Abstract
14-3-3σ has been implicated in the development of chemo and radiation resistance and in poor prognosis of multiple human cancers. While it has been postulated that 14-3-3σ contributes to these resistances via inhibiting apoptosis and arresting cells in G2/M phase of the cell cycle, the molecular basis of this regulation is currently unknown. In this study, we tested the hypothesis that 14-3-3σ causes resistance to DNA-damaging treatments by enhancing DNA repair in cells arrested in G2/M phase following DNA-damaging treatments. We showed that 14-3-3σ contributed to ionizing radiation (IR) resistance by arresting cancer cells in G2/M phase following IR and by increasing non-homologous end joining (NHEJ) repair of the IR-induced DNA double strand breaks (DSBs). The increased NHEJ repair activity was due to 14-3-3σ-mediated up-regulation of Poly(ADP-ribose) polymerase 1 (PARP1) expression that promoted the recruitment of DNA-PKcs to the DNA damage sites for repair of DSBs. On the other hand, the increased G2/M arrest following IR was due to 14-3-3σ-induced Chk2 expression.
Implications
These findings reveal an important molecular basis of 14-3-3σ function in cancer cell resistance to chemo/radiation therapy and in poor prognosis of human cancers.
Keywords: 14-3-3σ, DNA repair, PARP1, Chk2, radiation resistance
INTRODUCTION
14-3-3σ is a family member of 14-3-3 proteins (14-3-3β, ε, θ/τ, ζ, σ, γ and η) in human and has been implicated in the development of cancer and in treatment resistance and poor prognosis (1). While 14-3-3σ is thought to function as a tumor suppressor in mammary tissue, its expression has been found to up-regulate in drug resistant cancers of pancreas and breast and associates with poor prognosis (2–6). 14-3-3σ has also been found recently to regulate invasion of breast cancer cells (7) and EMT (8), which may contribute to poor cancer prognosis.
At the molecular level, 14-3-3σ was though to protect cancer cells against genotoxic treatments by regulating cell cycle progression and survival pathways (9,10). Somatic 14-3-3σ knockout led to mitotic catastrophe upon DNA damages (9). Following DNA damage, 14-3-3σ-sufficient cells are able to arrest in G2/M phase and survive while 14-3-3σ-deficient cells continue to progress through cell cycles and to cell death (11). It, thus, has been postulated that 14-3-3σ contributes to survival and DNA-damage resistance by arresting cells in G2/M phase (12). However, the molecular mechanism of 14-3-3σ action in this process remains unknown.
Radiation therapy is an important component of cancer treatments. IR impairs the survival of cancer cells mainly by causing double strand breaks (DSBs) in the DNA backbone. However, increased repair of DSBs would lead to IR resistance. Although DSBs are repaired by both homologous recombination (HR) and non-homologous end-joining (NHEJ) mechanisms, the latter directly ligates two DSB ends without the need of the template and, thus it functions throughout all phases of the cell cycle and is the predominant DSB repair pathway in mammalian cells while HR occurs mainly in mid-late S phases (13,14).
In this study, we tested the hypothesis that 14-3-3σ contributes to radiation resistance by up-regulating NHEJ repair while arresting cells in G2/M phase. We found that 14-3-3σ ectopic overexpression increased while its knockdown reduced IR resistance and NHEJ repair activity. We also showed that 14-3-3σ-induced increases in NHEJ repair activity was via up-regulating Chk2 and by increasing PARP1 expression via up-regulating its transcription and inhibiting caspase-mediated degradation of PARP1 protein. Furthermore, 14-3-3σ up-regulation of PARP1 increased DNA-PKcs recruitment to chromatin DNA, facilitating NHEJ repair of DSBs. These findings revealed an important molecular mechanism how 14-3-3σ contributes to chemo and radiation resistance and to poor prognosis of human cancers.
MATERIALS AND METHODS
Materials
Antibodies against 14-3-3σ, Chk1, Chk2 and DNA-PKcs were from EMD Millipore (Billerica, MA). The γ-H2AX antibody was from Enzo Biochem (New York, NY). 14-3-3σ siRNA pool and antibodies against Ku70 and Ku80 were from Santa Cruz Biotechnology (Dallas, TX). PARP1 and histone H3 antibodies were from Cell Signaling Technology (Danvers, MA). Adriamycin, mitoxantrone, and antibodies against GAPDH, β-Actin and α-Tubulin were from Sigma-Aldrich (St. Louis, MO). G418, pcDNA3.1(+) plasmid, and SYBR Green polymerase chain reaction (PCR) master mix were from Applied Biosystems (Grand Island, NY). The iScript cDNA synthesis kit, metafectene Pro transfection reagent, and gemcitabine were from Bio-Rad (Hercules, CA), Biontex (München, Germany), and Besse Medical (West Chester, OH), respectively. All other chemicals were purchased from Sigma-Aldrich or Fisher Scientific (Waltham, MA).
Cell lines and transfections
BxPC-3 cells with stable 14-3-3σ knockdown or harboring scrambled shRNA control were generated in a previous study (2) and cultured in RPMI1640 supplemented with 10% fetal bovine serum. MiaPaCa-2 cells with stable over-expression of ectopic 14-3-3σ and the control cells harboring vector control were also generated in a previous study (2) and cultured in (DMEM) supplemented with 10% fetal bovine serum and 2.5% horse serum. All cultures were at 37°C with 5% CO2. The cell lines were authenticated by analysis of tandem repeat sequences on 09/17/2013.
For transient knockdown, BxPC-3 cells were plated in 6-well plates at 2.0×105 cells/well and cultured overnight in complete media. About 60 pmol siRNAs targeting PARP1, Chk2, or control scrambled siRNA were diluted in serum-free RPMI1640 media and transiently transfected into cells using Metafectene Pro as previously described (15). For transient knockdown of 14-3-3σ, pSilencer-σ or scrambled shRNA construct engineered in a previous study (11) was transfected into BxPC-3 cells using Metafectene Pro transfection reagent as previously described (15). The sequences of siRNAs are shown in Supplemental Table S1.
Cytotoxicity assay
Cytotoxicity was determined using MTT and colony formation assays as previously described (2,16). MTT assay was used to determine cytotoxicity of anticancer drugs. Briefly, cells were seeded in 96-well plate at 3000 cells/well and cultured for 24 hrs followed by treatment with anticancer drugs and cultured continuously for 72 hrs at 37°C. Then, MTT (5 mg/mL) was added to the culture and incubated for another 4 hrs. The culture medium was then aspirated followed by addition of DMSO and absorption was determined using a 96-well plate reader. For colony formation assay, 100–200 cells/well were seeded in 6-well plate and cultured for 24 hrs before IR treatment. The cells were then cultured for 10 days before fixation and staining with crystal violent (0.005% in 20% methanol). The colonies were counted manually. Both the fitted sigmoidal dose response curve and IC50 were calculated using Graph Pad Prism Program.
Neutral comet assay
Neutral comet assay was performed using a kit from Trevigen (Gaithersburg, MD) according to the manufacturer’s instructions. Briefly, cells were harvested at different times following IR and single cell suspensions in PBS at 1×105 cells/ml were mixed with low temperature-melting agarose at 37°C and layered onto comet slides. The agarose on slides was allowed to solidify for 1 hour at 4°C, incubated in a pre-chilled lysis solution for 30 min at 4°C in the dark, and subjected to electrophoresis. The slides were then immersed twice in distilled water for 10 min, once in 70% ethanol for 5 min, dried completely at room temperature and stained with SYBR Green I (Trevigen). Comets were viewed and recorded using a Zeiss Axiovert 25 fluorescent microscope equipped with a camera and analyzed with CometScore V1.5 (TriTek, Sumerduck, VA). The Olive Tail Moment was determined by scoring at least 100 cells in each sample (17).
Cell lysate preparation, Western blot, and immunoprecipitation
Cell lysate preparation, Western blot analyses, and immunoprecipitation were performed as previously described (4,18). Briefly, cultured cells were harvested, washed with chilled PBS 3 times, and lysed in TNN lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5% Nonidet P-40, 50 mM NaF, 1 mM Na3VO4, 1 mM DTT, 0.1% SDS, and 2 mM PMSF) for 30 minutes on ice with agitation. The cell lysates were then sonicated briefly followed by centrifugation (16,000g at 4°C) for 15 minutes and the supernatant was collected with protein concentrations determined using Bradford assay. The cell lysates were then subjected to immunoprecipitation and Western blot analyses.
Immunofluorescence
Immunofluorescence imaging was performed as previously described (19). Briefly, 3×105 cells/well were cultured on coverslips in 24-well plate overnight followed by IR treatment. At different times after IR, cells were washed with chilled PBS, fixed with 50:50 (V:V) acetone/methanol for 10 min, blocked with 1% bovine serum albumin in PBS for 30 min, and incubated with anti-γ-H2AX antibody for 1 hr followed by washing and incubation with FITC-conjugated goat anti-mouse IgG for 30 min all at room temperature. The coverslips were counter-stained with DAPI before mounting on slides and viewing with an Olympus 2 confocal microscope.
Host cell reactivation-based NHEJ and HR assays
The host cell reactivation-based NHEJ assay was performed as previously described with modifications (20). Briefly, 5×104 cells/well were seeded in 24-well plates and cultured for 24 hrs before transfection with either linearized or intact pGL3-Luc plasmid with firefly luciferase reporter gene (400 ng/well) using Metafectene Pro. A pRL-TK (Promega) plasmid with renilla luciferase reporter gene was co-transfected (20 ng/well) to control transfection efficiency. Linearization of pGL3-Luc was achieved by HindIII digestion between CMV promoter and the luciferase gene and verified using agarose gel electrophoresis. Cells were harvested at 6 hrs after transfection and both the firefly and renilla luciferase activities were determined using a Dual-Luciferase Reporter Assay System on a luminometer. Relative NHEJ activity = F. Luclinear×100%/ F. Lucintact.
Host cell reactivation-based HR assay was performed using a kit following manufacturer’s instructions (Norgen Biotek Corp). Briefly, two different deletion mutants (dl-1 and dl-2) were engineered form the same parental reporter plasmid and transfected into cells individually or in combination. At 24 hrs after transfection, the reporter plasmids were recovered from these cells and used as template for real-time PCR analysis of production of wild type plasmid from HR reaction.
Real-time RT-PCR
Firstly, total RNAs were extracted from 1×107 fresh cells using the RNeasy Mini Kit (Qiagen) following the manufacturer's protocol. All RNA samples were treated with RNase-free DNase I (Promega) and quantified using a Nanodrop spectrophotometer (Fisher Scientific). For reverse transcription, 1 µg DNA-free RNA was used for cDNA synthesis using iScript cDNA synthesis kit (Bio-Rad) following the manufacturer's protocol exactly without any modification.
Real-time RT-PCR was performed using SYBR Green PCR master mix as previously described (Applied Biosystems) (21). Relative mRNA levels were quantified using the comparative Ct method by normalizing to the endogenous reference gene GAPDH and given by 2ΔΔCt where ΔCt=Ct(target gene)−Ct(GAPDH) and ΔΔCt=ΔCt(test sample)− ΔCt(control sample). Three independent experiments were performed with each in triplicate determinations. Supplemental Table S2 shows the primer sequences, which have been used to blast nucleotide database and no gene other than the targeted ones were found.
Subcellular fractionation
Subcellular fractionation was performed as previously described (22,23). Briefly, cells were harvested, washed with PBS, and lysed in buffer A (10 mM HEPES, pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 1 mM DTT and 2 mM PMSF) containing 0.1% Triton X-100 on ice for 10 min followed by centrifugation (1,300×g for 5 min at 4°C) to collect crude nuclei fraction (pellet). The supernatant was subjected to high-speed centrifugation (20,000×g for 5 min at 4°C) to collect the supernatant (cytosolic fraction). The low-speed pellet was subjected to lysis in buffer B (3 mM EDTA, 0.2 mM EGTA, 1 mM DTT and 2 mM PMSF) on ice for 30 min followed by centrifugation (1,700×g for 5 min at 4°C) to separate soluble nuclear (supernatant) and chromatin-bound (pellet) fractions.
Pulse labeling
Pulse labeling was performed as previously described (24). Briefly, 6×105 cells were seeded in 10-cm dish and cultured for 72 hrs. The cells were washed twice with PBS and once with RPMI1640 lacking methionine followed by incubation for 2 hrs in the same medium supplemented with 75 µCi/ml [35S]methionine. The labeled cells were then washed with PBS and harvested for lysate preparation and immunoprecipitation as described above. The immunoprecipitated proteins were separated by SDS-PAGE and signals of newly synthesized proteins were captured using X-ray films.
Cell cycle analysis
Cell cycle analysis was performed as previously described (25). Briefly, 2×105 cells were seeded in 6-well plates followed by IR treatment. Cells were collected at different times, fixed in cold 75% ethanol and stained with 50 µg/ml PI followed by analysis with FACS.
RESULTS
14-3-3σ contributes to cellular resistance to DNA-damaging drugs and IR
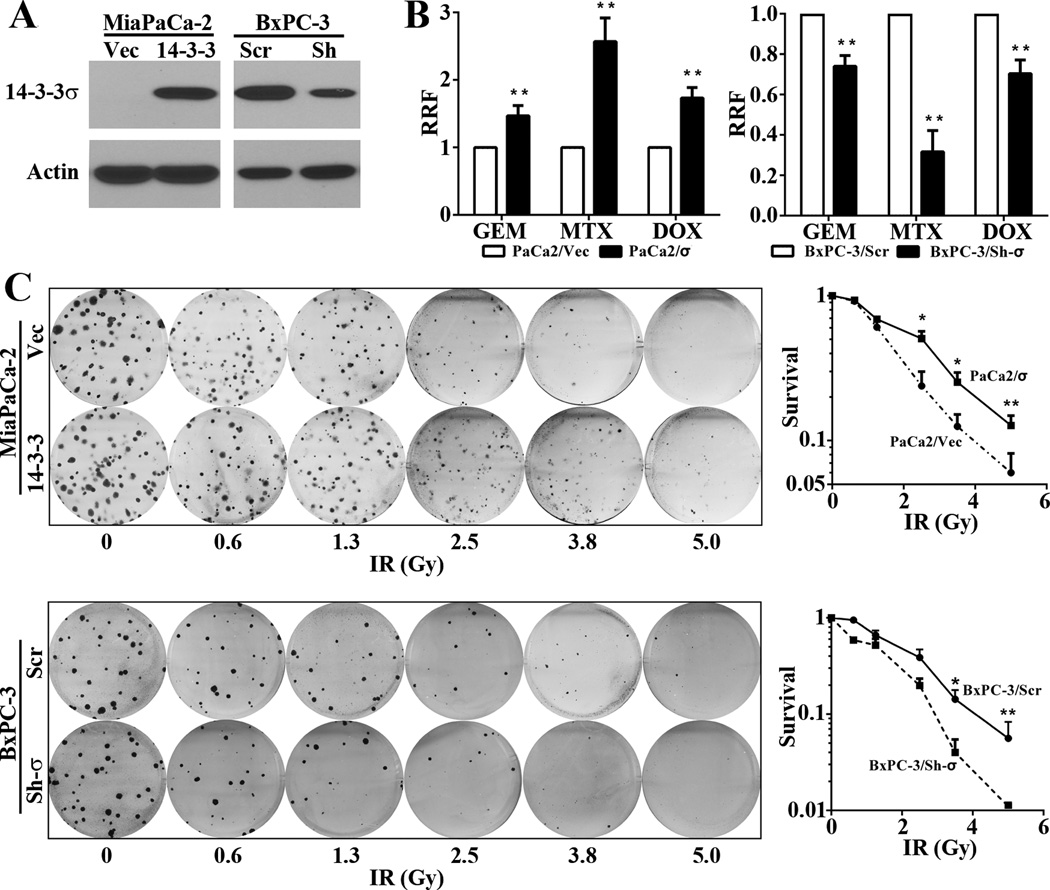
To determine the role of 14-3-3σ in cellular response to DNA-damaging drugs and IR, we took advantage of the previously established stable cell lines MiaPaCa-2/σ with over-expression of ectopic 14-3-3σ and BxPC-3/Sh-σ with knockdown of endogenous 14-3-3σ (Figure 1A). The parental BxPC-3 cells express high levels of endogenous 14-3-3σ, equivalent to that in human pancreatic cancer tissues (2) while MiaPaCa-2 cells lack endogenous 14-3-3σ (2), which are good models for knockdown and over-expression studies, respectively. Firstly, we subjected these cells to MTT survival assay following treatments with gemcitabine, mitoxantrone, or doxorubicin. As shown in Figure 1B, MiaPaCa-2/σ cells are significantly more resistant than the control vector-transfected MiaPaCa-2/Vec cells. Consistently, the stable BxPC-3/Sh-σ cells are significantly more sensitive to these drugs than the control BxPC-3/Scr cells. We next tested their response to IR treatments using colony formation survival assay. As shown in Figure 1C and supplemental Figure S1, higher expression level of 14-3-3σ also significantly increased cellular tolerance of IR.
Figure 1. 14-3-3σ contributes to cellular resistance to DNA-damaging drugs and IR.
(A) Western blot analysis of 14-3-3σ in stable MiaPaCa-2/Vec and MiaPaCa-2/σ and in BxPC-3/Scr and BxPC-3/Sh-σ cells. (B) MTT assay of survival against different therapeutic drugs. GEM, gemcitabine; MTX, mitoxantrone; DOX, doxorubicin. Relative resistance factor (REF)=IC50(MiaPaCa-2/σ)/IC50(MiaPaCa-2/Vec) or IC50(BxPC-3/Sh-σ)/IC50(BxPC-3/Scr). (C) Colony formation assay of survival against IR. (*p<0.05; **p<0.01; n=3).
We also tested another cell line, H1299 that expresses little endogenous 14-3-3σ (26), by stably over-expressing ectopic 14-3-3σ (H1299/σ) (Figure S2A). As shown in Figure S2B, H1299/σ was more resistant to IR than the control H1299/Vec cells. These studies suggest that 14-3-3σ contributes to cellular resistance to DNA-damaging drugs and IR in different human cancer cells.
14-3-3σ modulates γ-H2AX clearance and DSB accumulation
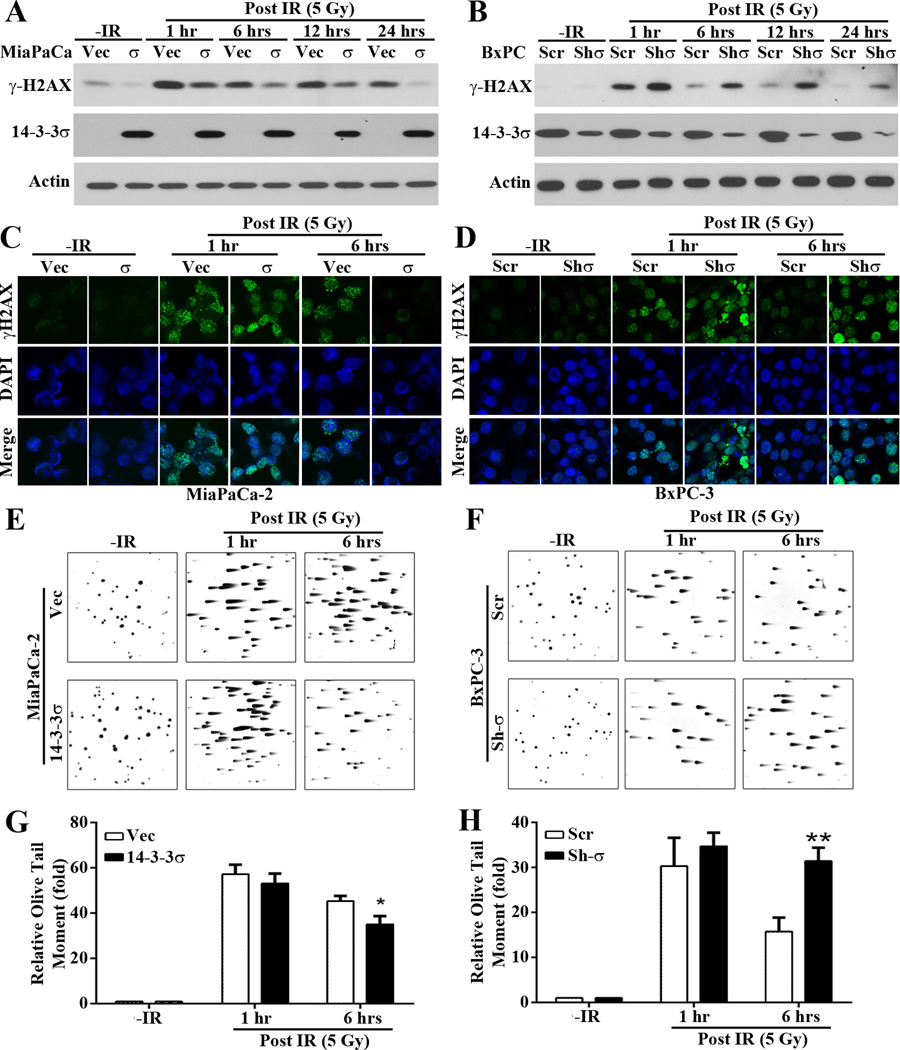
To determine if 14-3-3σ-mediated DNA-damage resistance is possibly due to its regulation of DNA repair, we first examined the accumulation of IR-induced DNA damages at different times following IR in these cells by examining γ-H2AX, a marker of DNA damages. As shown in Figure 2A, low basal level of γ-H2AX was detected similarly in both MiaPaCa-2/σ and the control MiaPaCa-2/Vec cells without IR treatment. At 1 hr post IR, γ-H2AX level dramatically increased in both cells. However, MiaPaCa-2/σ cells have much lower level of γ-H2AX than the control MiaPaCa-2/Vec cells, indicating that MiaPaCa-2/Vec cells have accumulated more DNA damages than the MiaPaCa-2/σ cells. While the γ-H2AX level continued to decrease with time in MiaPaCa-2/σ cells and reached the basal level at 24 hrs post IR, it remained at high levels in the MiaPaCa-2/Vec control cells at 24 hrs after IR. Similarly, the basal level of γ-H2AX was very low in both BxPC-3/Sh-σ and its control BxPC-3/Scr cells and the γ-H2AX level dramatically increased in both cells at 1 hr post IR (Figure 2B). However, BxPC-3/Sh-σ cells have more γ-H2AX than BxPC-3/Scr cells and it dissipated much slower in BxPC-3/Sh-σ than in BxPC-3/Scr cells. Consistently, we also found that at 1 hr post IR, both H1299/σ and H1299/Vec cells have much higher level of γ-H2AX than the control un-irradiated cells (Figure S3). However, at 24 hrs after IR, the γ-H2AX level returned to the basal level in H1299/σ cells while it remained high in the control H1299/Vec cells.
Figure 2. 14-3-3σ modulates IR-induced γ-H2AX and DSB accumulation.
(A–D) Western blot (A–B) and immuno-fluorescence staining (C–D) of γ-H2AX following IR treatment in in stable MiaPaCa-2/Vec and MiaPaCa-2/σ and in BxPC-3/Scr and BxPC-3/Sh-σ cells. (E-H) Comet assay of DSB accumulation in in stable MiaPaCa-2/Vec and MiaPaCa-2/σ (E, G) and in BxPC-3/Scr and BxPC-3/Sh-σ (F, H) cells. Olive Tail Moment=(tail mean–head mean)×(% of DNA in the tail) from 100 cells in each sample of 3 independent experiments scored by CometScore 1.5. (*p < 0.05; **p< 0.01).
To confirm above findings, we performed immunofluorescence staining of γ-H2AX following IR. As shown in Figure 2C–D, little staining was observed in the control untreated cells. At 1 hr post IR, punctate nuclear staining of γ-H2AX dramatically increased in all cells. However, at 6 hrs post IR the staining of H2AX dissipated in MiaPaCa-2/σ and BxPC-3/Vec cells, which express high levels of 14-3-3σ, while the staining remained high in MiaPaCa-2/Vec and BxPC-3/Sh-σ cells, which express little 14-3-3σ.
To further determine the role of 14-3-3σ in removing DNA damages, we performed neutral comet assay following IR. As shown in Figure 2E–H, similar levels of DNA damages were generated by IR in all cells at 1 hr post IR, indicated by Olive tail moment. However, at 6 hrs after IR MiaPaCa-2/σ cells have significantly less Olive tail moment than the control MiaPaCa-2/Vec cells while BxPC-3/Sh-σ cells have significantly more Olive tail moment than the control BxPC-3/Scr cells. Together, these findings suggest that 14-3-3σ may increase repair of IR-induced DSBs.
14-3-3σ promotes NHEJ repair of DSBs
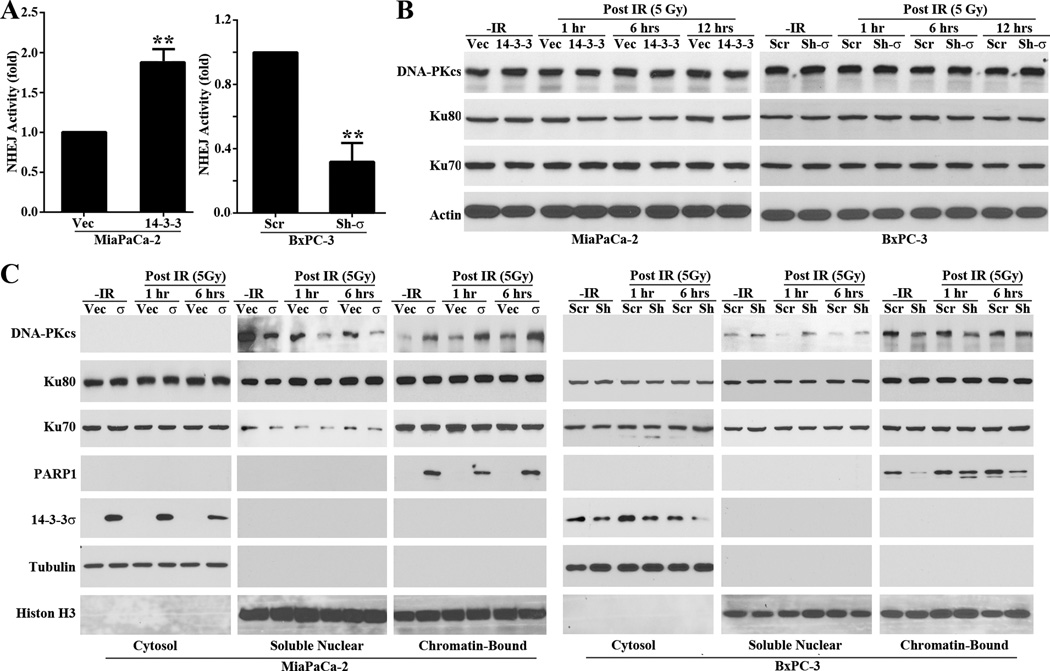
To assess directly if 14-3-3σ increases repair of IR-induced DSBs, we first examined the effect of 14-3-3σ on NHEJ activity by performing a host-cell-reactivation-based NHEJ assay in these cells. As shown in Figure 3A, cells expressing high levels of ectopic (MiaPaCa-2/σ) or endogenous (BxPC-3/Scr) 14-3-3σ had 2–3 fold higher NHEJ activity than the cells expressing little or no 14-3-3σ (BxPC-3/Sh-σ and MiaPaCa-2/Vec). Similarly, NHEJ repair activity was also significantly increased in H1299/σ cells compared with the control H1299/Vec cell (Figure S4A). However, 14-3-3σ may not affect HR repair as determined using host-cell-reactivation-based HR assay of the BxPC-3/Sh-σ and BxPC-3/Scr cell pair (Figure S4B). Interestingly, HR repair was significantly increased in H1299/σ cells compared with the control H1299/Vec cell (Figure S4C). Thus, 14-3-3σ positively regulates the NHEJ repair of DSBs and also possibly HR but in a cell line-dependent manner.
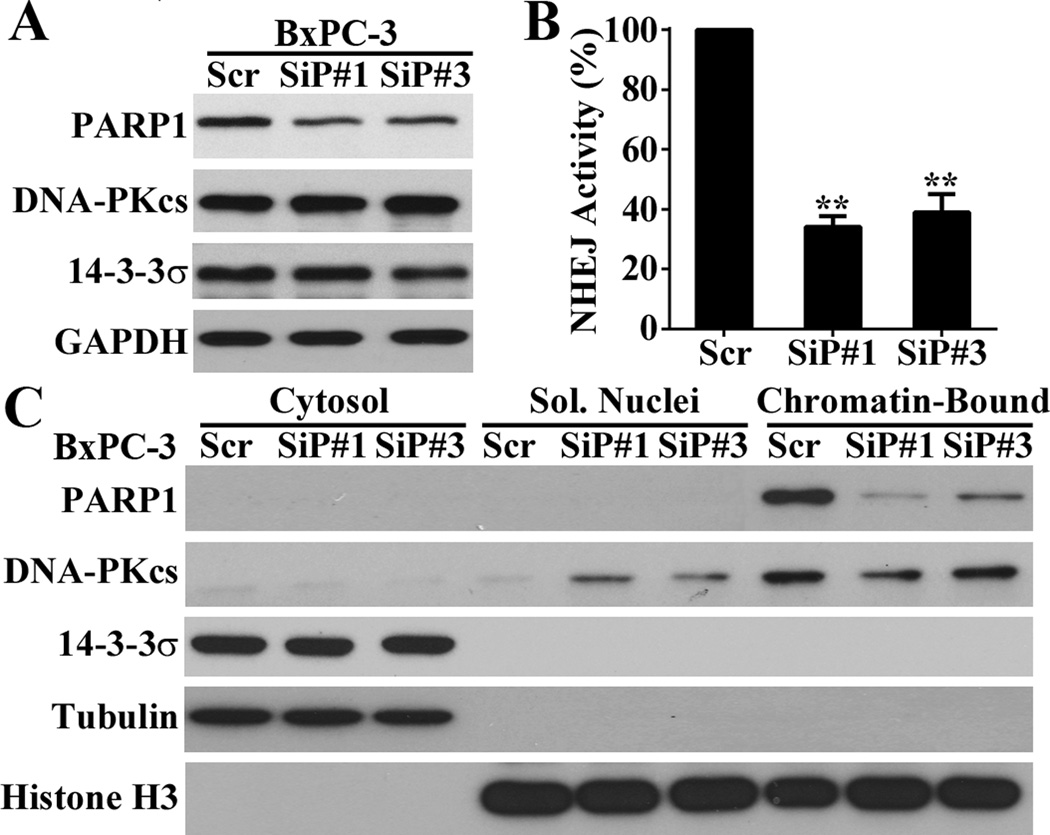
Figure 3. 14-3-3σ promotes NHEJ repair of DSBs via DNA-PKcs.
(A) Host cell reactivation assay of NHEJ activity. (**p<0.01; n=3). (B–C) Western blot analysis of Ku70, Ku80, DNA-PKcs expression (B) and recruitment to damaged chromatin DNA (C) following IR treatments. Tubulin and histone were used as cytosolic and nuclear fraction marker, respectively.
14-3-3σ promotes recruitment of DNA-PKcs
Because 14-3-3σ regulation of HR repair appears to be cell line-dependent, we decided to focus on NHEJ in the following studies. To understand the mechanism of 14-3-3σ action in regulating NHEJ, we first tested its effect on the expression of genes in the DNA-PK complex, Ku70, Ku80, and DNA-PKcs, which plays an important role in NHEJ. As shown in Figure 3B, 14-3-3σ had no discernible effect on the total level of Ku70, Ku80 and DNA-PKcs. Next, we determined the effect of 14-3-3σ on recruitment of the DNA-PK complex onto damaged chromatin following IR. For this purpose, we first isolated cytosolic, soluble nuclear and chromatin-bound proteins followed by Western blot analysis of Ku70, Ku80, and DNA-PKcs in these fractions. As shown in Figure 3C, the cells expressing high levels of 14-3-3σ (MiaPaCa-2/σ and BxPC-3/Scr) enhanced both the basal and IR-induced recruitment of DNA-PKcs to chromatin compared with the cells expressing little 14-3-3σ (MiaPaCa-2/Vec and BxPC-3/Sh-σ). However, the recruitment of Ku70 and Ku80 was not affected by altering 14-3-3σ expression. Thus, 14-3-3σ regulates NHEJ repair of DSBs possibly by promoting recruitment of DNA-PKcs to the damaged DNAs.
PARP1 mediates 14-3-3σ regulation of NHEJ
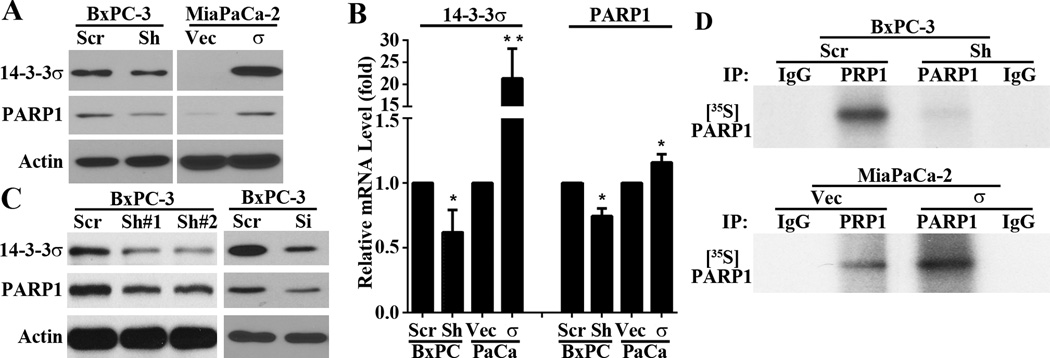
It has been reported that PARP1 participates in the NHEJ repair pathway (27) by PARylating and recruiting DNA-PK proteins (28). To test the possible role of PARP1 in mediating 14-3-3σ regulation of DNA-PKcs recruitment, we first determined if 14-3-3σ regulates PARP1 expression by testing the stable BxPC-3/Sh-σ and MiaPaCa-2/σ cells in comparison with their respective control cells using Western blot and real-time RT-PCR analysis. As shown in Figure 4A–B, BxPC-3/Sh-σ had reduced while MiaPaCa-2/σ had increased levels of PARP1 protein and mRNA compared with their respective control cells. Transient 14-3-3σ knockdown in BxPC-3 cells using two different shRNAs or a siRNA pool as describe previously (4) also successfully reduced PARP1 expression (Figure 4C).
Figure 4. 14-3-3σ regulation of PARP1 expression.
(A–C) Western blot (A and C) and real time RT-PCR (B) analyses of PARP1 expression in MiaPaCa-2 and BxPC-3 cells with altered 14-3-3σ expression. (**p<0.01; n=3). (D) PARP1 protein synthesis as determined using pulse-labeling with [35S]methionine, immunoprecipitation, and SDS-PAGE analysis.
The altered transcription of PARP1 by 14-3-3σ likely resulted in altered synthesis of PARP1 and change in PARP1 protein level. To test this possibility, we performed a pulse-labeling experiment in combination with immunoprecipitation of PARP1 followed by separation on SDS-PAGE and autoradiography. As shown in Figure 4D, indeed PARP1 protein synthesis was dramatically decreased in BxPC-3/Sh-σ cells and dramatically increased in MiaPaCa-2/σ cells compared with their respective control cells. Thus, 14-3-3σ regulates PARP1 expression, which in turn contributes to NHEJ repair of DSBs. However, it is noteworthy that the effect of 14-3-3σ on PARP1 appears to be more dramatic as determined using pulse labeling (Figure 4D) than Western blot (Figure 4A). While Western blot determines the steady state level of the protein, the pulse labeling shows the transient level of proteins that are labeled by [35S]methionine, which may amplify the difference between the control and 14-3-3σ-altered cells.
Next, we determined the effect of PARP1 knockdown on NHEJ. As shown in Figure 5A, PARP1 knockdown in BxPC-3 cells using two different siRNAs had not effect on the expression of DNA-PKcs or 14-3-3σ. However, the knockdown significantly reduced NHEJ activity (Figure 5B) and recruitment of DNA-PKcs to chromatin (Figure 5C). Consistently, PARP1 knockdown in MiaPaCa-2/σ cells with overexpression of ectopic 14-3-3σ using the same siRNA had not effect on the expression of DNA-PKcs or 14-3-3σ (Figure S5A), but reduced DNA-PKcs recruitment in these cells (Figure S5B). Thus, it is possible that PARP1 mediates 14-3-3σ regulation of DNA-PKcs recruitment.
Figure 5. Role of PARP1 in DNA-PKcs recruitment and NHEJ activity.
(A) Western blot analysis of PARP1 effect on DNA-PKcs expression. (B) Host cell reactivation assay of PARP1 effect on NHEJ activity. (*p< 0.05; **p< 0.01; n=3). (C) Western blot analysis of PARP1 effect on DNA-PKcs recruitment.
14-3-3σ inhibits IR-induced apoptosis
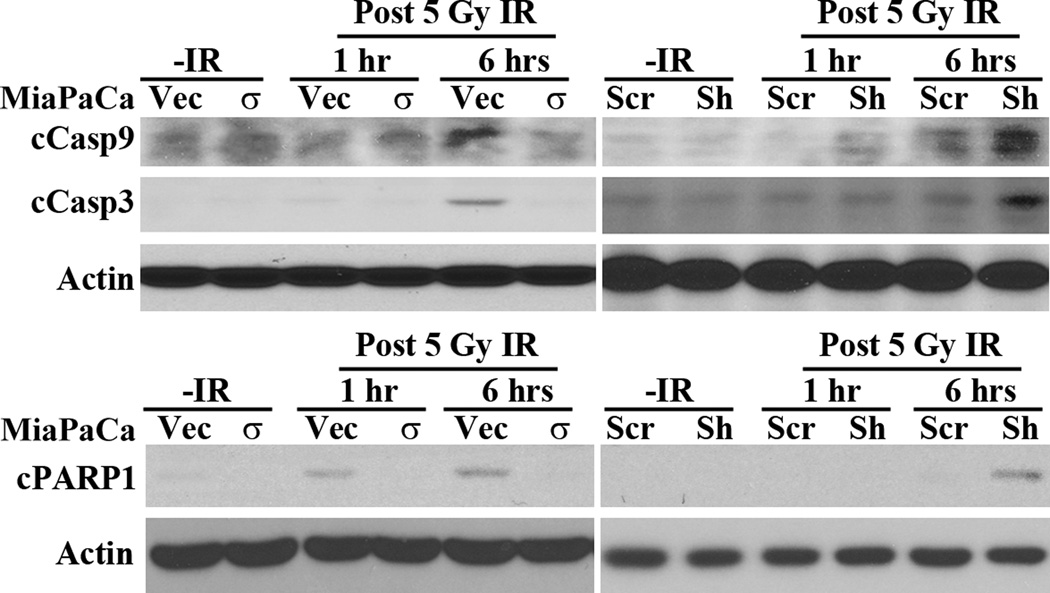
The above studies showed that 14-3-3σ might up-regulate NHEJ repair of IR-induced DSB by increasing PARP1 expression and DNA-PKcs recruitment. The increased DNA repair likely contributes to the increased cellular survival and resistance to IR. To test this possibility, we determined the effect of 14-3-3σ on IR-induced apoptosis. As shown in Figure 6A, activation and cleavage of caspase 9 and executioner caspase 3 induced by IR is dramatically inhibited in MiaPaCa-2/σ cells compared with the control MiaPaCa-2/Vec cells. Consistently, their activation and cleavage following IR are increased in BxPC-3/Sh-σ cells compared with the control BxPC-3/Scr cells. We also found that PARP1 cleavage by caspases, an indicator of apoptosis, was inhibited in MiaPaCa-2/σ and increased in BxPC-3/Sh-σ cells following IR. Thus, we conclude that 14-3-3σ protects cancer cells against IR-induced apoptosis by increasing the repair of IR-induced DNA damages.
Figure 6. 14-3-3σ inhibits IR-induced apoptosis.
Western blot analysis of caspase activation/cleavage (cCasp) and PARP1 cleavage (cPARP1) in MiaPaCa-2/Vec and MiaPaCa-2/σ cells prior to or after IR treatments.
14-3-3σ regulation of Chk2 expression in G2/M arrest for NHEJ repair
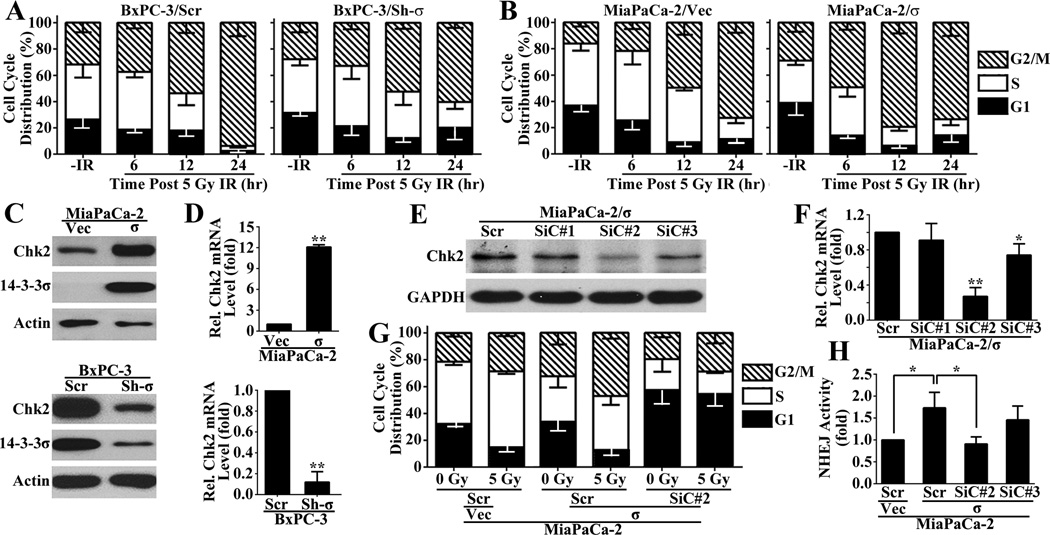
Previously, it has been suggested that 14-3-3σ plays an important role in regulating cell cycle and help arrest cells in G2/M phase following DNA damage to allow efficient repair of the DNA damages and to prevent mitotic catastrophe and apoptosis (29,30). Others and we have also shown that 14-3-3σ helps arrest cells in G2/M phase following DNA-damages (9,11,25). To determine how 14-3-3σ regulates cell cycle progression, we first analyzed cell cycle distribution of MiaPaCa-2/σ and BxPC-3/Sh-σ cells in comparison with their respective control cells following IR. As shown in Figure 7A–B, the untreated BxPC-3/Scr and MiaPaCa-2/σ cells with high level of 14-3-3σ had slightly more enrichment of G2/M population than MiaPaCa-2/Vec and BxPC-3/ Sh-σ cells with little 14-3-3σ, respectively. However, 14-3-3σ knockdown in BxPC-3 cells (BxPC-3/Sh-σ) caused remarkably less accumulation in G2/M population at 24 hrs post IR compared with the control BxPC-3/Scr cells (Figure 7A). For MiaPaCa-2/σ cells compared with the control MiaPaCa-2/Vec cells, the difference in G2/M population can be clearly observed at 6 and 12 hrs post IR treatment (Figure 7B).
Figure 7. 14-3-3σ regulation of G2/M arrest and Chk2 expression.
(A–B) Cell cycle distribution of MiaPaCa-2/σ and BxPC-3/Sh-σ in comparison with their respective control cells following IR. (C–D) Western blot (C) and real time RT-PCR (D) analysis of Chk2 expression in MiaPaCa-2/σ and BxPC-3/Sh-σ in comparison with their respective control cells. (E–F) Western blot (E) and real-time RT-PCR (F) analyses of Chk2 knockdown by siRNAs. (G) FACS analysis of Chk2 knockdown on cell cycle distribution following IR in MiaPaCa-2/σ cells. (H) Host cell reactivation assay of the effect of Chk2 knockdown on 14-3-3σ-induced NHEJ activity increase. (*p<0.05; **p<0.01; n=3).
To investigate how 14-3-3σ regulates G2/M arrest in response to IR, we determined the potential effect of 14-3-3σ on the expression of Chk2, which regulates several downstream effector molecules that are known ligand substrates of 14-3-3 proteins (12) and important for cell cycle progression following DNA damage (31). As shown in Figure 7C, Chk2 expression is up-regulated in MiaPaCa-2/σ and down-regulated in BxPC-3/Sh-σ cells compared with their respective control MiaPaCa-2/Vec and BxPC-3/Scr cells as determined using Western blot. Real-time RT-PCR analysis showed that the Chk2 mRNA is also increased in MiaPaCa-2/σ cells and decreased in BxPC-3/Sh-σ cells (Figure 7D). Similar findings were also observed with H1299/σ and H1299/Vec cell pair (Figure S6). Thus, 14-3-3σ likely up-regulates Chk2 expression in addition to up-regulating PARP1 expression.
To determine if Chk2 potentially mediates 14-3-3σ function in G2/M arrest following DNA damage, we first knocked known Chk2 expression in MiaPaCa-2/σ and MiaPaCa-2/Vec cells using three different siRNAs followed by treatment with IR before determining cell cycle distribution. As shown in Figure 7E–F, siRNA1 had no effect on Chk2 expression while siRNA#2 and siRNA#3 significantly knocked down Chk2 expression with different efficiency with siRNA#2 most effective, which was chosen for further study. As shown in Figure 7G, the scrambled siRNA-transfected MiaPaCa-2/σ cells have more G2/M population than MiaPaCa-2/Vec cells with or without IR treatment, consistent with the data shown in Figure 7B. However, Chk2 knockdown in MiaPaCa-2/σ cells eliminated the 14-3-3σ-induced increase in G2/M population. Thus, Chk2 may mediate 14-3-3σ-induced G2/M arrest following DNA damages.
To determine if 14-3-3σ-induced G2/M arrest enhances NHEJ repair of IR-induced DSB, we tested the effect of eliminating G2/M arrest by Chk2 knockdown on 14-3-3σ-induced NHEJ activity using the HCR assay as described above. As shown in Figure 7H, Chk2 knockdown in MiaPaCa-2/σ cells by siRNA#2 eliminated 14-3-3σ-induced NHEJ activity. It is also noteworthy that the siRNA#3 only slightly attenuated the 14-3-3σ-induced NHEJ activity, consistent with its lower activity in knocking down Chk2 expression (Figure 7E–F). Thus, we conclude that 14-3-3σ up-regulates Chk2 expression to arrest cells at G2/M phase following DNA damage, which may contribute to 14-3-3σ-induced NHEJ repair of IR-induced DSBs.
14-3-3σ regulation of Chk2 may be via PARP1
The above studies show that 14-3-3σ regulates both Chk2 and PARP1 expression. To determine if these regulations are independent events or have a linear relationship, we tested Chk2 and PARP1 expressions following their knockdowns. As shown in Figure S7A, PARP1 knockdown in BxPC-3 cells led to reduced Chk2 expression. However, Chk2 knockdown had no effect on PARP1 expression (Figure S7B). Thus, 14-3-3σ regulation of Chk2 expression may be via regulating PARP1.
DISCUSSION
Efficient DNA repair is considered as one of the major mechanisms that contribute to chemo and radiation resistance. While 14-3-3σ has been suggested to contribute to these resistances by arresting cells in G2/M phase for repair of DNA damages, this hypothesis has not been tested previously. Here, we showed that 14-3-3σ contributed IR resistance possibly by regulating cell cycle progression and NHEJ repair of IR-induced DSBs via regulating the expression of Chk2 and PARP1. These findings not only illustrate the molecular mechanisms of 14-3-3σ action in chemo and radiation resistance but also suggest that 14-3-3σ may be an upstream master regulator in chemo and radiation resistance and cancer cell survival. It is also noteworthy that 14-3-3σ has been shown to contribute to cisplatin resistance (1). Although the detailed mechanism for cisplatin resistance is unknown, it is tempting to speculate that 14-3-3σ may contribute to nucleotide excision repair of cisplatin-DNA adducts via NHEJ repair of DSB produced from these adducts.
The involvement of PARP1 in NHEJ repair (27) may be via PARylating and by increasing DNA-PKcs recruitment (28,32–34). Our finding that 14-3-3σ increase DNA-PKcs recruitment possibly via increasing PARP1 is consistent with these previous observations. Interestingly, 14-3-3σ has no effect on the expression or recruitment of Ku proteins. It remains unknown if Ku proteins are abundantly bound to chromatin and if DNA-PKcs may serve as a limiting factor for NHEJ repair of DSBs in BxPC-3 and MiaPaCa-2 cells. However, it has been reported previously that increased Ku70 expression promotes cellular resistance to IR in human cancer cells (35–37).
Previously, it has been suggested that 14-3-3σ binds to cdk2 and cdc25A, the downstream effectors of Chk2, and arrests them in cytoplasm, which results in G2/M arrest following DNA damages (38–40). Together with our finding that 14-3-3σ up-regulates Chk2 expression, we conclude that 14-3-3σ may act at multiple points in regulating cell cycle progression in response to DNA damages.
Although we have shown here that 14-3-3σ contributes to IR resistance by arresting cancer cells in G2/M phase and by increasing NHEJ repair via up-regulating Chk2 and PARP1 expression, it remains to be determined how 14-3-3σ regulates the expression of these genes. The finding that the mRNA levels of Chk2 and PARP1 were altered by 14-3-3σ suggests that 14-3-3σ may regulate the transcription of these genes. Although it has been shown that Chk2 expression is negatively regulated by p53 (41,42) and 14-3-3σ regulates p53 (43), p53 in BxPC-3 cells is inactive due to mutation and, thus, may not mediate 14-3-3σ regulation of Chk2 expression in these cells. However, PARP1 may mediate 14-3-3σ regulation of Chk2 expression. Recently, we have shown that the PARP1 gene promoter contains a NF-κB-binding element and that NF-κB negatively regulates PARP1 expression in another cell line, Panc-1 (20). It has also been shown that 14-3-3 proteins could bind to both p65 and IκBα and facilitate the nuclear export of IκBα-p65 complexes (44). Therefore, it is tempting to speculate that 14-3-3σ may bind to and arrest p65 in the cytoplasm and, thus, blocks p65 negative regulation of PARP1 promoter activity, resulting in increased PARP1 expression, which in turn regulates Chk2 expression.
It is of interest to note that PARP1 is also a substrate of apoptosis executioner caspases while participating in NHEJ repair of DSBs. In this study, we showed that 14-3-3σ up-regulates PARP1 transcription and prevents PARP1 cleavage due to apoptosis. The role of 14-3-3σ in resisting apoptosis has previously been shown and possibly via binding to and arresting pro-apoptotic proteins (10,45). The inhibition in PARP1 cleavage due to inhibition of apoptosis and the increased PARP1 expression by 14-3-3σ may form a feed-forward loop of DNA repairs for cancer cell survival against DNA-damaging drug and radiation treatments.
Although both NHEJ and HR mechanisms are responsible for repair of DSBs, it appears that the 14-3-3σ regulation of DSB repair via HR is cell-line-dependent. While it regulates HR in H1299 cells, it does not appear to do so in BxPC-3 cells, which are known to have wild type BRCA1 and BRCA2 (46). The cause of this cell line specificity in HR regulation is unknown. However, this finding is not surprising if 14-3-3σ regulation of DSB repair is via PARP-1 since the role of PARP-1 in regulating HR repair varies in different studies and PARP-1 may also controls the choice of the pathways for DSB repair (47). While PARP1 has been shown to activate HR repair (48), PARP-1 may also inhibit HR by preventing the HR machinery from recognizing and processing DNA lesions possibly by PARylating BRCA1 and inhibiting its activity, or by inhibiting the expression of BRCA1 and BRCA2 (49–54). Because NHEJ is associated with faster rate of repair compared to HR (55,56), consistent 14-3-3σ up-regulation of NHEJ repair in different cancer cells likely provides an efficient survival mechanism against DNA-damaging treatments to cancer cells.
Supplementary Material
Acknowledgments
Financial Support: This work was supported in part by a grant from NIH (R01 CA140582 to JTZ). Yifan Chen was supported in part by Sun Yet-sen University.
Footnotes
Conflict of Interest: The authors declare no conflict of interest.
REFERENCES
- 1.Li Z, Liu J-Y, Zhang J-T. 14-3-3σ, the double-edged sword of human cancers. American Journal of Translational Research. 2009;1(4):326–340. [PMC free article] [PubMed] [Google Scholar]
- 2.Li Z, Dong Z, Myer D, Yip-Schneider M, Liu J, Cui P, et al. Role of 14-3-3sigma in poor prognosis and in radiation and drug resistance of human pancreatic cancers. BMC Cancer. 2010;10:598. doi: 10.1186/1471-2407-10-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu Y, Liu H, Han B, Zhang JT. Identification of 14-3-3sigma as a contributor to drug resistance in human breast cancer cells using functional proteomic analysis. Cancer Res. 2006;66(6):3248–3255. doi: 10.1158/0008-5472.CAN-05-3801. [DOI] [PubMed] [Google Scholar]
- 4.Qin L, Dong Z, Zhang JT. Reversible epigenetic regulation of 14-3-3sigma expression in acquired gemcitabine resistance by uhrf1 and DNA methyltransferase 1. Mol Pharmacol. 2014;86(5):561–569. doi: 10.1124/mol.114.092544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simpson PT, Gale T, Reis-Filho JS, Jones C, Parry S, Steele D, et al. Distribution and significance of 14-3-3sigma, a novel myoepithelial marker, in normal, benign, and malignant breast tissue. J Pathol. 2004;202(3):274–285. doi: 10.1002/path.1530. [DOI] [PubMed] [Google Scholar]
- 6.Hustinx SR, Fukushima N, Zahurak ML, Riall TS, Maitra A, Brosens L, et al. Expression and Prognostic Significance of 14-3-3sigma and ERM Family Protein Expression in Periampullary Neoplasms. Cancer Biol Ther. 2005;4(5):596–601. doi: 10.4161/cbt.4.5.1748. [DOI] [PubMed] [Google Scholar]
- 7.Boudreau A, Tanner K, Wang D, Geyer FC, Reis-Filho JS, Bissell MJ. 14-3-3sigma stabilizes a complex of soluble actin and intermediate filament to enable breast tumor invasion. Proc Natl Acad Sci U S A. 2013;110(41):E3937–E3944. doi: 10.1073/pnas.1315022110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raychaudhuri K, Chaudhary N, Gurjar M, D'Souza R, Limzerwala J, Maddika S, et al. 14-3-3sigma Gene Loss Leads to Activation of the Epithelial to Mesenchymal Transition Due to the Stabilization of c-Jun Protein. J Biol Chem. 2016;291(31):16068–16081. doi: 10.1074/jbc.M116.723767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan TA, Hermeking H, Lengauer C, Kinzler KW, Vogelstein B. 14-3-3Sigma is required to prevent mitotic catastrophe after DNA damage. Nature. 1999;401(6753):616–620. doi: 10.1038/44188. [DOI] [PubMed] [Google Scholar]
- 10.Samuel T, Weber HO, Rauch P, Verdoodt B, Eppel JT, McShea A, et al. The G2/M regulator 14-3-3sigma prevents apoptosis through sequestration of Bax. J Biol Chem. 2001;276(48):45201–45206. doi: 10.1074/jbc.M106427200. [DOI] [PubMed] [Google Scholar]
- 11.Han B, Xie H, Chen Q, Zhang JT. Sensitizing hormone-refractory prostate cancer cells to drug treatment by targeting 14-3-3sigma. Mol Cancer Ther. 2006;5(4):903–912. doi: 10.1158/1535-7163.MCT-05-0393. [DOI] [PubMed] [Google Scholar]
- 12.Reinhardt HC, Yaffe MB. Phospho-Ser/Thr-binding domains: navigating the cell cycle and DNA damage response. Nat Rev Mol Cell Biol. 2013;14(9):563–580. doi: 10.1038/nrm3640. [DOI] [PubMed] [Google Scholar]
- 13.Burma S, Chen BP, Chen DJ. Role of non-homologous end joining (NHEJ) in maintaining genomic integrity. DNA Repair (Amst) 2006;5(9–10):1042–1048. doi: 10.1016/j.dnarep.2006.05.026. [DOI] [PubMed] [Google Scholar]
- 14.Karanam K, Kafri R, Loewer A, Lahav G. Quantitative live cell imaging reveals a gradual shift between DNA repair mechanisms and a maximal use of HR in mid S phase. Mol Cell. 2012;47(2):320–329. doi: 10.1016/j.molcel.2012.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu H, Liu Y, Zhang JT. A new mechanism of drug resistance in breast cancer cells: fatty acid synthase overexpression-mediated palmitate overproduction. Mol Cancer Ther. 2008;7(2):263–270. doi: 10.1158/1535-7163.MCT-07-0445. [DOI] [PubMed] [Google Scholar]
- 16.Qi J, Dong Z, Liu J, Peery RC, Zhang S, Liu JY, et al. Effective Targeting of the Survivin Dimerization Interface with Small-Molecule Inhibitors. Cancer Res. 2016;76(2):453–462. doi: 10.1158/0008-5472.CAN-15-1874. [DOI] [PubMed] [Google Scholar]
- 17.Olive PL, Wlodek D, Banath JP. DNA double-strand breaks measured in individual cells subjected to gel electrophoresis. Cancer research. 1991;51(17):4671–4676. [PubMed] [Google Scholar]
- 18.Dong Z, Liu Y, Zhang JT. Regulation of ribonucleotide reductase M2 expression by the upstream AUGs. Nucleic Acids Res. 2005;33(8):2715–2725. doi: 10.1093/nar/gki569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Q, Yang Y, Li L, Zhang JT. The amino terminus of the human multidrug resistance transporter ABCC1 has a U-shaped folding with a gating function. J Biol Chem. 2006;281(41):31152–31163. doi: 10.1074/jbc.M603529200. [DOI] [PubMed] [Google Scholar]
- 20.Wu X, Dong Z, Wang CJ, Barlow LJ, Fako V, Serrano MA, et al. FASN regulates cellular response to genotoxic treatments by increasing PARP-1 expression and DNA repair activity via NF-kappaB and SP1. Proc Natl Acad Sci U S A. 2016 doi: 10.1073/pnas.1609934113. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong Z, Liu Z, Cui P, Pincheira R, Yang Y, Liu J, et al. Role of eIF3a in regulating cell cycle progression. Exp Cell Res. 2009;315(11):1889–1894. doi: 10.1016/j.yexcr.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang W, Dong Z, Wang F, Peng H, Liu JY, Zhang JT. A Small Molecule Compound Targeting STAT3 DNA-Binding Domain Inhibits Cancer Cell Proliferation, Migration, and Invasion. ACS Chem Biol. 2014;9(5):1188–1196. doi: 10.1021/cb500071v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang W, Dong Z, Chen Y, Wang F, Wang CJ, Peng H, et al. Small-molecule inhibitors targeting the DNA-binding domain of STAT3 suppress tumor growth, metastasis and STAT3 target gene expression in vivo. Oncogene. 2016;35(6):783–792. doi: 10.1038/onc.2015.215. [DOI] [PubMed] [Google Scholar]
- 24.Dong Z, Zhang JT. EIF3 p170, a Mediator of Mimosine Effect on Protein Synthesis and Cell Cycle Progression. Mol Biol Cell. 2003;14(9):3942–3951. doi: 10.1091/mbc.E02-12-0784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Z, Peng H, Qin L, Qi J, Zuo X, Liu JY, et al. Determinants of 14-3-3sigma protein dimerization and function in drug and radiation resistance. J Biol Chem. 2013;288(44):31447–31457. doi: 10.1074/jbc.M113.467753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y, Chen Q, Zhang JT. Tumor suppressor gene 14-3-3sigma is down-regulated whereas the proto-oncogene translation elongation factor 1delta is up-regulated in non-small cell lung cancers as identified by proteomic profiling. J Proteome Res. 2004;3(4):728–735. doi: 10.1021/pr034127+. [DOI] [PubMed] [Google Scholar]
- 27.Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG. PARP inhibition: PARP1 and beyond. Nat Rev Cancer. 2010;10(4):293–301. doi: 10.1038/nrc2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.D'Silva I, Pelletier JD, Lagueux J, D'Amours D, Chaudhry MA, Weinfeld M, et al. Relative affinities of poly(ADP-ribose) polymerase and DNA-dependent protein kinase for DNA strand interruptions. Biochim Biophys Acta. 1999;1430(1):119–126. doi: 10.1016/s0167-4838(98)00278-7. [DOI] [PubMed] [Google Scholar]
- 29.Andersen SD, Keijzers G, Rampakakis E, Engels K, Luhn P, El-Shemerly M, et al. 14-3-3 checkpoint regulatory proteins interact specifically with DNA repair protein human exonuclease 1 (hEXO1) via a semi-conserved motif. DNA repair. 2012;11(3):267–277. doi: 10.1016/j.dnarep.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Su CH, Zhao R, Zhang F, Qu C, Chen B, Feng YH, et al. 14-3-3sigma exerts tumor-suppressor activity mediated by regulation of COP1 stability. Cancer research. 2011;71(3):884–894. doi: 10.1158/0008-5472.CAN-10-2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zannini L, Delia D, Buscemi G. CHK2 kinase in the DNA damage response and beyond. J Mol Cell Biol. 2014;6(6):442–457. doi: 10.1093/jmcb/mju045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruscetti T, Lehnert BE, Halbrook J, Le Trong H, Hoekstra MF, Chen DJ, et al. Stimulation of the DNA-dependent protein kinase by poly(ADP-ribose) polymerase. The Journal of biological chemistry. 1998;273(23):14461–14467. doi: 10.1074/jbc.273.23.14461. [DOI] [PubMed] [Google Scholar]
- 33.Spagnolo L, Barbeau J, Curtin NJ, Morris EP, Pearl LH. Visualization of a DNA-PK/PARP1 complex. Nucleic acids research. 2012;40(9):4168–4177. doi: 10.1093/nar/gkr1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chiruvella KK, Liang Z, Wilson TE. Repair of double-strand breaks by end joining. Cold Spring Harbor perspectives in biology. 2013;5(5):a012757. doi: 10.1101/cshperspect.a012757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen H, Schultz M, Kruh GD, Tew KD. Increased expression of DNA-dependent protein kinase confers resistance to adriamycin. Biochimica et biophysica acta. 1998;1381(2):131–138. doi: 10.1016/s0304-4165(98)00020-8. [DOI] [PubMed] [Google Scholar]
- 36.Shintani S, Mihara M, Li C, Nakahara Y, Hino S, Nakashiro K, et al. Up-regulation of DNA-dependent protein kinase correlates with radiation resistance in oral squamous cell carcinoma. Cancer science. 2003;94(10):894–900. doi: 10.1111/j.1349-7006.2003.tb01372.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luzhna L, Golubov A, Ilnytskyy S, Chekhun VF, Kovalchuk O. Molecular mechanisms of radiation resistance in doxorubicin-resistant breast adenocarcinoma cells. International journal of oncology. 2013;42(5):1692–1708. doi: 10.3892/ijo.2013.1845. [DOI] [PubMed] [Google Scholar]
- 38.Laronga C, Yang HY, Neal C, Lee MH. Association of the cyclin-dependent kinases and 14-3-3 sigma negatively regulates cell cycle progression. J Biol Chem. 2000;275(30):23106–23112. doi: 10.1074/jbc.M905616199. [DOI] [PubMed] [Google Scholar]
- 39.Falck J, Mailand N, Syljuasen RG, Bartek J, Lukas J. The ATM-Chk2-Cdc25A checkpoint pathway guards against radioresistant DNA synthesis. Nature. 2001;410(6830):842–847. doi: 10.1038/35071124. [DOI] [PubMed] [Google Scholar]
- 40.Vogel S, Herzinger T. The epithelium specific cell cycle regulator 14-3-3sigma is required for preventing entry into mitosis following ultraviolet B. Photodermatology, photoimmunology & photomedicine. 2013;29(6):300–310. doi: 10.1111/phpp.12071. [DOI] [PubMed] [Google Scholar]
- 41.Gottifredi V, Karni-Schmidt O, Shieh SS, Prives C. p53 down-regulates CHK1 through p21 and the retinoblastoma protein. Mol Cell Biol. 2001;21(4):1066–1076. doi: 10.1128/MCB.21.4.1066-1076.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matsui T, Katsuno Y, Inoue T, Fujita F, Joh T, Niida H, et al. Negative regulation of Chk2 expression by p53 is dependent on the CCAAT-binding transcription factor NF-Y. J Biol Chem. 2004;279(24):25093–25100. doi: 10.1074/jbc.M403232200. [DOI] [PubMed] [Google Scholar]
- 43.Yang HY, Wen YY, Lin YI, Pham L, Su CH, Yang H, et al. Roles for negative cell regulator 14-3-3sigma in control of MDM2 activities. Oncogene. 2007;26(52):7355–7362. doi: 10.1038/sj.onc.1210540. [DOI] [PubMed] [Google Scholar]
- 44.Aguilera C, Fernandez-Majada V, Ingles-Esteve J, Rodilla V, Bigas A, Espinosa L. Efficient nuclear export of p65-IkappaBalpha complexes requires 14-3-3 proteins. Journal of cell science. 2006;119(Pt 17):3695–3704. doi: 10.1242/jcs.03086. [DOI] [PubMed] [Google Scholar]
- 45.Subramanian RR, Masters SC, Zhang H, Fu H. Functional conservation of 14-3-3 isoforms in inhibiting bad-induced apoptosis. Exp Cell Res. 2001;271(1):142–151. doi: 10.1006/excr.2001.5376. [DOI] [PubMed] [Google Scholar]
- 46.Yuan J, Strack PR, Toniatti C, Pelletier M. A zinc finger nuclease assay to rapidly quantitate homologous recombination proficiency in human cell lines. Anal Biochem. 2013;434(1):96–98. doi: 10.1016/j.ab.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 47.Ko HL, Ren EC. Functional Aspects of PARP1 in DNA Repair and Transcription. Biomolecules. 2012;2(4):524–548. doi: 10.3390/biom2040524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bryant HE, Petermann E, Schultz N, Jemth AS, Loseva O, Issaeva N, et al. PARP is activated at stalled forks to mediate Mre11-dependent replication restart and recombination. EMBO J. 2009;28(17):2601–2615. doi: 10.1038/emboj.2009.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li D, Bi FF, Chen NN, Cao JM, Sun WP, Zhou YM, et al. A novel crosstalk between BRCA1 and poly (ADP-ribose) polymerase 1 in breast cancer. Cell Cycle. 2014;13(21):3442–3449. doi: 10.4161/15384101.2014.956507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang J, Bian C, Li J, Couch FJ, Wu K, Zhao RC. Poly(ADP-ribose) polymerase-1 down-regulates BRCA2 expression through the BRCA2 promoter. J Biol Chem. 2008;283(52):36249–36256. doi: 10.1074/jbc.M803693200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hu Y, Petit SA, Ficarro SB, Toomire KJ, Xie A, Lim E, et al. PARP1-driven poly-ADP-ribosylation regulates BRCA1 function in homologous recombination-mediated DNA repair. Cancer Discov. 2014;4(12):1430–1447. doi: 10.1158/2159-8290.CD-13-0891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Claybon A, Karia B, Bruce C, Bishop AJ. PARP1 suppresses homologous recombination events in mice in vivo. Nucleic Acids Res. 2010;38(21):7538–7545. doi: 10.1093/nar/gkq624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lindahl T, Satoh MS, Poirier GG, Klungland A. Post-translational modification of poly(ADP-ribose) polymerase induced by DNA strand breaks. Trends Biochem Sci. 1995;20(10):405–411. doi: 10.1016/s0968-0004(00)89089-1. [DOI] [PubMed] [Google Scholar]
- 54.Schultz N, Lopez E, Saleh-Gohari N, Helleday T. Poly(ADP-ribose) polymerase (PARP-1) has a controlling role in homologous recombination. Nucleic Acids Res. 2003;31(17):4959–4964. doi: 10.1093/nar/gkg703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shibata A, Conrad S, Birraux J, Geuting V, Barton O, Ismail A, et al. Factors determining DNA double-strand break repair pathway choice in G2 phase. Embo j. 2011;30(6):1079–1092. doi: 10.1038/emboj.2011.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Karanam K, Kafri R, Loewer A, Lahav G. Quantitative live cell imaging reveals a gradual shift between DNA repair mechanisms and a maximal use of HR in mid-S phase. Molecular cell. 2012;47(2):320–329. doi: 10.1016/j.molcel.2012.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.