZR1 retarded the proliferation of melanocytes. Control (shScr) or shFZR1 infected HPMs were subjected to clonogenic survival assays 5 days after viral infection. Crystal violet was used to stain the formed colonies.
(E–F) WT-FZR1, but not APC-binding deficient ΔC-box-FZR1, prevented senescence in FZR1-depleted melanocytes. HPMs stably expressing EV, WT- or ΔC-box-FZR1 were further infected with shScr or shFZR1 lentiviral constructs, the resulting cells were subjected to SA-β-gal staining assays 14 days after viral infection. (E) The pictures showed one representative experiment out of three independent experiments. (F) Quantification of β-gal positive cells of (E). Data are represented as mean ± SD, n=3. * p < 0.05, Student’s t test.
(G) WT-FZR1, but not APC-binding deficient ΔC-box-FZR1, suppressed the accumulation of CDK inhibitors in FZR1-depleted melanocytes. IB analysis of WCL derived from various HPMs generated in (E).
(H–I) Additional depletion of BRAF suppressed the onset of premature senescence upon FZR1 knockdown. HPMs were infected with the indicated lentiviral constructs, the resulting cells were subjected to SA-β-gal staining assays 14 days after viral infection. (H) The pictures showed one representative experiment out of three independent experiments. (I) Quantification of β-gal positive cells of (H). Data are represented as mean ± SD, n=3. * p < 0.05, Student’s t test.
(J) Expression of CDK inhibitors partly decreased in FZR1-depleted melanocytes upon further BRAF knockdown. IB analysis of WCL derived from various HPMs generated in (H).
(K–L) MEK inhibition reversed the senescence phenotype in FZR1-depleted melanocytes. Control (shScr) or shFZR1 infected HPMs were treated with or without 1 μM MEK inhibitor PD0325901 and subjected to SA-β-gal staining assays 14 days after viral infection. The pictures show one representative experiment (J) out of three independent experiments (K). Data are represented as mean ± SD, n=3. * p < 0.05, Student’s t test.
To gain further insight into FZR1-regulated melanocyte senescence, we found that in FZR1-depleted human primary melanocytes, re-introducing WT-FZR1 largely rescued the senescent phenotype (Fig. 2E–F), leading to escape from growth arrest phenotype (Supplementary Fig. S2F), in part by suppressing the elevated expression of p16INK4A and p21WAF1 that is associated with depleting FZR1 (Fig. 2G). Notably, this effect was not observed in the cells reintroduced the E3 ligase-deficient ΔC-box-FZR1 mutant (Fig. 2E–G and Supplementary Fig. S2F–G), which is unable to associate with the APC core complex (35). This result indicates an APC-dependent function of FZR1 in controlling BRAF signaling in primary melanocytes. Moreover, additional depletion of BRAF largely reversed the senescence phenotype in FZR1-depleted melanocytes (Fig. 2H–J and Supplementary Fig. S2H). Consistently, the MEK inhibitor PD0325901 (36) could also largely reverse FZR1-depletion induced senescence (Fig. 2K–L and Supplementary Fig. S2I), in part by suppressing FZR1 loss-induced upregulation of p16INK4A and p21WAF1 (Supplementary Fig. S2J). These results suggest that depletion of FZR1 triggers the onset of premature senescence in primary melanocytes mainly through activation of the BRAF/MEK/ERK signaling cascade.
BRAF is an APCFZR1 Ubiquitin Substrate in primary melanocytes
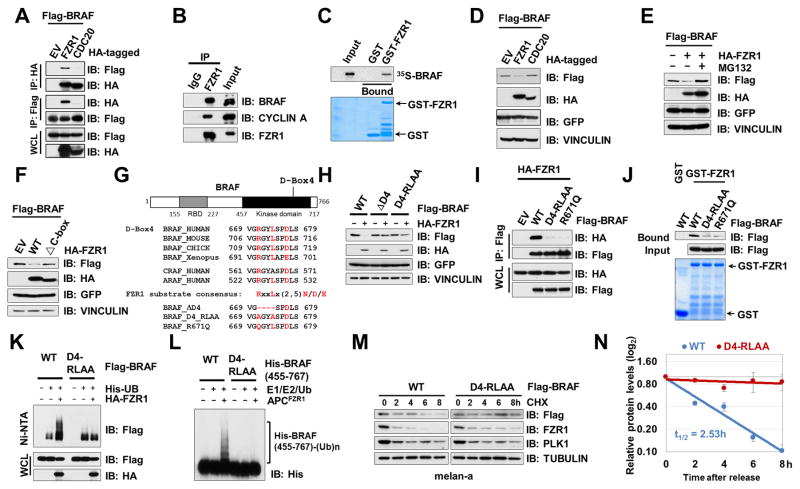
In support of APCFZR1 as a negative upstream regulator for BRAF, BRAF interacted with FZR1, the substrate recruiting subunit for the APCFZR1 E3 ligase complex, in cells (Fig. 3A–B and Supplementary Fig. S3A) and in vitro (Fig. 3C). Notably, BRAF was able to specifically interact with FZR1, but not CDC20 (Fig. 3A). Furthermore, FZR1 was co-immunoprecipitated with BRAF, but not CRAF, in cells (Supplementary Fig. S3B). Although FZR1 could also interact with ARAF in an ectopic expression condition, FZR1 only promoted the degradation of BRAF, but not ARAF (Supplementary Fig. S3C), supporting BRAF as a specific APCFZR1 ubiquitin substrate. Similar to other FZR1 substrates, BRAF specifically bound to the WD40 domain of FZR1 (37) (Supplementary Fig. S3D–E). Ectopic expression of FZR1, but not CDC20, led to BRAF downregulation (Fig. 3D), which could be largely abolished by the 26S proteasome inhibitor, MG132 (Fig. 3E). However, the E3 ligase activity-deficient mutant ΔC-box-FZR1 failed to promote BRAF degradation (Fig. 3F), indicating that APCFZR1 controls BRAF abundance probably through ubiquitination-mediated proteolysis.
Figure 3. APCFZR1 Promotes BRAF Ubiquitination in a D-box-Dependent Manner.
(A) BRAF specifically bound to FZR1, but not CDC20 in cells. Immunoblot (IB) analysis of whole cell lysates (WCL) and immunoprecipitates (IP) derived from 293T cells transfected with HA-FZR1 or HA-CDC20 together with the Flag-BRAF construct. 36 hours post-transfection, cells were pretreated with 10 μM MG132 for 10 hours before harvesting.
(B) Endogenous BRAF bound to endogenous FZR1. IB analysis of WCL and anti-FZR1 IP derived from HeLa cells.
(C) In vitro transcribed and translated BRAF (IVT-35S-BRAF) bound to purified recombinant GST-FZR1. Autoradiography of 35S-labelled BRAF bound to bacterially purified GST-FZR1, but not the GST recombinant protein.
(D) FZR1, but not CDC20, promoted the degradation of BRAF. IB analysis of WCL derived from 293 cells transfected with HA-FZR1 or HA-CDC20 with Flag-BRAF constructs. GFP serves as an internal transfection control.
(E) FZR1-mediated BRAF degradation could be blocked by the 26S proteasome inhibitor, MG132. IB analysis of WCL derived from 293 cells transfected with Flag-BRAF and EV or HA-FZR1 constructs. 10 μM MG132 was used to inhibit the 26S proteasome where indicated. GFP serves as an internal transfection control.
(F) APC-binding deficient ΔC-box-FZR1 failed to promote BRAF degradation. IB analysis of WCL derived from 293 cells transfected with Flag-BRAF and HA-tagged WT-FZR1 or E3 ligase activity deficient ΔC-box-FZR1 constructs. GFP serves as an internal transfection control.
(G) Sequence alignments of the putative D-boxes containing region between BRAF proteins from various species as well as a schematic representation of the various D-boxes deletion mutants generated and used in the following studies.
(H) D-box4-deleted or mutated BRAF mutants were resistant to FZR1-mediated degradation. IB analysis of WCL derived from 293 cells transfected with the indicated Flag-BRAF mutants with HA-FZR1 where indicated. GFP serves as an internal transfection control.
(I) D-box4 mutants of BRAF failed to bind FZR1. IB analysis of WCL derived from 293 cells transfected with the indicated Flag-tagged WT- or mutant BRAF constructs with HA-FZR1 where indicated. GFP serves as an internal transfection control.
(J) D-box4 mutated BRAF failed to bind FZR1 in vitro. GST pull down analysis to determine WT-, D4-RLAA, or R671Q mutant form of BRAF bound to the indicated GST fusion proteins.
(K) FZR1 promoted ubiquitination of WT-BRAF, but not D-box4 mutated BRAF, in cells. APCFZR1 promotes BRAF ubiquitination in vivo. IB analysis of WCL and subsequent His-tag pull-down in 6 M guanine-HCl containing buffer derived from 293 cells transfected with the indicated plasmids. Cells were pre-treated with 10 μM MG132 for 10 hours to block the proteasome pathway before harvesting.
(L) APCFZR1 promoted BRAF ubiquitination in vitro. Bacterially purified WT- and D4-RLAA-His-BRAF kinase domain (455–767) proteins were incubated with the APCFZR1 complex purified from G1 phase-arrested HeLa cell extract together with purified E1, E2 and ubiquitin as indicated at 30°C for 60 minutes before being resolved by SDS-PAGE and probed with the anti-His antibody.
(M–N) D-box4 mutated BRAF displayed an extended half-life compared with its WT counterpart. melan-a cells ectopically expressing WT- or D4-RLAA-BRAF were treated with 20 μg/ml cycloheximide (CHX) for the indicated time periods before harvesting. Equal amounts of whole cell lysates (WCL) were immunoblotted with the indicated antibodies (M). (N) Quantification of Flag-BRAF band intensities was plotted as mean ± SD from three independent experiments, Flag-BRAF bands were normalized to TUBULIN, then normalized to the t = 0 time point.
As most APCFZR1 substrates contain a destruction box (D-box) motif [RxxLx(2-5)N/D/E] (38), examination of the BRAF sequence revealed four putative D-boxes within its coding region (Fig. 3G and Supplementary Fig. S3F). Deletion of D-box 4 (ΔD4), and to a much lesser extent, D-box 3 (ΔD3), but not D-Box 1 or D-box 2, conferred a moderate resistance to FZR1-mediated BRAF degradation in cells (Supplementary Fig. S3G), indicating that D-box 4 is the primary degron motif. To minimize the impact of altering amino acids in the BRAF protein, we generated a D-box 4 mutated version of BRAF (D4-RLAA, Fig. 3G). Similar to ΔD4-BRAF, D4-RLAA-BRAF exhibited a noticeable resistance to FZR1-mediated degradation (Fig. 3H), in part due to impaired interaction with FZR1 (Fig. 3I–J).
In addition, we identified a cancer patient-derived mutation within D-box 4 (R671Q, mutation Id COSM159405, cancer.sanger.ac.uk and Fig. 3G), and further showed that analogous to D4-RLAA, the R671Q-BRAF mutant failed to interact with FZR1 (Fig. 3I–J), which might allow it to escape FZR1-mediated ubiquitination and subsequent degradation to favor tumorigenesis. In keeping with a critical role of D-box 4 in mediating BRAF ubiquitination by APCFZR1, compared to WT-BRAF, D4-RLAA-BRAF was insensitive to APCFZR1-promoted ubiquitination (Fig. 3K–L), thereby displaying an extended half-life (Fig. 3M–N) and relatively elevated abundance in the G1 phase (Supplementary Fig. S3H).
Notably, ultraviolet irradiation is a well-characterized risk factor for developing melanoma (39, 40). To this end, previous reports showed that acute UV radiation leads to FZR1 degradation and accumulation of APCFZR1 substrates (41, 42). Consistently, we found that UV exposure resulted in a reduction of FZR1 protein abundance in melan-a, HBL and A375 cells (Supplementary Fig. S3I–K). Of note, in normal melanocyte melan-a, UV-triggered FZR1 downregulation led to the accumulation of both BRAF and PLK1 (Supplementary Fig. S3I). In contrast, in melanoma cells HBL and A375 (Supplementary Fig. S3J–K), although FZR1 protein abundance decreased upon UV treatment, BRAF and PLK1 protein levels were largely unaffected. This result indicates that APCFZR1 might be more active in primary cells to target BRAF for proteolysis. Altogether, these data demonstrate that in the primary melanocyte setting, APCFZR1 controls the ubiquitination and subsequent degradation of BRAF in a D-box-dependent manner (Supplementary Fig. S3L).
Depletion of FZR1 in Cancer Cells Leads to ERK Activation Independent of APC
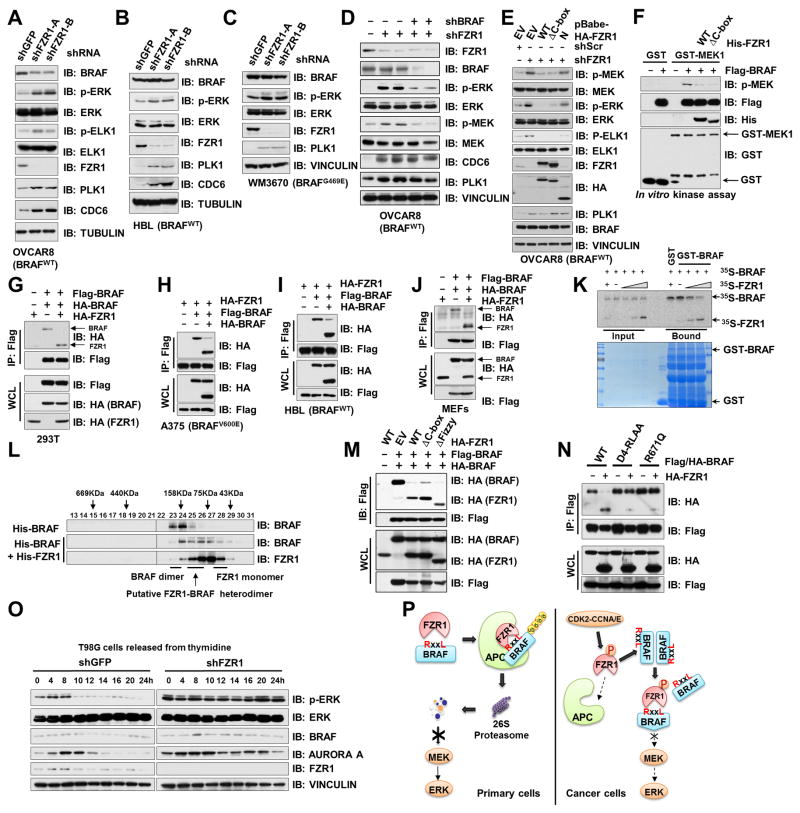
Consistent with results obtained in primary cells (Fig. 1A and Supplementary Fig. S1A), depletion of FZR1 in OVCAR8, HBL, WM3670, U2OS, HEK293 and H1755 cell lines, led to p-ERK upregulation (Fig. 4A–C and Supplementary Fig. S4A–C). However, unlike in normal cells, depletion of FZR1 did not significantly elevate BRAF abundance in these cancer or transformed cell lines (Fig. 4A–C and Supplementary Fig. S4A–C), which may be in part due to elevated CDK activity that has been shown to inhibit APCFZR1 E3 ligase activity (3–6). Additionally, compared to well-characterized APCFZR1 substrates such as PLK1, BRAF displayed reduced binding to both FZR1 and APC10 (Supplementary Fig. S4D–E), presumably due to lacking a canonical APC10 binding motif (43) (Supplementary Fig. S4F). Given that APC10 also participates in recognizing APC substrates (44) (Supplementary Fig. S4G), the lack of a strong interaction with APC10 indicates that BRAF might be a relatively weak, or distributive, rather than processive substrate for APCFZR1 (45). On the other hand, ARAF, which binds to FZR1 (Supplementary Fig. S3B) but not APC10 (Supplementary Fig. S4H), was resistant to FZR1-mediated degradation (Supplementary Fig. S3C), presumably due to its inaccessibility to the APC core complex. These results suggest that compared to other well-characterized strong APCFZR1 substrates, such as Hsl1, CYCLIN B1 and PLK1, BRAF is a weak APCFZR1 substrate likely due to the observation that recruitment of BRAF to the APC core complex for proteolysis is relatively inefficient, in part owing to its weaker interaction with APC10.
Figure 4. FZR1 Disrupts the BRAF dimerization Process to Inhibit BRAF Kinase Activity Independent of APC.
(A–C) Depletion of FZR1 in OVCAR8 (A), HBL (B) and WM3670 (C) cells led to ERK activation but not BRAF accumulation. Immunoblot (IB) analysis of whole cell lysates (WCL) derived from BRAFWT-expressing OVCAR8 (A) and HBL (B) or BRAFG469E-expressing WM3670 (C) cells, which were, infected with control (shScr) or shFZR1 lentiviral shRNA constructs. The infected cells were selected with 1 μg/ml puromycin for 72 hours before harvesting.
(D) Additional depletion of BRAF suppressed the ERK activation upon FZR1 knockdown. IB analysis of OVCAR8 cells infected with the indicated lentiviral shRNA constructs. The infected cells were selected with 1 μg/ml puromycin for 72 hours before harvesting.
(E) WT- or APC-binding deficient ΔC-box-FZR1, but not N-terminal FZR1, suppressed the activation of MEK/ERK signaling pathway upon FZR1 knockdown. OVCAR8 cells stably expressing EV, WT-, ΔC-box-, or N(1-174)-FZR1 were further infected with shScr or shFZR1 lentiviral constructs as indicated. The infected cells were selected with 1 μg/ml puromycin for 72 hours before harvesting.
(F) In vitro kinase assays showing that both WT-FZR1 and ΔC-box-FZR1 inhibited BRAF kinase activity towards phosphorylating GST-MEK1.
(G–I) FZR1 disrupted BRAF dimerization process in cells. IB analysis of WCL and immunoprecipitates (IP) derived from 293T (G), A375 (H) or HBL (I) cells transfected with both Flag-BRAF and HA-BRAF with HA-FZR1 as indicated. 36 hours post-transfection, cells were pretreated with 10 μM MG132 for 10 hours before harvesting.
(J) FZR1 was capable of disrupting BRAF dimerization in MEFs. IB analysis of WCL and IP derived from primary MEFs transfected with various HA-FZR1 constructs with Flag-BRAF and HA-BRAF. 36 hours post-transfection, cells were pretreated with 10 μM MG132 for 10 hours before harvesting.
(K) FZR1 disrupted BRAF dimerization process in vitro. Autoradiography of 35S-labelled BRAF bound to the indicated recombinant GST fusion proteins in the presence of increasing amounts of 35S-labelled FZR1.
(L) Gel filtration experiment to illustrate that FZR1 disrupts BRAF dimerization. Bacterially purified recombinant His-BRAF and His-FZR1 proteins were incubated as indicated before being separated by Superdex 200 gel filtration chromatography. Prior to running cell lysates, the molecular weight resolution of the column was first estimated by running native molecular weight markers (Thyroglobulin ~669KD, Ferritin ~440KD, Aldolase ~158KD, Conalbumin ~75KD and Ovalbumin ~44KD) and determining their retention times on coomassie-stained SDS-PAGE protein gels.
(M) WT-, APC-binding deficient ΔC-box- or ΔFizzy-FZR1 disrupted BRAF dimerization in cells. IB analysis of WCL and IP derived from 293T cells transfected with various HA-FZR1 constructs as well as Flag-BRAF and HA-BRAF. 36 hours post-transfection, cells were pretreated with 10 μM MG132 for 10 hours before harvesting.
(N) FZR1 failed to disrupt dimerization of BRAF D-box4-mutants. IB analysis of WCL and IP derived from 293T cells transfected with various Flag-BRAF and HA-BRAF constructs as indicated. 36 hours post-transfection, cells were pretreated with 10 μM MG132 for 10 hours before harvesting.
(O) ERK activity fluctuated during the cell cycle progression and depletion of FZR1 strongly activated ERK across the cell cycle. IB analysis of WCL derived from shScr- or shFZR1-infected T98G cells that were synchronized at the G1/S boundary by double-thymidine block then released back into the cell cycle for the indicated periods of time.
(P) A schematic illustration of the proposed models for FZR1-mediated inhibition of BRAF function via different mechanisms in different cellular contexts.
Interestingly, further depletion of endogenous BRAF in FZR1 knockdown OVCAR8 cells largely attenuated the upregulation of p-ERK levels (Fig. 4D), indicating an indispensable role for BRAF in mediating ERK activation upon loss of FZR1. Given that in tumor cells, BRAF is largely refractive to APCFZR1-mediated degradation, FZR1 might utilize an alternative mechanism to harness BRAF kinase activity, thereby restraining its downstream MEK/ERK oncogenic signaling. In support of this notion, we found that re-introducing WT-FZR1 or the APC-binding-deficient ΔC-box-FZR1, but not the non-BRAF-interacting N-terminal-FZR1, could effectively suppress p-MEK and p-ERK in FZR1-depleted OVCAR8 cells (Fig. 4E). Given that the APC core complex is indispensable for FZR1-mediated substrate ubiquitination and degradation (7), these results suggest that FZR1 might govern MEK/ERK activation independent of APC E3 ubiquitin ligase activity.
In keeping with this notion, bacterially purified His-WT- and ΔC-box-FZR1, both of which are APC-free, could efficiently inhibit the phosphorylation of GST-MEK1 by immuno-purified BRAF kinase in vitro (Fig. 4F). In addition, depletion of the core APC subunits, APC10, failed to influence the BRAF/MEK/ERK signaling cascade in HBL cells (Supplementary Fig. S4I). These results directed our efforts to further define the molecular mechanism by which FZR1 could possibly regulate BRAF activation through an APC-independent manner in the tumor cell settings (Supplementary Fig. S4J), where FZR1 mainly exists in an APC-free mode in part due to elevated CDK activities that promote phosphorylation of FZR1, blocking its association with the APC core complex (38) (Supplementary Fig. S4K).
FZR1 Disrupts BRAF Dimerization to Attenuate BRAF Activation in Tumor Cells
BRAF activation is under tight control by numerous mechanisms such as phosphorylation and dimerization (46). As our previous report suggested a scaffolding role for FZR1 in disrupting Smurf1 dimerization independent of its APC E3 ligase activity (9), we next sought to explore whether FZR1 could also regulate BRAF dimerization to control its activation. Notably, ectopic expression of FZR1 abolished BRAF dimerization both in cells (Fig. 4G–J) and in vitro (Fig. 4K), supporting a pivotal role for FZR1 in regulating BRAF dimerization independent of APC. In addition, utilizing gel filtration chromatography, single molecule kinetic analysis (47, 48) and chemical crosslinking, we further demonstrated a potent role for FZR1 in disrupting BRAF dimerization in vitro (Fig. 4L and Supplementary Fig. S4L) and in cells (Supplementary Fig. S4M). Furthermore, two FZR1 mutants, ΔC-box- and ΔFizzy-FZR1 (Supplementary Fig. S2G), both of which are deficient in interacting with the APC core complex (37), could disrupt BRAF dimer as effectively as WT-FZR1 (Fig. 4M). These results advocate a model that FZR1 largely disrupts the BRAF dimerization process in an APC-independent manner.
Unlike WT-BRAF, the dimerization of FZR1-interaction deficient mutants, D4-RLAA- and R671Q-BRAF (Fig. 3G and Supplementary Fig. 4N–O), could not be disrupted by FZR1 (Fig. 4N). In line with this finding, ectopic expression of FZR1 significantly suppressed p-ERK levels induced by WT-BRAF, but not CRAF, ARAF (Supplementary Fig. S4P), or the dimerization-deficient R509H-BRAF mutant that is relatively weak in activating ERK (49) (Supplementary Fig. S4Q). More importantly, depletion of FZR1 in T98G cells resulted in a stabilization of p-ERK levels across the cell cycle without a significant impact on BRAF protein abundance (Fig. 4O). Together, these data reveal a possible mechanism through which FZR1 suppresses BRAF activity by disrupting BRAF dimerization in cancer cells (Fig. 4P).
Examination of the crystal structure of BRAF kinase domain (50) revealed that the D-box4 motif of BRAF is located on a surface loop region of the C-lobe of BRAF kinase domain (Supplementary Fig. S4R). By docking the BRAF kinase domain onto the WD40 domain of yeast Fzr1 (Supplementary Fig. S4S) (51), we found that R671 of BRAF D-box4, which extrudes outwards the kinase domain, could potentially form hydrogen bonds with acidic residues of Fzr1-WD40 (Supplementary Fig. S4T–U). In addition, Y673 of BRAF D-box4 might interact with F286 of Fzr1-WD40 by aromatic stacking (Supplementary Fig. S4T). However, different from the Acm1 D-box in the resolved yeast Fzr1-WD40 structure (Supplementary Fig. S4S) (51), the side chain of L674 in the BRAF D-box4 faces backwards from Fzr1-WD40, indicating that to properly insert the BRAF D-box into the D-box binding pocket on the Fzr1-WD40, a conformational change on BRAF might be required. This hypothesis might partly explain why FZR1 could disrupt BRAF dimerization while the D-box4 motif is not located within the BRAF dimerization interface.
In further support of this notion, the D-box 4 motif of BRAF mediating interaction with FZR1 does not overlap with the BRAF-BRAF interacting surface (Supplementary Fig. S4R), indicating a possible mechanism that FZR1 interferes with BRAF dimerization through an interaction-induced conformational change, i.e. FZR1 might function as an allosteric inhibitor for the BRAF dimerization (52). In line with this notion, the constitutive dimerization of the BRAF-E586K mutant (53) could still be disrupted by FZR1 (Supplementary Fig. S4V–W). This observation thereby suggests an allosteric rather than a competitive regulation of BRAF dimerization by FZR1, which warrants further in-depth investigation.
In addition to V600E, many other BRAF oncogenic mutations or splicing variants have been identified in human cancers (49, 50, 54). Among them, mutations in the P-loop of the BRAF kinase domain (residues 464–469) have been categorized as dimerization-dependent mutations (21). In contrast to BRAFV600E-expressing A375 cells, depletion of FZR1 in BRAF G469 mutated melanoma and lung cancer cell lines led to an elevation of BRAF activity (Fig. 4C and Supplementary Fig. S4C), supporting a critical function for FZR1 in disrupting BRAF dimers (Supplementary Fig. S4X–Y).
Loss of FZR1 Contributes to Vemurafenib Resistance in BRAFV600E Melanoma Cells
It has been previously shown that RAF proteins utilize either homo-dimerization or hetero-dimerization to prime their activation (46, 55). However, the oncogenic BRAFV600E mutation at the activation segment of the BRAF kinase domain adopts an altered constitutive active conformation, which allows for full activation of the kinase without dimerization (24). Consistent with the dimerization-independent activation of BRAFV600E (24), we found that unlike its role in suppressing WT-BRAF activity, FZR1 failed to suppress p-ERK levels when co-expressed with BRAFV600E in vivo (Supplementary Fig. S5A) or in an in vitro kinase assay (Supplementary Fig. S5B). Furthermore, unlike depleting FZR1 in melanoma cells harboring WT-BRAF (Fig. 4B), depletion of FZR1 in BRAFV600E melanoma cells did not induce ERK activation (Supplementary Fig. S5C), although FZR1 could still efficiently bind to and disrupt dimer formation of the BRAFV600E mutant (Supplementary Fig. S5D–E).
Although monomeric BRAFV600E is fully active, recent studies have demonstrated critical roles for BRAFV600E dimerization in contributing to vemurafenib (PLX4032) resistance (49, 56). For instance, several BRAF splicing variants have been identified in vemurafenib resistant melanoma cells that lack exons encoding the RAS binding domain. As such, these BRAFV600E variants exhibit increased dimerization in the absence of RAS binding to confer PLX4032 resistance (49). Additionally, vemurafenib has been found to induce the trans-activation of WT-BRAF or CRAF through hetero-dimerization between vemurafenib-bound BRAFV600E and WT-BRAF or CRAF (53, 56). These studies suggest that further understanding of the molecular mechanisms governing BRAFV600E dimerization could have great clinical significance in overcoming PLX4032 resistance.
Intriguingly, we found that in keeping with its ability to disrupting WT-BRAF or BRAFV600E homodimers, FZR1 could also inhibit the hetero-dimerization of BRAFV600E with CRAF (Supplementary Fig. S5F), or with ARAF (Supplementary Fig. S5G). Moreover, depletion of FZR1 in A375 and HBL cells resulted in an increased BRAF-CRAF heterodimerization (Supplementary Fig. S5H–I). This finding prompted us to further explore the role of FZR1 in regulating vemurafenib resistance in BRAFV600E melanoma cells. Notably, in A375 melanoma cells harboring homozygous BRAFV600E, depletion of FZR1 led to a moderate resistance to PLX4032 treatment as evidenced by elevated p-MEK and p-ERK signals (Supplementary Fig. S5J) as well as increased cell viability under drug challenge (Supplementary Fig. S5K). These findings therefore indicate that loss or reduced FZR1 abundance might contribute to vemurafenib resistance in melanoma patients.
Phosphorylation of FZR1 N-terminus by ERK and CYCLIN D1/CDK4 Inhibits APCFZR1 E3 Ligase Activity
In contrast to BRAFWT-expressing cancer cells (Fig. 4A), in A375 melanoma cell line harboring the BRAFV600E oncogenic mutation, protein levels of most APCFZR1 substrates examined including PLK1, AURORA A, GEMININ, CYCLIN B and CDC20, were barely affected by depletion of endogenous FZR1 (Supplementary Fig. S5C), suggesting that in these BRAF hyper-active melanoma cells, the ubiquitin E3 ligase activity of APCFZR1 is largely attenuated.
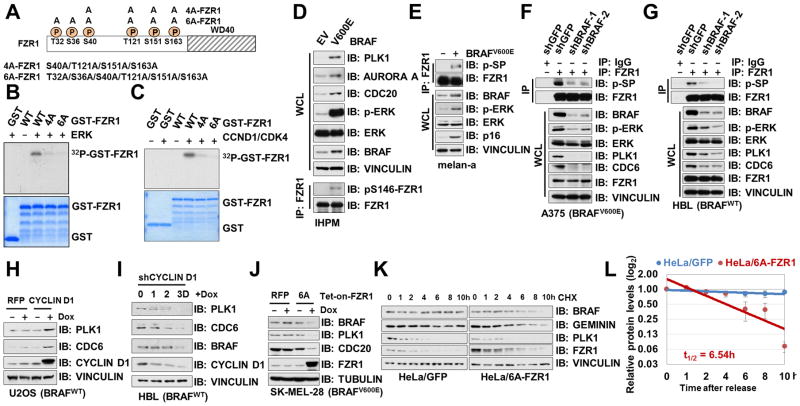
We and others have previously pinpointed serine/threonine residues within the N-terminal domain of FZR1 as CYCLIN A2 (also named CCNA2)/CDK2 or CYCLIN E1 (also named CCNE1)/CDK2 target sites (Fig. 5A) (3–6), phosphorylation of which abolishes interaction between FZR1 and the APC core complex (3, 43). Since elevation of RAS/RAF/MEK/ERK and its downstream CYCLIN D1 (also named CCND1)/CDK4 signaling pathways have been found in most melanoma patients (57), we next sought to examine whether these CDK2 target sites within FZR1 could also be phosphorylated by ERK and/or CYCLIN D1/CDK4. Notably, in vitro kinase assays revealed that purified ERK or CYCLIN D1/CDK4 could phosphorylate WT-FZR1, but not 4A- or 6A-FZR1 in which the previous identified CDK2 sites were mutated to alanines (Fig. 5B–C). Our finding is consistent with a recent report that FZR1 could be phosphorylated by CDK4 at its N-terminus (58).
Figure 5. Phosphorylation of FZR1 N-terminus by ERK and CYCLIN D1/CDK4 Inhibits the APCFZR1 E3 Ligase Activity.
(A) A schematic illustration of previously identified serine/threonine sites that were phosphorylated by CDK kinases, as well as the 4A-FZR1 and 6A-FZR1 mutants used in the following studies.
(B–C) In vitro kinase assays showing that bacterially purified WT- but not 4A- or 6A-GST-FZR1 could be phosphorylated by ERK (B) or CYCLIN D1/CDK4 (C).
(D–E) Ectopic expression of BRAFV600E led to the elevation of FZR1 phosphorylation and the accumulation of various APCFZR1 ubiquitin substrates in immortalized human primary melanocytes. Immunoblot (IB) analysis of whole cell lysates (WCL) and anti-FZR1 immunoprecipitates (IP) derived from EV or BRAFV600E expressing hTERT/p53DD/CDK4R24C human melanocytes (IHPMs, D) or melan-a cells (E). Cells were pretreated with 10 μM MG132 for 10 hours before harvesting.
(F–G) Depletion of BRAF led to a hypo-phosphorylated, more active FZR1 in cells. IB analysis of WCL and anti-FZR1 IP derived from control or shBRAF-infected A375 (F) or HBL (G) cells. Cells were pretreated with 10 μM MG132 for 10 hours before harvesting.
(H) Doxycycline-induced expression of CYCLIN D1 led to the accumulation of APCFZR1 substrate CDC6 in U2OS cells. U2OS cells were infected with pTRIPZ lentiviral vectors that allow the ectopic expression of either RFP (as a negative control) or CYCLIN D1 under the control of doxycycline. The infected cells were selected with 1 μg/ml puromycin for 72 hours. Afterwards, 300 ng/mL doxycycline were added for 24 hours before harvesting to induce the expression of CYCLIN D1.
(I) Doxycycline-induced depletion of CYCLIN D1 led to the decrease of APCFZR1 substrate CDC6, PLK1 and BRAF in HBL cells. HBL cells were infected with the pLKO-Tet-on lentiviral vector that allows the depletion of CYCLIN D1 under the control of doxycycline. The infected cells were selected with 1 μg/ml puromycin for 72 hours. Afterwards, 1 μg/mL doxycycline was added for the indicated time periods before harvesting.
(J) Doxycycline-induced expression of non-phosphorylatable 6A-FZR1 led to the decrease of APCFZR1 substrate PLK1 and CDC20 in BRAFV600E and CDK4R24C-expressing melanoma cells. BRAFV600E expressing SK-MEL-28 melanoma cells were infected with pTRIPZ lentiviral vectors that allow ectopic expression of either RFP (as a negative control) or 6A-FZR1 cDNA under the control of doxycycline. The infected cells were selected with 1 μg/ml puromycin for 72 hours. Afterwards, 300 ng/mL doxycycline were added for 24 h before harvesting.
(K–L) Ectopic expression of non-phosphorylatable 6A-FZR1 in HeLa cells destabilized BRAF. HeLa cells stably expressing GFP (as a negative control) or HA-6A-FZR1 were treated with 20 μg/ml cycloheximide (CHX) for the indicated time periods before harvesting. Equal amounts of WCL were immunoblotted with the indicated antibodies (K). (L) Quantification of BRAF band intensities was plotted as mean ± SD from three independent experiments, BRAF bands were normalized to VINCULIN, then normalized to the t = 0 time point.
To further evaluate the function of these phosphorylation on FZR1, compared with empty vector (EV)-infected parental cells, increased phosphorylation of FZR1 (5) and elevation of various FZR1 substrates were observed in BRAFV600E-expressing IHPM (Fig. 5D) and melan-a cells (Fig. 5E), indicating that in BRAFV600E expressing cells, increased phosphorylation of FZR1 might lead to inactivation of its E3 ligase activity. In support of this notion, depletion of BRAF in A375 (Fig. 5F) and HBL cells (Fig. 5G) led to a significant reduction of FZR1 phosphorylation as well as steady state level of APCFZR1 substrates including PLK1 and CDC6. Furthermore, induced expression of CYCLIN D1, led to an elevation in protein abundance of known APCFZR1 substrates (Fig. 5H), whereas depletion of CYCLIN D1 gradually suppressed the levels of various APCFZR1 substrates including BRAF (Fig. 5I).
To further determine a causal relationship between hyper-activation of ERK and/or CDK4 in BRAFV600E expressing cells and phosphorylation-dependent inactivation of APCFZR1 (Fig. 5A), we demonstrated that compared with EV or WT-FZR1 expressing cells, ectopic expression of the non-phosphorylatable 6A-FZR1 mutant led to a more dramatic decrease in APCFZR1 substrates such as PLK1, and to a lesser extent, BRAF (Fig. 5J–L). Furthermore, the number of senescent cells was decreased when introducing 6A-FZR1 in BRAFV600E-expressing melan-a cells (Supplementary Fig. S5L–M), suggesting that the non-phosphorylatable FZR1 mutant were more efficient in suppressing BRAFV600E-mediated premature senescence (29). These findings indicate that 4A- or 6A-FZR1, which are deficient in ERK- or CDK-mediated phosphorylation (Fig. 5B–C), displays elevated APCFZR1 E3 ligase activity even in BRAFV600E expressing cells with hyper-active ERK, presumably due to its higher affinity towards the APC core complex regardless of CDK and ERK activation status (43).
Interestingly, although N-terminal phosphorylation suppresses the E3 ligase activity of FZR1 (8), it barely affected FZR1’s function in disrupting BRAF dimerization (Supplementary Fig. S5N), a process mainly requires the WD40 domain of FZR1 to bind BRAF (Supplementary Fig. S3D–E). However, how APCFZR1-mediated BRAF ubiquitination is suppressed in different cancer settings warrants further in-depth studies.
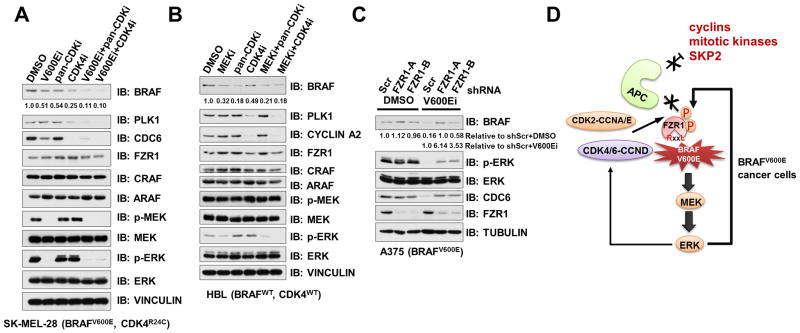
Pharmacologically Inhibiting BRAF/ERK and CDK4 Restores the APCFZR1 E3 Ligase Activity
Given the pivotal role of BRAF and CYCLIN D1/CDK4 signaling pathways in driving melanomagenesis (57), inhibitors targeting BRAFV600E, MEK or CDK4/6 are either approved or under clinical trials (59), which include the combinational treatment of melanoma patients with CDK4/6 inhibitor (LEE011) and BRAF inhibitor (LGX818) (ClinicalTrials.gov identifiers: 01777776 and 01820364). However, mechanistic studies to reveal the benefit of CDK4/6 inhibitors are largely absent. As our findings pinpointed both ERK and CYCLIN D1/CDK4 as FZR1 upstream kinases to inhibit APCFZR1 E3 ligase activity, next we challenged multiple melanoma cells with BRAFV600E inhibitor PLX4032 (60), MEK inhibitor PD0325901, pan-CDK inhibitor mimosine (61) or CDK4/6 inhibitor PD0332991 (62). Consistent with frequent aberrancies in CDK4, but not CDK2, signaling in the melanoma disease setting, we found that CDK4/6 inhibitor is more potent than pan-CDK inhibitor, which mainly targets CDK2, in restoring APCFZR1 activity in both BRAFV600E and BRAFWT genetic backgrounds (Fig. 6A–B and Supplementary Fig. S6A–B). Furthermore, we also observed a co-operative role for PLX4032 and PD0332991 in reactivating APCFZR1 E3 ligase activity in BRAFV600E expressing cells as evidenced by the downregulation of various APCFZR1 substrates including BRAF (Fig. 6A and Supplementary Fig. S6A–B).
Figure 6. Pharmacologically Inhibiting BRAF/ERK and CDK4 Restores the APCFZR1 E3 Ligase Activity.
(A) Protein levels of BRAF and other APCFZR1 substrates decreased upon BRAFV600E and CDK4/6 inhibition in melanoma cells. IB analysis of BRAFV600E and CDK4R24C expressing SK-MEL-28 melanoma cells treated with either 1 μM BRAFV600E inhibitor PLX4032 (V600Ei), 10 μM pan-CDK inhibitor mimosine (pan-CDKi), 1 μM CDK4/6 inhibitor PD0332991 (CDK4i), 1 μM PLX4032+10 μM mimosine, 1 μM PLX4032+1 μM PD0332991 or DMSO as a negative control for 24 h before harvesting. BRAF band intensities were quantified using ImageJ, normalized to corresponding TUBULIN band intensities, and then normalized to DMSO control lane.
(B) Protein levels of BRAF and other APCFZR1 substrates reduced upon MEK and CDK4/6 inhibition in melanoma cells. IB analysis of BRAFWT expressing HBL melanoma cells treated with either 1 μM MEK inhibitor PD0325901 (MEKi), 10 μM pan-CDK inhibitor mimosine (pan-CDKi), 1 μM CDK4/6 inhibitor PD0332991 (CDK4i), 1 μM PD0325901+10 μM mimosine, 1 μM PD0325901+1 μM PD0332991 or DMSO as a negative control for 24 h before harvesting. BRAF band intensities were quantified using ImageJ, normalized to corresponding Tubulin band intensities, and then normalized to DMSO control lane.
(C) Depletion of FZR1 in BRAFV600E-inhibited melanoma cells led to the upregulation of BRAF and PLK1. IB analysis of BRAFV600E expressing A375 melanoma cells, which were, infected with the control (shScr) or the indicated shFZR1 lentiviral shRNA constructs. The infected cells were selected with 1 μg/ml puromycin for 72 hours to eliminate the non-infected cells before harvesting. Prior to the harvest, cells were treated with DMSO (as a negative control) or 1 μM BRAFV600E inhibitor PLX4032 (V600Ei) for 24 hours as indicated. BRAF band intensities were quantified using ImageJ, normalized to corresponding Tubulin band intensities, and then normalized to DMSO control lane (upper row) or normalized to shScr+V600Ei lane (lower row).
(D) A schematic illustration of the proposed model for the putative role of FZR1 in suppressing BRAF dimerization-mediated transactivation of downstream MEK/ERK signaling to bypass PLX4032 triggered BRAFV600E inhibition in melanoma cells, as well as how mechanistically hyperactive ERK and/or CYCLIN D1/CDK4-mediated phosphorylation of FZR1 inhibits APCFZR1 E3 ligase activity in BRAFV600E melanoma cells.
In supporting our hypothesis that inhibition of BRAF/MEK and/or CDK activities in cancer cells reactivates APCFZR1, we further found that although depletion of FZR1 did not affect BRAF or CDC6 protein abundance in A375 cells, further PLX4032 treatment resulted in an apparent upregulation of BRAF and CDC6 levels compared with PLX4032-treated shScr-A375 cells (Fig. 6C). Similarly, in BRAFWT-expressing OVCAR8 cells, combinational treatment of PD0325901 with mimosine led to an increase of BRAF protein levels in shFZR1-OVCAR8 cells compared to control cells (Supplementary Fig. S6C).
Moreover, BRAFV600E or MEK inhibitor treatment led to an increase of ubiquitinated species of endogenous CDC20, a well-characterized APCFZR1 substrate (63) (Supplementary Fig. S6D–E), supporting the notion that the observed downregulation of various APCFZR1 substrates upon BRAFV600E or MEK inhibitor treatment is at least in part through restoring APCFZR1 E3 ligase activity. Although CDK4/6 inhibitor treatment of melanoma cells led to an enrichment in G1 phase, MEK or BRAFV600E inhibitor has a minimal effect on cell cycle profiles of various melanoma cell lines we examined (Supplementary Fig. S6F–H). These results indicate that the observed downregulation of APCFZR1 substrates upon MEK or BRAFV600E inhibitor treatment (Fig. 6A–B and Supplementary Fig. S6A–B) are mainly through restoring APCFZR1 activity rather than simply altering the cell cycle progression status. Moreover, the elevated APCFZR1 activity might be partly due to the increased interaction between FZR1 and the APC core complex (Fig. 6D and Supplementary Fig. S6I).
To gain further genetic insight into the critical role of APCFZR1 deficiency in promoting melanoma development, we found that in the Skin Cutaneous Melanoma (TCGA, Provisional) dataset with 287 cases (cbioportal.org) (64, 65), there are 23.0% mutation or deletion rate for FZR1 and 14 APC subunits (Supplementary Table S1 and Fig. S6J), 13% mutation or amplification rate for CYCLIN D1 and CDK4, and 80% mutation or amplification rate for BRAF and NRAS (Supplementary Fig. S6J). Notably, the genetic alterations of FZR1 and APC subunits do not significantly overlap with the genetic alterations of CCND1 and CDK4 (Supplementary Fig. S6J), while the wild-type BRAF/NRAS genetic status displays a significant association with the amplification of CCND1/CDK4 (χ2=6.877, p<0.01 by Pearson’s Chi-squared test). Consistent with the genetic analysis, we found that melanoma-derived FZR1 mutants displayed a reduced binding with BRAF and subsequently attenuated activity in promoting BRAF ubiquitination (Supplementary Fig. S6K–M). These analysis suggest that the genetic inactivation of APCFZR1 via mutation or deletion, together with phosphorylation-dependent inactivation of APCFZR1, lead to the deficiency of APCFZR1 in a vast majority of melanoma patients, which in turn elevates a cohort of oncogenic substrates of APCFZR1 to drive melanomagenesis.
Depletion of FZR1 Cooperates with AKT Activation to Promote Melanomagenesis in vitro
Having pinpointed that FZR1 deficiency leads to hyper-activation of the BRAF/ERK signaling pathway via either an APC-dependent or APC-independent mechanism in different cellular contexts (Fig. 4P), we next sought to explore whether depletion of FZR1 could promote melanomagenesis, in which BRAF activation plays a pivotal role (66). To this end, we took advantage of a widely utilized in vitro melanomagenesis model (67) to evaluate the contribution of FZR1 deficiency in transforming melanocytes.
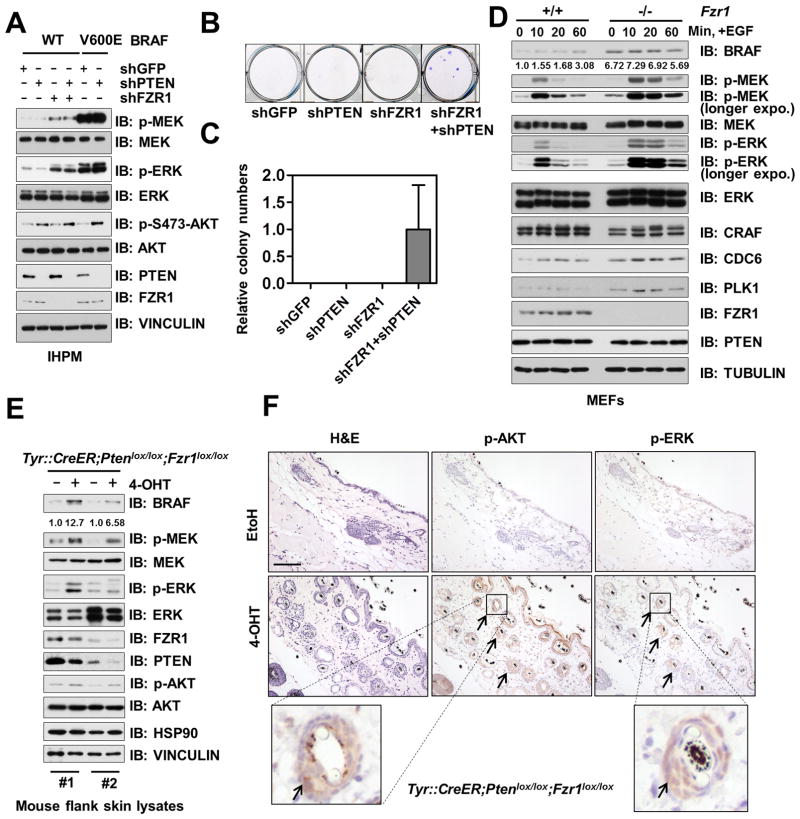
Activation of both ERK and AKT signaling pathways has been shown to facilitate melanoma development (68, 69). Notably, simultaneous depletion of FZR1 and PTEN in IHPM cells led to the upregulation of both ERK and AKT oncogenic signaling (Fig. 7A). Furthermore, consistent with previous reports (67, 70), complete withdrawal of growth factors such as TPA abolished the proliferation of the parental IHPM cells (Fig. 7B–C), whereas co-depletion of FZR1 and PTEN conferred growth factor-independent growth to IHPM cells (Fig. 7B–C and Supplementary Fig. S7A). Moreover, co-depletion of FZR1 and PTEN favored anchorage-independent growth of IHPM cells in soft agar (Supplementary Fig. S7B). These results support the synergetic role of FZR1 depletion-induced ERK activation, and PTEN loss-induced AKT activation in transforming primary melanocytes to drive melanomagenesis in vitro.
Figure 7. Depletion of FZR1 Co-operates with PTEN deficiency to Promote Co-activation of BRAF/ERK and Akt Oncogenic Signaling Both in vitro and in vivo.
(A) Co-depletion of FZR1 and PTEN activated both ERK and AKT. Immunoblot (IB) analysis of hTERT/p53DD/CDK4R24C human melanocytes (IHPMs) infected with the indicated lentiviral constructs.
(B–C) IHPM cells could proliferate independent of TPA upon co-depletion of FZR1 and PTEN. IHPM cells generated in (A) were subjected to clonogenic survival assays in RPMI-1640 media supplemented with 10% FBS without the essential growth factor, TPA for 21 days. Crystal violet was used to stain the formed colonies (B) and the colony numbers were counted from three independent experiments. The colony numbers were calculated as mean ± SD (C).
(D) BRAF was accumulated and ERK was activated in Fzr1−/− MEFs. IB analysis of WT and Fzr1−/− MEFs treated with 100 ng/ml EGF for the indicated period of time after 16 h serum deprivation.
(E) BRAF was accumulated and ERK was activated in mouse skin lysates derived from melanocyte conditional Fzr1 knockout mice. IB analysis of lysates from flank skin tissue of the engineered Tyr::CreER;Ptenlox/lox;Fzr1lox/lox mice, treated with EtOH (as a negative control) or 4-OHT. Flank skin tissues were harvested 21 days post-treatment for IB analysis.
(F) Elevation of both p-AKT and p-ERK was found in mouse skin samples derived from melanocyte conditional Fzr1 knockout mice. H&E staining and immunohistochemistry analysis of flank skin tissues from (E) using anti-p-AKT and anti-p-ERK antibodies as indicated. Arrows indicate the positively stained cells around hair follicles that are putative melanocytes. Scale bar: 100 μm.
Genetic Ablation of Fzr1 Synergizes with Pten Loss to Promote co-Activation of BRAF/ERK and AKT Oncogenic Signaling in vivo
Although complete deletion of both Fzr1 (Fzr1−/−) alleles in mice led to embryonic lethality (71), mouse embryonic fibroblasts (MEFs) obtained from Fzr1−/− mice embryo displayed elevated BRAF and p-ERK levels compared with WT-MEFs (Fig. 7D and Supplementary Fig. S7B–C), which further strengthens our finding that BRAF is an APCFZR1 substrate. Moreover, in keeping with the fact that RAF/MEK/ERK signaling pathway is rapidly stimulated by growth factor treatment, we found that, compared to WT-MEFs, Fzr1−/− MEFs were more responsive to EGF treatment and displayed extended activation kinetics (Fig. 7D).
Given that the BRAF/MEK/ERK oncogenic pathway plays a pivotal role in governing melanocyte proliferation, differentiation, as well as melanomagenesis (17), to further examine the physiological role of the FZR1-BRAF signaling axis in the melanocyte setting, melanocyte-specific Fzr1 and Pten conditional knockout mice (Tyr::CreER;Ptenlox/lox;Fzr1lox/lox) (68, 72) were generated. Notably, we found that topical application of 4-hydroxytamoxifen (4-OHT) on one side of the mouse flank (ethanol was used on the other side as a negative control), which induced the expression of the Cre-recombinase specifically in melanocytes to delete endogenous Fzr1 and Pten, led to a marked increase of BRAF and its downstream signals as evidenced by both immunoblot (Fig. 7E) and immunohistochemistry (Fig. 7F and Supplementary Fig. S7D) analyses.
More importantly, we found that the elevation of both p-AKT and p-ERK mainly occurred near hair follicles (Fig. 7F and Supplementary Fig. S7D), where most melanocytes reside in the postnatal mice (26). This observation is in agreement with previous studies demonstrating that Tyr::CreERT2 mainly targets melanoblasts and melanocytes within hair follicles (73), which might be the major reason accounting for the phenotype of pigmented observed in 4-OHT treated mouse skin (Supplementary Fig. S7E). Although it will be critical to further determine whether the elevated BRAF and AKT signaling could eventually drive melanoma development after long term 4-OHT treatment, these in vivo mice genetic data, however, coherently demonstrate that FZR1 might function as a tumor suppressor in vivo in part by suppressing the activation of the BRAF/MEK/ERK oncogenic pathway. As such, loss of the FZR1 tumor suppressor synergizes with PTEN deficiency, leading to a concomitant elevation of AKT and ERK signaling in vivo.
DISCUSSION
Deregulation of the RAS/RAF/MEK/ERK oncogenic signaling cascade is considered a hallmark for driving tumorigenesis in various human cancers (19). However, how different isoforms of RAF proteins are restrained from becoming hyperactive in normal tissue and how these negative regulations are attenuated during cancer progression still remains largely undefined. To date, several proteins have been identified to negatively regulate this important signaling pathway. For instance, binding of 14-3-3 to phospho-serines at both the N-terminus and the C-terminus of RAF proteins locked their closed inhibitory conformation (74), and phosphorylation of CRAF by PKA at Ser43 inhibited CRAF activation (75). Ubiquitination-mediate proteolysis has also been shown to negative regulate RAF kinases. SEL-10, a C. elegans homologue of human FBW7 controls the turnover of LIN-45, the C. elegans homologue of RAF kinases (76). Furthermore, in human cells, ring finger protein 149 (RNF149) targets RAF proteins for ubiquitination and subsequent destruction (77). These studies suggested that the RAF/MEK signaling was tightly regulated through various post-translational modifications including phosphorylation, ubiquitination and scaffolding.
Our findings here have demonstrated that FZR1, which is a key cell cycle regulator with a potential tumor suppressor role (8), could negatively regulate BRAF protein abundance and kinase activation through both APC-dependent BRAF proteolysis and APC-independent disruption of BRAF dimerization. Importantly, we have found that the mechanisms by which FZR1 regulates BRAF are different in normal cells and cancer cells (Supplementary Fig. S7F). In non-transformed cells, such as human primary fibroblasts, melanocytes or MEFs, APCFZR1 governs BRAF proteolysis in a cell cycle and D-box dependent manner, which retains BRAF abundance to prevent hyper-activation of MEK and ERK downstream of BRAF. Our finding therefore identifies APCFZR1 as a novel upstream regulator of BRAF abundance in human somatic cells, serving to suppress unscheduled cell proliferation, an important step towards neoplasia.
In cancer cells, however, our data suggest that APCFZR1-mediated BRAF degradation was largely attenuated (Fig. 4A–C and Supplementary Fig. S4A–C), which might be attributed to several mechanisms that lead to reduced APCFZR1 E3 ligase activity as well as escape from APCFZR1-mediated ubiquitination of various known APCFZR1 substrates. Previous studies have revealed that phosphorylation of FZR1 by CYCLIN A/E-CDK2 in late G1/S phases or by CYCLIN B1/CDK1 in G2/M phases dissociates FZR1 from the APC core complex (1) to partly inactivate APCFZR1 in cancer cells. Furthermore, numerous reports have identified that phosphorylation could disrupt the interaction between FZR1 and its substrates, hence facilitating the escape from APCFZR1-mediated ubiquitination and degradation in cancer cells (78–80). Notably, inhibiting MEK/ERK signaling by PLX4032, PD0325901, or suppressing CDK activity by mimosine, PD0332991, could repress BRAF abundance in various cancer cell lines (Fig. 6A–B and Supplementary Fig. S6A–B).
Recent structural and biochemical studies have shed light on the molecular mechanisms by which APC catalyzes poly-ubiquitination of its substrates (81). It has been demonstrated that poly-ubiquitination of substrates by APC displays distinct processivities due to different binding affinities between FZR1 and its substrates (45). In support of these reports, compared to well characterized APCFZR1 substrate PLK1, BRAF displayed attenuated affinity to both FZR1 and APC10, both of which are required for substrate interaction (Supplementary Fig. S4D–E) (44), presumably due to the lacking of the APC10 interacting motif within the D-box4 degron of BRAF (Supplementary Fig. S4F–G) (44). In support of this notion, a replacement of BRAF D-box4 (RGYLSPDLSK) with Hsl1 D-box sequence (RAALSDITN) (termed Opti-D4, Supplementary Fig. S4F) resulted in an increased interaction between APC10 and the generated BRAF chimera protein (Supplementary Fig. S4N), leading to more efficient degradation of the chimera protein by APCFZR1 (Supplementary Fig. S4O).
Although APCFZR1 displays marginal effect in controlling BRAF turnover in cancer cells, our studies have revealed that APC-free FZR1 in BRAFWT cancer cells could still suppress BRAF/ERK signaling, largely by disrupting BRAF dimerization without affecting BRAF protein abundance. This finding further advocates for the tumor suppressive role of FZR1 towards BRAF, especially when it is largely APC-free in most cancer cells. FZR1-mediated disruption of BRAF dimerization was also observed in immortalized MEFs (Fig. 4J), in which deletion of FZR1 led to the accumulation of BRAF (Fig. 7D). These results suggest a model that FZR1 might harness BRAF oncogenic function via both APC-dependent and APC-independent mechanisms in normal cells, similar to the reported dual suppressive role of SOCS proteins by inhibiting the JAK kinases and targeting them for degradation (82).
Taken together, our findings reveal a reciprocal suppression mechanism between FZR1 and BRAF in controlling tumorigenesis, and further suggest that FZR1 might exert its tumor suppressor role in part by functioning as a novel upstream inhibitor of the BRAF oncogenic signaling pathway (Fig. 4P and Supplementary Fig. S7F).
METHODS
Plasmids and Antibodies
Cell lines and their culturing conditions, plasmids and antibodies, experimental procedures for cell synchronization and FACS analysis could be found in the Extended Methods section of the Supplementary Information available online.
Cell Culture, Transfection and Infection
HeLa, HEK293, HEK293T, U2OS and T98G cells were cultured in DMEM containing 10% FBS (HyClone), 100 mg/mL penicillin-streptomycin as described previously (9), which were obtained from Dr. William G. Kaelin, Jr. in June, 2006. OVCAR8 ovarian cancer cell line was obtained from Dr. Marc W. Kirschner in March 2011. WM266.4, SK-MEL-28, A375, B16 melanoma cell lines, human primary melanocytes (HPM), mouse primary melanocytes (melan-a) and hTERT/p53DD/R24C-CDK4 immortalized human melanocytes (IHPM) have been described previously (83). HPM cells were isolated from normal discarded foreskins as described before (84), which were cultured in Medium 254 (Gibco). melan-a cells were obtained from the Wellcome Trust Functional Genomics Cell Bank at University of London in June 2013. IHPM cells were engineered with stable expression of human telomerase reverse transcriptase (hTERT), dominant negative p53 (p53DD) and a constitutively active CDK4 mutant (R24C-CDK4), allowing the bypass of premature senescence under oncogenic stresses (67), which were obtained from Dr. Hans Widlund in September 2012. A375 and HBL melanoma cell lines have been described previously (85), which were obtained from Dr. David Fisher in June 2008. WM266.4 and SK-MEL-28 cells were obtained from Dr. Richard Marais in August 2012. WM3670 melanoma cell line was obtained from Dr. Keiran S. Smalley in November 2016. H1755 NSCLC cell line was obtained from Dr. Eric B. Haura in November 2016. All cell lines were obtained between 2006 and 2016 and routinely tested negative for Mycoplasma. Cell line authentication was not routinely performed.
Cell culture transfection, lentiviral shRNA virus packaging and subsequent infection of various cell lines were performed according to the protocol described previously (86).
The purposes of using different cell lines in the manuscript could be found in the Extended Methods section of the Supplementary Information available online.
BrdU, SA-β-Gal Assays, and Crystal Violet Staining
Lentivirus-infected human primary melanocytes or murine melanocytes melan-a were subjected to SA-β-gal staining, BrdU labeling or crystal violet staining assays 14 days after viral infection. The experimental procedures for BrdU labeling and SA-β-Gal and crystal violet staining were described previously (83, 87).
Immunoblots and Immunoprecipitation
Cells were lysed in EBC buffer (50 mM Tris pH 7.5, 120 mM NaCl, 0.5% NP-40) supplemented with protease inhibitors (Complete Mini, Roche) and phosphatase inhibitors (phosphatase inhibitor cocktail set I and II, Calbiochem). The protein concentrations of the lysates were measured using the Bio-Rad protein assay reagent on a Beckman Coulter DU-800 spectrophotometer. The lysates were then resolved by SDS-PAGE and immunoblotted with indicated antibodies. For immunoprecipitation, 800 μg lysates were incubated with the appropriate antibody (1–2 μg) for 3–4 hours at 4 °C followed by 1 hour incubation with Protein A sepharose beads (GE Healthcare). Immuno-complexes were washed five times with NETN buffer (20 mM Tris, pH 8.0, 100 mM NaCl, 1 mM EDTA and 0.5% NP-40) before being resolved by SDS-PAGE and immunoblotted with indicated antibodies.
In Vitro Kinase Assay
BRAF in vitro kinase assays were performed as described (42). Briefly, BRAF was immune-purified from 293T cells transfected with Flag-BRAF constructs. GST-MEK1 and His-FZR1 were expressed in BL21 E.coli and purified using Glutathione Sepharose 4B media (GE Healthcare Life Sciences) and Ni-NTA agarose (Qiagen), respectively. BRAF kinase was incubated with 0.2 μg of GST-MEK in the absence or presence of His-FZR1 in kinase assay buffer (10 mM HEPES pH 8.0, 10 mM MgCl2, 1 mM dithiothreitol, 0.1 mM ATP). The reaction was initiated by the addition of GST-MEK in a volume of 30 μl for 15 min at 30 °C followed by the addition of SDS-PAGE sample buffer to stop the reaction before resolved by SDS-PAGE.
In Vivo Ubiquitination Assay
Denatured in vivo ubiquitination assays were performed as described (88). Briefly, 293 cells were transfected with Flag-BRAF, His-ubiquitin and HA-FZR1. 36 hours after transfection, 10 μM MG132 was added to block proteasome degradation, and cells were harvested in denatured buffer (6M guanidine-HCl, 0.1 M Na2HPO4/NaH2PO4, 10 mM imidazole), followed by Ni-NTA (Ni-nitrilotriacetic acid) purification and Immunoblot analysis.
In Vitro Ubiquitination Assay
APCFZR1 in vitro ubiquitination assays were performed as described previously (89). Briefly, 8 μg of anti-CDC27 antibodies coupled to 80 μL of protein A-agarose (Sigma) were incubated with 0.8 ml extracts from nocodazole arrested and released G1 (3 hours post release) HeLa cells, and mixed for 2 hours at 4 °C. The beads were washed three times with 1 ml swelling buffer (SB) (25 mM HEPES, pH7.5, 1.5 mM MgCl2, 5 mM KCl) supplemented with 0.05% Tween 20 and twice with SB. Finally, the beads were resuspended in 40 μl of SB and aliquoted into 8 tubes (5 μl for each tube). 0.15 μM E1, 1.5 μM E2, 1 mg/ml Ubiquitin, energy mix (7.5 mM creatine phosphate, 1 mM ATP, 1 mM MgCl2, 0.1 mM EGTA), and 1 U creatine phosphokinase were mixed in UBAB buffer (25 mM Tris/HCl, pH7.5, 50 mM NaCl, 10 mM MgCl2) in a final volume of 8 μl, i.e., reaction mix. Reactions were started by the addition of bacterially purified various GST-BRAF proteins as substrates, incubated for 60 minutes at 30 °C, resolved by SDS-PAGE and immunoblotted with the indicated antibodies.
Gel Filtration Chromatography Analysis
For gel filtration experiment using purified His-BRAF and His-FZR1, recombinant His-tagged proteins were purified using Ni-NTA Agarose (Cat. No. 30210, QIAGEN) according to manufacturer’s instructions. His-tagged proteins were further dialyzed with PBS supplemented with 0.1 M NaHCO3 (PBSC) and subjected to Superdex 200 10/300 GL column (GE Lifesciences Cat. No. 17-5175-01). Chromatography was performed on the AKTA-FPLC (GE Lifesciences Cat. No. 18-1900-26) with EBC buffer as described previously (9). One column volume of elutes were fractionated with 500 ml in each fraction, at the elution speed of 0.5 mL/min. 30 μl aliquots of each fraction were loaded onto SDS-PAGE gels and detected with indicated antibodies.
For gel filtration experiment using cell lysates, cells were washed with phosphate-buffered saline, lysed in 0.5 ml of EBC buffer (50 mM Tris pH 7.5, 120 mM NaCl, 0.5% NP-40) containing protease inhibitors (Complete Mini, Roche) and phosphatase inhibitors (phosphatase inhibitor cocktail set I and II, Calbiochem), and filtered through a 0.45 μm syringe filter. Total protein concentration was then adjusted to 8 mg/ml with EBC buffer and 500 μl of the lysate was loaded onto a Superdex 200 10/300 GL column as described above.
GST or His recombinant protein purification and in Vitro Binding Assay
Purification of GST- or His-tag-fused recombinant proteins and GST-pull down analyses were performed as described previously (27, 90).
Clonogenic Survival and Soft Agar Assays
The clonogenic survival and soft agar assays for hTERT/p53DD/R24C-CDK4 melanocytes (IHPM) were performed as described previously (83). Briefly, for growth factor independent clonogenic survival experiments, IHPM cells were cultured in 10% FBS containing RPMI-1640 media before plating into 6-well plate at 3,000 cells per well. 3 weeks later, cells were stained with crystal violet and the colony numbers were counted.
For soft agar assays, IHPM cells (30,000 per well) were seeded in 0.5% low-melting-point agarose in RPMI-1640 with 10% FBS, layered onto 0.8% agarose in RPMI-1640 with 10% FBS. The plates were kept in the cell culture incubator for 80 days after which the colonies >50 μm were counted under a light microscope.
Single-molecule Analysis of Transient Protein-protein Interactions
Coverslip passivation, TIRF microscope configuration and image analysis were described previously (47, 48). 50 nM biotinylated anti-Flag antibody (M2) was added to cell lysate expressing FLAG-BRAF, incubated at room temperature for 20 minutes. Cell lysate-antibody mix was applied to streptavidin-functionalized coverslip right before the experiment to immobilize Flag-BRAF. After 3 minute incubation, cell lysate on coverslip was washed off, replaced with buffer A (25 mM Tris-HCl pH 7.5, 100 mM NaCl, 20 mM imidazole, 5 mg/mL BSA) containing 10 nM BRAF or FZR1 labeled with Dylight550-NHS, and various concentrations of unlabeled BRAF or FZR1 as a competitor. Time series were acquired at 5 frames/second for 30 seconds. Binding constants of the competitor (KI~Kd) was calculated from the titration curve of the total number of binding events.
, N is the number of binding events; Nmax is the number of binding events in the absence of competitor; [C] is the concentration of competitor.
Mouse Models
All animal experiments were approved by the Beth Israel Deaconess Medical Center IACUC Committee on Animal Research. The Tyr::Cre-ERT2 transgenic mice, Ptenflox/flox mice and Fzr1flox/flox mice have been described previously (72, 91). Ptenflox/flox mice were first crossed with Tyr::Cre-ERT2 mice. The resulting compound mice or Tyr::Cre-ERT2 transgenic mice were then crossed with Fzr1flox/flox mice to generate conditional knockout mouse models of Pten and/or Fzr1. To activate the Tyr::Cre-ERT2 transgene to delete Pten and/or Fzr1 gene in the mouse melanocyte, the adult mice (6–8 weeks) were treated topically with 20 mg/mL 4-hydroxytamoxifen (4-OHT) in 100% ethanol for up to 7 months at the right ear, flank, paw and tail. Mouse tissues were fixed in 4% PFA. Normal and tumor tissues were embedded in paraffin, sectioned, and hematoxylin and eosin (H&E) stained for pathological evaluation.
Immunoblot Analysis of Mice Skin Tissues
Mice skin tissues from EtOH or 4-hydroxytamoxifen (4-OHT) treated flanks of Tyr::CreER;Ptenlox/lox;Fzr1lox/lox mice were lysed with RIPA buffer following the sonication. The lysates were subjected to immunoblot analysis with the indicated antibodies.
Histology and Immunohistochemical Analysis of Mice Skin Tissues
Mice skins were dissected and fixed in 4% paraformaldehyde for histology and IHC. For staining, the tissues were embedded in paraffin in according with standard procedures. 5 μm sections were cut and processed for H&E staining or stained for FZR1 (34-2000, 1:100), p-ERK (20G11, 1:100) and p-AKT (D9E, 1:100). The stained slides were visualized by a bright-field microscope.
Statistical Analysis
All quantitative data were presented as the mean ± SEM or the mean ± SD as indicated of at least three independent experiments by Student’s t test for between group differences. The p < 0.05 was considered statistically significant.
Supplementary Material
STATEMENT OF SIGNIFICANCE.
FZR1 inhibits BRAF oncogenic functions via both APC-dependent proteolysis and APC-independent disruption of BRAF dimers, while hyper-activated ERK and CDK4 reciprocally suppresses APCFZR1 E3 ligase activity. Aberrancies in this newly defined signaling network might account for BRAF hyper-activation in human cancers, suggesting that targeting CYCLIN D1/CDK4, alone or in combination with BRAF/MEK inhibition, can be an effective anti-melanoma therapy.
Acknowledgments
Financial support: L.W. is supported by CA183914. W.W. is supported by R01CA177910, GM089763 and R01GM094777.
We thank Brian North, Alan W. Lau, Jianping Guo, Naoe Nihira and Wenjian Gan for critical reading of the manuscript, William C. Hahn, Peter K. Jackson, Hans R. Widlund, Keiran S. Smalley and Eric B. Haura for providing reagents and members of the Wei, Kirschner, Cui and Pandolfi labs for useful discussions.
GRANT SUPPORT
W. Wei is a Leukemia and Lymphoma Society Scholar and ACS Research Scholar. This work was supported in part by the NIH grants (W. Wei, GM089763, GM094777 and CA177910; L. Wan, CA183914) and in part by Skin SPORE (1P50CA168536) Developmental Research Program (L. Wan).
Abbreviations
- KD
kinase domain
- APC/C
anaphase promoting complex/cyclosome
- FZR1
fizzy related protein 1, also called Cdh1
Footnotes
Conflict of interest: None
AUTHORS’ CONTRIBUTIONS
Conception and design: L. Wan, M. Chen, J. Cao, R. Cui, P.P. Pandolfi, W. Wei
Development of methodology: L. Wan, M. Chen, J. Cao, S. Song, Y. Lu, H. Inuzuka
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): L. Wan, M. Chen, J. Cao, X. Dai, Q. Yin, S. Song, Y. Lu, J. Liu, H. Inuzuka, J.M. Katon, K. Berry, J. Fung, C. Ng, J. Zhang, P. Liu, M.S. Song, L. Xue, R.T. Bronson
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): L. Wan, M. Chen, J. Cao, X. Dai, S. Song, Y. Lu, R.T. Bronson
Writing, review, and/or revision of the manuscript: L. Wan, M. Chen, J. Cao, X. Dai, Q. Yin, R. Cui, P.P. Pandolfi, W. Wei
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): J.M. Katon, K. Berry, J. Fung, C. Ng, R.T. Bronson
Study supervision: L. Wan, M. Chen, J. Cao, M.W. Kirschner, R. Cui, P.P. Pandolfi, W. Wei
References
- 1.Pines J. Cubism and the cell cycle: the many faces of the APC/C. Nat Rev Mol Cell Biol. 2011;12:427–38. doi: 10.1038/nrm3132. [DOI] [PubMed] [Google Scholar]
- 2.Manchado E, Eguren M, Malumbres M. The anaphase-promoting complex/cyclosome (APC/C): cell-cycle-dependent and -independent functions. Biochem Soc Trans. 2010;38:65–71. doi: 10.1042/BST0380065. [DOI] [PubMed] [Google Scholar]
- 3.Lukas C, Sorensen CS, Kramer E, Santoni-Rugiu E, Lindeneg C, Peters JM, et al. Accumulation of cyclin B1 requires E2F and cyclin-A-dependent rearrangement of the anaphase-promoting complex. Nature. 1999;401:815–8. doi: 10.1038/44611. [DOI] [PubMed] [Google Scholar]
- 4.Keck JM, Summers MK, Tedesco D, Ekholm-Reed S, Chuang LC, Jackson PK, et al. Cyclin E overexpression impairs progression through mitosis by inhibiting APC(Cdh1) The Journal of cell biology. 2007;178:371–85. doi: 10.1083/jcb.200703202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fukushima H, Ogura K, Wan L, Lu Y, Li V, Gao D, et al. SCF-mediated Cdh1 degradation defines a negative feedback system that coordinates cell-cycle progression. Cell Rep. 2013;4:803–16. doi: 10.1016/j.celrep.2013.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lau AW, Inuzuka H, Fukushima H, Wan L, Liu P, Gao D, et al. Regulation of APC(Cdh1) E3 ligase activity by the Fbw7/cyclin E signaling axis contributes to the tumor suppressor function of Fbw7. Cell Res. 2013;23:947–61. doi: 10.1038/cr.2013.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peters JM. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol. 2006;7:644–56. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- 8.Zhang J, Wan L, Dai X, Sun Y, Wei W. Functional characterization of Anaphase Promoting Complex/Cyclosome (APC/C) E3 ubiquitin ligases in tumorigenesis. Biochimica et biophysica acta. 2014;1845:277–93. doi: 10.1016/j.bbcan.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wan L, Zou W, Gao D, Inuzuka H, Fukushima H, Berg AH, et al. Cdh1 regulates osteoblast function through an APC/C-independent modulation of Smurf1. Molecular cell. 2011;44:721–33. doi: 10.1016/j.molcel.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Geley S, Kramer E, Gieffers C, Gannon J, Peters JM, Hunt T. Anaphase-promoting complex/cyclosome-dependent proteolysis of human cyclin A starts at the beginning of mitosis and is not subject to the spindle assembly checkpoint. The Journal of cell biology. 2001;153:137–48. doi: 10.1083/jcb.153.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindon C, Pines J. Ordered proteolysis in anaphase inactivates Plk1 to contribute to proper mitotic exit in human cells. The Journal of cell biology. 2004;164:233–41. doi: 10.1083/jcb.200309035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petersen BO, Wagener C, Marinoni F, Kramer ER, Melixetian M, Lazzerini Denchi E, et al. Cell cycle- and cell growth-regulated proteolysis of mammalian CDC6 is dependent on APC-CDH1. Genes & development. 2000;14:2330–43. doi: 10.1101/gad.832500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujita T, Liu W, Doihara H, Wan Y. Regulation of Skp2-p27 axis by the Cdh1/anaphase-promoting complex pathway in colorectal tumorigenesis. Am J Pathol. 2008;173:217–28. doi: 10.2353/ajpath.2008.070957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bassermann F, Frescas D, Guardavaccaro D, Busino L, Peschiaroli A, Pagano M. The Cdc14B-Cdh1-Plk1 axis controls the G2 DNA-damage-response checkpoint. Cell. 2008;134:256–67. doi: 10.1016/j.cell.2008.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia-Higuera I, Manchado E, Dubus P, Canamero M, Mendez J, Moreno S, et al. Genomic stability and tumour suppression by the APC/C cofactor Cdh1. Nat Cell Biol. 2008;10:802–11. doi: 10.1038/ncb1742. [DOI] [PubMed] [Google Scholar]
- 16.Cao J, Dai X, Wan L, Wang H, Zhang J, Goff PS, et al. The E3 ligase APC/CCdh1 promotes ubiquitylation-mediated proteolysis of PAX3 to suppress melanocyte proliferation and melanoma growth. Science signaling. 2015;8:ra87. doi: 10.1126/scisignal.aab1995. [DOI] [PubMed] [Google Scholar]
- 17.Wellbrock C, Karasarides M, Marais R. The RAF proteins take centre stage. Nat Rev Mol Cell Biol. 2004;5:875–85. doi: 10.1038/nrm1498. [DOI] [PubMed] [Google Scholar]
- 18.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–54. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 19.Roberts PJ, Der CJ. Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene. 2007;26:3291–310. doi: 10.1038/sj.onc.1210422. [DOI] [PubMed] [Google Scholar]
- 20.Bollag G, Tsai J, Zhang J, Zhang C, Ibrahim P, Nolop K, et al. Vemurafenib: the first drug approved for BRAF-mutant cancer. Nature reviews Drug discovery. 2012;11:873–86. doi: 10.1038/nrd3847. [DOI] [PubMed] [Google Scholar]
- 21.Holderfield M, Deuker MM, McCormick F, McMahon M. Targeting RAF kinases for cancer therapy: BRAF-mutated melanoma and beyond. Nature reviews Cancer. 2014;14:455–67. doi: 10.1038/nrc3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tap WD, Gong KW, Dering J, Tseng Y, Ginther C, Pauletti G, et al. Pharmacodynamic characterization of the efficacy signals due to selective BRAF inhibition with PLX4032 in malignant melanoma. Neoplasia. 2010;12:637–49. doi: 10.1593/neo.10414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flaherty KT, Infante JR, Daud A, Gonzalez R, Kefford RF, Sosman J, et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. The New England journal of medicine. 2012;367:1694–703. doi: 10.1056/NEJMoa1210093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poulikakos PI, Rosen N. Mutant BRAF melanomas--dependence and resistance. Cancer cell. 2011;19:11–5. doi: 10.1016/j.ccr.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 25.Catalanotti F, Solit DB. Will Hsp90 inhibitors prove effective in BRAF-mutant melanomas? Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18:2420–2. doi: 10.1158/1078-0432.CCR-12-0626. [DOI] [PubMed] [Google Scholar]
- 26.Yoshida H, Kunisada T, Kusakabe M, Nishikawa S, Nishikawa SI. Distinct stages of melanocyte differentiation revealed by anlaysis of nonuniform pigmentation patterns. Development. 1996;122:1207–14. doi: 10.1242/dev.122.4.1207. [DOI] [PubMed] [Google Scholar]
- 27.Gao D, Inuzuka H, Korenjak M, Tseng A, Wu T, Wan L, et al. Cdh1 regulates cell cycle through modulating the claspin/Chk1 and the Rb/E2F1 pathways. Mol Biol Cell. 2009;20:3305–16. doi: 10.1091/mbc.E09-01-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Engelbert D, Schnerch D, Baumgarten A, Wasch R. The ubiquitin ligase APC(Cdh1) is required to maintain genome integrity in primary human cells. Oncogene. 2008;27:907–17. doi: 10.1038/sj.onc.1210703. [DOI] [PubMed] [Google Scholar]
- 29.Pollock PM, Harper UL, Hansen KS, Yudt LM, Stark M, Robbins CM, et al. High frequency of BRAF mutations in nevi. Nat Genet. 2003;33:19–20. doi: 10.1038/ng1054. [DOI] [PubMed] [Google Scholar]
- 30.Sackton KL, Dimova N, Zeng X, Tian W, Zhang M, Sackton TB, et al. Synergistic blockade of mitotic exit by two chemical inhibitors of the APC/C. Nature. 2014;514:646–9. doi: 10.1038/nature13660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jacob K, Quang-Khuong DA, Jones DT, Witt H, Lambert S, Albrecht S, et al. Genetic aberrations leading to MAPK pathway activation mediate oncogene-induced senescence in sporadic pilocytic astrocytomas. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17:4650–60. doi: 10.1158/1078-0432.CCR-11-0127. [DOI] [PubMed] [Google Scholar]
- 32.Zhu J, Woods D, McMahon M, Bishop JM. Senescence of human fibroblasts induced by oncogenic Raf. Genes & development. 1998;12:2997–3007. doi: 10.1101/gad.12.19.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Michaloglou C, Vredeveld LC, Soengas MS, Denoyelle C, Kuilman T, van der Horst CM, et al. BRAFE600-associated senescence-like cell cycle arrest of human naevi. Nature. 2005;436:720–4. doi: 10.1038/nature03890. [DOI] [PubMed] [Google Scholar]
- 34.Haqq C, Nosrati M, Sudilovsky D, Crothers J, Khodabakhsh D, Pulliam BL, et al. The gene expression signatures of melanoma progression. Proc Natl Acad Sci U S A. 2005;102:6092–7. doi: 10.1073/pnas.0501564102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwab M, Neutzner M, Mocker D, Seufert W. Yeast Hct1 recognizes the mitotic cyclin Clb2 and other substrates of the ubiquitin ligase APC. The EMBO journal. 2001;20:5165–75. doi: 10.1093/emboj/20.18.5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barrett SD, Bridges AJ, Dudley DT, Saltiel AR, Fergus JH, Flamme CM, et al. The discovery of the benzhydroxamate MEK inhibitors CI-1040 and PD 0325901. Bioorg Med Chem Lett. 2008;18:6501–4. doi: 10.1016/j.bmcl.2008.10.054. [DOI] [PubMed] [Google Scholar]
- 37.Kraft C, Vodermaier HC, Maurer-Stroh S, Eisenhaber F, Peters JM. The WD40 propeller domain of Cdh1 functions as a destruction box receptor for APC/C substrates. Molecular cell. 2005;18:543–53. doi: 10.1016/j.molcel.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 38.Harper JW, Burton JL, Solomon MJ. The anaphase-promoting complex: it’s not just for mitosis any more. Genes & development. 2002;16:2179–206. doi: 10.1101/gad.1013102. [DOI] [PubMed] [Google Scholar]
- 39.Miller AJ, Mihm MC., Jr Melanoma. The New England journal of medicine. 2006;355:51–65. doi: 10.1056/NEJMra052166. [DOI] [PubMed] [Google Scholar]
- 40.Viros A, Sanchez-Laorden B, Pedersen M, Furney SJ, Rae J, Hogan K, et al. Ultraviolet radiation accelerates BRAF-driven melanomagenesis by targeting TP53. Nature. 2014;511:478–82. doi: 10.1038/nature13298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu W, Li W, Fujita T, Yang Q, Wan Y. Proteolysis of CDH1 enhances susceptibility to UV radiation-induced apoptosis. Carcinogenesis. 2008;29:263–72. doi: 10.1093/carcin/bgm251. [DOI] [PubMed] [Google Scholar]
- 42.Zhang BH, Guan KL. Activation of B-Raf kinase requires phosphorylation of the conserved residues Thr598 and Ser601. The EMBO journal. 2000;19:5429–39. doi: 10.1093/emboj/19.20.5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chang L, Zhang Z, Yang J, McLaughlin SH, Barford D. Atomic structure of the APC/C and its mechanism of protein ubiquitination. Nature. 2015;522:450–4. doi: 10.1038/nature14471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chang L, Zhang Z, Yang J, McLaughlin SH, Barford D. Molecular architecture and mechanism of the anaphase-promoting complex. Nature. 2014;513:388–93. doi: 10.1038/nature13543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rape M, Reddy SK, Kirschner MW. The processivity of multiubiquitination by the APC determines the order of substrate degradation. Cell. 2006;124:89–103. doi: 10.1016/j.cell.2005.10.032. [DOI] [PubMed] [Google Scholar]
- 46.McKay MM, Morrison DK. Integrating signals from RTKs to ERK/MAPK. Oncogene. 2007;26:3113–21. doi: 10.1038/sj.onc.1210394. [DOI] [PubMed] [Google Scholar]
- 47.Lu Y, Lee BH, King RW, Finley D, Kirschner MW. Substrate degradation by the proteasome: a single-molecule kinetic analysis. Science. 2015;348:1250834. doi: 10.1126/science.1250834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu Y, Wang W, Kirschner MW. Specificity of the anaphase-promoting complex: a single-molecule study. Science. 2015;348:1248737. doi: 10.1126/science.1248737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Poulikakos PI, Persaud Y, Janakiraman M, Kong X, Ng C, Moriceau G, et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E) Nature. 2011;480:387–90. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wan PT, Garnett MJ, Roe SM, Lee S, Niculescu-Duvaz D, Good VM, et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116:855–67. doi: 10.1016/s0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- 51.He J, Chao WC, Zhang Z, Yang J, Cronin N, Barford D. Insights into degron recognition by APC/C coactivators from the structure of an Acm1-Cdh1 complex. Molecular cell. 2013;50:649–60. doi: 10.1016/j.molcel.2013.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Arkin MR, Wells JA. Small-molecule inhibitors of protein-protein interactions: progressing towards the dream. Nature reviews Drug discovery. 2004;3:301–17. doi: 10.1038/nrd1343. [DOI] [PubMed] [Google Scholar]
- 53.Hatzivassiliou G, Song K, Yen I, Brandhuber BJ, Anderson DJ, Alvarado R, et al. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature. 2010;464:431–5. doi: 10.1038/nature08833. [DOI] [PubMed] [Google Scholar]
- 54.Solit DB, Rosen N. Towards a unified model of RAF inhibitor resistance. Cancer discovery. 2014;4:27–30. doi: 10.1158/2159-8290.CD-13-0961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roskoski R., Jr RAF protein-serine/threonine kinases: structure and regulation. Biochem Biophys Res Commun. 2010;399:313–7. doi: 10.1016/j.bbrc.2010.07.092. [DOI] [PubMed] [Google Scholar]
- 56.Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 2010;464:427–30. doi: 10.1038/nature08902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–63. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.The I, Ruijtenberg S, Bouchet BP, Cristobal A, Prinsen MB, van Mourik T, et al. Rb and FZR1/Cdh1 determine CDK4/6-cyclin D requirement in C. elegans and human cancer cells. Nat Commun. 2015;6:5906. doi: 10.1038/ncomms6906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jang S, Atkins MB. Treatment of BRAF-mutant melanoma: the role of vemurafenib and other therapies. Clin Pharmacol Ther. 2014;95:24–31. doi: 10.1038/clpt.2013.197. [DOI] [PubMed] [Google Scholar]
- 60.Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. The New England journal of medicine. 2010;363:809–19. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu P, Begley M, Michowski W, Inuzuka H, Ginzberg M, Gao D, et al. Cell-cycle-regulated activation of Akt kinase by phosphorylation at its carboxyl terminus. Nature. 2014;508:541–5. doi: 10.1038/nature13079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fry DW, Harvey PJ, Keller PR, Elliott WL, Meade M, Trachet E, et al. Specific inhibition of cyclin-dependent kinase 4/6 by PD 0332991 and associated antitumor activity in human tumor xenografts. Mol Cancer Ther. 2004;3:1427–38. [PubMed] [Google Scholar]
- 63.Huang JN, Park I, Ellingson E, Littlepage LE, Pellman D. Activity of the APC(Cdh1) form of the anaphase-promoting complex persists until S phase and prevents the premature expression of Cdc20p. The Journal of cell biology. 2001;154:85–94. doi: 10.1083/jcb.200102007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer discovery. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science signaling. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tsao H, Chin L, Garraway LA, Fisher DE. Melanoma: from mutations to medicine. Genes & development. 2012;26:1131–55. doi: 10.1101/gad.191999.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–22. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- 68.Dankort D, Curley DP, Cartlidge RA, Nelson B, Karnezis AN, Damsky WE, Jr, et al. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nat Genet. 2009;41:544–52. doi: 10.1038/ng.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vredeveld LC, Possik PA, Smit MA, Meissl K, Michaloglou C, Horlings HM, et al. Abrogation of BRAFV600E-induced senescence by PI3K pathway activation contributes to melanomagenesis. Genes & development. 2012;26:1055–69. doi: 10.1101/gad.187252.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Van Raamsdonk CD, Bezrookove V, Green G, Bauer J, Gaugler L, O’Brien JM, et al. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature. 2009;457:599–602. doi: 10.1038/nature07586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Garcia-Higuera I, Manchado E, Dubus P, Canamero M, Mendez J, Moreno S, et al. Genomic stability and tumour suppression by the APC/C cofactor Cdh1. Nat Cell Biol. 2008;10:802–11. doi: 10.1038/ncb1742. [DOI] [PubMed] [Google Scholar]
- 72.Bosenberg M, Muthusamy V, Curley DP, Wang Z, Hobbs C, Nelson B, et al. Characterization of melanocyte-specific inducible Cre recombinase transgenic mice. Genesis. 2006;44:262–7. doi: 10.1002/dvg.20205. [DOI] [PubMed] [Google Scholar]
- 73.Harris ML, Pavan WJ. Postnatal lineage mapping of follicular melanocytes with the Tyr::CreER(T) (2) transgene. Pigment cell & melanoma research. 2013;26:269–74. doi: 10.1111/pcmr.12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dougherty MK, Muller J, Ritt DA, Zhou M, Zhou XZ, Copeland TD, et al. Regulation of Raf-1 by direct feedback phosphorylation. Molecular cell. 2005;17:215–24. doi: 10.1016/j.molcel.2004.11.055. [DOI] [PubMed] [Google Scholar]
- 75.Ramstad C, Sundvold V, Johansen HK, Lea T. cAMP-dependent protein kinase (PKA) inhibits T cell activation by phosphorylating ser-43 of raf-1 in the MAPK/ERK pathway. Cell Signal. 2000;12:557–63. doi: 10.1016/s0898-6568(00)00097-8. [DOI] [PubMed] [Google Scholar]
- 76.de la Cova C, Greenwald I. SEL-10/Fbw7-dependent negative feedback regulation of LIN-45/Braf signaling in C. elegans via a conserved phosphodegron. Genes & development. 2012;26:2524–35. doi: 10.1101/gad.203703.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hong SW, Jin DH, Shin JS, Moon JH, Na YS, Jung KA, et al. Ring finger protein 149 is an E3 ubiquitin ligase active on wild-type v-Raf murine sarcoma viral oncogene homolog B1 (BRAF) J Biol Chem. 2012;287:24017–25. doi: 10.1074/jbc.M111.319822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gao D, Inuzuka H, Tseng A, Chin RY, Toker A, Wei W. Phosphorylation by Akt1 promotes cytoplasmic localization of Skp2 and impairs APCCdh1-mediated Skp2 destruction. Nat Cell Biol. 2009;11:397–408. doi: 10.1038/ncb1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lin HK, Wang G, Chen Z, Teruya-Feldstein J, Liu Y, Chan CH, et al. Phosphorylation-dependent regulation of cytosolic localization and oncogenic function of Skp2 by Akt/PKB. Nat Cell Biol. 2009;11:420–32. doi: 10.1038/ncb1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mailand N, Diffley JF. CDKs promote DNA replication origin licensing in human cells by protecting Cdc6 from APC/C-dependent proteolysis. Cell. 2005;122:915–26. doi: 10.1016/j.cell.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 81.Sivakumar S, Gorbsky GJ. Spatiotemporal regulation of the anaphase-promoting complex in mitosis. Nat Rev Mol Cell Biol. 2015;16:82–94. doi: 10.1038/nrm3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ungureanu D, Saharinen P, Junttila I, Hilton DJ, Silvennoinen O. Regulation of Jak2 through the ubiquitin-proteasome pathway involves phosphorylation of Jak2 on Y1007 and interaction with SOCS-1. Molecular and cellular biology. 2002;22:3316–26. doi: 10.1128/MCB.22.10.3316-3326.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cao J, Wan L, Hacker E, Dai X, Lenna S, Jimenez-Cervantes C, et al. MC1R is a potent regulator of PTEN after UV exposure in melanocytes. Molecular cell. 2013;51:409–22. doi: 10.1016/j.molcel.2013.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dunham WR, Klein SB, Rhodes LM, Marcelo CL. Oleic acid and linoleic acid are the major determinants of changes in keratinocyte plasma membrane viscosity. The Journal of investigative dermatology. 1996;107:332–5. doi: 10.1111/1523-1747.ep12363174. [DOI] [PubMed] [Google Scholar]
- 85.Ghosh S, Rosenthal R, Zajac P, Weber WP, Oertli D, Heberer M, et al. Culture of melanoma cells in 3-dimensional architectures results in impaired immunorecognition by cytotoxic T lymphocytes specific for Melan-A/MART-1 tumor-associated antigen. Annals of surgery. 2005;242:851–7. doi: 10.1097/01.sla.0000189571.84213.b0. discussion 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Boehm JS, Hession MT, Bulmer SE, Hahn WC. Transformation of human and murine fibroblasts without viral oncoproteins. Molecular and cellular biology. 2005;25:6464–74. doi: 10.1128/MCB.25.15.6464-6474.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Banito A, Rashid ST, Acosta JC, Li S, Pereira CF, Geti I, et al. Senescence impairs successful reprogramming to pluripotent stem cells. Genes Dev. 2009;23:2134–9. doi: 10.1101/gad.1811609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wan L, Tan M, Yang J, Inuzuka H, Dai X, Wu T, et al. APC(Cdc20) suppresses apoptosis through targeting Bim for ubiquitination and destruction. Developmental cell. 2014;29:377–91. doi: 10.1016/j.devcel.2014.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wu T, Merbl Y, Huo Y, Gallop JL, Tzur A, Kirschner MW. UBE2S drives elongation of K11-linked ubiquitin chains by the anaphase-promoting complex. Proc Natl Acad Sci U S A. 2010;107:1355–60. doi: 10.1073/pnas.0912802107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wei W, Ayad NG, Wan Y, Zhang GJ, Kirschner MW, Kaelin WG., Jr Degradation of the SCF component Skp2 in cell-cycle phase G1 by the anaphase-promoting complex. Nature. 2004;428:194–8. doi: 10.1038/nature02381. [DOI] [PubMed] [Google Scholar]
- 91.Song MS, Carracedo A, Salmena L, Song SJ, Egia A, Malumbres M, et al. Nuclear PTEN regulates the APC-CDH1 tumor-suppressive complex in a phosphatase-independent manner. Cell. 2011;144:187–99. doi: 10.1016/j.cell.2010.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.