Abstract
The bacterial flagellum contains a specialized secretion apparatus in its base that pumps certain protein subunits through the growing structure to their sites of installation beyond the membrane. A related apparatus functions in the injectisomes of gram-negative pathogens to export virulence factors into host cells. This mode of protein export is termed type-III secretion (T3S). Details of the T3S mechanism are unclear. It is energized by the proton gradient; here, a mutational approach was used to identify proton-binding groups that might function in transport. Conserved proton-binding residues in all the membrane components were tested. The results identify residues R147, R154, and D158 of FlhA as most critical. These lie in a small, well conserved cytoplasmic domain of FlhA, located between trans-membrane segments 4 and 5. Two-hybrid experiments demonstrate self-interaction of the domain, and targeted cross-linking indicates that it forms a multimeric array. A mutation that mimics protonation of the key acidic residue (D158N) was shown to trigger a global conformational change that affects the other, larger cytoplasmic domain that interacts with the export cargo. The results are discussed in the framework of a transport model based on proton-actuated movements in the cytoplasmic domains of FlhA.
Keywords: membrane transport, motility, pathogenesis, bioenergetics
Abbreviated Summary
A key proton-interacting site is identified in the flagellar protein FlhA and is shown to modulate conformation of the cargo-engaging parts of the protein. These findings give clues to the mechanism of the remarkably rapid protein export process known as type-III secretion.
Introduction
Assembly of the bacterial flagellum begins with structures in the vicinity of the cytoplasmic membrane and proceeds through steps that add the exterior structures in a proximal-to-distal sequence (Minamino & Namba, 2004, Macnab, 2003). Assembly of the rod, hook, and filament requires the action of the flagellar type III secretion (T3S) apparatus, which delivers the needed protein subunits into an internal channel that conducts them to the appropriate sites of installation in the growing structure (Macnab, 2004, Aizawa, 2006). Export by the T3S apparatus can be very fast; in the early stages of filament growth, flagellin protein (MW ~55 kDa) is transported at a rate of several subunits per second (Iino, 1974). The flagellar export system includes an ATPase, called FliI (Fan & Macnab, 1996), that for some time was believed to provide the energy for export. More recent work has shown that trans-membrane cargo movement does not require ATP hydrolysis but is instead energized by the membrane proton gradient (Wilharm et al., 2004, Paul et al., 2008, Minamino & Namba, 2008, Galperin et al., 1982). The requirement for ATPase activity can be bypassed by secondary mutations that increase the levels of export substrate or the magnitude of the protonmotive force (Erhardt et al., 2014). The T3S apparatus is thus a proton-energized protein pump.
The membrane part of the flagellar export apparatus is located within the MS-ring of the basal body (Katayama et al., 1996, Macnab, 2003, Liu et al., 2009) and is formed from the proteins FlhA, FlhB, FliP, FliQ and FliR (Minamino et al., 1994, Ohnishi et al., 1997). FliO also contributes to optimal export function but a recent study indicates that fliO-deletion strains retain some ability to assemble flagella (Barker et al., 2010). FliH, FliI and FliJ are non-membrane proteins that function in the cytoplasm to deliver cargo to the base of the apparatus (Vogler et al., 1991, Fan & Macnab, 1996, Minamino & Macnab, 2000b, Fraser et al., 2003a, Minamino et al., 2000). Export of some cargo subunits is further assisted by the substrate-specific chaperones FlgN, FliS, and FliT (Aldridge et al., 2003, Aldridge et al., 2006, Auvray et al., 2001, Bennett & Hughes, 2001, Cornelis, 2006). Core components of the flagellar export apparatus are closely related to proteins of the ‘injectisome,’ an apparatus that functions in many gram-negative pathogens to export virulence factors into host cells. Given these resemblances in the functionally critical proteins, the export mechanisms in the two systems are likely to be similar.
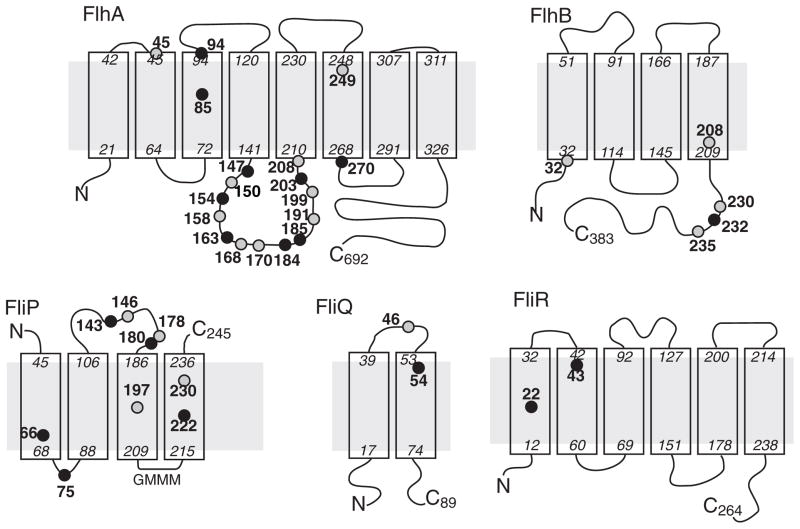
The probable topology and approximate relative sizes of the integral membrane components of the apparatus are illustrated in Fig. 1. Several lines of evidence support the hypothesized location of the T3S apparatus within the MS-ring: FliP and FliR have been found in purified basal bodies (Fan et al., 1997), intergenic suppression studies identified an interaction between FlhA and FliF (Kihara et al., 2001), and a feature seen in electron cryo-tomographs of the Shigella injectisome has a size and shape that would fit the cytoplasmic domains of nine subunits of MxiA (an FlhA homolog) (Abrusci et al., 2013). Hara et al. (Hara et al., 2011) reported additional instances of intergenic suppression that provide further evidence of interactions among FlhA, FlhB, and FliR (Hara et al., 2011). FlhB and FliR are likely to occur near each other and in equal numbers, because some Clostridia contain a naturally occurring fliR-flhB genetic fusion (Nolling et al., 2001), and an engineered FliR-FlhB fusion was shown to retain function in Salmonella (Nolling et al., 2001, Van Arnam et al., 2004). The MS-ring is large enough to fit multiple copies of each membrane component; for FlhA and its injectisome homologs there is strong evidence indicating nine copies (Morimoto et al., 2014, Abrusci et al., 2013, Zilkenat et al., 2016). FlhA and FlhB have sizable carboxyl-terminal domains on the cytoplasmic side of the membrane that engage the FliH/FliI/FliJ cargo-delivery complex (Fraser et al., 2003a, Minamino & Macnab, 2000c, Minamino et al., 2003, Zhu et al., 2002, Minamino et al., 1994, McMurry et al., 2004, Minamino et al., 2010, Saijo-Hamano et al., 2010, Fraser et al., 2003b, Hirano et al., 1994). The apparatus first exports ‘early-class’ substrates that form the hook and basal body and then, once the hook has reached its normal length, switches to export of the ‘late-class’ proteins that form the filament (Fraser et al., 2003b, Hirano et al., 1994, Minamino & Macnab, 2000a). This specificity switch is in large part regulated by FlhB.
Fig. 1.
Consensus topology predictions for the membrane components of the flagellar export apparatus. Approximate endpoints of the TM segments are indicated, using numbering appropriate to the Salmonella proteins. Positions of conserved acidic (gray circles) and basic residues (black circles) are indicated (see Table S1 for a breakdown by individual topology-prediction method, and Fig. S1 for sequence alignments). While all methods predict two TM segments for FliQ they are less definite the location of the termini (whether both are in the cytoplasm or the periplasm); this uncertainty does not affect the conclusions of the present study. The eight-segment topology for FlhA was predicted by only one of the methods, but the other methods indicated a quite long TM-7 segment, with highly variable endpoints, that would be consistent with the presence of two short TM segments. The eight-segment topology is in accord with the known cytoplasmic location of the C-terminal domain.
Although the membrane proteins of the T3S apparatus were identified several years ago (Minamino et al., 1994, Ohnishi et al., 1997), the details of their organization and the molecular mechanism of substrate translocation remain unclear. In contrast to the soluble components FliH, FliI, and FliJ, which are evolutionarily related to components of the ATP synthase (Pallen et al., 2006, Ibuki et al., 2011), the membrane components do not appear to be related to any other, more fully characterized transport system. Other proton-energized systems can nevertheless provide guidance: Because export is fueled by the membrane proton gradient, some component(s) of the apparatus should contain critical proton-binding groups, such as have been identified in the ATP synthase (Hoppe et al., 1982, Hoppe & Sebald, 1984, Miller et al., 1990, Imada et al., 2007, Imada et al., 2016), the E. coli lactose transporter (Abramson et al., 2003), or the flagellar rotary motor (Zhou et al., 1998). The membrane proteins of the flagellar export apparatus contain many conserved acidic and basic residues that might serve to interact with the energizing protons. Hara et al. (Hara et al., 2011) undertook a mutational analysis of eight conserved protonatable residues in the trans-membrane segments of FlhA and found that FlhA residue D208 is important for function. A recent mutational analysis by Barker et al. (Barker et al., 2016) was focused on a cytoplasmic domain of FlhA, and although the question of proton coupling was not addressed explicitly, several potentially proton-binding residues were found to be important. In the present study, we sought to identify functionally important proton-binding residues of the T3S apparatus by systematic mutational analysis of the conserved acidic and basic residues in all of the membrane components. The results implicate FlhA in coupling to the gradient; proton-binding residues in the other membrane components (FlhB, FliP, FliQ, and FliR) all proved dispensable for function in one or more of the assays for transport. Three charged residues of FlhA--R147, R154, and D158--proved most critical. These are in a relatively small, highly conserved cytoplasmic domain of FlhA (sometimes termed the FHIPEP domain; (McMurry et al., 2004)). This domain was shown to interact with itself in experiments using the bacterial adenylate cyclase two-hybrid system, and targeted disulfide crosslinking showed that it forms a multi-subunit array, presumably a ring. Limited-proteolysis experiments showed that a protonation-mimicking mutation in the critical aspartate residue (D158N) triggered a substantial conformational change that affected the other, larger cytoplasmic domain of FlhA that is known to interact with the exported cargo. The results are discussed in the framework of a model for the transport mechanism based on proton-regulated conformational changes in the cytoplasmic domains of FlhA.
Results
Mutational analysis of potentially proton-binding residues
The essential membrane proteins of the flagellar export apparatus are FliP, FliQ, FliR, FlhA, and FlhB. A summary of their topologies, based on predictions from a number of web-based tools (Claros & vonHeijne, 1994, Hirokawa et al., 1998, Hofmann & Stoffel, 1993, Melen et al., 2003, Tusnady & Simon, 2001), is presented in Fig. 1. The predictions are in reasonably close agreement overall (for details of individual predictions, see Table S1) and indicate four trans-membrane (TM) segments in FliP, two in FliQ, six in FliR, four in FlhB, and seven or eight in FlhA. A study using tandem LacZ-PhoA fusions to probe topology in the Xanthomonas T3S apparatus (Berger et al., 2010) indicated the presence of just single TM segments in both HrcS (a FliR paralog) and HrcT (a FliQ paralog), in sharp contrast to all the predictions here. The FliQ and FliR topologies in Fig. 1 are based on the presence of multiple, strongly hydrophobic segments, and were therefore adopted in preference to the single-segment topologies. Sequence alignments of the FliP, FliQ, FliR, FlhB, and FlhA proteins from about 50 species (Fig. S1) were used to identify conserved acidic and basic residues, on the assumption that residues that function in coupling to the protonmotive force are likely to be capable of binding protons. Conserved acidic and basic residues were individually replaced with alanine by mutagenesis of the genes cloned in salicylate-regulated plasmids. Mutant plasmids were transformed into the corresponding deletion strains, and function was assayed on soft-agar motility plates.
In FliP, four acidic residues and five basic residues are either invariant or strongly conserved (Fig. 1). Alanine replacements at most of these positions had only minor effects on function as assayed on motility plates (Table 2). Function was decreased but not eliminated by alanine replacement of E178 (near the periplasmic end of TM-3) or K222 (in TM-4). Function was eliminated upon replacement of D197 with alanine, asparagine, or proline. A glycine replacement retained significant function, however (about 20% of wild type). Thus, although E178, D197, and K222 are important for optimal function of the flagellar T3S apparatus, none are indispensable. To determine whether the defective FliP proteins exert dominant-negative effects, several variants, including the three non-functional D197 replacements, were expressed in wild-type cells. Non-functioning mutant proteins might be expected to inhibit motility if they can replace the wild-type protein in the apparatus; such dominant-negative effects were noted, for example, for mutations in the critical residue D32 of MotB in the flagellar motor (Zhou et al., 1998). Migration of wild-type cells was unaffected (Fig. S2), suggesting that the mutant FliP proteins are unstable or do not compete effectively with the wild-type protein for installation into the apparatus.
Table 2.
Effects of mutations in FliP, FliQ, FliR, and FlhB
| Mutation | Migration Ratea | |
|---|---|---|
| FliP | D146A | 1.02 (± .03) |
| E178A | 0.22 (± .05) | |
| D197A | 0.0 | |
| D197N | 0.0 | |
| D197P | 0.0 | |
| D197G | 0.17 (± .04) | |
| D197E | 0.97 (± .02) | |
| D230A | 0.88 (± .04) | |
| R66A | 0.79 (± .02) | |
| R75A | 0.75 (± .04) | |
| R143A | 1.00 (± .04) | |
| K180A | 0.88 (± .03) | |
| K222A | 0.21 (± .04) | |
| FliQ | E46A | 0.0 |
| K54A | 0.0 | |
| Q39A | 0.78 (± .09) | |
| T42A | 0.80 (± .05) | |
| Q43A | 1.05 (± .06) | |
| T48A | 1.02 (± .05) | |
| FliR | R22A | 0.90 (± .05) |
| K43A | 1.0 (± .05) | |
| FlhB | E32A | 0.98 (± .14) |
| D208Ab | 0.05 (± .05) | |
| E230A | 0.85 (± .10) | |
| K232A | 0.81 (± .08) | |
| E235A | 0.90 (± .07) |
Swarming rate of strains with chromosomal deletions complemented with the corresponding plasmid-borne genes, induced with 2.5 μM Na-salicylate. Rates are relative to control strains expressing the wild-type genes present on the same motility plates. Values are averages of six biological replicates (± s.e.m.)
Motility was restored to nearly wild-type levels upon over-expression of FlhA (see Fig. S3).
FliQ has one conserved acidic residue (E46) and one conserved basic residue (K54) (Fig. 1). Motility was eliminated by alanine replacement of either residue (Table 2). The E46A and K54A variants did not affect motility when expressed in wild-type cells (Fig. S2). This non-dominance might indicate a role for these residues in stabilizing the protein or facilitating its installation into the apparatus. In addition to the conserved E46 and K54, FliQ contains some well-conserved residues with polar character positioned near the periplasmic end of TM-1 (Q39, T42, Q43, and T48) that would presumably not bind protons directly but that might help form an aqueous proton-conducting channel. Alanine replacements of these residues had fairly minor effects (Table 2).
FliR contains no invariant acidic residues. It has one invariant basic residue located in TM-1 (R22), and basic character is also conserved at position 43, in TM-2. Motility was not significantly affected by alanine replacements of R22 or R43. Basic character is also conserved at position 206 (arginine in the protein of Salmonella) but this conservation does not extend to certain injectisome homologs (EscT of E. carotovora or HrcT of P. syringae), so R206 was not tested.
FlhB contains one invariant basic residue (K232), three invariant acidic residues (D208, E230, and E235), and one acidic residue (E32) that is only moderately conserved but was included due to its position near the inner end of a TM segment (a location similar to that of the putative proton-binding residue of MotB, D32) (Fig. 1). Motility was not significantly affected by alanine replacement of E32, E230, K232, or E235. Replacement of D208 of FlhB caused a severe motility reduction, but this defect was rescued upon moderate overexpression of FlhA (Fig. S3).
FlhA contains the largest number of conserved proton-binding residues, nine each acidic and basic (Fig. 1). Motility was eliminated by alanine replacement of the acidic residues D158, D170, D199, D208, or D249, or the basic residues R94, R147, R154, R185, or K203 (Table 3). All but two of these (R94 and D249) lie within or adjacent to a highly conserved, approximately 65-residue cytoplasmic domain between TM-4 and TM-5 (Fig. 1), hereafter termed CD-1 (to contrast with the larger ‘CD-2’ domain at the C-terminus). Hara et al. (Hara et al., 2011) previously noted motility defects in R94A, D208A, and K203A FlhA mutants. Attention was directed particularly to residue D208, which lies near the inner end of TM-segment 5, just adjacent to CD-1. We tested additional replacements of D208, and also residue D158, which exhibited distinctive properties in other experiments (below). At position 208, motility was eliminated by replacement of the native aspartate with asparagine, glutamine, serine, histidine, or glutamate (Table 3). At position 158, motility was eliminated by replacements of the native aspartate with asparagine, glutamine, serine, histidine, or proline. A D158E replacement gave motility like wild-type even though glutamate does not appear to occur naturally at this position (Fig. S1).
Table 3.
Effects of mutations in FlhA
| Mutation | Migration Ratea |
|---|---|
| D45A | 0.62 (± .07) |
| E150A | 0.88 (± .06) |
| D158A | 0.0 |
| D158N | 0.0 |
| D158Q | 0.0 |
| D158S | 0.0 |
| D158H | 0.0 |
| D158P | 0.0 |
| D158E | 0.94 (± .02) |
| D168A | 0.24 (± .08) |
| D170A | 0.0 |
| E191A | 0.85 (± .02) |
| D199A | 0.0 |
| D208A | 0.0 |
| D208N | 0.0 |
| D208Q | 0.0 |
| D208S | 0.0 |
| D208E | 0.0 |
| D208H | 0.0 |
| D249A | 0.0 |
| D249N | 0.0 |
| R85A | 0.86 (± .02) |
| R94A | 0.0 |
| R147A | 0.0 |
| R147Q | 0.0 |
| R154A | 0.0 |
| R154Q | 0.0 |
| K163A | 1.01 (± .03) |
| R184A | 1.06 (± .03) |
| R185A | 0.0 |
| K203A | 0.0 |
| K203Q | 0.98 (± .02) |
| R270A | 0.87 (± .06) |
Swarming rate of the ΔflhA strain transformed with plasmids expressing FlhA with the indicated mutations, induced with 2.5 μM Na-salicylate. Rates are relative to control strains expressing wild-type flhA present on the same plates (mean ± s.e.m.; n=6).
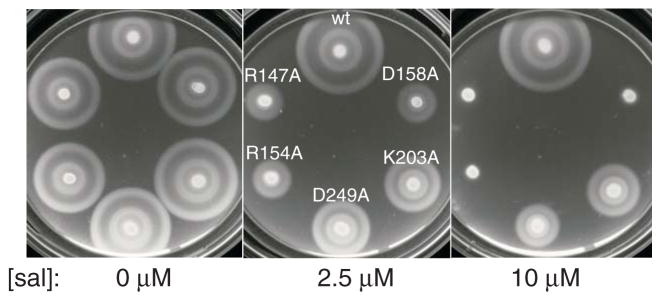
Dominant-negative effects were examined by expressing the mutant FlhA proteins in wild-type cells. All of the nonfunctional alanine-replacements caused a measurable motility inhibition. Dominant-negative effects were strongest with the R147A, R154A, and D158A mutations (Fig. 2; for cases of weaker dominance see Fig. S2). These strong dominant-negative effects give a first indication that residues R147, R154, and D158 might constitute a functionally distinct group. The dominance presumably indicates that the mutant proteins are stable enough to accumulate in cells; stability of the mutant proteins was confirmed on immunoblots using anti-FlhA antibody (Fig. S4). The motility impairment was not due to discharge of the membrane potential or other nonspecific effects on cellular physiology, because growth rates were the same in cells expressing the wild-type and dominant-negative mutant FlhA proteins (Fig. S4).
Fig. 2.
Dominant-negative effects of FlhA mutations. Results are shown for five mutations that eliminated FlhA function in a soft-agar motility assay. FlhA proteins were expressed in the wild-type Salmonella strain LT2 from plasmids based on pMS122 (Table 1), induced with salicylate at the levels indicated. Plates contain 50 μg/ml chloramphenicol. The control (‘wt’) expressed wild-type flhA from the plasmid. Plates were inoculated with 3 μl from overnight cultures and photographed after 5.5 h at 32 °C.
Intragenic suppression of motility defects in FlhA mutants
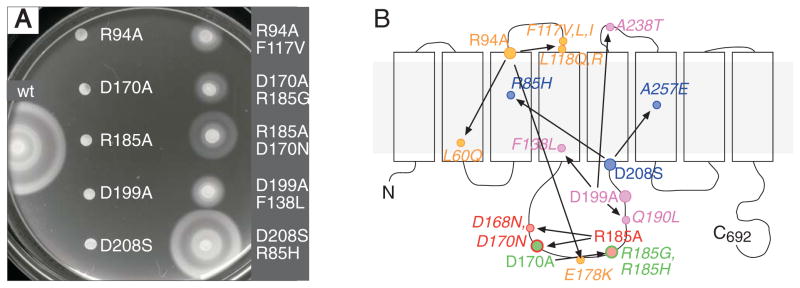
The soft-agar motility assay provides a convenient means for isolating variants that regain motility as the result of further mutation(s). To examine rescue-ability of the FlhA charged-residue mutants, cultures of the ten nonfunctional FlhA mutants were inoculated in streaks on soft agar, and the plates were monitored for the appearance of “flares” indicative of motile cells. When motile flares were observed, plasmids were isolated from the motile cells and re-introduced into the ΔflhA strain to determine whether the motility rescue was associated with the plasmid, then the plasmids were sequenced to identify the mutation(s). In this way, intragenic suppressors were found for the mutations R94A, D170A, R185A, D199A, D208S, and D208E. The starting alleles and suppressing mutations are summarized on a diagram of the FlhA topology in Fig. 3, which also shows examples of rescued-motility phenotypes. An implication of this suppression analysis is that FlhA variants lacking a protonatable side-chain at positions 94, 170, 185, 199, or 208 can function fairly well provided that certain additional mutations are present elsewhere in the protein. In most cases, the suppressing mutations are quite distant in the protein structure (because they are at very different positions relative to the membrane), and in no case would the rescuing mutation restore a proton-binding group in the neighborhood of the originally mutated residue. Motile flares were not observed for the mutants with alanine replacements of R147, R154, D158, K203, or D249.
Fig. 3.
Phenotypic suppression of certain FlhA mutations by spontaneously arising second mutations elsewhere in the protein. The background was the flhA-deletion strain TH12642. Cells contained derivatives of plasmid pMS122 expressing the indicated flhA variants from a salicylate-regulated promoter. Left: Examples of the immotility of starting mutants and partial motility rescue by the suppressing mutations. Plates contained 50 μg/ml chloramphenicol and 2.5 μM sodium salicylate. The plate was inoculated with 3 μl from overnight cultures and photographed after 5 h at 32° C. Right: summary of isolated mutation/suppressor pairs, mapped onto the predicted FlhA membrane topology.
Other replacements at the potentially important positions
The five positions that failed to yield suppressors might require a protonatable side-chain for function or, alternatively, might require a side-chain more polar than alanine. As described above, polar but non-protonatable replacements of D158 (S, N, H, Q) were nonfunctional, suggesting a requirement for an acidic group at this position. To test whether polar character is sufficient for function at the other four positions, a polar but non-protonatable replacement (N or Q) was tested at each. The K203Q mutant was highly motile. R147Q, R154Q, and D249N mutants were immotile.
Assays of export using a flgE-bla (Hook-β-lactamase) fusion
Lee and Hughes (Lee & Hughes, 2006) have developed a sensitive method for monitoring flagellar export based on translocation of a FlgE-Bla (Flagellar hook–β lactamase) fusion protein. If the central rod of the basal body is disrupted by mutation, the fusion protein is transported into the periplasm, and measurements of ampicillin (Amp) resistance (such as IC50) can provide a measure of transport activity. This assay was used with the collection of charged-residue mutations in FliP, FliQ, and FlhA, using a set of tester strains that each contained a chromosomally encoded flgE-bla fusion gene, a deletion of the rod genes flgBC, and a deletion in one of the export-apparatus genes (fliP, fliQ, or flhA). Plasmids encoding the corresponding export-apparatus gene, either wild-type or mutant, were transformed into these deletion backgrounds, and transport was assayed using the ampicillin IC50 assay. Briefly, assays employed an initial 2.5-h culturing step in presence of inducer (sodium salicylate) to stimulate synthesis of the plasmid-encoded export-apparatus protein, followed by a 3.5-h culturing step in presence of various concentrations of ampicillin (with continued induction by salicylate). Growth at each Amp concentration was measured by OD600 to determine the values that reduced growth to 50% of the uninhibited value. The assay was used for all of the mutations that eliminated motility in the soft-agar assays and, in the case of FliP, certain mutations that severely impaired motility. Results are summarized in Fig. 4.
Fig. 4.

Effects of mutations on transport of a FlgE-Bla (Flagellar hook/β-lactamase) fusion protein. The protein confers ampicillin resistance when exported to the periplasm. Transport activity was assayed by measuring IC50 for ampicillin (the concentration that reduced growth to half of the uninhibited level). Box plots represent data for at least six biological replicates. Mutants shown here were all nonfunctional in the soft-agar motility assay, with the exception of the FliP E178A and K222A mutants, which were weakly motile (Tables 2 and 3).
In FliP, measurements on the negative control strain gave IC50 values of approximately 6 μg/ml, and the positive control (expressing wild-type FliP) had an average value above 400 μg/ml. Three FliP mutations that impaired motility severely (E178A, D197G, K222A) all showed IC50 values in the 50–150 μg/ml range. The significant Amp resistance remaining in these mutants reinforces the conclusion that these invariant residues of FliP are not essential for transport.
In FliQ, negative controls gave IC50 values in the 7 μg/ml range and positive controls were approximately 300 μg/ml. Both of the mutations that eliminated motility (E46A and K54A) retained significant transport activity in the Flg-Bla assay, with average IC50 values near 40 μg/ml. This result, like the recessive character of the mutations (Fig. S2), argues against an essential role for these FliQ residues in the transport mechanism.
Several of the FlhA mutations fully eliminated transport activity in the IC50 assays, in most cases agreeing with the motility assays. Important exceptions included the D199A, D208N, and D249N mutants, which retained significant transport activity in the FlgE-Bla assays in spite of being totally immotile. The activity of the D199A and D208N mutants reinforces the suppression results (Fig. 3) in arguing against the essentiality of proton-binding side-chains at these positions; activity in the D249N mutant provides a first indication that transport does not require a protonatable side-chain at this position. Residues R147, R154, and D158 proved as critical in the Hook-Bla transport assays as in the motility measurements. All tested replacements of D158 (A, N, S, H, or Q) reduced IC50 values to the level of negative controls, as did replacements of R147 or R154 with either A or Q.
Inter-subunit complementation
The export apparatus contains several, most likely nine, copies of FlhA (Abrusci et al., 2013, Morimoto et al., 2014). Mutations that prevent motility when they are the only allele present might be tolerated when present in only some of the subunits, and two such mutations might complement each other if they affect distinct functions of the protein. To test for inter-subunit complementation between various pairs of flhA mutations, certain of the nonfunctional alleles were transferred onto a kanamycin-resistance plasmid and tested in combination with other nonfunctioning flhA alleles carried on chloramphenicol-resistance plasmids. In motility tests on soft-agar plates, several pairs of flhA mutations conferred fair motility in spite of being nonfunctional by themselves (Fig. 5). In these cases, motility was also seen in a large proportion of cells grown in liquid culture, indicating that it was not the result of recombination to restore the wild-type gene (an event that can be selected for fairly rapidly in motility plates). The R94A mutation in particular showed effective inter-subunit complementation when paired with several other mutations, including all of the acidic-residue replacements except D170A. The D170A mutation was instead complemented by R185A, echoing the mutual phenotypic suppression observed with replacements at positions 170 and 185 (Fig. 3).
Fig. 5.
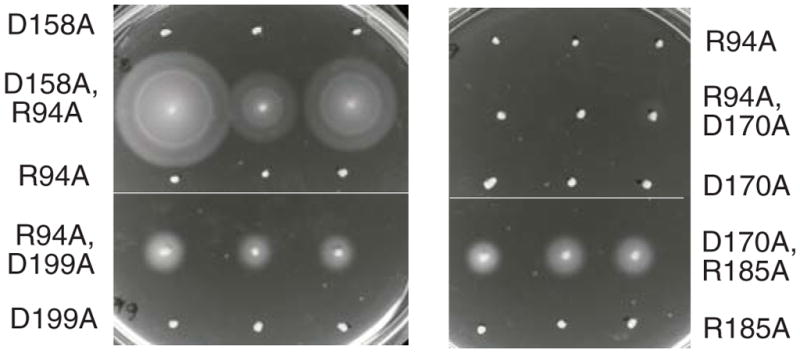
Inter-subunit complementation observed with certain pairs of FlhA mutations. Mutant variants of FlhA were expressed from plasmids with CmR or KmR markers. Both plasmids used a salicylate-inducible promoter. The background was the ΔflhA strain TH12642. Plates contained each antibiotic at 25 μg/ml, and 5 μM sodium salicylate. Single-allele controls contained just one of the flhA mutations, together with a control plasmid to provide the other antibiotic resistance. Plates were inoculated with picks from different single colonies (3 for each strain) and incubated for 7 h at 32 °C. An example of a non-complementing mutation pair (R94A + D170A) is also shown. (Lighting arrangements necessitated use of two photos to capture each plate; thin horizontal lines are boundaries between the photos.)
Positions for which a complementing allele was not found in this survey were K203 (which tolerates Q replacement and is thus not a candidate for an essential proton-binding function), and R147 and R154, two of the three residues that appear most critical overall. A mutation in the third critical residue, D158A, showed effective inter-subunit complementation with R94A. Thus, although D158 is the most critical acidic residue in the apparatus, function does not require D158 to be present in every subunit.
Self-interaction of CD-1
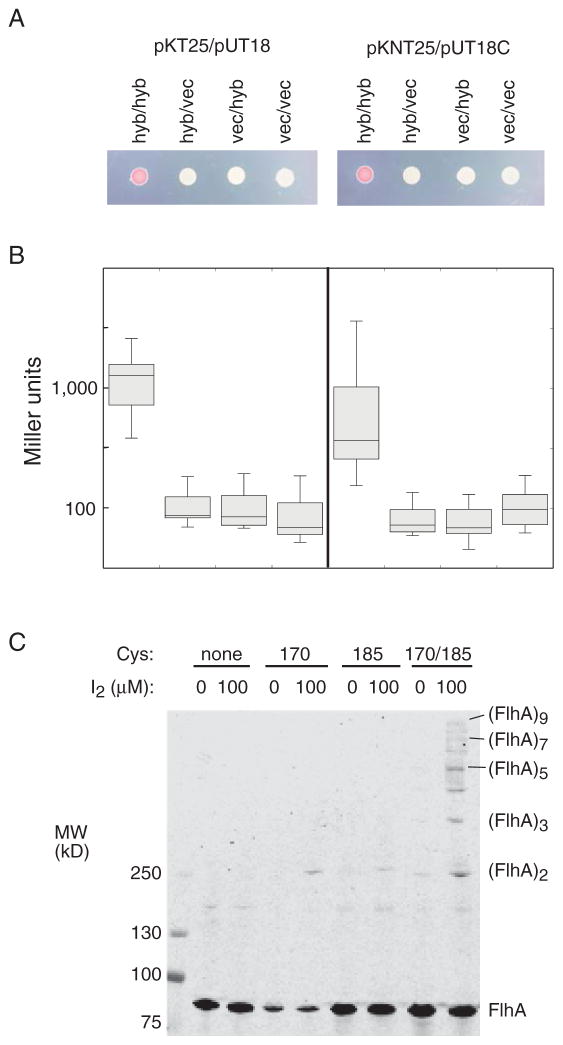
The mutual suppression and inter-subunit complementation observed for mutations of D170 and R185 point to an interaction between adjacent CD-1 domains in the apparatus. To more directly test for ‘self’ (homotypic) interactions between CD-1 domains, we used the bacterial adenylate cyclase two-hybrid (BACTH) system (Karimova et al., 1998). Hybrid constructs contained flhA codons 143–209 fused to sequences encoding N- or C-terminal fragments of adenylate cyclase. Homotypic interactions can be tested in four ways corresponding to the four possible fusions of adenylate cyclase N- and C-terminal domains to either the N- or C-termini of the test protein. Two of the four combinations showed strong color on MacConkey plates and β-galactosidase activities significantly above negative controls (Fig. 6). The clean negative controls as well as the absence of any detectable interaction with two of the hybrid-protein combinations argue that the binding is not the result of non-specific interactions. The CD-1 domain thus appears to interact with other copies of itself. If FlhA subunits are arranged with axial symmetry, as is suggested by the arrangement of C-terminal domains seen in some crystal structures (Abrusci et al., 2013, Saijo-Hamano et al., 2010), then several (probably nine) interacting CD-1 domains would form a ring. To test for the presence of CD-1 multimers, we introduced cysteine residues in place of D170 and R185 (the positions exhibiting mutual suppression) and examined disulfide crosslinking induced by I2. To enable detection on immunoblots, the protein carried an N-terminal FLAG tag, which permitted nearly normal function in soft-agar plates. As with the Ala replacements, the single Cys replacements abolished motility (Fig. S5), and proteins with only a single Cys replacement also accumulated to lower levels in cells (as judged from anti-FLAG immunoblots of whole-cell extracts). A double-Cys mutant retained some motility (Fig. S5) and accumulated to more nearly normal levels. Mutants with one position changed to Cys and the other to Ala also retained significant motility (Fig. S5), and were therefore used as the single-Cys controls for cross-linking experiments. On treatment with iodine, the D170C/R185C protein formed a ladder of cross-linked products extending to nonamer; products as large as pentamer were observed even prior to treatment with oxidant (Fig. 6). The crosslinking must occur within a pre-existing FlhA complex rather than through collision of diffusing subunits, because products as large as heptamer persisted (while dimer and trimer became stronger) when the membrane was dissolved by detergent (Triton X-100) (Fig. S6). The FlhA complex evidently forms and is stable in the absence of any other flagellar proteins, because a similar product ladder was observed in the ΔflhDC background (Fig. S6). Crosslinking was also studied using the bifunctional reagent bis-maleimidohexane. On treatment with BMH, the double-Cys protein again formed a ladder of products up to nonamer. The single-Cys proteins formed only dimer, which could result either from protein motions within the complex or collisional crosslinking between individual subunits (Fig. S5).
Fig. 6.
Self-interaction of the small FlhA cytoplasmic domain. (A) CD-1/CD-1 interaction observed in the bacterial adenylate cyclase two-hybrid (BACTH) system (Karimova et al., 1998). The BACTH vectors used in each experiment are indicated above (hyb = hybrid protein; vec = vector control). The MacConkey-agar plate was spotted with 2 μL of overnight cultures and incubated for 24 h at 32 °C. (B) β-galactosidase activities for the construct pairs shown in panel A. The box plots summarize results for 12 biological replicates. (C) Disulfide cross-linking of 170C/185C FlhA. Single-Cys controls had one Cys (at the position indicated) and the other residue replaced by Ala. FlhA carried an N-terminal FLAG tag. Products were resolved on a 4%-20% polyacrylamide gel and detected using anti-FLAG antibody.
Conformational change affecting the large cytoplasmic domain
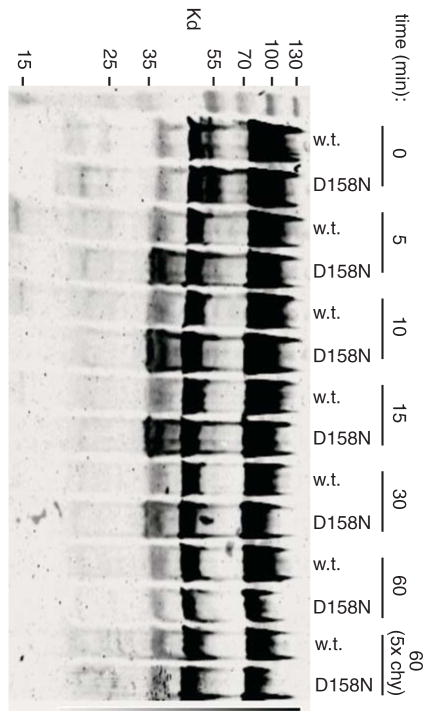
Several studies have shown that the other, larger cytoplasmic domain of FlhA (here termed CD-2) interacts with export substrate and chaperones and with the FliH/I/J cargo-delivery complex (Minamino & Macnab, 2000c, Bange et al., 2010, McMurry et al., 2004, Minamino et al., 2012, Stone et al., 2010, Hartmann & Büttner, 2013). If coupling to the proton gradient involves CD-1 as we propose, then the transport mechanism is likely to involve some linkage between CD-1 and CD-2. We conjectured that the conformation of the large cytoplasmic domain might be responsive to the state of the R147/R154/D158 site. This was tested in limited-proteolysis experiments comparing wild-type FlhA to the protein with a protonation-mimicking mutation (D158N) in the functionally critical acidic group. Experiments used N-terminally tagged FlhA. Cells expressing wild-type or D158N FlhA were converted into inside-out vesicles using a French press at relatively low pressure, then treated with chymotrypsin for various times. Results are shown in Fig. 7. At intermediate times of digestion, a band at about 35-kDa appeared at significantly higher level in the D158N mutant than in the wild type. Changes were also seen in weaker bands near 20 kDa (stronger in the mutant) and 15 kDa (stronger in the wild-type). The band near 35 kDa would result from cleavage in the vicinity of the linker connecting membrane-embedded parts of the protein to the globular C-terminal domain (i.e., near the beginning of CD-2). In a similar experiment done in the ΔflhDC background where no other flagellar genes are expressed, the pattern of proteolysis was different than in the ΔflhA background, and was little affected by the D158N mutation (Fig. S6). The modulation of protease sensitivity by D158 thus requires other flagellar proteins in addition to FlhA, presumably the other proteins of the export apparatus. We conclude that the critical residue D158 in CD-1 regulates a functionally relevant conformational change that extends to the cargo-engaging CD-2 domain.
Fig. 7.
Conformational change in FlhA induced by mutation of D158 in CD-1. Inside-out membrane vesicles (total protein concentration 1.4 mg/ml) prepared from cells expressing N-terminally FLAG-tagged FlhA were subjected to limited proteolysis with chymotrypsin (28 μg/ml for most lanes; 5x this for the last two lanes) at room temperature for the times indicated. Progress of FlhA digestion was monitored by immunoblotting with anti-FLAG antibody. FlhA had either the native D158 residue or the D158N mutation, as indicated.
Discussion
A functionally critical cluster of charged residues in FlhA
Both FliP and FliQ show strong conservation in certain segments in or near the membrane, and both proteins contain invariant, potentially proton-binding residues (Fig. S1). However, none of the conserved acidic or basic residues of FliP or FliQ proved critical in all assays of transport function. The same applies to FliR and FlhB, which are also less well conserved. FlhA, on the other hand, has three functionally important charged residues, including an acidic residue that could engage in rapid proton binding/dissociation reactions. FlhA thus appears the best candidate for coupling type-III secretion to the proton gradient. A protein that functions to harness the membrane gradient would presumably also contain a channel to conduct the energizing protons. Patterns of residue conservation in the FlhA TM segments appear consistent with such a function: Several positions in TM-2 through TM-6 (N51, T82, N89, T93, N137, N219, Q254, and S261, in Salmonella) exhibit nearly invariant polar character that might reflect their involvement in forming a relatively polar (and probably well-hydrated) path for protons. TM-5 contains the invariant acidic residue D208 near its inner end, and TM-3 has an invariant charged residue near its middle (R85) that gave rise to suppressors of D208 mutations (discussed further below).
The functionally critical residues fall in CD-1, the relatively small cytoplasmic domain of FlhA between TM-4 and TM-5. This domain is well conserved and has numerous charged groups (22 of 65 residues are charged in the protein of Salmonella, for example) (Fig. S1). Most of these charged residues are invariant, eight did not tolerate Ala replacement in motility assays, and R147, R154, and D158 proved essential for transport. Mutational replacements at these positions, whether with alanine or with a polar non-protonatable residue, eliminated function in both the motility and hook-Bla transport assays, and the motility defect in these mutants has (so far) not been rescued by mutations elsewhere in the protein. Other positions can be classed as less critical either because suppressing mutations were readily isolated (for mutations of R94, D170, R185, D199, and D208), because a non-protonatable side-chain allowed significant function (K203Q), or because non-protonatable variants function well in the hook-Bla transport assay (D199A, D208N, and D249N). Sequence-based algorithms (Drozdetskiy et al., 2015) are ambiguous regarding the probable secondary structure of the segment containing residues 147–158. It could be helical, in which case it would have substantial amphipathic character and would display the three critical groups on the same face.
Barker et al. recently carried out a mutational study targeted to the CD-1 domain (Barker et al., 2016). Their study explored the probable role of the domain in regulating the sorting of substrates into the export pore, and although the focus was not on the mechanism of coupling to the gradient, their observations are consistent with the present results and with the proposal that the R147/R154/D158 group is involved in coupling. Residue V151, which would lie on the same (hypothetical) helix surface as R147/R154/D158, gave complex phenotypes and was suggested to have an especially important role in substrate sorting.
Networks of intragenic suppression
Though D208 of FlhA was identified as particularly important in a previous mutational study (Hara et al., 2011), we found that the D208S motility defect was suppressible by mutations in R85 or A257. One of the suppressing mutations here (R85H) was also found in the recent study of Barker et al. (Barker et al., 2016). This implies that a non-protonatable group is not essential at position 208, provided certain other mutations are present some distance away (R85 and A257 are both predicted to lie near the middles of membrane segments) (Fig. 3). We further note that the FlhA protein of T. pallidum has asparagine in place of D208 and glycine in place of R85 (thus resembling a mutation/suppressor pair found here, with respect to the positions involved) (Fig. S1). We suggest that D208 is not a primary site of coupling to the proton gradient. Nevertheless, both the present results and the study of Hara et al. (Hara et al., 2011) point to an important role for this residue. Given its location near the inner end of TM-5, we suggest that D208 might function (in species other than T. pallidum) to facilitate proton transfers in the lower part of the channel, possibly delivering protons to the site of coupling in the cytoplasmic domain. In this hypothesis, the suppressing mutations in R85 and A257 (Fig. 3) would act by altering the structure or dynamics of the channel, possibly enlarging it so that the requirement for an acidic residue at the lower end is relaxed.
Other mutation/suppression pairs point to the importance of electrostatic properties of CD-1. The R185A replacement removing a positive charge is suppressed by D168N or D170N mutations removing a negative charge; D170A eliminating a negative charge is suppressed by R185G or R185H that remove positive charge. Charged residues in the domain might be important for stabilizing its structure or/and enabling its interactions with neighboring subunits. The inter-subunit complementation seen with D170 and R185 mutations support the latter possibility (discussed further below). Structural characterization of the domain will be needed to clarify the details of these suppression effects. Some other mutation/suppressor linkages, such as those involving R94 or D199, involve several sites that are widely separated in the protein. These far-reaching genetic connections might reflect conformational interdependencies between different regions of the protein.
Special cases distinguished by the FlgE-Bla transport assay
The FlgE-Bla export assay gave results that were in general agreement with motility assays but with evidently greater sensitivity to low-level export. Three FliP variants with severe motility impairments (E178A, D197G, K222A) retained substantial transport function in the ampicillin IC50 assays. Two FliQ mutants that were completely immotile (E46A and K54A) had IC50 values indicating significant residual transport activity, arguing against a critical proton-coupling role for these FliQ residues. Similarly, the D249N variant, though immotile, retained significant transport activity in the FlgE-Bla assays. The dispensability of an acidic group at position 249 is in agreement with the findings of Hara et al. (Hara et al., 2011), who reported that a lysine replacement there supports motility even though alanine does not. While both the aspartate and lysine side-chains can bind protons, they differ so greatly in structure and proton affinity that the motility of a D249K mutant can be taken as evidence against a critical proton-coupling role for D249. A final distinctive case was that of the D199A mutant, which supported substantial export in the IC50 assay in spite of being immotile. We suggest that export might be slowed in the D199A mutant to a rate that is sufficient to confer Amp resistance but not enough to produce filaments of useful length. Consistent with this, Barker et al. (Barker et al., 2016) found that mutations of D199 displayed complex phenotypes indicating that transport was perturbed but not prevented.
The apparatus tolerates a subset of nonfunctional subunits
The several instances of inter-subunit complementation imply that most non-functioning FlhA mutations (R94A, D158A, D170A, R185A, D199A, D208A, D249A) can be tolerated if they are present in only some of the subunits. R94A in particular supported significant function when combined with several of the other nonfunctioning proteins, indicating that R94 provides a distinct function that does not need to be present in every FlhA subunit. Moreover, the results indicate that even the critically important residue Asp158 does not need to be wild-type in all copies of FlhA present in the apparatus.
The inter-subunit complementation observed with the D170A and R185A mutations (Fig. 5) recapitulates the mutual rescue seen with these residues in the suppressor screen (Fig. 3) and suggests that they are present at an interface between adjacent subunits. This provided guidance for the cross-linking experiment to probe the organization of the CD-1 domains.
A membrane-proximal ring formed by the small cytoplasmic domain
Experiments using the bacterial two-hybrid system gave further evidence that adjacent CD-1 domains interact, and targeted cross-linking experiments showed that residue 170 in one subunit is near residue 185 of the adjacent subunit. The large multimer formed upon oxidative crosslinking indicates that many copies of CD-1 are present together in a regular array. Crystallographic studies of the CD-2 domain (Saijo-Hamano et al., 2010, Bange et al., 2010, Abrusci et al., 2013) show multiple, most likely nine, subunits arranged with roughly axial symmetry. We suggest that the nine copies of CD-1 also form a ring. Because CD-1 is closely flanked by hydrophobic, putatively membrane-spanning segments, the CD-1 ring must lie close to the membrane. The CD-2 ring, by contrast, is joined to the membrane regions by a fairly long linker and could have greater freedom of movement.
Communication between CD-1 and CD-2
A possible functional linkage between the CD-1 and CD-2 domains was suggested previously by the work of Hartmann and Büttner, who studied interactions of the FlhA-homolog HrcV of Xanthomonas (Hartmann & Büttner, 2013). They identified several interaction partners of CD-2 and found that certain of these interactions were perturbed by mutations in CD-1, including some mutations in the putative coupling-site identified here. Their findings can be readily rationalized in terms of a conformational change in CD-2 modulated by the state of the 147/154/158 site in CD-1 and are germane to a specific proposal for the transport mechanism described below.
Implications for the transport mechanism
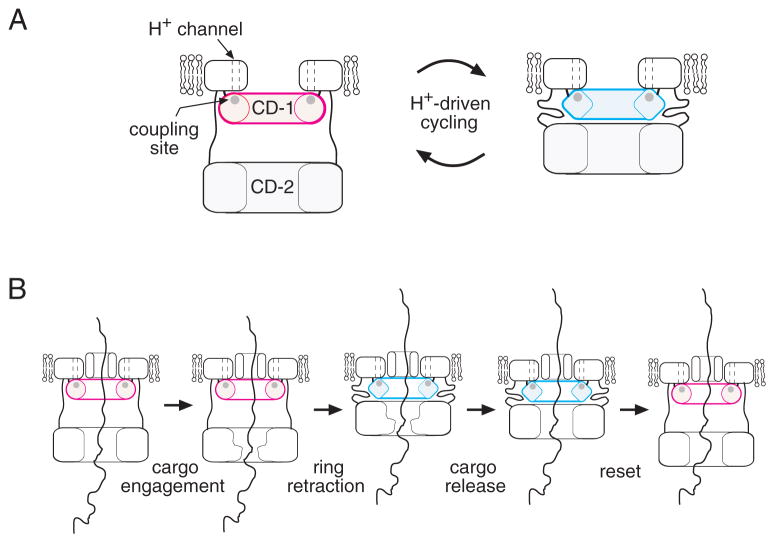
We suggest that the 147/154/158 site in FlhA is a key site of energy transduction in the T3S apparatus. Whether the membrane gradient consists primarily of ΔpH or ΔΨ, gradient protons will be at a high effective concentration (low pH) as they reach the cytoplasm. The acidic residue D158 would then be best suited to accepting gradient protons and subsequently releasing them rapidly into the cytoplasm (with proton-actuated transport events occurring in between). A proposal for the transport mechanism takes shape if we make the further assumption that the CD-1 ring interacts with the larger CD-2 ring, a linkage that is plausible given the D158-regulated conformational change (Fig. 7) and the results of Hartmann and Büttner (Hartmann & Büttner, 2013). The CD-2 ring interacts with the cargo (Minamino & Macnab, 2000c, Bange et al., 2010, McMurry et al., 2004, Minamino et al., 2012, Stone et al., 2010, Hartmann & Büttner, 2013), and was assigned to a feature some distance (~6 nm) from the membrane in the electron microscopic study of Abrusci et al. (Abrusci et al., 2013). We suggest that proton-actuated conformational changes in the CD-1 ring could modulate its interactions with CD-2 to drive cyclical movement of the cargo-engaging ring toward the membrane (to bind CD-1) and away (when released from CD-1). Such a motion, appropriately synchronized to cycles of cargo binding and release, could drive the transport of sizable segments of protein with each cycle. This hypothesis is illustrated in Fig. 8.
Fig. 8.
Model for the action of FlhA in flagellar export. (A) Hypothesized proton-driven conformational cycling in the small cytoplasmic domain of FlhA (CD-1). Several copies of the CD-1 domain are proposed to form a ring, positioned close to the membrane and on the axis of the apparatus. The CD-2 forms a larger ring, which can lie either some distance from the membrane (tethered by a linker with extended secondary structure) or near the membrane (held there by interaction with CD-1). Proton-actuated conformational changes in the CD-1 ring are suggested to modulate its interaction with CD-2 to drive cyclical movement of the CD-2 ring between the membrane-proximal and -distal positions. (B) Substrate transport will occur if the cyclical motion of the CD-2 ring is linked to cycles of cargo binding and release. This would presumably involve conformational changes in the CD-2 ring, which could be induced by its cyclical interactions with CD-1. A trans-membrane conduit for the cargo, formed by a protein other than FlhA, is also shown.
Experimental Procedures
Strains, plasmids and mutagenesis
Experiments used strains of Salmonella enterica (serovar Typhimurium) listed in Table 1. The fliP, fliQ, fliR, flhA, and flhB genes were cloned into the salicylate-inducible expression vector pKG116, a gift from J. S. Parkinson (University of Utah), using NdeI and KpnI sites. For experiments testing intragenic complementation, a particular allele of flhA (R94A) was cloned into a variant of pKG116 that had Cmr replaced by Kmr. Function of some FlhB mutants was tested in the presence of over-expressed wild-type FlhA. These experiments used a second plasmid encoding wild-type flhA, based on the IPTG-inducible, Apr plasmid pRR48 (a gift from J. S. Parkinson). Mutations were made using the QuikChange method (Stratagene) and confirmed by Sanger sequencing (carried out by GeneWiz® or by core facilities at the University of Utah).
Table 1.
Strains and plasmids
| Strain | Relevant genotype or property | Source or reference |
|---|---|---|
| LT2 | Wild type Salmonella enterica serovar Typhimurium | K. T. Hughes |
| TH12642 | ΔflhA7452 | This study |
| TH12644 | ΔflhB7454 | This study |
| TH10549 | ΔfliP6709 | This study |
| TH10550 | ΔfliQ6710 | This study |
| TH10551 | ΔfliR null strain | This study |
| TH12643 | ΔflgBC6557flgE6569::blaΔflhA7453 | This study |
| TH12731 | ΔflgBC6557flgE6569::blaΔfliP7457 | This study |
| TH15930 | ΔflgBC6557flgE6569::blaΔfliQ6710 | This study |
| Plasmid | ||
| pKG116 | Salicylate-inducible vector; Cmr | J. S. Parkinson |
| pRR48 | IPTG-inductible vector; Apr | J. S. Parkinson |
| pMS10 | fliP in pKG116 | This study |
| pMS11 | fliQ in pKG116 | This study |
| pMS12 | fliR in pKG116 | This study |
| pMS122 | flhA in pKG116 | This study |
| pMS123 | flhB in pKG116 | This study |
| pMM108 | pTrc99A-His-FLAG-flhA | (Saijo-Hamano et al., 2004) |
| pEK1007 | flhA in pRR48 | This study |
| pDB1002 | Kmr control plasmid | This study |
| pDB1012 | flhA-D170A in Kmr version of pKG116 | This study |
| pDB1015 | flhA-R94A in Kmr version of pKG116 | This study |
Function of mutated proteins
For assays of function, salicylate-inducible plasmids encoding the mutated proteins were transformed into corresponding deletion strains, and rates of migration in soft-agar tryptone plates (0.27% Bacto-agar, 10 g L−1 tryptone, 5 g L−1 NaCl) were measured as described previously (Tang & Blair, 1995), with induction by salicylate at the levels indicated in the Figures and Tables (in most cases 2.5 μM). Rates are relative to that of wild-type controls present on the same plates. For tests of dominance, the mutant plasmids were introduced into the wild-type strain LT2, and migration in soft agar was measured and compared to that of a control strain expressing the wild-type protein from the plasmid. Stability of selected FlhA proteins (R147A, R154A, D158A) was examined on immunoblots using polyclonal anti-FlhA antibody, a gift of T. Minamino (Minamino et al., 2010). Monoclonal anti-DnaK antibody (Abcam) was used as loading control.
Hook-β-lactamase secretion assay
Assays of secretion based on IC50 values for ampicillin (antibiotic concentration giving 50% growth-inhibition) were adapted from the procedure described by Lee and Hughes (Lee & Hughes, 2006). Strains contained a deletion of the rod genes flgBC and a chromosomally encoded fusion of the hook gene flgE to the bla gene encoding β-lactamase. Each tester strain was additionally deleted for one of the export-apparatus genes flhA, fliP, or fliQ, and were transformed with plasmids encoding the corresponding export-apparatus gene, either wild-type or mutant. Cells were cultured from single colonies at 37 °C overnight in LB (10 g L−1 tryptone, 5 g L−1 yeast extract, 5 g L−1 NaCl) plus chloramphenicol (Cm) (50 μg ml−1), then diluted 50-fold in LB containing Cm plus 2.5 μM sodium salicylate to induce expression of the plasmid-encoded gene. Following 2.5 h of growth at 37 °C, cultures were again diluted 50-fold into LB/Cm/salicylate containing various concentrations of ampicillin (0, 10, 20, 50, 100, 200, 500, or 1,000 μg mL−1), and grown for an additional 3.5 h at 37 °C. Optical density at 600 nm was measured, and curves of relative growth vs. [Amp] were generated by normalizing to growth in zero ampicillin. The concentration of Amp that permitted growth at 50% of the control (no-Amp) level was determined by interpolation.
Prediction of membrane topologies
Trans-membrane segments were predicted using the web-based tools MEMSAT-SVM (Nugent & Jones, 2009), HMMTOP (Tusnady & Simon, 2001), SOSUI (Hirokawa et al., 1998), TMHMM (Melen et al., 2003), TMPRED (Hofmann & Stoffel, 1993), and TopPred (Claros & vonHeijne, 1994). Segment endpoints shown in Fig. 1 are averages of the values obtained from the various methods.
Isolation of suppressors of flhA mutations
Cells of the flhA-deletion strain transformed with plasmids expressing non-functional FlhA variants (having mutations in residues Arg 94, Arg 147, Arg 154, Asp 158, Asp170, Arg185, Asp199, Lys203, Asp208, or Asp249) were grown overnight in LB/Cm and inoculated in streaks onto soft-agar tryptone plates containing Cm (50 μg mL−1) and sodium salicylate (10 μM). Plates were incubated at 32 °C to allow the outgrowth of motile variants. Certain of the Arg-94, Asp170, Arg185, Asp199, and Asp-208 mutants displayed motile ‘flares’ within 24–36 h. Cells from the edges of flares were colony purified and cultured to isolate the flhA-encoding plasmid. Plasmids were re-introduced into the flhA-deletion strain to determine whether they encoded changes responsible for the motility rescue. Plasmids conferring motility were sequenced to determine the nature of the suppressing mutations.
Inter-subunit complementation
Selected flhA mutations were transferred onto a plasmid derived from pKG116 with the CmR-marker replaced by KmR. A control plasmid expressed KmR but no cloned gene. The ΔflhA strain was transformed with CmR and KmR plasmids encoding various pairs of flhA mutations, and fresh transformants were picked onto soft-agar motility plates containing both antibiotics and 5 μM sodium salicylate for induction. Plates were incubated at 32 °C and photographed at the times indicated in the figure legend. In parallel, cells were cultured at 32 °C in LB with antibiotics and inducer, and motility was scored under the microscope.
Two-hybrid measurements of domain interactions
Interactions of the small cytoplasmic domain of FlhA were examined using the bacterial adenylate cyclase two hybrid (BACTH) system (Karimova et al., 1998). Codons 143–209 of flhA were cloned into each of the four BACTH vectors (pkT25, pkNT25, pUT18 pUT18c), and suitable pairs of constructs (i.e., one encoding the adenylate cyclate N-terminal domain and another encoding the C-terminal domain) were transformed into the adenylate cyclase-deficient strain BTH101. Transformants were cultured overnight at 32 °C in LB media containing antibiotics (50 μg ml−1 Km, 100 μg ml−1 Amp), and IPTG (0.5 mM) to induce expression of the hybrid constructs, then spotted onto MacConkey plates containing the same additives. Plates were incubated at 32 °C and photographed at the times indicated in the fig. legend. Measurements of β-galactosidase activity followed the protocol of Zhang and Bremer (Zhang & Bremer, 1995). For these assays, cells were cultured overnight in LB (with antibiotics and IPTG), then OD600 was measured (for later calculation of activity) and 10 μl of cells were mixed with 90 μL of substrate solution (Zhang & Bremer, 1995) for use in the assay.
Crosslinking
C-terminally FLAG-tagged FlhA was expressed from an IPTG-inducible promoter in plasmid p-His-FLAG-flhA, in the ΔflhA strain TH12642. Cells were cultured overnight at 32 °C in LB containing 100 μg ml−1 Amp. Overnight cultures were diluted 100-fold into 10 ml of the same medium additionally containing 10 μM IPTG and cultured at 32 °C until OD600 reached 1.2–1.4. OD600 was measured and used to equalize numbers of cells (by taking suitable sample volumes), then cells were collected by centrifugation and resuspended in 2 mL XL buffer (20 mM Na-phosphate pH 7.5, 150 mM NaCl) and incubated on ice for 10 min. Samples were divided into experimental and control tubes (0.5 ml each). For oxidative crosslinking by iodine, I2 was added to experimental samples to final concentration of 0.1 mM from a 20 mM stock in ethanol. Controls received an equal volume of ethanol. Crosslinking was allowed to proceed for 3–5 minutes at room temperature (the color associated with I2 was dissipated in the early part of this incubation, and results were not affected by incubation for longer times). Sulfhydryls were blocked with 10 mM NEM (5 μL from a 1 M stock). Proteins were resolved on SDS-PAGE gels (either 4% - 20% gradient gels or 7.5% gels using a 70:1 acrylamide:bis-acrylamide ratio) and visualized on immunoblots with monoclonal anti-FLAG antibody (Sigma) used at 1:1000 dilution. For cross-linking by bis-maleimidohexane (BMH), cells were resuspended in 40 mM MOPS, 20 mM Na-acetate, 1 mM EDTA, pH 7.3, and treated with 0.2 mM BMH (added from a 20 mM stock in DMSO) and incubated for 1 h at 22° C. Reaction was quenched with 10 mM NEM. Following crosslinking, cells were pelleted and resuspended in 1 mL lysis buffer (50 mM Tris pH 7.4, 100 mM NaCl, 5 mM EDTA, 1 mg ml−1 lysozyme) and incubated at 37° C for 30 min. Samples were sonicated briefly (Branson model 450, power 3, duty cycle 50%, 15 s), pelleted, and resuspended and boiled in 100 μL non-reducing gel buffer. Proteins were resolved on 4%-20% gradient gels and visualized as described above.
Limited proteolysis
Strain 12642 (ΔflhA) transformed with plasmid pMM108 were cultured overnight at 32 °C in LB-Amp, diluted 100-fold into 250 mL of the same medium containing 10 μM IPTG, and grown at 32 °C to OD600 of 1.2–1.4. Inside-out vesicles were prepared using a French pressure cell at relatively low pressure (~4,000 psi), as described by Kojima and Blair (Kojima & Blair, 2001). Membranes (1.4 mg protein mL−1) were treated with chymotrypsin (28 μg mL−1) at 22° C for the times indicated. Reaction was quenched with protease-inhibitor cocktail (Sigma) and addition to an equal volume of preheated 6X reducing-gel buffer. Samples were boiled, loaded on gels, and products were visualized on anti-FLAG immunoblots as described above.
Supplementary Material
Acknowledgments
We thank Tohru Minamino and Keiichi Namba for the gift of pMM108, Niquele Nunes Almeida for assistance with motility measurements, Runkun Wang for assistance with IC50 assays, and Sandy Parkinson for discussions and strains. Supported by grants R01-GM64664 from the National Institute of General Medical Sciences of the NIH (to D.F.B.), R01- GM056141 (to K.T.H.), and GM087260Z (to D.F.B. and K.T.H.) from the National Institute of General Medical Sciences of the NIH, the Helmholtz Association Young Investigator grant VH-NG-932 (to M.E.), and the People Programme (Marie Curie Actions) of the European Union Seventh Framework Programme grant 334030 (to M.E.).
Footnotes
Author contributions
M.E., K.T.H., and D.F.B. contributed to conception, design and analysis of experiments, and drafting the paper; P.W., E.AK., M.E., D.F.B., T.H., Y.Z., and M.K.S. contributed to strain construction, conducting experiments, and data analysis and interpretation.
References
- Abramson J, Smirnova I, Kasho V, Verner G, Kaback HR, Iwata S. Structure and mechanism of the lactose permease of Escherichia coli. Science. 2003;301:610–615. doi: 10.1126/science.1088196. [DOI] [PubMed] [Google Scholar]
- Abrusci P, Vergara-Irigaray M, Johnson S, Beeby MD, Hendrixson DR, Roversi P, Friede ME, Deane JE, Jensen GJ, Tang CM, Lea SM. Architecture of the major component of the type III secretion system export apparatus. Nat Struct & Molec Biol. 2013;20:99–104. doi: 10.1038/nsmb.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aizawa SI. Bacterial flagella and type III secretion systems. FEMS Microbiol Letters. 2006;202:157–164. doi: 10.1111/j.1574-6968.2001.tb10797.x. [DOI] [PubMed] [Google Scholar]
- Aldridge P, Karlinsey JE, Hughes KT. The type III secretion chaperone FlgN regulates flagellar assembly via a negative feedback loop containing its chaperone substrates FlgK and FlgL. Mol Microbiol. 2003;49:1333–1345. doi: 10.1046/j.1365-2958.2003.03637.x. [DOI] [PubMed] [Google Scholar]
- Aldridge PD, Karlinsey JE, Aldridge C, Birchall C, Thompson D, Yagasaki J, Hughes KT. The flagellar-specific transcription factor, σ28, is the Type III secretion chaperone for the flagellar-specific anti-σ28 factor FlgM. Genes Dev. 2006;20:2315–2326. doi: 10.1101/gad.380406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auvray F, Thomas J, Fraser GM, Hughes C. Flagellin polymerization control by a cytosolic export chaperone. J Mol Biol. 2001;308:221–229. doi: 10.1006/jmbi.2001.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bange G, Kümmerer N, Engel C, Bozkurt G, Wild K, Sinning I. FlhA provides the adaptor for coordinated delivery of late flagella building blocks to the type III secretion system. Proc Natl Acad Sci USA. 2010;107:11295–11300. doi: 10.1073/pnas.1001383107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker CS, Inoue T, Meshcheryakova SK, Samatey FA. Function of the conserved FHIPEP domain of the flagellar type III export apparatus, protein FlhA. Mol Microbiol. 2016;100:278–288. doi: 10.1111/mmi.13315. [DOI] [PubMed] [Google Scholar]
- Barker CS, Meshcheryakova IV, Kostyukova AS, Samatey FA. FliO regulation of FliP in the formation of the Salmonella enterica flagellum. PLoS Genetics. 2010:6. doi: 10.1371/journal.pgen.1001143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett JCQ, Hughes C. Substrate complexes and domain organization of the Salmonella flagellar export chaperones FlgN and FliT. Mol Microbiol. 2001;39:781–791. doi: 10.1046/j.1365-2958.2001.02268.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger C, Probin G, Bonas U, Koebnik R. Membrane topology of conserved components of the type III secretion system frm the plant pathogen Xanthomonas campestris pv. vesicatoria. Microbiology. 2010;156:1963–1974. doi: 10.1099/mic.0.039248-0. [DOI] [PubMed] [Google Scholar]
- Claros M, vonHeijne G. TopPred II: An improved software for membrane protein structure predictions. Comput Appl Biosci. 1994;10:685–686. doi: 10.1093/bioinformatics/10.6.685. [DOI] [PubMed] [Google Scholar]
- Cornelis GR. The type III secretion injectisome. Nat Rev Microbiol. 2006;4:811–825. doi: 10.1038/nrmicro1526. [DOI] [PubMed] [Google Scholar]
- Drozdetskiy A, Cole C, Procter J, Barton GJ. JPred4: a protein secondary structure prediction server. Nucl, Acids Res. 2015;(43):W389–W394. doi: 10.1093/nar/gkv332. [DOI] [PMC free article] [PubMed]
- Erhardt M, Mertens ME, Fabiani FD, Hughes KT. ATPase-independent Type-III protein secretion in Salmonella enterica. PloS Genet. 2014;10:e1004800. doi: 10.1371/journal.pgen.1004800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan F, Macnab RM. Enzymatic characterization of FliI: an ATPase involved in flagellar assembly in Salmonella typhimurium. J Biol Chem. 1996;271:31981–31988. doi: 10.1074/jbc.271.50.31981. [DOI] [PubMed] [Google Scholar]
- Fan F, Ohnishi K, Francis NR, Macnab RM. The FliP and FliR proteins of Salmonella typhimurium, putative components of the flagellar export apparatus, are located in the flagellar basal body. Mol Microbiol. 1997;26:1035–1046. doi: 10.1046/j.1365-2958.1997.6412010.x. [DOI] [PubMed] [Google Scholar]
- Fraser GM, Gonzalez-Pedrajo B, Tame JR, Macnab RM. Interactions of FliJ with the Salmonella type III flagellar export apparatus. J Bacteriol. 2003a;185:5546–5554s. doi: 10.1128/JB.185.18.5546-5554.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser GM, Hirano T, Ferris HU, Devgan LL, Kihara M, Macnab RM. Substrate specificity of type III flagellar protein export in Salmonella is controlled by subdomain interactions in FlhB. Mol Microbiol. 2003b;48:1043–1057. doi: 10.1046/j.1365-2958.2003.03487.x. [DOI] [PubMed] [Google Scholar]
- Galperin M, Dibrov PA, Glagolev AN. ΔμH+ is required for flagellar growth in Escherichia coli. FEBS Letters. 1982;143:319–322. doi: 10.1016/0014-5793(82)80125-7. [DOI] [PubMed] [Google Scholar]
- Hara N, Namba K, Minamino T. Genetic characterization of conserved charged residues in the bacterial flagellar type III export protein FlhA. PLoS one. 2011;6:e22417. doi: 10.1371/journal.pone.0022417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann N, Büttner D. The inner membrane protein HrcV from Xanthomonas spp. is involved in substrate docking during type III secretion. Molec Plant-Microbe Interactions. 2013;26:1176–1189. doi: 10.1094/MPMI-01-13-0019-R. [DOI] [PubMed] [Google Scholar]
- Hirano T, Yamaguchi S, Oosawa K, Aizawa SI. Roles of FliK and FlhB in determination of flagellar hook length in Salmonella typhimurium. J Bacteriol. 1994;176:5439–5449. doi: 10.1128/jb.176.17.5439-5449.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa T, Boon-Chieng S, Mitaku S. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics. 1998;14:378–379. doi: 10.1093/bioinformatics/14.4.378. [DOI] [PubMed] [Google Scholar]
- Hofmann K, Stoffel W. TMbase-a database of membrane spanning protein segments. Biol Chem Hoppe-Seyler. 1993;347:166. [Google Scholar]
- Hoppe J, Schairer HU, Friedl P, Sebald W. An Asp-Asnsubstitution in the proteolipid subunit of the ATP-synthase from Escherichia coli leads to a non-functional proton channel. FEBS Letters. 1982;145:21–29. doi: 10.1016/0014-5793(82)81198-8. [DOI] [PubMed] [Google Scholar]
- Hoppe J, Sebald W. The proton conducting Fo-part of bacterial ATP synthase. Biochim Biophys Acta. 1984;768:1–27. doi: 10.1016/0304-4173(84)90005-3. [DOI] [PubMed] [Google Scholar]
- Ibuki T, Imaga K, Minamino T, Kato T, Miyata T, Namba K. Common architecture of the flagellar type III protein export apparatus and F- and V-type ATPases. Nat Struct Mol Biol. 2011;18:277–282. doi: 10.1038/nsmb.1977. [DOI] [PubMed] [Google Scholar]
- Iino T. Assembly of Salmonella flagellin in vitro and in vivo. J Supramol Struct. 1974;2:372–384. doi: 10.1002/jss.400020226. [DOI] [PubMed] [Google Scholar]
- Imada K, Minamino T, Tahara A, Namba K. Structural similarity between the flagellar type III ATPase FliI and F1-ATPase subunits. Proc Natl Acad Sci USA. 2007;104:485–490. doi: 10.1073/pnas.0608090104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imada K, Minamino T, Uchida Y, Kinoshita M, Namba K. Insight into the flagella type III export revealed by the complex structure of the type III ATPase and its regulator. Proc Natl Acad Sci USA. 2016;113:3633–3638. doi: 10.1073/pnas.1524025113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimova G, PIdoux J, Ullmann A, Ladant D. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci USA. 1998;95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama E, Shiraishi T, Oosawa K, Baba N, Aizawa SI. Geometry of the flagellar motor in the cytoplasmic membrane of Salmonella typhimurium as determined by stereo-photogrammetry of quick-freeze deep-etch replica images. J Mol Biol. 1996;255:458–475. doi: 10.1006/jmbi.1996.0038. [DOI] [PubMed] [Google Scholar]
- Kihara M, Minamino T, Yamaguchi S, Macnab RM. Intergenic suppression between the flagellar MS ring protein FliF of Salmonella and FlhA, a membrane component of its export apparatus. J Bacteriol. 2001;183:1655–1662. doi: 10.1128/JB.183.5.1655-1662.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S, Blair DF. Conformational change in the stator of the bacterial flagellar motor. Biochemistry. 2001;40:13041–13050. doi: 10.1021/bi011263o. [DOI] [PubMed] [Google Scholar]
- Lee HJ, Hughes KT. Posttranscriptional control of the Salmonella enterica flagellar hook protein FlgE. J Bacteriol. 2006;188:3308–3316. doi: 10.1128/JB.188.9.3308-3316.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, LT, Botkin DJ, McCrum E, Winkler H, Norris SJ. Intact flagellar motor of Borrelia burgdorferi revealed by cryo-electron tomography: Evidence for stator ring curvature and rotor/C-ring assembly flexion. J Bacteriol. 2009;191:5026–5036. doi: 10.1128/JB.00340-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macnab RM. How bacteria assemble flagella. Ann Rev Microbiol. 2003;57:77–100. doi: 10.1146/annurev.micro.57.030502.090832. [DOI] [PubMed] [Google Scholar]
- Macnab RM. Type III flagellar protein export and flagellar assembly. Biochim Biophys Acta. 2004;1694:207–217. doi: 10.1016/j.bbamcr.2004.04.005. [DOI] [PubMed] [Google Scholar]
- McMurry JL, Van Arnam JS, Kihara M, Macnab RM. Analysis of the cytoplasmic domains of Salmonella FlhA and interactions with components of the flagellar export machinery. J Bacteriol. 2004;186:7586–7592. doi: 10.1128/JB.186.22.7586-7592.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melen K, Krogh A, von Heijne G. Reliability measures for membrane protein topology prediction algorithms. J Mol Biol. 2003;327:735–744. doi: 10.1016/s0022-2836(03)00182-7. [DOI] [PubMed] [Google Scholar]
- Miller MJ, Oldenberg M, Fillingame RH. The essential carboxyl group in subunit c of F1Fo ATP synthase can be moved and H+ translocating function retained. Proc Natl Acad Sci USA. 1990;87:4900–4904. doi: 10.1073/pnas.87.13.4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino T, Chu R, Yamaguchi S, Macnab RM. Role of FliJ in flagellar protein export in Salmonella. J Bacteriol. 2000;182:4207–4215. doi: 10.1128/jb.182.15.4207-4215.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino T, Gonzalez-Pedrajo B, Kihara M, Namba K, Macnab RM. The ATPase FliI can interact with the type III flagellar protein export apparatus in the absence of its regulator, FliH. J Bacteriol. 2003;185:5546–5554. doi: 10.1128/JB.185.13.3983-3988.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino T, Iino T, Kutsukake K. Molecular characterization of the Salmonella typhimurium flhB operon and its protein products. J Bacteriol. 1994;176:7630–7637. doi: 10.1128/jb.176.24.7630-7637.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino T, Kinoshita M, Hara N, Takeuchi S, Hida A, Koya S, Glenwright H, Imaga K, Aldridge PD, Namba K. Interaction of a bacterial flagellar chaperone FlgN with FlhA is required for efficient export of its cognate substrates. Mol Microbiol. 2012;83:775–788. doi: 10.1111/j.1365-2958.2011.07964.x. [DOI] [PubMed] [Google Scholar]
- Minamino T, Macnab RM. Domain structure of Salmonella FlhB, a flagellar export component responsible for substrate-specificity switching. J Bacteriol. 2000a;182:4906–4914. doi: 10.1128/jb.182.17.4906-4914.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino T, Macnab RM. FliH, a soluble component of the type III flagellar export apparatus of Salmonella, forms a complex with FliI and hinhibits its ATPase activity. Mol Microbiol. 2000b;37:1494–1503. doi: 10.1046/j.1365-2958.2000.02106.x. [DOI] [PubMed] [Google Scholar]
- Minamino T, Macnab RM. Interactions among components of the Salmonella flagellar export apparatus and its substrates. Mol Microbiol. 2000c;35:1052–1064. doi: 10.1046/j.1365-2958.2000.01771.x. [DOI] [PubMed] [Google Scholar]
- Minamino T, Namba K. Self-assembly and type III protein export of the bacterial flagellum. J Mol Microbiol Biotechnol. 2004;7:5–17. doi: 10.1159/000077865. [DOI] [PubMed] [Google Scholar]
- Minamino T, Namba K. Distinct roles of the FliI ATPase and proton motive force in bacterial flagellar protein export. Nature. 2008;451:485–488. doi: 10.1038/nature06449. [DOI] [PubMed] [Google Scholar]
- Minamino T, Shimada M, Okabe M, Saijo-Hamano Y, Imada K, Kihara M, Namba K. Role of the C-terminal cytoplasmic domain of FlhA in bacterial flagellar type III protein export. J Bacteriol. 2010;192:1929–1936. doi: 10.1128/JB.01328-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto YV, Ito M, Hiraoka KD, Che YS, Bai F, Kami-Ike N, Namba K, Minamino T. Assembly and stoichiometry of FliF and FlhA in Salmonella flagellar basal body. Mol Microbiol. 2014;91:1214–1226. doi: 10.1111/mmi.12529. [DOI] [PubMed] [Google Scholar]
- Nolling J, Breton G, Omelchenko MV, Makarova KS, Zeng Q, Gibson R, Lee HM, Dubois J, Qiu D, Hitti J. Genome sequence and comparative analysis of the solvent-producing bacterium Clostridium acetobutylicum. J Bacteriol. 2001;183:4823–4838. doi: 10.1128/JB.183.16.4823-4838.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent T, Jones DT. Transmembrane protein topology prediction using support vector machines. BMC Bioinformatics. 2009:10. doi: 10.1186/1471-2105-10-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi K, Fan F, Schoenhals GJ, Kihara M, Macnab RM. The FliO, FliP, FliQ, and FliR proteins of Salmonella typhimurium: Putative components for flagellar assembly. J Bacteriol. 1997;179:6092–6099. doi: 10.1128/jb.179.19.6092-6099.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallen MJ, Bailey CM, Beatson SA. Evolutionary links between FliH/YscL-like proteins from bacterial type III secretion systems and second-stalk components of the FoF1 and vacuolar ATPases. Protein Sci. 2006;15:935–941. doi: 10.1110/ps.051958806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul K, Erhardt M, Hirano T, Blair DF, Hughes KT. Energy source of flagellar type III secretion. Nature. 2008;451:489–492. doi: 10.1038/nature06497. [DOI] [PubMed] [Google Scholar]
- Saijo-Hamano Y, Imada K, Minamino T, Kihara M, Shimada M, Kitao A, Namba K. Structure of the cytoplasmic domain of FlhA and implication for flagellar type III export. Mol Microbiol. 2010;76:260–268. doi: 10.1111/j.1365-2958.2010.07097.x. [DOI] [PubMed] [Google Scholar]
- Saijo-Hamano Y, Minamino T, Macnab RM, Namba K. Structural and functional analysis of the C-terminal cytoplasmic domain of FlhA, an integral membrane component of the Type III flagellar protein export apparatus in Salmonella. J Mol Biol. 2004;343:457–466. doi: 10.1016/j.jmb.2004.08.067. [DOI] [PubMed] [Google Scholar]
- Stone CB, Bulir DC, Gilchrist JD, Toor RK, Mahony JB. Interactions between flagellar and type III secretion proteins in Chlamydia pneumoniae. BMC Microbiol. 2010:10. doi: 10.1186/1471-2180-10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Blair DF. Regulated underexpression of the FliM protein of Escherichia coli and evidence for a location in the flagellar motor distinct from the MotA/MotB torque generators. J Bacteriol. 1995;177:3485–3495. doi: 10.1128/jb.177.12.3485-3495.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusnady GE, Simon I. The HMMTOP transmembrane topology prediction server. Bioinformatics. 2001;17:849–850. doi: 10.1093/bioinformatics/17.9.849. [DOI] [PubMed] [Google Scholar]
- Van Arnam JS, McMurry JL, Kihara M, Macnab RM. Analysis of an engineered Salmonella flagellar fusion protein, FliR-FlhB. J Bacteriol. 2004;186:2495–2498. doi: 10.1128/JB.186.8.2495-2498.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogler AP, Homma M, Irikura VM, Macnab RM. Salmonella typhimurium mutants defective in flagellar filament regrowth and sequence similarity of FliI to F0F1, vacuolar, and archaebacterial ATPase subunits. J Bacteriol. 1991;173:3564–3572. doi: 10.1128/jb.173.11.3564-3572.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilharm G, Lehmann V, Krauss K, Lehnert B, Richter S, Ruckdeschel K, Heesemann J, Trulzsch K. Yersinia enterocolitica type III secretion depends on the proton motive force but not on the flagellar motor components MotA and MotB. Infect and Immun. 2004;72:4004–4009. doi: 10.1128/IAI.72.7.4004-4009.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Bremer H. Control of the Escherichia coli rrnB P1 promoter strength by ppGpp. J Biol Chem. 1995;270:11181–11189. doi: 10.1074/jbc.270.19.11181. [DOI] [PubMed] [Google Scholar]
- Zhou J, Sharp LL, Tang HL, Lloyd SA, Billings S, Braun TF, Blair DF. Function of protonatable residues in the flagellar motor of Escherichia coli: a critical role for Asp 32 of MotB. J Bacteriol. 1998;180:2729–2735. doi: 10.1128/jb.180.10.2729-2735.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu K, Gonzalez-Pedrajo B, Macnab RM. Interactions among membrane and soluble components of the flagellar export apparatus of Salmonella. Biochemistry. 2002;41:9516–9524. doi: 10.1021/bi0203280. [DOI] [PubMed] [Google Scholar]
- Zilkenat S, Franz-Wachtal S, Stierhof YD, Galan JE, Macek B, Wagner S. Determination of the stoichiometry of the complete bacterial type III secretion needle complex using a combined quantitative proteomic approach. Mol Cell Proteomics. 2016;15:1598–1609. doi: 10.1074/mcp.M115.056598. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.