Abstract
Background
Studies of colorectal cancer (CRC) screening by multi-target stool DNA (MT-sDNA) show false positive (FP) rates of 7–13%. It is unclear whether FP patients are at increased long-term risk of adverse outcomes.
Methods
We compared subsequent clinical events among patients with apparent FP MT-sDNA to those in patients reported as true negative (TN). This was a retrospective cohort study of participants in pre-FDA approval MT-sDNA studies having non-advanced or negative baseline colonoscopy findings from a single referral-center. Per-protocol and calibrated cutoffs defined FP and TN groups. From the time of stool collection, we measured differences between FP and TN groups in time to death, subsequent cancer diagnosis and onset of alarm symptoms.
Results
Of 1050 eligible patients, only 6 were lost to follow-up. Median age was 65.6 years (IQR, 56.8, 72.3); 54% were female. Median follow-up time was 4 years (IQR, 3.5-5.3). Eight aerodigestive (lung & gastrointestinal tract) cancers occurred. FP status by calibrated, but not per-protocol cut-offs was associated with subsequent aerodigestive cancer; however, cumulative incidence did not exceed SEER expectations from the general population. By any cut-off method, FP status was not associated with mortality or alarm symptoms.
Conclusions
While FP status was associated with long-term aerodigestive cancers, new cases were not temporally related and did not exceed incidence estimates from general population.
Impact
These observations do not justify aggressive follow-up evaluation for patients with FP MT-sDNA at this time. Larger studies are needed to confirm these early findings.
Keywords: Colorectal Neoplasms/prevention and control, DNA methylation, predictive value of tests, meta-analysis, tumor biomarkers
INTRODUCTION
Colorectal cancer (CRC) is the second leading cause of cancer death in the United States (1). Because CRC deaths are preventable (2–4), the U.S. Preventive Services Task Force (USPSTF) currently recommends that all adults aged 50 years to 75 years and select adults aged 76 years to 85 years undergo screening (5). In their updated guidelines, the USPSTF now includes the new multi-target stool DNA test (MT-sDNA) as a fully-endorsed screening option; MT-sDNA has also been endorsed by the American College of Gastroenterology (6) and the American Cancer Society (7). More recently, the National Committee for Quality Assurance has included MT-sDNA among its Healthcare Effectiveness Data and Information Set for 2017 (8).
The MT-sDNA test is now available for general CRC screening (ColoGuard™, Exact Sciences, Madison WI). In August 2014, this noninvasive test was jointly approved by the US Food and Drug Administration (FDA) and the Centers for Medicare and Medicaid Services (CMS) for CRC screening in average-risk patients after demonstration of significantly greater sensitivity for detecting CRC and advanced precancerous lesions compared to fecal immunochemical testing (FIT) alone (9). In the cross-sectional screening studies (9, 10), MT-sDNA sensitivities for CRC were 92-100% and for adenomas ≥1cm, >1cm, ≥2cm, and ≥3cm were 40–42%, 51–52%, 62–66%, and 68–80%, respectively. In these same screen-setting studies (9, 10), CRC or advanced precancerous lesions (advanced adenoma or sessile serrated polyps ≥1 cm) were absent in 7–13% of follow-up colonoscopies after positive MT-sDNA tests (9–12).
The MT-sDNA test incorporates both DNA (methylated BMP3 & NDRG4 and mutant KRAS, all normalized by β-actin) and hemoglobin markers. A logistic algorithm converts quantitative data from these component assays into a binomial “positive” or “negative” result, and a FP rate approximating 10% was pre-specified in development studies (11, 12). The biological and clinical contributions underlying these apparent FPs remain to be established. Molecular pre-neoplastic field changes in the colorectum may exist in the absence of gross lesions and could potentially be detected by stool testing. Colorectal lesions could have been overlooked on colonoscopic evaluation of positive MT-sDNA results, as neoplasm misses by colonoscopy are well-documented (13, 14). And, theoretically, neoplasms above the colon could bleed or exfoliate markers that are excreted in stool. DNA abnormalities among the MT-sDNA panel have been observed in tissues of other airway and digestive tract (collectively, aerodigestive (AD)) neoplasms (15–17). Yet, minimal decreases in MT-sDNA specificity have been observed as a result of cross-reactivity with AD neoplasms in cross-sectional experiments (18). Furthermore, supra-colonic neoplasms were not detected by additional endoscopic and cross-sectional imaging testing in a previous small cohort study, evaluating patients with positive stool DNA tests results (19).
It is uncertain whether or not significant long-term adverse outcomes are associated with FP by the now available MT-sDNA test. Specifically, are FP MT-sDNA patients at sufficiently increased risk of mortality or subsequent cancer diagnosis to justify additional follow-up testing or intensification of cancer surveillance? This area of uncertainty is an important concern for patients and providers, notably the American Academy of Family Physicians (AAFP).(20). As such, we aimed to address this question by examining long-term outcomes among participants from three pre-approval studies of MT-sDNA (9, 11, 21) who had either non-advanced adenomas only or negative findings on baseline colonoscopy. We measured differences in mortality rate, subsequent cancer incidence, and development of alarm symptoms among patients with apparent FP and true negative (TN) MT-sDNA results.
MATERIALS AND METHODS
Study Design and Population
This study was conducted after institutional review board approval. Methods and results are reported in accordance with the STROBE (Strengthening the Reporting of Observational Studies in Epidemiology) guidelines (22). We identified a retrospective cohort of patients at Mayo Clinic (Rochester MN, Scottsdale AZ and Jacksonville FL) who participated in any of 3 MT-sDNA pre-approval clinical studies and whose study colonoscopy showed neither CRC nor advanced precancerous lesions (Table 1).
Table 1.
Design and assays used in pre-approval studies
| Study 1 | Study 2 | Study 3 | |
|---|---|---|---|
| Study name | Specificity | Cutoff | DeeP-C |
| Design | Cohort | Case-control | Cohort |
| Participants | Low risk patients, recruited after negative colonoscopy |
Cases referred for surveillance or known colorectal neoplasm, asymptomatic average-risk controls |
Average risk patients due for CRC screening |
| Methylated DNA markers |
BMP3* NDRG4* TFPI2 Vimentin |
BMP3* NDRG4* |
BMP3* NDRG4* |
| Mutant DNA markers |
- | KRAS | KRAS |
| Normalizing DNA marker |
β-actin* | β-actin* | β-actin* |
| FIT | - | Yes | Yes |
FIT, fecal immunochemical test;
Markers used in calibrated analysis
Study 1 (“Specificity”) cross-sectionally sampled asymptomatic patients at average risk for CRC between February 2010 and August 2010 (21). Study 2 (“Cutoff”) was a case-control sampling of higher-risk patients referred for known colorectal neoplasia and asymptomatic average-risk control patients who were scheduled for screening at the time of study enrollment, between August 2011 and December 2011 (11). Study 3 (“Deep-C”) was a cross sectional cohort sample of asymptomatic, average-risk patients who were scheduled to undergo screening colonoscopy at the time of study enrollment, between June 2011 and November 2012 (ClinicalTrials.gov number, NCT01397747)(9). At the time of enrollment in each of the original studies, patients were excluded for personal history of cancer in the lungs or gastro-intestinal tract, family history of CRC in a 1st degree relative, a comorbid inflammatory bowel disease diagnosis or a known genetic cancer syndrome. Additionally, exposures to tobacco and alcohol were collected on all patients. Prototype molecular marker panels performed in each study are listed in Table 1.
Data Collection
After an Accurint database (LexisNexis, Dayton OH) search to determine patients’ vital status, patients or next-of-kin were invited by mail for structured telephone interview to document new cancer or precancer diagnoses, subsequent esophagogastroduodenoscopy (EGD) or colonoscopy results, and the development of alarm symptoms (anemia, unintentional weight loss, dysphagia, early satiety, bowel habit change or gross GI bleeding), since study participation (Supplemental Information). Chart review was performed on all who declined interview and those who did not respond; chart review was also performed on a random subset of those interviewed to measure potential reporting bias. Patient follow-up was censored at the time of death, loss to follow-up, or finalization of the study database on December 31, 2015. Deaths were ascertained from the Accurint database, next of kin interview and chart review. Subsequent cancer diagnoses were confirmed, wherever possible, by review of endoscopic, radiographic and histopathologic reports.
Determination of false positive rate
Because each of the 3 studies used a different MT-sDNA prototype marker panel, baseline FP MT-sDNA results were determined by two methods. First, the FP and TN rates were measured according to per-protocol definitions, reported separately for each study. Briefly, 90th percentile values for each marker panel were applied to all patients in the “Specificity” study and “Cutoff” study controls to optimize panel specificity to a goal of 90%, whereas “DeeP-C” used a pre-specified logistic-regression algorithm, with a calculated score of ≥183 indicating a positive result (9). In the second method, the results of MT-sDNA across each of the three studies were calibrated using three DNA markers, methylated BMP3, methylated NDRG4 and β-actin, measured in common for all patients. To scale all three markers, raw data for each was standardized to the mean and standard deviation within the study of origin. Then a smoothing spline logistic regression model was used to calibrate marker values from “Specificity” and “Cutoff” to “DeeP-C” test results. From this calibrated data set, 90th and 95th percentile rates for all 3 markers combined, were used to establish 10% and 5% FP rates across all 3 studies.
Statistical analysis
Assuming MTsDNA test specificity of 90%, 850 patients were anticipated to provide 80% power to detect a difference between a 6% cumulative event rate in the FP group from a 1% event rate in the TN group, at the 5% significance level. Primary study end-points included rates of mortality, subsequent AD cancer diagnosis, any subsequent cancer diagnosis and development of alarm symptoms, assessed from the time of stool collection, using the Kaplan-Meier method. Association of FP status with each of these rates was assessed by proportional hazards, reported as hazard ratios (HR) with 95% confidence intervals (CI). Cumulative incidence of AD cancers in FP and TN groups were also compared with expected Surveillance, Epidemiology, and End Results (SEER) Program cumulative incidence (23). Potential differences in proportions or continuous distributions of baseline characteristics between patients in each study were assessed with chi-square, Fisher’s exact or Wilcoxon rank-sum tests, where applicable. Geographic distance from the site of study participation to each patient’s primary address was measured as a surrogate for ongoing access to the study site for clinical follow-up. A distance of 100 miles or less was used as a surrogate for access to primary care at the study site.
RESULTS
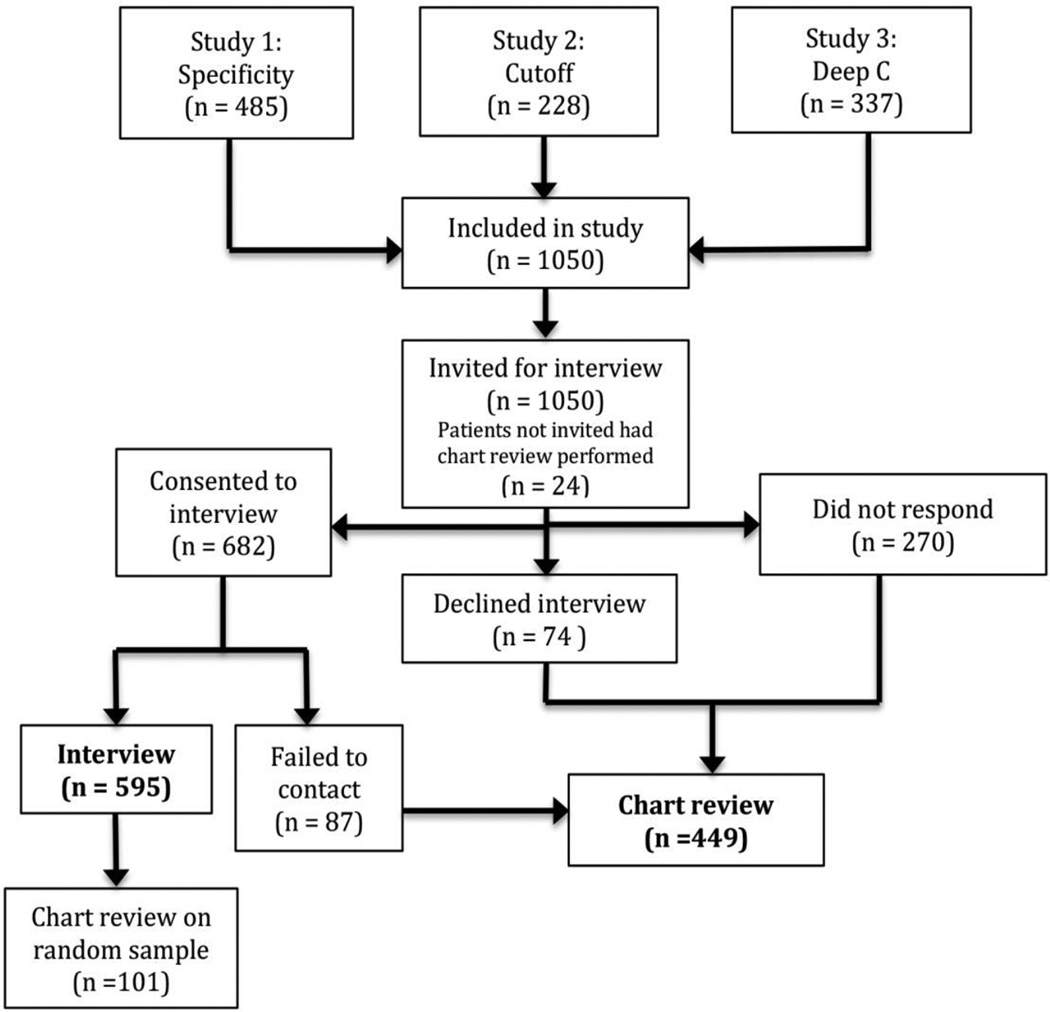
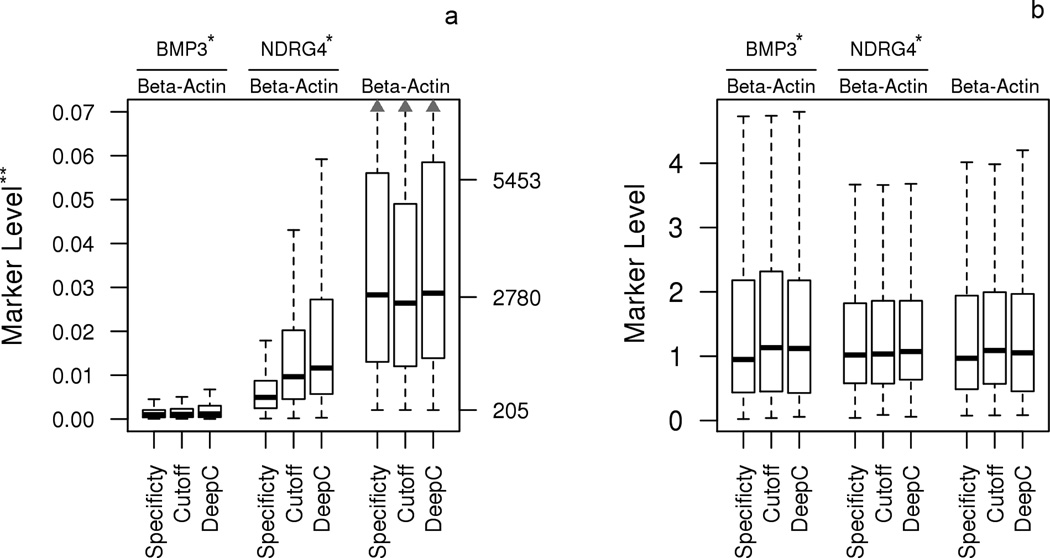
There were 1050 eligible participants. Of these 595 (57%) participated in the structured interview. Six patients declined both interview and chart review; however, their vital status was publically available (Figure 1). Using the per-protocol method, MT-sDNA was FP in 160 (15%) and TN in 890 (85%) patients. The stool biomarkers common to all three studies, before and after calibration are shown in Figure 2. Using the calibrated method, MT-sDNA was falsely positive in 113 (11%) and 51 (5%) patients at 90th and 95th percentile thresholds, respectively, and this was not significantly different by study (Table 2) Baseline demographic and clinical features of all patients are shown in Table 2. Median age of the “Deep-C” cohort was 69 years (IQR, 65-74); this was significantly older than the median age in the other two studies (p < 0.0001). While sex was similar across all three studies, the “Specificity” study participants were more racially diverse. Of the 1050 patients, 570 (54%) were geographically defined as having access to primary care at Mayo Clinic. This was similar across MT-sDNA test result groups: 490/890 (55%) of TN vs 80/160 (50%) of FP (p=0.26), using the per protocol cutoff; 514/937 (55%) of TN vs 56/113 (50%) of FP (p=0.32), using the calibrated 90% cutoff; and, 544/999 (54%) of TN vs 26/51 (51%) of FP (p=0.67), using the calibrated 95% cutoff. There was also no difference in geographic variation when distance to Mayo Clinic was analyzed as a continuous variable (p=0.06), using the per protocol cutoff.
Figure 1.
Study flow diagram.
Figure 2.
Distributions of stool DNA markers (BMP3, NDRG4 and β-actin) by study, (A) standardized to the mean and standard deviation within the study of origin and (B) after center and scaling the data.
* Copy numbers of BMP3 and NDRG4 were standardized by beta-actin
** Left and right Y-axes show ratios for the standardized markers and copy numbers for beta-actin PCR products, respectively
Table 2.
Demographic and Clinical Factors in Study Cohort
| Variable | Overall Cohort |
Study 1 “Specificity” |
Study 2 “Cutoff” |
Study 3 “Deep-C” |
P-value | |
|---|---|---|---|---|---|---|
| Patients, n | 1050 | 485 | 228 | 337 | ||
| Median age (IQR), years |
65.6 (56.8, 72.3) | 64.1 (56.6, 71.8) | 62.4 (53.0, 70.9) | 68.5 (65.0, 73.8) | <0.0001 | |
| Women, n (%) | 563 (54) | 257 (53) | 119 (52) | 187 (55) | 0.70 | |
| White race, n (%) | 981 (93) | 437 (90) | 216 (95) | 328 (97) | <0.0001 | |
| Patient status by specificity cut-off |
||||||
| Per-protocol (90%) | ||||||
| FP, n (%) | 160(15) | 65 (13) | 36 (16) | 59 (18) | 0.27 | |
| TN, n (%) | 890 (85) | 420 (87) | 192 (84) | 278 (82) | ||
| Calibrated (90%) | ||||||
| FP, n (%) | 113 (11) | 53 (11) | 25 (11) | 35 (10) | 1.00 | |
| TN, n (%) | 937 (89) | 432 (89) | 203 (89) | 302 (90) | ||
| Calibrated (95%) | ||||||
| FP n (%) | 51 (5) | 25 (5) | 10 (4) | 16 (5) | 0.93 | |
| TN, n (%) | 999 (95) | 460 (95) | 218 (96) | 321 (95) | ||
IQR, Interquartile Range; FP, False Positive; TN, True Negative
Median follow-up time was 4 years (IQR, 3.5-5.3) and was similar between FP and TN groups (p=0.10, per-protocol, p > 0.9, calibrated). The proportion of patients who underwent a subsequent invasive gastrointestinal diagnostic procedure (any of colonoscopy, flexible sigmoidoscopy or EGD) during follow-up was: 187/885 (21%) of TN vs 36/159 (23%) of FP (p=0.67), using the per protocol cutoff; 199/932 (21%) of TN vs 24/112 (21%) of FP (p=1.00), using the calibrated 90% cutoff; and, 215/993 (22%) of TN vs 8/51 (16%) of FP (p=0.38), using the calibrated 95% cutoff.
During study follow-up a total of 25 patients were reported deceased. Of these, all were confirmed by chart review or telephone interview with next of kin. A random sample of patients who had both interview and chart review performed were analyzed to assess agreement on cancer events (yes vs no) between the two methods of data abstraction. There was agreement in 51 out of the 53 patients assessed. The 2 that were not in agreement had noted skin cancer on the interview but not in the chart review. Of 48 total subsequent cancer events, 36 were reported during interview and 12 additional events were found on chart review. All cancer events, except non-melanoma skin neoplasms, were confirmed by histopathology review. Subsequent alarm signs were common, occurring in 13% of the cohort after 5 years of follow-up.
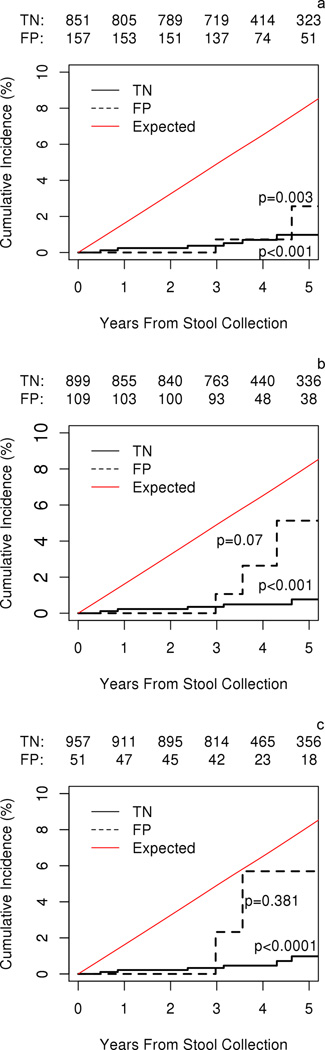
By either cut-off method, FP status was not associated with mortality, subsequent cancer of all types or alarm symptoms (Table 3). FP status by calibrated, but not per-protocol, cut-offs was associated with subsequent AD cancer (Table 3). There were 8 incident AD cancers (1 CRC, 3 lung, 3 pancreatic, 1 bile duct) in the study cohort (Supplemental Table 1). One patient considered false positive by all three cut-off definitions, subsequently developed CRC; this individual’s multiple sub-centimeter adenoma lesions at index colonoscopy did not meet pre-specified advanced colorectal neoplasia endpoint criteria (9). By any cut-off method, cumulative incidence of AD cancer in the FP group fell below the expected age- and gender-specific SEER incidence, and all AD cancers discovered on follow-up were diagnosed beyond 3 years from MT-sDNA testing (Figure 3).
Table 3.
Hazard ratios, calibrated and age-adjusted
| Outcome | 5 Yr. Cumulative |
Hazard ratio (95% CI) for false positive MT-sDNA† |
||
|---|---|---|---|---|
| Incidence % (95% CI) |
Per Protocol Cutoffs |
Calibrated 90th Percentile |
Calibrated 95th Percentile |
|
| All-cause mortality | 3 (2–4) | 0.9 (0.4–2.7) | 1.7 (0.6–4.5) | 2.3 (0.7–7.6) |
| Subsequent aerodigestive cancer* |
1 (0.3–2) | 1.3 (0.3–6.7) | 4.1 (1.0–17.6) | 5.5 (1.1–27.5) |
| Any subsequent cancer** |
5 (4–7) | 1.1 (0.6–2.3) | 0.8 (0.3–2.1) | 1.1 (0.4–3.7) |
| Any subsequent alarm signs |
13 (10–17) | 0.8 (0.4–1.5) | 0.6 (0.3–1.4) | 0.6 (0.2–2.1) |
Age-adjusted
Included 1 colorectal, 3 pancreatic, 3 lung, 1 bile duct
Above plus 16 skin, 12 genitourinary, 8 breast, 2 hematological, 1 ophthalmological, 1 glioblastoma
Figure 3.
Cumulative incidence of aerodigestive cancers in FP and TN groups versus expected SEER cumulative incidence (1953 / 100,000 person-years) by A) per-protocol results, B) calibrated 90% specificity, and C) calibrated 95% specificity. Aerodigestive cancers occurred in FP patients at a rate of 278, 698, and 1015 per 100,000 person-years in per protocol, calibrated 90% specificity and calibrated 95% specificity cut-offs, respectively. FP = false positive cohort; TN = true negative cohort; SEER = Surveillance, Epidemiology, and End Results Program of the National Cancer Institute
DISCUSSION
This is the first study on the long-term follow-up of patients who underwent MT-sDNA testing for CRC screening. Five-year survival is similar between patients with FP and TN MT-sDNA results. Only 8 subsequent AD cancer events were observed in the cohort of 1043 patients, a rate far below that expected based on SEER incidence data. By per-protocol assignments, these were not associated with FP MT-sDNA tests. However, after calibrated cut-offs were applied, the AD cancer rates were statistically increased among those with FP MT-sDNA. However, temporal association of these events with MT-sDNA appears to be lacking as the earliest AD cancer among FP patients presented 3 years after MT-sDNA testing. The first event, an early-stage CRC, would have been detected at 93% sensitivity by MT-sDNA at the subsequent recommended 3-year screening interval. The other AD cancers were detected over 4 years after MT-sDNA testing. Given the rapid growth rate of lung cancers, current guidelines recommend annual screening of patients at increased risk (24). There is no population-level screening for other AD cancers, but even those at risk for familial pancreatic cancer might be offered only annual surveillance (25). Therefore, it is doubtful that the positive MT-sDNA test was attributable to a pre-clinical AD cancer.
For patient inclusion in each study, index colonoscopies were performed by endoscopists whose adenoma detection rates are regularly monitored. Study procedures also required documented evidence of cecal intubation, annotation of withdrawal time and bowel preparation sufficient for examination of ≥90% of the mucosal surface. These standards of care for screening colonoscopy are supported by jointly issued guidelines from the American Society for Gastrointestinal Endoscopy and American College of Gastroenterology (26).
For follow-up of other non-invasive CRC screening methods, little data is available data on the yield of additional testing, beyond colonoscopy. Despite high cumulative false positive rates of programmatic FOBT screening (27), there is insufficient evidence to recommend additional invasive testing (28). While there are no U.S. guidelines to direct management in this subgroup of patients, a recent microsimulation study (29) concluded that these individuals could resume screening with annual FOBT, 10 years after negative follow-up colonoscopy (30).
Extrapolation of screen-setting observations to a theoretical population of 10,000 persons revealed that 16% of MT-sDNA tests will come back positive, with 45.4% of these having negative colonoscopy results (9). Based on the SEER incidence and studies in stools of patients with liver, pancreatic and gynecologic cancers, it was estimated that specificity of MT-sDNA for colorectal cancer would only be reduced by 0.02% by cross-reactivity to these neoplasms (18).
We have opportunistically utilized a ready pool of patients from three pre-approval studies performed sufficiently long ago to calculate 5-year event rates. The strengths of this study include the large sample size, high response rate, high agreement between survey and chart review, and low loss to follow up. The majority of patients in our cohort were local to the Mayo Clinic system, optimizing the chances of capturing subsequent cancer events. The Kaplan-Meier analysis method also was used to account for variable rates of follow-up during the study period. Hard outcomes like cancer incidence can the difficult to ascertain as there is no national-level cancer registry to completely enumerate cancer events. We feel that our method of event ascertainment by structured survey and electronic medical records review was adequate, but could have also missed events. As with all telephone interviews, recall bias has to be considered. However, it is unlikely that our primary outcomes, death and subsequent cancer diagnosis, would be affected. This was confirmed by measurement of agreement between chart review and survey for a randomly selected sub-set of patients. The development and reporting of subsequent alarm signs may have been influenced by recall bias. This secondary outcome was measured to avoid missing potentially un-diagnosed cancers and was not significantly different between groups.
We acknowledge that the total sample size limited the ability to measure between-group differences in event rates of <5%. A multivariate analysis of subsequent cancer events in larger sets of patients will also be needed to measure for interaction of false-positive MT-sDNA results and exposures to other cancer risk factors, especially smoking. Additionally, the now FDA-approved version of MT-sDNA test was only used in 337 patients, representing 32% of our overall combined cohort. To account for this, data were logistically calibrated using 3 markers common to all studies. While this method also permitted assessment of more stringent hypothetical specificity thresholds, direct extrapolation of these findings to the test specificity of the FDA-approved MT-sDNA test, which uses a larger panel of markers, is potentially limited. Also, these observations may not be applicable to patient populations at higher risk of cancer. As the MT-sDNA test becomes more widely used in the population, additional study of clinical outcomes of patients screened by MT-sDNA is warranted.
Clinical MT-sDNA test specificity in clinical use may differ from that observed in clinical trials. This difference may be attributable to patient factors. In the largest screen-setting study of MT-sDNA, sub-group analyses showed that specificity was 94% in participants age 65 years or less but 87% in those over 65 (p < 0.0001) (9). Moreover, MT-sDNA specificity was 89.8% (95% CI, 88.9 to 90.7) in the sub-set of patients with completely negative colonoscopies; this was higher than the observed specificity of 86.6% (95% CI, 85.9 to 87.2) from the entire study cohort which included colonoscopy findings of non-advanced adenomas or other non-neoplastic pathology among “negative” results (9). Provider factors may also be influence MT-sDNA yield. Early observations from the first year of clinical use in a referral-center practice, suggest that polyp detection rates and colonoscopy withdrawal times were significantly higher among endoscopists aware of a positive MT-sDNA results in clinical practice compared to those blinded in pre-approval studies (31). Specificity also needs to be examined over the duration of a screening program. Used every 3 years, MT-sDNA is expected to generate fewer FP results and therefore fewer follow-up colonoscopies than annual FIT screening (32) or other modalities over a lifetime (33). This directly influences the risk to benefit ratio of programmatic screening. A recent paper analyzing the comparative and cost effectiveness of CRC screening methods using a Markov model demonstrated that colonoscopy and FIT were more effective and less costly than MT-sDNA, assuming equal adherence (34). However, the same study acknowledged that consistent participation in yearly CRC screening by FIT is only 15%; if MT-sDNA yielded CRC participation rates more than 1.7-fold relative to FIT then MT-sDNA every 3 years cost less than $100,000 per quality-adjusted life-year gained compared with yearly FIT. Additionally, another recent study commissioned by the USPSTF, showed that, MT-sDNA performed at 3 year intervals yielded the greatest number of life-years gained and averted the most colorectal cancer deaths relative to the harms from complications of screening and follow-up tests generated by all-other USPSTF-endorsed screening approaches (33).
In summary, patients with positive MT-sDNA tests are unlikely to benefit from additional follow-up evaluations after negative high-quality colonoscopy. We observed similar 5-year mortality and subsequent all-cancer event rates between those with positive and negative MT-sDNA, following a negative study colonoscopy. False-positive tests were associated with a low, but statistically significant increase in AD cancers between groups, all of which presented after 3–5 years of follow-up. The clinical significance of this observation appears questionable, as events do not show temporal association and fall below SEER estimates of AD cancer incidence in the US population. Larger studies are needed to confirm these observations.
Supplementary Material
Acknowledgments
Financial Support: This research was supported by a grant from the Maxine and Jack Zarrow Family Foundation of Tulsa Oklahoma and the Paul Calabresi Program in Clinical-Translational Research (NCI CA90628). Additional partial support was provided by the Carol M. Gatton endowment for Digestive Diseases Research. These funding sources had no role in the funding, design or conduct of the Study.
Footnotes
Conflict of Interest Statement: Conflicts of Interest Statement: Mayo Clinic is a minor equity holder in Exact Sciences (Madison WI) and has an intellectual property agreement with Exact Sciences under which inventors, such as Drs. Kisiel and Ahlquist and Mr. Mahoney could receive royalties in accordance with Mayo Clinic policy. Exact Sciences had no role in the funding, design or conduct of the study. TG Cotter, KN Burger, ME Devens, JA Simonson, KL Lowrie, DH Johnson and RI Heigh have no relevant conflict of interest. Portions of this manuscript were presented in abstract form at Digestive Diseases Week, May 23, 2016, San Diego CA.
References
- 1.Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64(2):104–117. doi: 10.3322/caac.21220. [DOI] [PubMed] [Google Scholar]
- 2.Atkin WS, Edwards R, Kralj-Hans I, Wooldrage K, Hart AR, Northover JM, et al. Once-only flexible sigmoidoscopy screening in prevention of colorectal cancer: a multicentre randomised controlled trial. Lancet. 2010;375(9726):1624–1633. doi: 10.1016/S0140-6736(10)60551-X. [DOI] [PubMed] [Google Scholar]
- 3.Shaukat A, Mongin SJ, Geisser MS, Lederle FA, Bond JH, Mandel JS, et al. Long-term mortality after screening for colorectal cancer. N Engl J Med. 2013;369(12):1106–1114. doi: 10.1056/NEJMoa1300720. [DOI] [PubMed] [Google Scholar]
- 4.Nishihara R, Ogino S, Chan AT. Colorectal-cancer incidence and mortality after screening. N Engl J Med. 2013;369(24):2355. doi: 10.1056/NEJMc1313116. [DOI] [PubMed] [Google Scholar]
- 5.US Preventive Services Task Force. Screening for Colorectal Cancer: US Preventive Services Task Force Recommendation Statement. JAMA. 2016;315(23):2564–2575. doi: 10.1001/jama.2016.5989. [DOI] [PubMed] [Google Scholar]
- 6.Rex DK, Johnson DA, Anderson JC, Schoenfeld PS, Burke CA, Inadomi JM, et al. American College of Gastroenterology guidelines for colorectal cancer screening 2009 [corrected] Am J Gastroenterol. 2009;104(3):739–750. doi: 10.1038/ajg.2009.104. [DOI] [PubMed] [Google Scholar]
- 7.Colorectal cancer prevention and early detection. [Accessed December 2, 2016];American Cancer Society website. Available at http://www.cancer.org/cancer/colonandrectumcancer/moreinformation/colonandrectumcancerearlydetection/colorectal-cancer-early-detection-screening-tests-used. Updated April 24, 2016.
- 8.National Committee for Quality Assurance (NCQA) [Accessed December 2, 2016];HEDIS®1 2017 Volume 2: Technical Update. Available at http://www.ncqa.org/Portals/0/HEDISQM/HEDIS2017/HEDIS%202017%20Volume%202%20Technical%20Update.pdf?ver=2016-10-03-114902-317 Updated October 3, 2016.
- 9.Imperiale TF, Ransohoff DF, Itzkowitz SH, Levin TR, Lavin P, Lidgard GP, et al. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med. 2014;370(14):1287–1297. doi: 10.1056/NEJMoa1311194. [DOI] [PubMed] [Google Scholar]
- 10.Redwood DG, Asay ED, Blake ID, Sacco PE, Christensen CM, Sacco FD, et al. Stool DNA Testing for Screening Detection of Colorectal Neoplasia in Alaska Native People. Mayo Clin Proc. 2016;91(1):61–70. doi: 10.1016/j.mayocp.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 11.Lidgard GP, Domanico MJ, Bruinsma JJ, Light J, Gagrat ZD, Oldham-Haltom RL, et al. Clinical performance of an automated stool DNA assay for detection of colorectal neoplasia. Clin Gastroenterol Hepatol. 2013;11(10):1313–1318. doi: 10.1016/j.cgh.2013.04.023. [DOI] [PubMed] [Google Scholar]
- 12.Ahlquist DA, Zou H, Domanico M, Mahoney DW, Yab TC, Taylor WR, et al. Next-generation stool DNA test accurately detects colorectal cancer and large adenomas. Gastroenterology. 2012;142(2):248–256. doi: 10.1053/j.gastro.2011.10.031. quiz e25-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brenner H, Hoffmeister M, Arndt V, Stegmaier C, Altenhofen L, Haug U. Protection from right- and left-sided colorectal neoplasms after colonoscopy: population-based study. J Natl Cancer Inst. 2010;102(2):89–95. doi: 10.1093/jnci/djp436. [DOI] [PubMed] [Google Scholar]
- 14.Kahi CJ, Hewett DG, Norton DL, Eckert GJ, Rex DK. Prevalence and variable detection of proximal colon serrated polyps during screening colonoscopy. Clin Gastroenterol Hepatol. 2011;9(1):42–46. doi: 10.1016/j.cgh.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 15.Dai Z, Popkie AP, Zhu WG, Timmers CD, Raval A, Tannehill-Gregg S, et al. Bone morphogenetic protein 3B silencing in non-small-cell lung cancer. Oncogene. 2004;23(20):3521–3529. doi: 10.1038/sj.onc.1207441. [DOI] [PubMed] [Google Scholar]
- 16.Liu Z, Zhang J, Gao Y, Pei L, Zhou J, Gu L, et al. Large-scale characterization of DNA methylation changes in human gastric carcinomas with and without metastasis. Clin Cancer Res. 2014;20(17):4598–4612. doi: 10.1158/1078-0432.CCR-13-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hashimoto D, Arima K, Yokoyama N, Chikamoto A, Taki K, Inoue R, et al. Heterogeneity of KRAS Mutations in Pancreatic Ductal Adenocarcinoma. Pancreas. 2016 doi: 10.1097/MPA.0000000000000624. [DOI] [PubMed] [Google Scholar]
- 18.Exact Sciences Corporation. Summary of Effectiveness and Safety Data 2014. Accessed at Food and Drug Administration (FDA) website at http://www.accessdata.fda.gov/cdrh_docs/pdf13/P130017b.pdf on September 9, 2015.
- 19.Ahlquist DASD, Loprinzi CL, Goldberg RM, Rex DK, Levin TR, Ahnen DJ, Knigge KL, Lance MP, Lawson MJ, Allison JE, Devens ME, Stella PJ, Hillman SL. False-positive stool DNA results on colorectal cancer screening yield from supracolonic evaluation. Gastroenterology. 2004;126:A-58. [Google Scholar]
- 20.Clinical Preventive Service Recommendation: Colorectal Cancer. [Accessed December 2, 2016];American Academy of Family Physicians website. Available at http://www.aafp.org/patient-care/clinical-recommendations/all/colorectal-cancer.html Updated August 31, 2016.
- 21.Ahlquist DA, Taylor WR, Yab TC, Devens ME, Mahoney DW, Boardman LA, et al. Aberrantly Methylated Gene Marker Levels in Stool: Effects of Demographic, Exposure, Body Mass, and Other Patient Characteristics. J Mol Biomark Diagn. 2012;3(133) [Google Scholar]
- 22.von Elm E, Altman DG, Egger M, Pocock SJ, Gotzsche PC, Vandenbroucke JP, et al. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: guidelines for reporting observational studies. Ann Intern Med. 2007;147(8):573–577. doi: 10.7326/0003-4819-147-8-200710160-00010. [DOI] [PubMed] [Google Scholar]
- 23.National Cancer Institute. [Accessed September 9, 2015];Surveillance, Epidemiology, and End Results (SEER) Program: Incidence - SEER 9 Regs Research Data, Nov 2014 Sub (1973-2012) Available at https://seer.cancer.gov/data/seerstat/nov2014/
- 24.U.S. Preventive Services Task Force. [Accessed on December 2, 2016];Final Recommendation Statement: Lung Cancer: Screening. Available at https://www.uspreventiveservicestaskforce.org/Page/Document/RecommendationStatementFinal/lung-cancer-screening Updated October 2014.
- 25.Canto MI, Harinck F, Hruban RH, Offerhaus GJ, Poley JW, Kamel I, et al. International Cancer of the Pancreas Screening (CAPS) Consortium summit on the management of patients with increased risk for familial pancreatic cancer. Gut. 2013;62(3):339–347. doi: 10.1136/gutjnl-2012-303108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rex DK, Schoenfeld PS, Cohen J, Pike IM, Adler DG, Fennerty MB, et al. Quality indicators for colonoscopy. Am J Gastroenterol. 2015;110(1):72–90. doi: 10.1038/ajg.2014.385. [DOI] [PubMed] [Google Scholar]
- 27.Hubbard RA, Johnson E, Hsia R, Rutter CM. The cumulative risk of false-positive fecal occult blood test after 10 years of colorectal cancer screening. Cancer Epidemiol Biomarkers Prev. 2013;22(9):1612–1619. doi: 10.1158/1055-9965.EPI-13-0254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Allard J, Cosby R, Del Giudice ME, Irvine EJ, Morgan D, Tinmouth J. Gastroscopy following a positive fecal occult blood test and negative colonoscopy: systematic review and guideline. Can J Gastroenterol. 2010;24(2):113–120. doi: 10.1155/2010/516363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frazier AL, Colditz GA, Fuchs CS, Kuntz KM. Cost-effectiveness of screening for colorectal cancer in the general population. JAMA. 2000;284(15):1954–1961. doi: 10.1001/jama.284.15.1954. [DOI] [PubMed] [Google Scholar]
- 30.Haug U, Knudsen AB, Kuntz KM. How should individuals with a false-positive fecal occult blood test for colorectal cancer be managed? A decision analysis. Int J Cancer. 2012;131(9):2094–2102. doi: 10.1002/ijc.27463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson D, Kisiel JB, Burger KN, Mahoney DW, Devens ME, Ahlquist DA, Sweetser S. Knowledge of a Positive Cologuard™ Result Improves Yield and Quality of Colonoscopy. Gastroenterology. 2016;150(4):S454. [Google Scholar]
- 32.Kisiel JB, Ahlquist DA. Stool DNA screening for colorectal cancer: opportunities to improve value with next generation tests. J Clin Gastroenterol. 2011;45(4):301–308. doi: 10.1097/MCG.0b013e3181f0f028. [DOI] [PubMed] [Google Scholar]
- 33.Knudsen AB, Zauber AG, Rutter CM, Naber SK, Doria-Rose VP, Pabiniak C, et al. Estimation of Benefits, Burden, and Harms of Colorectal Cancer Screening Strategies: Modeling Study for the US Preventive Services Task Force. JAMA. 2016;315(23):2595–2609. doi: 10.1001/jama.2016.6828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ladabaum U, Mannalithara A. Comparative Effectiveness and Cost Effectiveness of a Multitarget Stool DNA Test to Screen for Colorectal Neoplasia. Gastroenterology. 2016;151(3):427–439. doi: 10.1053/j.gastro.2016.06.003. e6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.