Abstract
Purpose
Several molecular glioma markers (including isocitrate dehydrogenase 1 [IDH1] mutation, amplification of the epidermal growth factor receptor [EGFR] and methylation of the O6-methylguanine-DNA methyltransferase [MGMT] promoter) have been associated with glioblastoma survival. In this study, we examined the association between tumoral amino acid uptake, molecular markers, and overall survival in patients with IDH1 wild-type (primary) glioblastoma.
Materials and Methods
Twenty-one patients with newly-diagnosed IDH1 wild-type glioblastomas underwent presurgical MRI and PET scanning with alpha[C-11]-L-methyl-tryptophan (AMT). MRI characteristics (T2 and T1 contrast volume), tumoral tryptophan uptake, PET-based metabolic tumor-volume and kinetic variables were correlated with prognostic molecular markers (EGFR and MGMT) and overall survival.
Results
EGFR amplification was associated with lower T1 contrast volume (P=.04) as well as lower T1-contrast/T2 volume (P=.04) and T1-contrast/PET volume ratios (P=.02). Tumors with MGMT promoter methylation showed lower metabolic volume (P=.045) and lower tumor/cortex AMT unidirectional uptake ratios than those with unmethylated MGMT promoter (P=.009). While neither EGFR amplification nor MGMT promoter methylation was significantly associated with survival, high AMT tumor/cortex uptake ratios on PET were strongly prognostic for longer survival (hazard ratio: 30; P=.002). Estimated mean overall survival was 26 months in patients with high vs. 8 months in those with low tumoral AMT uptake ratios.
Conclusions
The results demonstrate specific MRI and amino acid PET imaging characteristics associated with EGFR amplification and MGMT promoter methylation in patients with primary glioblastoma. High tryptophan uptake on PET may identify a subgroup with prolonged survival.
Keywords: Glioblastoma, IDH1 mutation, amino acid PET, molecular markers, survival
Introduction
Gliomas represent approximately 30% of all central nervous system tumors and 80% of all malignant brain tumors.1 Glioblastomas are the most common malignant primary brain tumors in adults and carry a dismal prognosis with a median survival of 15 months,1,2 however, a small percentage of patients remains stable for a prolonged period of time after initial treatment. The majority of glioblastomas (90%) is primary and affects older patients compared to secondary glioblastomas which develop from WHO grade II gliomas.3
Main prognostic factors for survival of glioblastomas include age, performance status, radiologic features, and extent of initial tumor resection.3 In addition, several molecular markers were shown to provide prognostic information in these tumors.4 Among these, high Ki-67 nuclear labeling index (characterizing tumoral proliferative activity) and overexpression of epidermal growth factor receptor (EGFR) carry an unfavorable prognosis, while isocitrate dehydrogenase 1 (IDH1) mutation is associated with longer survival. IDH1 mutation is more frequent in WHO grade II and III gliomas and secondary glioblastoma (70–80%), while less than 5% in primary glioblastoma (IDH1 wild-type). O6-methylguanine-DNA methyltransferase (MGMT) promoter methylation in high-grade gliomas is associated with favorable response to the alkylating chemotherapeutic agent temozolomide and longer survival.5–9
Recent studies have demonstrated the association of molecular glioma markers with specific tumoral imaging characteristics. Most studies were performed with MRI and showed a number of radiophenotypes associated with glioblastoma genetic biomarkers.10 In contrast, radiogenomic studies utilizing metabolic and molecular imaging markers have been scarce. In one study, MGMT promoter methylation was associated with high methionine uptake on positron emission tomography (PET) in patients with grade II–III gliomas.11 However, no similar studies have been reported for glioblastomas. A study using the amino acid PET tracer [F-18]-fluoro-ethyl-tyrosine (FET) in previously-treated glioblastomas demonstrated that the PET-assessed recurrence pattern was influenced by MGMT promoter methylation status.12 A recent FET-PET study also showed that PET-defined biological tumor volume in newly-diagnosed glioblastomas was a prognostic imaging biomarker for survival, independent of MGMT promoter methylation.13 Nonetheless, the association between glioblastoma amino acid uptake, genetic biomarkers, and survival remains to be determined.
Alpha[C-11]-L-methyl-tryptophan (AMT) is an amino acid PET tracer utilized to study brain tryptophan uptake and metabolism.14 AMT-PET can detect and differentiate various primary and metastatic brain tumors, including glioblastomas.15–17 We have also demonstrated that high tryptophan uptake in the suspected recurrence site was a strong prognostic imaging marker for 1-year and overall survival in patients with previously-treated glioblastoma independent of MRI and clinical prognostic factors.18 However, PET findings were not correlated with molecular prognostic markers, and the potential prognostic value of AMT-PET in pre-treatment glioblastomas remained to be determined.
The present study had two main goals: (i) to evaluate if prognostic molecular markers in primary (IDH1 wild-type) glioblastomas are associated with a specific pattern of amino acid uptake or metabolism on PET imaging and/or MRI variables; and (ii) to determine if pre-treatment tryptophan uptake measured by PET has a prognostic value for overall survival in the same group.
Materials and Methods
Subjects
The study included 21 patients (14 males, mean age: 62 years; Table 1) with a suspected glioblastoma who underwent presurgical imaging including dynamic AMT-PET following tumor resection and standard postsurgical chemoradiation. In all patients, histopathology confirmed WHO grade IV glioma with wild-type IDH1. Thirteen patients had gross total tumor resection and 8 had subtotal resection at initial surgery. Five patients underwent a second resection after tumor recurrence, and two patients received additional chemotherapy (bevacizumab and bevacizumab with irinotecan in one case each; Table 1). No patients received NovoTTF treatment that could prolong overall survival.19 The study was approved by the Wayne State University Institutional Review Board with written informed consent obtained from all participants.
Table 1.
Clinical data of the 21 patients with IDH1 wild-type glioblastoma
| Patient No. |
Gender | Age (Years) |
1st Surgery (Total or Subtotal Resection) |
2nd Surgery (Yes/No) |
Additional Treatment | Survival (Days) |
|---|---|---|---|---|---|---|
| 1 | F | 70 | subtotal | no | none | 282 |
| 2 | F | 54 | total | yes | none | 596* |
| 3 | M | 47 | subtotal | yes | bevacizumab | 1067 |
| 4 | F | 70 | subtotal | no | none | 444 |
| 5 | M | 79 | total | no | none | 486* |
| 6 | M | 40 | subtotal | no | none | 185 |
| 7 | M | 65 | total | no | none | 1650* |
| 8 | M | 54 | total | yes | none | 760 |
| 9 | M | 61 | total | yes | none | 860 |
| 10 | M | 68 | subtotal | no | none | 217 |
| 11 | F | 60 | total | no | none | 859 |
| 12 | M | 67 | total | no | none | 262 |
| 13 | F | 82 | subtotal | no | none | 127 |
| 14 | F | 65 | total | no | none | 167 |
| 15 | M | 67 | total | no | none | 445 |
| 16 | M | 62 | total | no | none | 310 |
| 17 | M | 66 | subtotal | no | none | 961 |
| 18 | F | 76 | subtotal | no | bevacizumab/irinotecan | 815 |
| 19 | M | 54 | total | yes | none | 670 |
| 20 | M | 46 | total | yes | none | 291 |
| 21 | M | 59 | total | no | none | 391 |
alive at last follow-up
MRI acquisition
Diagnostic MRIs with routine T1, T2, fluid attenuated inversion recovery (FLAIR), and post-contrast T1 (T1-Gad) sequences acquired closest in time to the AMT-PET were used in this study. MRI was performed on a Siemens MAGNETOM Trio TIM 3.0 Tesla scanner (Siemens Medical Solutions, Malvern, Pennsylvania), a GE Signa HDxt 3.0 Tesla scanner (GE Medical Systems, Milwaukee, Wisconsin), or a Philips Achieva TX 3.0 Tesla scanner (Philips Medical Systems Inc., Da Best, the Netherlands), using similar parameters on all scanners.
AMT-PET scanning protocol
PET studies were performed using a Siemens EXACT/HR whole-body positron emission tomograph (Siemens Medical Systems, Knoxville, TN) or GE Discovery STE PET/CT scanner (GE Medical Systems, Milwaukee, WI). The PET image in plane resolution was 7.5±0.4 mm at full-width half-maximum (FWHM) and 7.0±0.5 mm FWHM in the axial direction, with a slice thickness of 3.125 mm. The AMT tracer was synthesized by using a high-yield procedure as outlined before.20 The procedure for AMT-PET scanning has been described previously.15,16,18,21 In brief, after 6 h of fasting, AMT (1 mCi/kg=37 MBq/kg) was injected intravenously. For collection of timed blood samples, a second venous line was established. In the initial 20 min of the scan following tracer injection, a dynamic PET scan of the heart was performed to obtain the blood input function from the left cardiac ventricle noninvasively. The blood input function was continued beyond these initial 20 min by using venous blood samples (0.5 mL/sample, collected at 20, 30, 40, 50, and 60 min after AMT injection). At 25 min after tracer injection, a dynamic emission scan of the brain (7×5 min) was acquired. Measured attenuation correction, scatter, and decay correction was applied to all PET images. For visualization of AMT uptake, averaged activity images 30–55 min post-injection were created and converted to an AMT standardized uptake value (SUV) image.
PET and MRI processing and analysis
For image analysis, the 3D Slicer software22 version 3.6.3 (www.slicer.org), was used as described previously.15 First, pre-treatment AMT-PET and MR images were co-registered using the Fast Rigid Registration module, and a transformation matrix was created.23 This transformation matrix was then applied to the summed AMT-PET image and to the dynamic AMT-PET images loaded via the 4D image module of 3D Slicer. Following fusion of the summed AMT-PET with MR images, regions of interest (ROIs) were drawn on the tumor mass in tumor regions with MRI contrast enhancement and/or T2/FLAIR signal changes, and the ROIs were then applied on the co-registered AMT PET images. As a reference (background) region, at least 3 ROIs were drawn on the homotopic cortex contralateral to the tumor, and the values from these ROIs were averaged. For quantification of the AMT net transport and metabolism, Patlak graphical analysis16,24 was performed, which yielded AMT volume of distribution (VD) and K values, as described previously.16,25 VD is an estimate of the volume of distribution of the tracer in the free precursor pool. We have previously shown that tumoral VD is increased when the blood-brain barrier is compromised16 and tumoral VD was also a good estimate of tumor proliferative activity in newly-diagnosed gliomas.26 The AMT K value reflects the unidirectional uptake of tracer into tissue,27,28 which is thought to be higher with increased metabolism of tryptophan via the serotonin (in normal brain) and/or the kynurenine pathway.14,16
Tumor volumes on MRI were defined as described previously: (i) thresholds slightly above the highest signal of normal white matter contralateral to the tumor were used for T2 images; and (ii) the area of contrast enhancement was used for post-contrast images.29 Contrast-enhancing volumes were created by segmentation of post-contrast abnormalities semi-automatically, while T2 images were segmented manually to avoid erroneous inclusion of cerebrospinal fluid in the volume of interest.
Tissue processing
Freshly-resected tumor specimens were divided and fragments were flash-frozen in liquid nitrogen or formalin-fixed and paraffin-embedded (FFPE). Flash-frozen tissues were stored at −80°C until DNA extraction. FFPE tissues were sectioned at 5 µM and mounted on slides for immunohistochemical staining or collected for DNA extraction. Genomic DNA was extracted from all samples with the ZR Genomic DNA-Tissue MiniPrep (Zymo Research, Irvine, CA) according to the manufacturer’s protocol for flash-frozen or FFPE tissues. DNA was spin-column purified and eluted in DNase-free water. DNA samples were quantified with a NanoDrop 2000 spectrophotometer (ThermoFisher Scientific, Wilmington, DE). The 260/280 ratios for all of the samples were 1.91±0.06.
Ki-67 labeling index
Immunohistochemical staining for Ki-67 (using CONFIRM™ anti-Ki-67 rabbit monoclonal antibody, clone 30-9, Ventana Medical Systems, Inc., Tucson, AZ) was performed to assess glioma proliferative activity. The Ki-67 labeling index was determined by identifying the areas of greatest tumor cellularity, examining at least 10 fields at 400× (high-power) magnification on an Olympus microscope, and determining the ratio between the number of Ki-67-positive tumor cell nuclei and the total number of tumor cell nuclei in each high-power field. The proliferation index was expressed as a percent range (lowest to highest) in the 10 high-power fields (e.g., 5–10%). The middle value of the defined range was then used in correlations with the PET variables.
MGMT promoter methylation
The MGMT promoter region analyzed was −20 to +178, which contains some of the CpG sites that have been shown to correlate most highly with levels of MGMT mRNA.30 For each sample tested, 500 ng of DNA were bisulfite-converted with the Cells-to-CpG™ Bisulfite Conversion Kit (ThermoFisher, Carlsbad, CA). Bisulfite-converted DNA was quantified (Nanodrop) and 20 ng was used in the PCR reaction with primers 5’-GTTTTTTTGTTTTTTTTAGGTTTT and 5’-AAACRACCCAAACACTCACC and Power SYBR® Green PCR Master Mix (ThermoFisher).30 PCR reactions were performed with a StepOnePlus® real-time PCR thermocycler (ThermoFisher). The resulting PCR products were cleaned up with the DNA Clean & Concentrator™ (Zymo Research) prior to Sanger sequencing. All sequencing reactions were performed by ACGT, Inc. (Wheeling, IL) with primers 5’-GGTTTYGTTTYGTTTTAGATT and 5’-AGGATATGTTGGGATAGTT.30 Sequence data were analyzed with Chromas 2.6 (Technelysium Pty Ltd., South Brisbane, Australia). The MGMT promoter was considered methylated if greater than 28% of the CpG sites assayed were methylated.30
IDH1 mutation
To detect mutations associated with arginine 132 (base pairs 394–395), exon 4 of IDH1 was PCR amplified from 10 ng of genomic DNA with the primers 5’-TGAGAAGAGGGTTGAGGAGTT and 5’-AACATGCAAAATCACATTATTGCC. PCR reactions were performed with and Power SYBR® Green PCR Master Mix (ThermoFisher). PCR reactions were performed with a StepOnePlus® real-time PCR thermocycler (ThermoFisher). The resulting PCR products were cleaned up with the DNA Clean & Concentrator™ (Zymo Research) prior to Sanger sequencing. All sequencing reactions were performed by ACGT, Inc. using the same primers. Sequence data were analyzed with Chromas 2.6 (Technelysium Pty Ltd., South Brisbane, Australia).
EGFR copy number variation
Levels of EGFR amplification in the tumor DNA was determined using the TaqMan® Copy Number Assay (Assay ID Hs01646307_cn; ThermoFisher). Data were normalized using the TaqMan® Copy Number Reference Assay for RNase P. For each sample, 20 ng of genomic DNA was amplified using TaqMan® Universal PCR Master Mix (ThermoFisher). Data were analyzed with CopyCaller® Software v2.0 (ThermoFisher) to determine the degree of EGFR amplification in each tumor DNA sample. Samples with 3 or more copies of the EGFR gene were considered positive for amplification.
Statistical analysis
Group comparisons of clinical and imaging variables of patients with various molecular markers were performed using the Mann-Whitney U-test. Univariate correlations were performed using the Spearman’s rank correlation. After identifying the PET variable(s) with a significant correlation with overall survival, a receiver-operating characteristic (ROC) analysis was performed to identify the optimal threshold for differentiating patients who were alive at 1-year follow-up from those who had died. Using this cutoff threshold, a Cox-regression analysis was done to obtain a hazard ratio (HR) for survival in patients having above- vs. below-threshold values. Statistical analysis was carried out using SPSS Statistics 23.0 software. A P value <.05 was considered to be significant.
Results
Imaging correlates of molecular markers
Mean Ki-67 labeling index was 30% (range: 10–70%), MGMT promoter methylation was present in 7/19 (37%), while EGFR amplification was detected in 6/20 tumors (30%). EGFR amplification was associated with higher T1-Gad/PET volume ratio (P=.02), lower T1-Gad/T2 volume ratio (P=.04) and lower T1-Gad volume (P=.04; Table 2). Tumors with EGFR amplifications also had markedly higher Ki-67 labeling index (41±17% vs. 20±6%, respectively, P=.001). Tumors with MGMT promoter methylation showed lower tumor/cortex AMT K-ratios (mean: 1.3±0.4 vs. 2.05±0.6, P=.009) and lower AMT-PET-defined metabolic volume (mean: 23.2±14.9 vs. 48.2±29.1 cm3, P=.045) than those with unmethylated MGMT. None of the molecular markers showed an association with age, performance status or survival (Table 2).
Table 2.
Relation of clinical and imaging variables to molecular tumor markers. For EGFR and MGMT promoter methylation, mean values in positive vs. negative subgroups are listed; the P value refers to the Mann-Whitney U test. For Ki-67 labeling, correlation coefficients (Spearman’s) and related P values are listed. Statistically significant values (P<.05) are in bold.
| Variables | EGFR | MGMT | Ki-67 | |||||
|---|---|---|---|---|---|---|---|---|
| Positive | Negative |
P value |
Positive | Negative |
P value |
r value |
P value |
|
| Age (years) | 64 | 62 | 0.8 | 67 | 59 | 0.1 | 0.21 | 0.4 |
|
Performance status |
83 | 82 | 0.6 | 86 | 80 | 0.9 | 0.07 | 0.8 |
| Survival (days) | 651 | 506 | 0.2 | 746 | 493 | 0.4 | 0.2 | 0.3 |
|
T1-Gad volume (cm3) |
6 | 21.3 | 0.04 | 11.7 | 16.1 | 0.5 | −0.58 | 0.005 |
| T2 volume (cm3) | 36.4 | 44.8 | 0.9 | 48.8 | 40.9 | 0.1 | −0.1 | 0.6 |
| T1/T2 volume | 0.14 | 0.52 | 0.04 | 0.19 | 0.49 | 0.2 | −0.47 | 0.03 |
|
PET volume (cm3) |
52.7 | 41.3 | 0.6 | 23.2 | 48.3 | 0.045 | −0.2 | 0.5 |
| T1/PET volume | 0.14 | 0.56 | 0.02 | 0.51 | 0.34 | 0.9 | −0.5 | 0.02 |
| PET SUV-ratio | 2.4 | 2.1 | 0.2 | 2 | 2.3 | 0.3 | 0.04 | 0.9 |
| PET VD-ratio | 3.6 | 2.4 | 0.1 | 2.8 | 2.8 | 0.7 | 0.1 | 0.7 |
| PET K-ratio | 1.7 | 1.9 | 0.7 | 1.3 | 2 | 0.009 | −0.1 | 0.7 |
Abbreviation: PET= positron emission tomography, EGFR= epidermal growth factor receptor, MGMT=O6-methylguanine-DNA methyltransferase, SUV= standardized uptake value, VD= volume of distribution, T1-Gad= post-contrast T1
Prognostic value of pre-treatment AMT-PET for survival
The median survival time in our study group was 14.8 months (445 days), and 13 patients (62%) had >1-year survival (Table 1). Overall survival showed a significant positive correlation with tumor/cortex SUV-ratios (r=0.49; P=.023), indicating longer survival in those with higher pre-treatment tumoral tryptophan uptake (Figure 1). No other clinical, molecular or imaging variables correlated with survival.
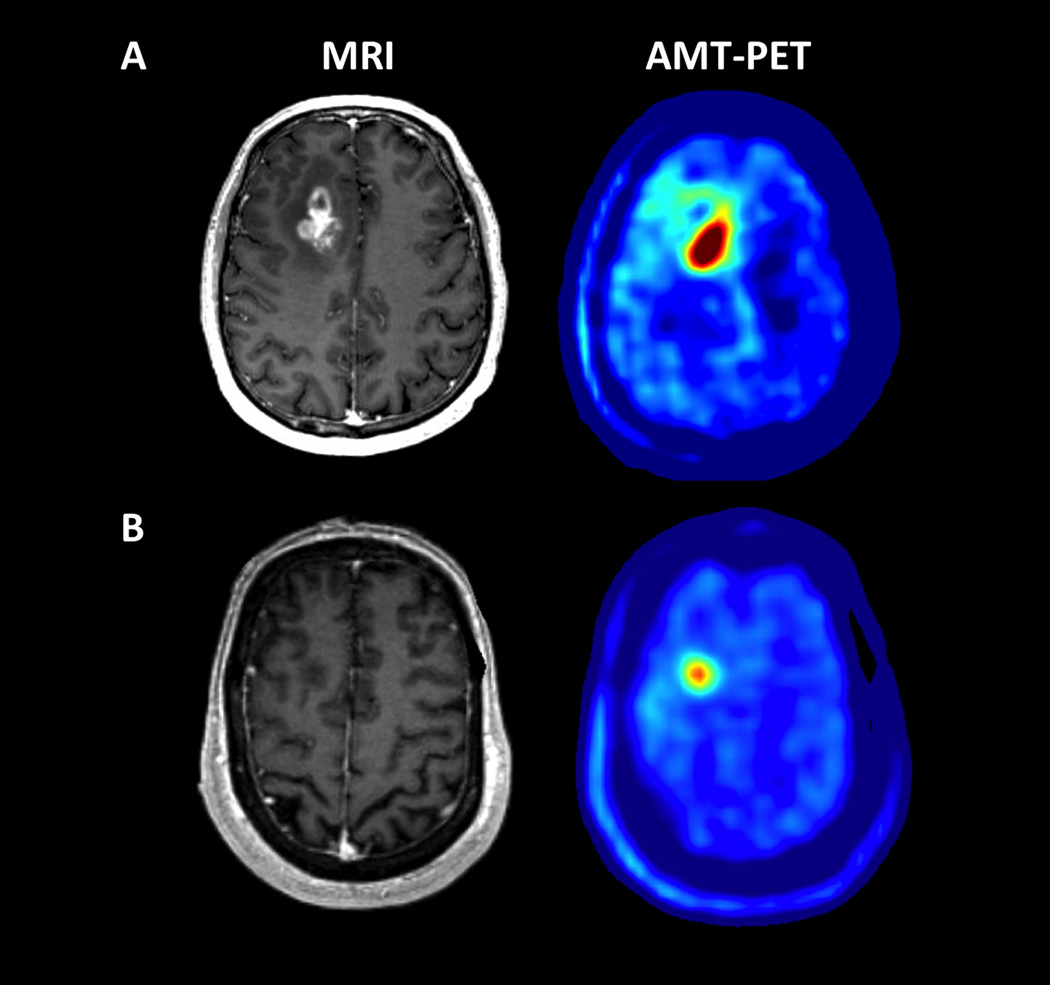
Figure 1.
Representative examples of AMT-PET in a patient with high (A) vs. low (B) uptake associated with different survival. (A) A 54 year-old male (patient #8 in Table 1) with a right frontal glioblastoma, measured with a 2.59 tumor/cortex AMT SUV-ratio, above the ROC-defined threshold of 1.94. He survived for more than 2 years after the PET scan. (B) A 68 year-old male (patient #10 in Table 1) with a right frontal glioblastoma showing a below-threshold tumor/cortex AMT SUV-ratio (1.56). He survived for only 7 months.
The ROC analysis showed high area under the curve (AUC) for AMT SUV-ratios (0.94; P=.001). Using a 1.94 tumor/cortex SUV-ratio as the cutoff threshold, 1-year survival was correctly predicted with 100% sensitivity and 88% specificity. Cox-regression analysis showed that AMT SUV-ratios above 1.94 were highly predictive for long survival (hazard ratio: 30.2 [95% CI: 3.5–259]; P=.002). Estimated mean overall survival was 26 [SEM: 3] vs. 8 [SEM: 1] months in patients with above- vs. those with below-threshold AMT uptake ratios, respectively (Figure 2).
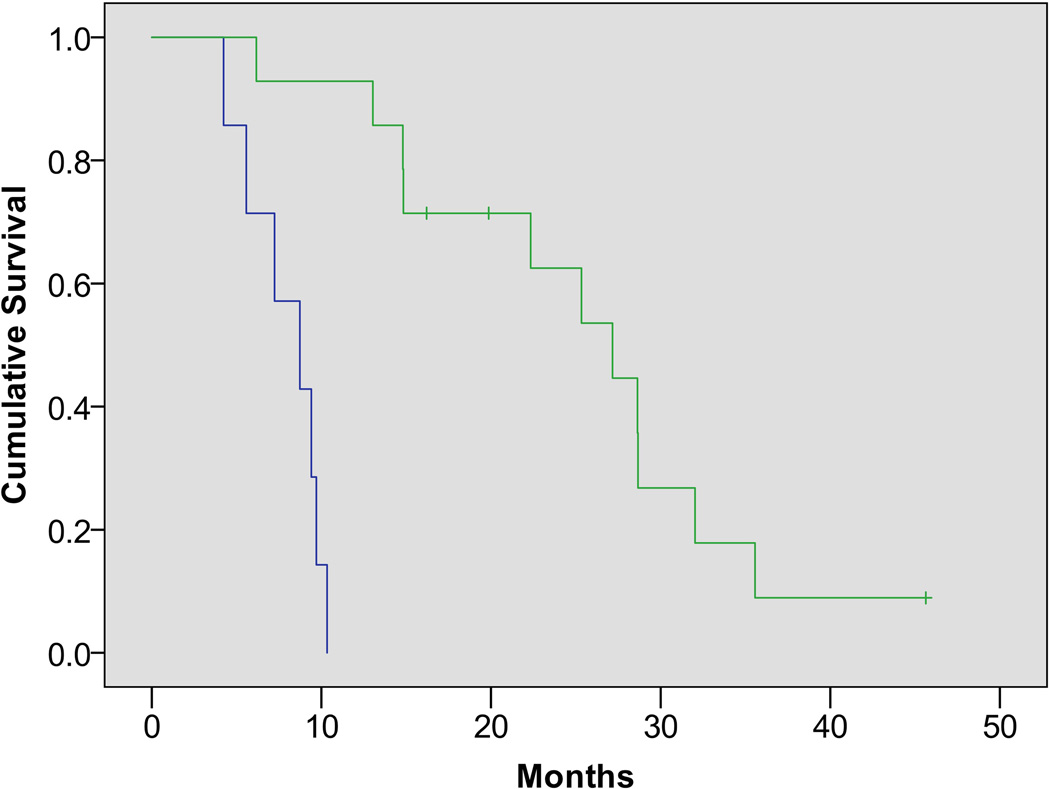
Figure 2.
Kaplan-Meijer survival curves in patients with high (green) vs. low (blue) AMT uptake ratios (based on the cutoff threshold of 1.94 tumor/cortex SUV ratio, determined by an ROC analysis based on 1-year survival). Patients with above-threshold ratios had a substantially longer cumulative survival (hazard ratio: 30.2 [95% CI: 3.5–259]; P=.002).
Discussion
This is the first study to demonstrate a link between prognostic genetic biomarkers and tumoral amino acid uptake in IDH1 wild-type glioblastomas. MGMT promoter methylation was associated with lower AMT-PET-based metabolic volumes and K-ratios, while EGFR amplification was associated with lower T1-Gad/PET and lower T1-Gad/T2 volume ratios. In addition, while neither EGFR nor MGMT promoter methylation status was associated with survival, high AMT SUV tumor/cortex ratio was a strong prognostic imaging marker associated with a markedly longer survival in this patient group.
In previous studies, high tumoral amino acid uptake was generally associated with poor prognosis in patients with malignant glioma. For example, high [C-11]-methionine (MET) accumulation was found to be an independent, significant prognostic factor for short survival in glioma patients when various tumor grades were included.31 Several MET-PET studies also demonstrated that high metabolic tumor volume was associated with shorter progression-free and/or overall survival.32–34 Also, in a recent FET-PET study, higher biological tumor volume before radiotherapy was the most important imaging marker for shorter survival independent of MGMT promoter methylation and clinical prognostic factors.13
Our present data, showing that high pre-treatment AMT uptake actually predicts longer overall survival, appear to be at odds with the above-mentioned PET studies. This difference could be explained by several factors. First, unlike previous studies, our patient group included only IDH1 wild-type glioblastomas, a selective glioma group with particularly dismal prognosis. Despite this, patients with above-threshold (>1.94) tumor/cortex uptake ratios showed a relatively prolonged survival, with 8/14 patients (57%) achieving >2-year survival. In contrast, all 7 patients with below-threshold values died within 1 year. This was a very robust difference, especially when considering that no other clinical or molecular markers showed a significant association with survival in this relatively small cohort. It is compelling to speculate that this unexpected finding may be related to potential differences in tumoral metabolism between AMT and other amino acid PET tracers. While most clinically-tested amino acid tracers (including AMT) are transported by the same (L-type) transport system, they show important differences in their metabolic fate (reviewed in Juhász et al.35). Specifically, while FET or [F-18]-Fluoro-DOPA are not metabolized in glioma tissue, both MET and AMT can undergo at least partial tumoral metabolism. Specifically, AMT uptake, trapping, and breakdown may be affected by tryptophan metabolism via the immunosuppressive kynurenine pathway.14,15 Rate-limiting enzymes of this pathway, such as indoleamine 2,3 dioxygenase (IDO) 1 and 2, as well as tryptophan 2,3 dioxygenase (TDO) can be upregulated in various cancers, including brain tumors,36–38 and generate metabolites that suppress T-cell mediated immune responses.39 As a result, high activity of these enzymes has been associated with poor survival in several cancer types including malignant gliomas.37,40 In addition to IDO1/2 and TDO, recent studies have also shown high expression and activity of downstream enzymes of the kynurenine pathway in gliomas (such as kynurenine 3-monooxygenase and kynureninase).38,41 As a result, tryptophan and its derivatives may undergo breakdown and elimination from tumor tissue, and this may be manifested as lower AMT SUV measured beyond 30 min after tracer injection. Whether lower late AMT accumulation is indeed due to enhanced kynurenine pathway activity, will require further studies evaluating if in vivo uptake of AMT or other tryptophan derivative PET tracers can be modulated (increased) by emerging kynurenine pathway enzyme inhibitors.42 In this respect, recently developed F-18 labeled tryptophan analogs, such as 1-(2-[F-18]fluoroethyl)-L-tryptophan,43 or others, should be also explored because of their potential for more widespread clinical application as compared to AMT. Initial in vivo studies in patient-derived xenograft mouse models have been encouraging.44
The association between EGFR amplification, high Ki-67 labeling index, and low T1-Gad/PET (and also low T1-gad/T2) volume ratios is generally consistent with the notion that EGFR amplification leads to EGFR overexpression that promotes angiogenesis and aggressive tumor growth leading to glioma cell infiltration.45 Low T1-Gad/T2 ratio has been reported in EGFR-amplified glioblastomas,46 consistent with extensive infiltrative edema as a result of enhanced angiogenesis and tumor cell invasion in the non-contrast-enhancing brain. Our data are in keeping with these results and also demonstrate larger relative areas with high tryptophan uptake beyond the contrast-enhancing tumor mass in EGFR-amplified glioblastomas. Increased amino acid uptake outside the contrast-enhancing tumor mass can mark glioma cell infiltration, as demonstrated by previous studies employing PET-image-guided biopsy.29,47 In addition, both lower tryptophan unidirectional uptake ratios and lower metabolic volumes were associated with MGMT promoter methylation. A previous study reported that MGMT promoter methylation was associated with higher methionine uptake on PET in patients with grade II–III gliomas.11 However, no similar results have been reported for glioblastomas. Whether the differences between the present and the previous methionine PET study is due to the different glioma grades or different amino acid tracers used (or both), will need to be addressed in future studies. Nevertheless, the studies demonstrate that MGMT promoter methylation may affect tumoral amino acid metabolism that can be captured by PET imaging.
In summary, our data demonstrate specific MRI and AMT-PET characteristics associated with prognostic molecular markers in IDH1 wild-type glioblastoma. Despite the limited number of patients, high AMT uptake ratios on PET were found to be a robust prognostic imaging marker, regardless of other, molecular or clinical prognostic factors. Therefore, molecular imaging of tryptophan metabolism is worth further studies for prognostic imaging in patients with newly-diagnosed glioblastoma.
Acknowledgments
We are grateful to the entire staff at the PET Center, Children’s Hospital of Michigan, who provided invaluable technical help in performing the PET scans. We also thank Drs. Natasha Robinette and Alit Amit-Yousif who performed the clinical interpretation of the MR images.
Funding: The study was supported by a grant (CA123451 to C.J. and S.M.) from the National Cancer Institute; a grant from the Fund for Medical Research and Education, Wayne State University School of Medicine (to S.M.); and Strategic Research Initiative Grant from the Karmanos Cancer Institute (to S.M. and C.J.). The Biobanking and Correlative Sciences Core is supported, in part, by NIH Center Grant P30 CA022453 to the Karmanos Cancer Institute at Wayne State University.
Footnotes
Conflict of Interest: None of the authors report any conflict of interest or disclosure.
REFERENCES
- 1.Omuro A, DeAngelis LM. Glioblastoma and other malignant gliomas: a clinical review. JAMA. 2013;310:1842–1850. doi: 10.1001/jama.2013.280319. [DOI] [PubMed] [Google Scholar]
- 2.Stupp R, Hegi ME, Mason WP, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 3.Delgado-Lopez PD, Corrales-Garcia EM. Survival in glioblastoma: a review on the impact of treatment modalities. Clin Transl Oncol. 2016;18:1062–1071. doi: 10.1007/s12094-016-1497-x. [DOI] [PubMed] [Google Scholar]
- 4.Xie Q, Mittal S, Berens ME. Targeting adaptive glioblastoma: an overview of proliferation and invasion. Neuro Oncol. 2014;16:1575–1584. doi: 10.1093/neuonc/nou147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 6.van den Bent MJ, Dubbink HJ, Sanson M, et al. MGMT promoter methylation is prognostic but not predictive for outcome to adjuvant PCV chemotherapy in anaplastic oligodendroglial tumors: a report from EORTC Brain Tumor Group Study 26951. J Clin Oncol. 2009;27:5881–5886. doi: 10.1200/JCO.2009.24.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wick W, Hartmann C, Engel C, et al. NOA-04 randomized phase III trial of sequential radiochemotherapy of anaplastic glioma with procarbazine, lomustine, and vincristine or temozolomide. J Clin Oncol. 2009;27:5874–5880. doi: 10.1200/JCO.2009.23.6497. [DOI] [PubMed] [Google Scholar]
- 8.Weller M, Stupp R, Reifenberger G, et al. MGMT promoter methylation in malignant gliomas: ready for personalized medicine? Nat Rev Neurol. 2010;6:39–51. doi: 10.1038/nrneurol.2009.197. [DOI] [PubMed] [Google Scholar]
- 9.Appin CL, Brat DJ. Molecular genetics of gliomas. Cancer J. 2014;20:66–72. doi: 10.1097/PPO.0000000000000020. [DOI] [PubMed] [Google Scholar]
- 10.ElBanan MG, Amer AM, Zinn PO, et al. Imaging genomics of Glioblastoma: state of the art bridge between genomics and neuroradiology. Neuroimaging Clin N Am. 2015;25:141–153. doi: 10.1016/j.nic.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 11.Okita Y, Nonaka M, Shofuda T, et al. 11C-methinine uptake correlates with MGMT promoter methylation in nonenhancing gliomas. Clin Neurol Neurosurg. 2014;125:212–216. doi: 10.1016/j.clineuro.2014.08.004. [DOI] [PubMed] [Google Scholar]
- 12.Niyazi M, Schnell O, Suchorska B, et al. FET-PET assessed recurrence pattern after radio-chemotherapy in newly diagnosed patients with glioblastoma is influenced by MGMT methylation status. Radiother Oncol. 2012;104:78–82. doi: 10.1016/j.radonc.2012.04.022. [DOI] [PubMed] [Google Scholar]
- 13.Suchorska B, Jansen NL, Linn J, et al. Biological tumor volume in 18FET-PET before radiochemotherapy correlates with survival in GBM. Neurology. 2015;84:710–719. doi: 10.1212/WNL.0000000000001262. [DOI] [PubMed] [Google Scholar]
- 14.Chugani DC, Muzik O. Alpha[C-11]methyl-L-tryptophan PET maps brain serotonin synthesis and kynurenine pathway metabolism. J Cereb Blood Flow Metab. 2000;20:2–9. doi: 10.1097/00004647-200001000-00002. [DOI] [PubMed] [Google Scholar]
- 15.Bosnyák E, Kamson DO, Guastella AR, et al. Molecular imaging correlates of tryptophan metabolism via the kynurenine pathway in human meningiomas. Neuro Oncol. 2015;17:1284–1292. doi: 10.1093/neuonc/nov098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Juhasz C, Chugani DC, Muzik O, et al. In vivo uptake and metabolism of alpha-[11C]methyl-L-tryptophan in human brain tumors. J Cereb Blood Flow Metab. 2006;26:345–357. doi: 10.1038/sj.jcbfm.9600199. [DOI] [PubMed] [Google Scholar]
- 17.Kamson DO, Mittal S, Buth A, et al. Differentiation of glioblastomas from metastatic brain tumors by tryptophan uptake and kinetic analysis: a positron emission tomographic study with magnetic resonance imaging comparison. Mol Imaging. 2013;12:327–337. [PMC free article] [PubMed] [Google Scholar]
- 18.Kamson DO, Mittal S, Robinette NL, et al. Increased tryptophan uptake on PET has strong independent prognostic value in patients with a previously treated high-grade glioma. Neuro Oncol. 2014;16:1373–1383. doi: 10.1093/neuonc/nou042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stupp R, Taillibert S, Kanner AA, et al. Maintenance Therapy With Tumor-Treating Fields Plus Temozolomide vs Temozolomide Alone for Glioblastoma: A Randomized Clinical Trial. JAMA. 2015;314:2535–2543. doi: 10.1001/jama.2015.16669. [DOI] [PubMed] [Google Scholar]
- 20.Chakraborty PK, Mangner TJ, Chugani DC, et al. A high-yield and simplified procedure for the synthesis of alpha-[11C]methyl-L-tryptophan. Nucl Med Biol. 1996;23:1005–1008. doi: 10.1016/s0969-8051(96)00127-8. [DOI] [PubMed] [Google Scholar]
- 21.Bosnyak E, Kamson DO, Robinette NL, et al. Tryptophan PET predicts spatial and temporal patterns of post-treatment glioblastoma progression detected by contrast-enhanced MRI. J Neurooncol. 2016;126:317–325. doi: 10.1007/s11060-015-1970-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kikinis R, Pieper S. 3D Slicer as a tool for interactive brain tumor segmentation. Conf Proc IEEE Eng Med Biol Soc. 2011;2011:6982–6984. doi: 10.1109/IEMBS.2011.6091765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mattes D, Haynor DR, Vesselle H, et al. PET-CT image registration in the chest using free-form deformations. IEEE Trans Med Imaging. 2003;22:120–128. doi: 10.1109/TMI.2003.809072. [DOI] [PubMed] [Google Scholar]
- 24.Patlak CS, Blasberg RG, Fenstermacher JD. Graphical evaluation of blood-to-brain transfer constants from multiple-time uptake data. J Cereb Blood Flow Metab. 1983;3:1–7. doi: 10.1038/jcbfm.1983.1. [DOI] [PubMed] [Google Scholar]
- 25.Alkonyi B, Barger GR, Mittal S, et al. Accurate differentiation of recurrent gliomas from radiation injury by kinetic analysis of alpha-11C-methyl-L-tryptophan PET. J Nucl Med. 2012;53:1058–1064. doi: 10.2967/jnumed.111.097881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Juhasz C, Chugani DC, Barger GR, et al. Quantitative PET imaging of tryptophan accumulation in gliomas and remote cortex: correlation with tumor proliferative activity. Clin Nucl Med. 2012;37:838–842. doi: 10.1097/RLU.0b013e318251e458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chugani DC, Muzik O, Chakraborty P, et al. Human brain serotonin synthesis capacity measured in vivo with alpha-[C-11]methyl-L-tryptophan. Synapse. 1998;28:33–43. doi: 10.1002/(SICI)1098-2396(199801)28:1<33::AID-SYN5>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 28.Muzik O, Chugani DC, Chakraborty P, et al. Analysis of [C-11]alpha-methyl-tryptophan kinetics for the estimation of serotonin synthesis rate in vivo. J Cereb Blood Flow Metab. 1997;17:659–669. doi: 10.1097/00004647-199706000-00007. [DOI] [PubMed] [Google Scholar]
- 29.Kamson DO, Juhasz C, Buth A, et al. Tryptophan PET in pretreatment delineation of newly-diagnosed gliomas: MRI and histopathologic correlates. J Neurooncol. 2013;112:121–132. doi: 10.1007/s11060-013-1043-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Everhard S, Tost J, El Abdalaoui H, et al. Identification of regions correlating MGMT promoter methylation and gene expression in glioblastomas. Neuro Oncol. 2009;11:348–356. doi: 10.1215/15228517-2009-001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim S, Chung JK, Im SH, et al. 11C-methionine PET as a prognostic marker in patients with glioma: comparison with 18F-FDG PET. Eur J Nucl Med Mol Imaging. 2005;32:52–59. doi: 10.1007/s00259-004-1598-6. [DOI] [PubMed] [Google Scholar]
- 32.Galldiks N, Dunkl V, Kracht LW, et al. Volumetry of [11C]-methionine positron emission tomographic uptake as a prognostic marker before treatment of patients with malignant glioma. Mol Imaging. 2012;11:516–527. [PubMed] [Google Scholar]
- 33.Yoo MY, Paeng JC, Cheon GJ, et al. Prognostic Value of Metabolic Tumor Volume on 11C-Methionine PET in Predicting Progression-Free Survival in High-Grade Glioma. Nucl Med Mol Imaging. 2015;49:291–297. doi: 10.1007/s13139-015-0362-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kobayashi K, Hirata K, Yamaguchi S, et al. Prognostic value of volume-based measurements on 11C-methionine PET in glioma patients. Eur J Nucl Med Mol Imaging. 2015;42:1071–1080. doi: 10.1007/s00259-015-3046-1. [DOI] [PubMed] [Google Scholar]
- 35.Juhasz C, Dwivedi S, Kamson DO, et al. Comparison of amino acid positron emission tomographic radiotracers for molecular imaging of primary and metastatic brain tumors. Mol Imaging. 2014;13 doi: 10.2310/7290.2014.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uyttenhove C, Pilotte L, Theate I, et al. Evidence for a tumoral immune resistance mechanism based on tryptophan degradation by indoleamine 2,3-dioxygenase. Nat Med. 2003;9:1269–1274. doi: 10.1038/nm934. [DOI] [PubMed] [Google Scholar]
- 37.Mitsuka K, Kawataki T, Satoh E, et al. Expression of indoleamine 2,3-dioxygenase and correlation with pathological malignancy in gliomas. Neurosurgery. 2013;72:1031–1038. doi: 10.1227/NEU.0b013e31828cf945. discussion 1038–1039. [DOI] [PubMed] [Google Scholar]
- 38.Adams S, Teo C, McDonald KL, et al. Involvement of the kynurenine pathway in human glioma pathophysiology. PLoS One. 2014;9:e112945. doi: 10.1371/journal.pone.0112945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Munn DH, Mellor AL. Indoleamine 2,3-dioxygenase and tumor-induced tolerance. J Clin Invest. 2007;117:1147–1154. doi: 10.1172/JCI31178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Opitz CA, Litzenburger UM, Sahm F, et al. An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature. 2011;478:197–203. doi: 10.1038/nature10491. [DOI] [PubMed] [Google Scholar]
- 41.Guastella AR, Michelhaugh SK, Klinger NV, et al. Tryptophan PET Imaging of the Kynurenine Pathway in Patient-Derived Xenograft Models of Glioblastoma. Mol Imaging. 2016;15:1–11. doi: 10.1177/1536012116644881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sheridan C. IDO inhibitors move center stage in immuno-oncology. Nat Biotechnol. 2015;33:321–322. doi: 10.1038/nbt0415-321. [DOI] [PubMed] [Google Scholar]
- 43.Henrottin J, Lemaire C, Egrise D, et al. Fully automated radiosynthesis of 1N-[18F]fluoroethyl-tryptophan and study of its biological activity as a new potential substrate for indoleamine 2,3-dioxygenase PET imaging. Nucl Med Biol. 2016;43:379–389. doi: 10.1016/j.nucmedbio.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 44.Michelhaugh SK, Guastella AR, Klinger NV, et al. Assessment of tryptophan uptake and kinetics using 1-(2-[18F]fluoroethyl)-L-tryptophan and α-[11C]-methyl-L-tryptophan PET imaging in mice implanted with patient-derived brain tumor xenografts. J Nucl Med. 2016 Oct 20; doi: 10.2967/jnumed.116.179994. pii: jnumed.116.179994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Furnari FB, Cloughesy TF, Cavenee WK, et al. Heterogeneity of epidermal growth factor receptor signalling networks in glioblastoma. Nat Rev Cancer. 2015;15:302–310. doi: 10.1038/nrc3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aghi M, Gaviani P, Henson JW, et al. Magnetic resonance imaging characteristics predict epidermal growth factor receptor amplification status in glioblastoma. Clin Cancer Res. 2005;11:8600–8605. doi: 10.1158/1078-0432.CCR-05-0713. [DOI] [PubMed] [Google Scholar]
- 47.Kracht LW, Miletic H, Busch S, et al. Delineation of brain tumor extent with [11C]L-methionine positron emission tomography: local comparison with stereotactic histopathology. Clin Cancer Res. 2004;10:7163–7170. doi: 10.1158/1078-0432.CCR-04-0262. [DOI] [PubMed] [Google Scholar]