Abstract
Pancreatic ductal adenocarcinoma (PDAC) is referred to as a silent killer due to the lack of clear symptoms, a lack of early detection methods, and a high frequency of metastasis at diagnosis. In addition, pancreatic cancer is remarkably resistant to chemotherapy, and clinical treatment options remain limited. The tumor microenvironment (TME) and associated factors are important determinants of metastatic capacity and drug resistance. Here, oncostatin M (OSM), an IL-6 cytokine family member, was identified as an important driver of mesenchymal and cancer stem cell (CSC) phenotypes. Furthermore, the generation of cells that harbor mesenchymal/CSC properties following OSM exposure resulted in enhanced tumorigenicity, increased metastasis, and resistance to gemcitabine. OSM induced the expression of ZEB1, Snail (SNAI1), and OSM receptor (OSMR), engaging a positive feedback loop to potentiate the mesenchymal/CSC program. Suppression of JAK1/2 by Ruxolitinib prevented STAT3-mediated transcription of ZEB1, SNAI1, and OSMR, as well as the emergence of a mesenchymal/CSC phenotype. Likewise, ZEB1 silencing, by shRNA-mediated knockdown, in OSM-driven mesenchymal/CSC reverted the phenotype back to an epithelial/non-CSC state. Importantly, the generation of cells with mesenchymal/CSC properties was unique to OSM, and not observed following IL-6 exposure, implicating OSMR and downstream effector signaling as a distinct target in PDAC. Overall, these data demonstrate the capacity of OSM to regulate an epithelial-mesenchymal transition (EMT)/CSC plasticity program that promotes tumorigenic properties.
Keywords: Pancreatic cancer, Oncostatin-M, Cancer stem cells, STAT3, Zeb1
INTRODUCTION
Pancreatic ductal adenocarcinoma (PDAC) has been referred to as a silent killer due to the lack of clear symptoms, and the lack of early detection methods. In the great majority of cases, patients present with advanced disease that has already metastasized. In addition, pancreatic cancer cells are remarkably resistant to chemotherapy, and additional treatment options remain limited. As a result, the 5 year survival rate is ~5% (1), with nearly as many patients dying each year of pancreatic cancer as are diagnosed. The problem lies, in part due to the early cellular dissemination of PDAC cells to distant organs, which can precede primary tumor formation (2). This highly invasive characteristic of PDAC (3) is largely associated with the gain of mesenchymal properties by the ductal epithelial cells, indicative of an epithelial-mesenchymal transition (EMT) (2). Expression of key transcription factors capable of driving EMT, such as ZEB1, Snail (SNAI1), and TWIST1/2, initiate the repression of tight cell-cell protein interactions (claudin, occludin, ZO1, and E-Cadherin), ultimately releasing cells to invade neighboring tissue (4–6). These invasive and motile properties allow the tumor cells to escape a primary site of disease, systemically disseminate, and seed metastases at secondary sites. Concomitant with this transition to an invasive, mesenchymal phenotype is the acquisition of cancer stem cells (CSC) properties responsible for enhanced tumorigenicity and acquired resistance to therapeutic drugs (7–11).
Emerging evidence suggests a pivotal role for the tumor microenvironment (TME) in promoting epithelial-mesenchymal plasticity (E-M plasticity) and the acquisition of CSC properties (12–15). The TME is composed of secreted factors emanating from infiltrating immune components, fibroblasts, and other stromal cells. Notably, severe immune infiltration and stromal fibrosis (desmoplasia) is observed in early and late stages of PDAC (16, 17), and enhances tumor growth/survival (18). Identifying key mediators within the TME that engage mesenchymal/CSC plasticity will be important for developing effective therapeutic targets for the treatment of PDAC.
Oncostatin M (OSM), an IL-6 family cytokine, is elevated in the serum of PDAC patients relative to healthy controls (19). Moreover, tumor-associated macrophages (at both primary and metastatic tumors) exhibit increased secretion of OSM in murine PDAC models (20). OSM signals through a gp130/OSM receptor (OSMR) complex, which activates the Janus kinase/ Signal Transducer and Activator of Transcription 3 (JAK/STAT3) pathway. STAT3 promotes gene expression both as a transcription factor as well as by epigenetic mechanisms, and is critical in inducing inflammation and cancer. Furthermore, constitutive activation of oncogenic STAT3 signaling promotes tumorigenesis and is associated with poor outcomes in PDAC (21, 22). Importantly, OSM can more potently activate STAT3 relative to other IL-6 family members (23, 24). However, very little is known about the biological effects of OSM-induced STAT3 in PDAC. Here, we examine the ability of OSM to promote the emergence of mesenchymal/CSC properties in PDAC. We demonstrate that OSM is capable of inducing a potent EMT/CSC program that enhances tumorigenic potential, cell motility, invasiveness and metastasis. Importantly, OSM conferred resistance to gemcitabine, a current front line therapy for PDAC. We propose that targeting OSM within the TME or OSM signaling within the tumor is likely to reduce the aggressive, metastatic and tumorigenic properties associated with PDAC, thereby improving patient response to therapy and extending patient survival.
MATERIALS AND METHODS
Cell Culture
All PDAC cell lines were obtained from ATCC and grown in a humidified atmosphere containing 5% CO2 at 37°C. HPAC cells and human fibroblasts were grown in DMEM/F12 (#10-092-CV; Corning) with 10% FCS (#S11150; Atlanta Biologicals), 0.005 mg/mL of human transferrin (#T2252; Sigma Aldrich), 10 ng/mL of Human Epidermal Growth Factor (#01-107; Millipore), 0.002 mg/mL of Human Insulin (#I9278; Sigma Aldrich), and 40 ng/mL of Hydrocortisone (#H4001; Sigma Aldrich). Panc 04.03 and Panc 08.13 cells were grown in RPMI-1640 (#SH30027.01; Hyclone) with 15% FBS (#S11150; Atlanta Biologicals). Panc 05.04 cells were grown in RPMI-1640 (#SH30027.02; Hyclone) with 15% FBS (#S11150; Atlanta Biologicals) and 0.2 U/mL of Human Insulin (#I9278; Sigma Aldrich). Treatments were either with 10 ng/mL human recombinant Oncostatin M (OSM; #OSM01-13; DAPCEL), 10 ng/ml IL-6 or 10 μM Ruxolitinib (#R-6688; LC Laboratories). All short-term treatments were performed as denoted in the figure legend; all long-term treatments were given at each medium change unless denoted otherwise (~48 hours). Human Fibroblast cultures were established from the digestion medium filtrate of primary reduction mammoplasty tissue as previously described (25). The fibroblast-containing filtrate was plated into DMEM (#10-013-CV; Corning) with 10% FBS (#S11150; Atlanta Biologicals) and immortalized using pBabe-Puro-hTERT.
Microscopy, Western blot analysis, and Quantitative Real-Time RT-PCR (qPCR)
Bright-field images were captured at 40X & 200X on a Nikon Eclipse TE2000-S using MetaMorph (Molecular Devices, Sunnyvale, CA). Western blots were conducted as described previously (12). Primary antibodies used were Actin (#MS-1295-P; Thermo Scientific), E-Cadherin (#sc-27191; Santa Cruz, #3195; Cell Signaling), Snail (#3879; Cell Signaling), phosphorylated STAT3Y705 (#9145; Cell Signaling), STAT3 (#9139; Cell Signaling), and ZEB1 (#3396; Cell Signaling). Secondary antibodies used were HRP-linked Anti-Mouse (#7076; Cell Signaling) and HRP-linked Anti-Rabbit (#7074; Cell Signaling). For qPCR, total RNA was isolated as previously described (12). RNA (1 μg) was reverse transcribed by iScript cDNA Synthesis Kit (#170-8891; Bio Rad). Gene expression was and identified using iQ SYBR Green Supermix (#170-8880; Bio Rad) and a CFX96 thermocycler (Bio Rad). All samples were normalized to GAPDH expression, error bars represent ± SEM for a representative experiment performed in triplicate. A two tailed unpaired student’s t-Test was performed in order to determine significance; * = p-value ≤ 0.05, ** = p-value ≤ 0.01, *** = p-value ≤ 0.001, **** = p-value ≤ 0.0001. Primer sequences were as follows: OSMR Forward: 5′-TCCCAATACCACAAGCACAG-3′; OSMR Reverse: 5′-GCAAGTTCCTGAGAGTATCCTG-3′; SNAI1 Forward: 5′-GGAAGCCTAACTACAGCGAG-3′; SNAI1 Reverse: 5′-CAGAGTCCCAGATGAGCATTG-3′; ZEB1 Forward: 5′-ACCCTTGAAAGTGATCCAGC-3′; ZEB1 Reverse: 5′-CATTCCATTTTCTGTCTTCCGC-3′; GAPDH Forward: 5′-TGCACCACCAACTGCTTAGC-3′; GAPDH Reverse: 5′-GGCATGGACTGTGGTCATGAG-3′; CDH1 Forward: 5′-CCCAATACATCTCCCTTCACAG-3′; CDH1 Reverse: 5′-CCACCTCTAAGGCCATCTTTG-3′.
Flow Cytometry, Migration and Growth Assays
For flow cytometry and FACS, HPAC cells (~1–2 x 106) were stained with anti-human CD44 APC (clone BJ18; #338806; BioLegend), anti-human CD24 PE (clone ML5; #311106; BioLegend), and anti-human CD133 VioBright FITC (clone AC133; #130-105-225; Miltenyi Biotec). Cells were analyzed using a BD LSRII and FACSDiva software version 6.2. Migration assays were performed using the live cell IncuCyte Zoom imaging system. Briefly, HPAC cells (1000 cells/well) were suspended in DMEM/F-12 containing 0.5% FBS were seeded onto 96-well ClearView-Chemotaxis plates with 8mm pores. Wherever indicated, plates were coated with 50 μg/ml matrigel, 5 μg/ml collagen or 5 μg/ml Fibronectin, prior to the seeding of the cells. The plates were incubated and imaged over the indicated time points. Cells migrating to the bottom chamber across the pores were imaged and quantified. Similarly, growth assays were performed using 96-well tissue culture dishes seeded with HPAC-VEC, HPAC-OSM, parental HPAC or OSM-treated HPAC (day7) cells (2000 cells/well). Each well was imaged at regular intervals to determine confluence over time.
Mouse Xenografts
Athymic NCr (nude) and NSG mice were bred and maintained at the Athymic Animal and Xenograft Core facility at the Case Western Reserve University Case Comprehensive Cancer Center. For tumorigenicity assays, 1 x 106 of HPAC-VEC or HPAC-OSM cells were resuspended in 50 μl of 1:1 mix of matrigel:media mix. Orthotopic pancreatic injections were performed on 8–12 week old male or female, Nude or NSG mice anesthetized using isoflurane. The mouse was laid on its right side and after appropriate sterilization of the surgical site, a small incision was made slightly medial to the splenic silhouette. The pancreas was externalized and 50 μl of cells were slowly injected close to the tail of the pancreas. The pancreas was then internalized and abdominal wall and incision site closed with sterile sutures. Tail vein injections were performed by slightly warming the tail under IR lamp and injecting 100 μl of cell suspension containing 1x 106 tumor cells. The in vivo, co-inoculation studies (using human fibroblasts and HPAC cells) were performed as follows. HPAC cells were co-cultured at a 1:3 ratio with FB-VEC or FB-OSM for 7 days. GFP positivity (from the GFP-expressing HPAC cells) was used to assess the ratio of HPAC:FB in both groups and the ratio was normalized to 4:1 prior to orthotopic injection of 1.2 x 106 cells into the pancreas of nude mice. Limiting dilution assays were performed by injecting the indicated number of tumor cells in 100 μl matrigel-media mix into the flanks of the mice. For all experiments tumor growth was monitored by bioluminescence imaging after injecting luciferin 10 min prior to imaging. Tumor volume was also measured manually using calipers and tumor weight assessed following euthanasia and tumor retrieval. All animals were used in compliance with the guidelines approved by the Case Western Reserve University Institutional Animal Care and Use Committee.
Viral Constructs and virus production
Lentiviruses were packaged and used to infect target cells as previously described (26). PLK0.1 vectors containing shRNA’s targeting ZEB1 (TRC Version: TRCN0000017565; Clone Name: NM_030751.2-572s1c1 and TRC Version: TRCN0000017566; Clone Name: NM_030751.2-70s1c1) were purchased from Sigma Aldrich. PLK0.1 vector containing shRNA-targeting GFP has been previously described (26). The SORE6 cancer stem cell reporter construct was a kind gift from Dr. Lalage Wakefield (27). Lentiviral pLenti-CMV-GFP was obtained from Addgene; Plasmid #17447. pBABE-puro-hTERT was purchased from addgene (plasmid #1771). pLenti-Neo-VEC was generated using gateway cloning and recombining the multiple cloning site of PCR8 topo vector (Invitrogen; Cat#46-0899) into lentiviral vector pLenti-CMV-Dest (Addgene; Plasmid#17451). pLenti-Neo-OSM as generated by sub-cloning OSM cDNA (OriGene; Cat# SC-121421) into gateway entry vector pLenti-ENTR4 (Addgene; Plasmid# 17424) and recombined into lentiviral destination vector (Addgene; Plasmid#17451). pFLUG-GFP-LUC (Firefly) fusion construct was a kind gift from Dr. Huiping Liu (28).
RESULTS
Elevated OSM and OSMR induce EMT in PDAC cells
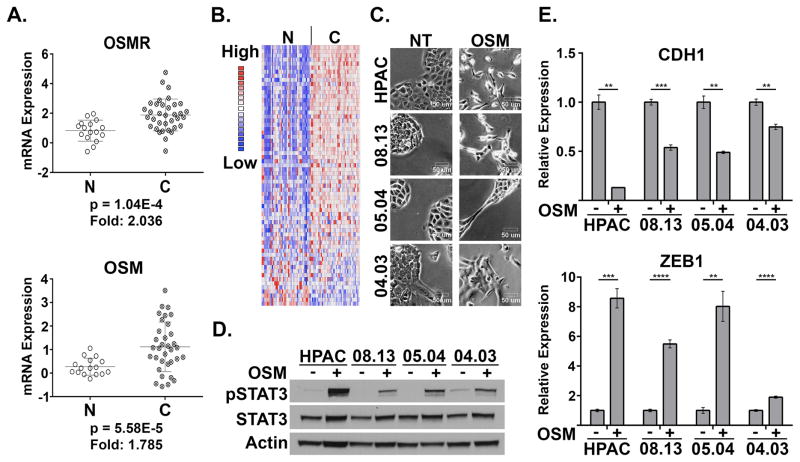
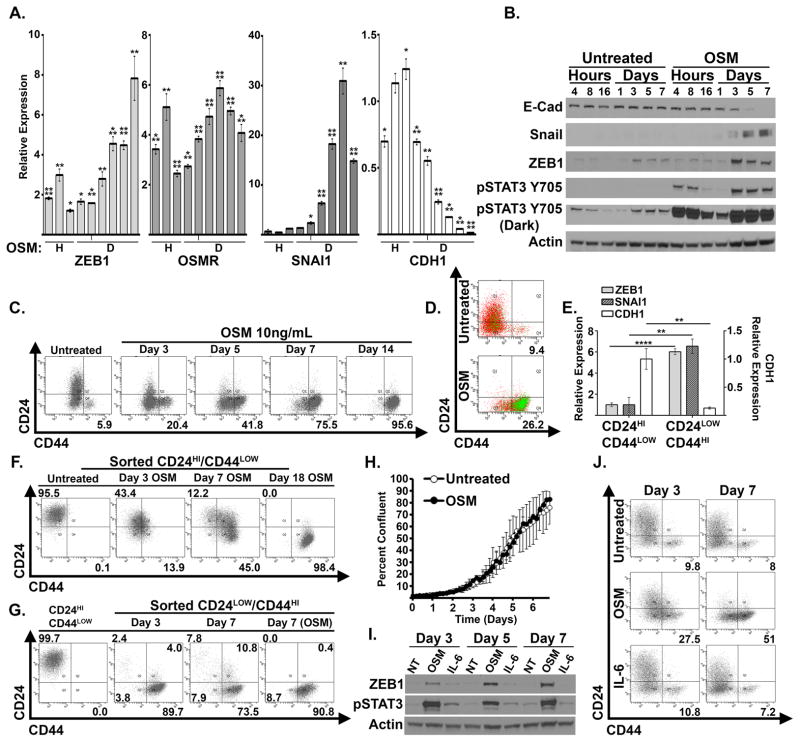
Utilizing publically available gene expression data sets, we interrogated the levels of OSM and OSMR in invasive pancreatic adenocarcinomas. PDAC tissue exhibited a significant increase in OSM and OSMR gene expression relative to normal pancreas (Figure 1A), which correlates with an elevated STAT3 gene expression signature (Figure 1B). Additional data sets also demonstrated elevated OSM and OSMR expression in PDAC tissue (Supplementary Figure S1 A–D) (29–32). Moreover, studies documenting the presence of OSM within primary and metastatic lesions in murine models, and elevated levels of OSM in the serum of PDAC patients who respond poorly to treatment provide further rationale for examining OSM in the biology of PDAC (19, 20). The elevated level of OSM, OSMR, and STAT3 target genes support our further investigation into the biological role of OSM in PDAC. For this, we treated four PDAC cell lines with recombinant human OSM, and observed an emergent mesenchymal morphology consistent with an epithelial-mesenchymal transition (EMT), concomitant with STAT3 activation (Figure 1C and 1D). The mesenchymal morphology was accompanied by the repression of epithelial junctional protein, E-cadherin (CDH1; Figure 1E), and induction of ZEB1 (Figure 1E), an EMT transcription factor capable of repressing E-cadherin expression. Further analysis of OSM-induced EMT confirmed the time-dependent induction of ZEB1 and Snail (SNAI1; another key EMT transcription factor), which correlated with the upregulation of CD44, a cell surface marker used to enrich for CSCs from various cancer types (Figure 2A–C). Moreover, exposure to OSM increases expression of the OSM receptor (OSMR), creating a positive feedback loop capable of enhancing the impact of TME OSM (Figure 2A). Conversely, the repression of E-cadherin correlated with the loss of CD24 expression, indicating that the emergent mesenchymal cells have acquired a CD24LOW/CD44HI phenotype, a well-described CSC phenotype across multiple tumor types, including pancreatic cancer (Figure 2C) (10, 28, 33).
Figure 1. Elevated OSM and OSMR in PDAC drive EMT.
(A) Oncomine data of whole tumor tissue vs. normal pancreas tissue examining OSM and OSMR mRNA levels in PDAC and normal pancreas (60). (B) Expression of a STAT3 gene expression signature (generated by filtering Pancreatic Cancer vs. Normal analysis; then applying filter “genes differentially expressed in melanoma cells in response to STAT3 expression”) in PDAC and normal pancreas. (C–E) A panel of PDAC cells lines were treated with recombinant OSM for one week and analyzed for morphological changes using bright-field microscopy (D; images shown at 200X). STAT3 phosphorylation (Tyr 705), total STAT3, and Actin were assessed using Western blot analysis (D), and E-Cadherin (CDH1) and ZEB1 were assessed using qRT-PCR (E).
Figure 2. OSM-induces mesenchymal/CSC plasticity.
(A–C) HPAC cells were exposed to recombinant OSM for 4, 8, 16 hours (H) and 1, 3, 5, 7, 10, 14 days (D). qRT-PCR analysis was used to quantify expression of the indicated genes. (A). Western blot analysis was used to assess the indicated proteins (B). CD24 and CD44 surface expression was assessed by flow cytometry (C). (D) HPAC cells were infected with lentiviral particles encoding a CSC-specific SORE6 GFP-reporter construct (HPAC-SORE6), and left untreated or treated with recombinant OSM for 8 days. Flow cytometry was performed for CD24/CD44 surface expression, as well as GFP intensity (numbers represent the percentage of cells that were GFP positive). (E–G) HPAC cells were sorted using FACS for CD24HI/CD44LOW and CD24LOW/CD44HI populations. qRT-PCR analysis was used to quantify the expression of the indicated genes (E). CD24HI/CD44LOW cells were treated with OSM for 3, 7 and 18 days and CD24 and CD44 expression was assessed by flow cytometry (F). CD24HI/CD44LOW cells were sorted from native HPAC populations and cultured in basal HPAC medium for 7 days (far left panel); CD24LOW/CD44HI sorted cells were cultured for 3 and 7 days in the presence or absence of OSM (adjacent three panels) and CD24 and CD44 expression was assessed by flow cytometry (G). (H) HPAC cells were treated with recombinant OSM for 7 days and subsequently were monitored for growth over a period of four days. The graph shows percent confluence of cells at the indicated times. (I–J) HPAC cells were treated for 7 days with recombinant OSM or IL-6.Western blot analysis (I) and flow cytometry (J) were performed after 3 and 7 days of OSM treatment.
Additionally, exposure of HPAC cells to recombinant OSM increased activity of a GFP-reporter construct containing OCT4/SOX2 response elements (SORE6), which has been previously described to identify cancer stem cell populations (27) (Figure 2D). To confirm the connection between a CD24LOW/CD44HI cell surface profile and a mesenchymal phenotype, the small fraction of CD24LOW/CD44HI cells present in parental HPAC population were separated by Fluorescence-Activated Cell Sorting (FACS; Supplementary Figure S2). Indeed, the CD24LOW/CD44HI population expressed elevated levels of SNAI1 and ZEB1, as well as reduced levels of CDH1 when compared to the CD24HI/CD44LOW population (Figure 2E). To further confirm that OSM converts cells into a CD24LOW/CD44HI state rather than selecting for a pre-existing CD44HI population, sorted CD24HI/CD44LOW cells were exposed to OSM for various times. Again, OSM induced the acquisition of CD44 and the loss of CD24 (Figure 2F), while untreated CD24HI/CD44LOW cells retained their epithelial, CD24HI/CD44LOW state, indicating the epithelial/non-CSC state is quite stable (Supplementary Figure S3). Conversely, the CD24LOW/CD44HI-sorted population spontaneously generated differentiated CD24HI cells (Figure 2G). The ability to produce differentiated progeny is a property of CSC. Notably, the de novo generation of a differentiated CD24HI population was prevented by adding OSM to the medium following the sort for CD44HI cells, indicating that OSM can suppress the differentiation of the CD44HI population. Finally, OSM generated the emergence of a CD44HI/CD133HI population (Supplementary Figure S4), which possess significant tumor initiating capabilities and metastatic potential within PDAC (11, 34). Importantly, analysis of cell confluency using the Incucyte Zoom imaging system or analysis of cell number determined that OSM does not alter proliferation at the doses used in our experiments (Figure 2H; Supplementary Figure S5B), further indicating that OSM is not selecting for pre-existing CD24LOW/CD44HI cells. In contrast to OSM, IL-6 was unable to convert parental HPAC into CD24LOW/CD44HI, likely due to the weaker induction of STAT3Y705 phosphorylation and ZEB1 expression (Figure 2I & J). This further implies that OSM may be a more potent driver of aggressive phenotypes in PDAC in certain contexts. While we do not observe comparable phenotypic response to IL-6 in our studies, we note that IL-6 has also been demonstrated to play a significant role in PDAC initiation and progression in transgenic murine mouse models(22, 35).
OSM induces motility, gemcitabine resistance, and increased tumorigenic potential
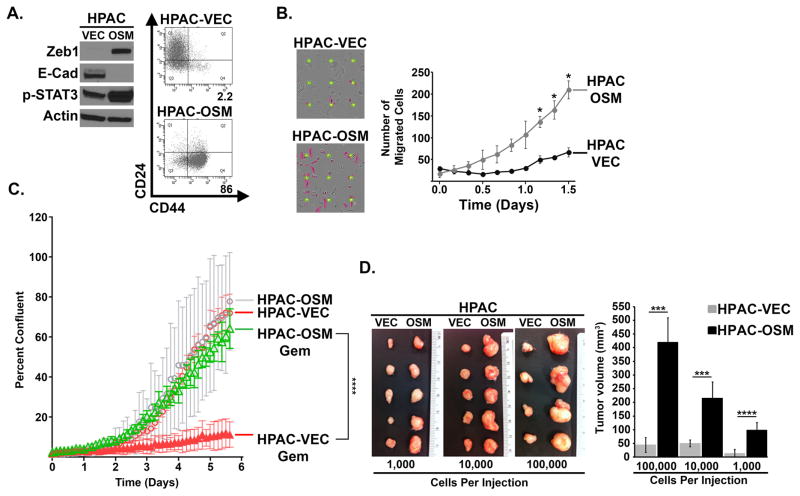
Our observation that OSM induces key EMT transcription factors, generates cells with a CD24LOW/CD44HI phenotype, and activates a CSC-reporter prompted us to assess whether OSM exposure enhanced biological characteristics consistent with mesenchymal/CSC. For this, OSM was exogenously expressed in HPAC cells by lentiviral transduction (HPAC-OSM) so as to enrich for a stable CD24LOW/CD44HI population. An empty lentivirus was used as a control (HPAC-VEC). One week post-infection, HPAC-OSM cells exhibited increased STAT3Y705 phosphorylation, increased ZEB1 expression, a loss of E-cadherin expression, and a conversion from CD44LOW to CD44HI (Figure 3A). As observed with recombinant OSM, exogenous OSM expression did not impact the proliferation of HPAC cells (Supplementary Figure S5). However, HPAC-OSM cells were significantly more migratory on multiple matrices (including fibronectin and matrigel, as well as uncoated wells, but not collagen; Figures 3B and Supplementary Figure S6A–C).
Figure 3. OSM Induces Properties of CSC and Increases Cell Motility.
HPAC cells (expressing GFP-LUC fusion protein) were infected with lentiviruses encoding OSM (HPAC-OSM) or control vector (HPAC-VEC). (A) One week post-infection, Western analysis and flow cytometry was performed, as indicated. (B) Migration of HPAC-VEC and HPAC-OSM cells was assessed using a live cell IncuCyte Zoom imaging system. Graph shows number of migrated cells (purple colored cells in the adjacent images), error bars represent ± SEM for a representative experiment performed in triplicate. A two tailed paired t-test was performed to determine significance; * = p-value ≤ 0.01. (C) HPAC-VEC and HPAC-OSM cells were cultured in the presence or absence of Gemcitabine (30 nM) and cell number was monitored using the IncuCyte Zoom live cell imaging system. Graph shows percent confluence of cells at the indicated times, error bars represent ± SEM for a representative experiment performed in triplicate; a 2-way ANOVA was performed in order to determine significance; **** = p-value ≤ 0.001. (D) HPAC-VEC and HPAC-OSM populations were injected subcutaneously at limiting dilutions of 100,000, 10,000, and 1000 cells/injection. Images of resected primary tumors and quantification of primary tumor volume are presented. p-values were calculated using a two tailed Student’s t test; *** = p-value ≤ 0.01, **** = p-value ≤ 0.01.
Next, we assessed the ability of OSM to induce therapeutic resistance and tumorigenic potential, key characteristics of mesenchymal/CSC (36–38). HPAC-OSM and HPAC-VEC cells were treated with gemcitabine; a frontline therapy for PDAC, and cell growth was monitored over time. HPAC-OSM cells were significantly less sensitive to gemcitabine (Figure 3C), when compared to HPAC-VEC cells, suggesting that elevated OSM in the TME may promote gemcitabine resistance. Finally, limiting dilutions of HPAC-VEC and HPAC-OSM cells were subcutaneously injected into the flank of NSG mice (Supplementary Figure S7). Importantly, HPAC-OSM cells formed significantly larger tumors (~5–10 times larger) at each of the cell numbers tested, implicating the OSM-induced CD44HI population as highly tumorigenic (Figure 3D). The existence of the small number of CD44HI cells in the HPAC-VEC population (ranging from ~2–12%) is likely contributing to the small tumors generated from this population. Taken together, our data support a role for OSM in generating PDAC cells with mesenchymal/CSC properties that are highly migratory, tumorigenic, and resistant to gemcitabine.
OSM increases tumor burden and metastases in vivo
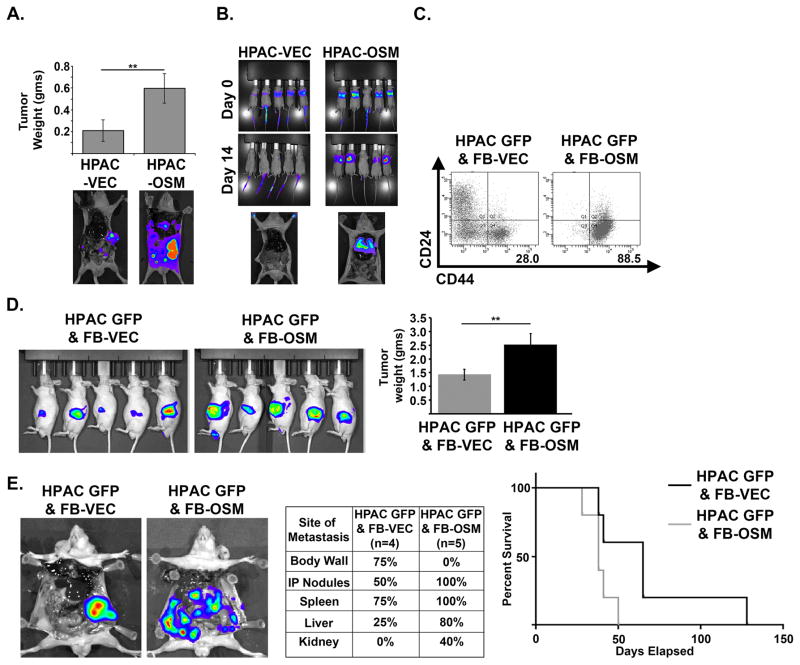
We next assessed whether the increased motility and tumorigenicity induced by OSM impacted tumor invasiveness and metastatic spread using orthotopic injection of HPAC-VEC and HPAC-OSM. OSM expression conferred a significant increase in primary tumor burden as well as increased intraperitoneal metastatic spread (Figure 4A), a common feature in patients with PDAC. Furthermore, HPAC-OSM demonstrated a greater capacity to colonize the lung following tail-vein injection, when compared to HPAC-VEC (Figure 4B). In order to better represent a tumor microenvironment with high levels of OSM, we co-cultured GFP-labeled HPAC cells (HPAC-GFP) with control human fibroblasts (FB-VEC), or fibroblasts expressing OSM (FB-OSM). HPAC cells co-cultured with FB-OSM showed a nearly three-fold increase in the CD24LOW/CD44HI population (Figure 4C). Moreover, orthotopic co-injection of HPAC and FB-OSM into the pancreas of mice yielded greater tumor burden and metastatic dissemination when compared to HPAC/FB-VEC co-injection (Figure 4D & E). HPAC/FB-OSM co-injections spread more often to every site except the body wall, resulting in more rapid lethality (Figure 4E). This suggests that a stroma high in OSM will impart a more invasive and aggressive phenotype within the tumor by eliciting a mesenchymal/CSC program.
Figure 4. OSM increases tumor burden and metastases in vivo.
(A) HPAC-VEC and HPAC-OSM cells were injected orthotopically into the pancreas of athymic, nude mice. Tumor weight and representative BLI images at the end of the experiment are shown (23 days post injection). A two tailed Student’s t-test was used to determine significance; ** = p ≤ 0.01. (B) HPAC-VEC and HPAC-OSM cells were injected via the tail vein in athymic, nude mice. BLI images showing lung colonization at 14 days post injection are shown. (C–E) HPAC-GFP were co-cultured with control fibroblasts (FB-VEC) or fibroblasts expressing OSM (FB-OSM). Following one week of co-culture, CD24 and CD44 surface expression on HPAC-GFP was assessed via flow cytometry by gating on the GFP+ cells (C). HPAC-GFP were orthotopically co-implanted with FB-OSM or FB-VEC into nude mice (D). BLI images at 21 days post injection and quantification of primary tumor weight at the time animals were sacrificed. A two tailed Student’s t-test was used to determine significance; ** = p ≤ 0.01. Representative BLI images, site and frequency of metastasis, and Kaplan-Meier survival curve of mice that received HPAC GFP & FB-VEC or HPAC GFP & FB-OSM cells (E).
OSM-induced mesenchymal/CSC plasticity requires JAK/STAT3 signaling
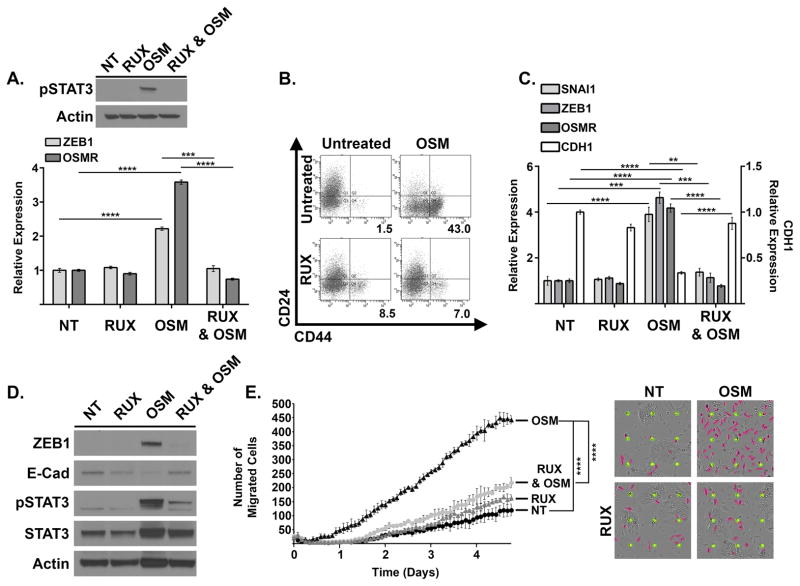
OSM induces activation of the heterodimeric receptor gp130/OSMR, which activates JAK1/2 mediated STAT3 phosphorylation, dimerization, and subsequent STAT3-mediated transcription (of Zeb1, Snail, OSMR, as shown in Figure 2A). To assess whether suppression of JAK/STAT3 activation prevents OSM-induced mesenchymal/CSC plasticity, HPAC cells were treated with recombinant OSM in the presence or absence of a JAK1/2 inhibitor, Ruxolitinib (RUX). Indeed, RUX inhibited STAT3Y705 phosphorylation and suppressed ZEB1 and OSMR expression induced by a 4-hour treatment with OSM (Figure 5A). Furthermore, upon longer-term treatment, RUX prevented CD44 acquisition, transcription of OSMR and key EMT transcription factors ZEB1 and Snail, and E-Cadherin (CDH1) repression (Figure 5B–D). Finally, RUX inhibited OSM-induced cell motility (Figure 5E). Taken together, the invasive and stem-like properties induced by OSM require JAK-activated STAT3, thus opening up avenues of therapy targeting the intracellular signaling required for OSM-induced mesenchymal/CSC plasticity in PDAC.
Figure 5. OSM-induced mesenchymal/CSC plasticity requires activated STAT3.
(A) HPAC cells were treated with recombinant OSM, Ruxolitinib, or a combination of both for four hours. Western blot (upper panel) and qRT-PCR (lower panel) were performed as indicated. (B–D) HPAC cells were treated with recombinant OSM, Ruxolitinib, or a combination of both for 8 days. Flow cytometry (B), qRT-PCR (C) and Western blot analysis (D) were performed as indicated. The percentage of CD24LOW/CD44HI cells is indicated by the number below the flow cytometry scan in panel B. (E) Migration of HPAC cells treated as indicated was assessed using a live cell IncuCyte Zoom imaging system. The graph (left panel) and pictures (right panel) show the number of migrated cells at the indicated times; error bars represent ± SEM for a representative experiment performed in triplicate; a 2-way ANOVA was performed in order to determine significance; **** = p-value ≤ 0.001.
STAT3-activated ZEB1 drives OSM-induced mesenchymal/CSC plasticity
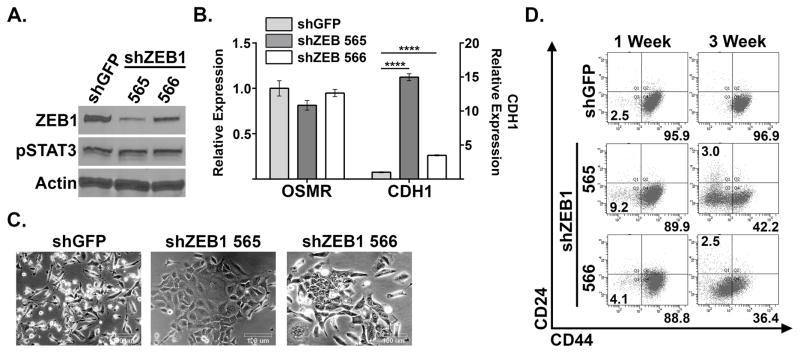
Since ZEB1 has been shown to be a master regulator of EMT, and a direct target of OSM-activated STAT3, we hypothesized that ZEB1 was a key component of OSM-induced mesenchymal/CSC plasticity. To test this hypothesis, HPAC-OSM, which have a 95.9% CD24LOW/CD44HI profile, were infected with lentiviruses encoding short hairpin RNAs targeting ZEB1 (or shGFP as a control), and knockdown of ZEB1 was confirmed by Western blot (Figure 6A). Importantly, knockdown of ZEB1 did not alter STAT3Y705 phosphorylation (Figure 6A) or downstream transcriptional target OSMR (Figure 6B). ZEB1 knockdown increased E-Cadherin expression (CDH1; Figure 6B), though to a lesser extent in the population with less ZEB1 knockdown (shZEB1-566). The increase in CDH1 correlated with a more epithelial morphology compared to shGFP control (Figure 6C). Most importantly, the suppression of ZEB1 in HPAC-OSM cells led to a marked reduction in the number CD44HI cells at both 1 week and 3 weeks following infection with the ZEB1 shRNAs (from ~96% in shGFP controls to 42.2% for shZEB1-565 and 36.4% for shZEB1-566). These data support our hypothesis that ZEB1 is a crucial JAK/STAT3 activated gene required for OSM-induced mesenchymal/CSC plasticity in PDAC cells.
Figure 6. ZEB1 is crucial to OSM-induced mesenchymal/CSC plasticity.
CD24LOW/CD44HI HPAC-OSM cells were infected with lentiviruses encoding two short hairpin RNAs targeting ZEB1 (shZEB1-765 and shZEB1-766) or a control (shGFP). Following selection, Western blot analysis (A), qRT-PCR (B), bright-field microscopy (C) and flow cytometry (D) were performed as indicated.
DISCUSSION
Patients diagnosed with pancreatic cancer currently have dismal survival rates, with 75% of patients succumbing to their disease within one year of diagnosis. The observation that transformed pancreatic epithelial cells can disseminate to distant organs even before a frank malignancy is detectable at the primary site underscores the difficulty in managing patients with aggressive PDAC (2). Our understanding of how OSM, (39) present in the PDAC TME, impacts mesenchymal/CSC plasticity and gives rise to metastatic and therapy-resistant PDAC growth is currently limited and would provide important insights into how PDAC may be targeted.
The cancer stem cell (CSC) hypothesis places cells with self-renewal capacity and the ability to differentiate at the top of the tumor cell hierarchy. Importantly, CSC harboring a mesenchymal phenotype display a decreased sensitivity to chemo- and radiation-therapy, resulting in an enrichment of CSC following treatment. Evidence is emerging that suggests that, non-CSC can be induced into a transient CSC-like, drug-tolerant state when exposed to chemotherapies. Our studies support such a model, whereby OSM exposure induces the conversion of epithelial/non-CSC into mesenchymal/CSC (Figure 7). Mechanistically, we demonstrate that gp130/OSMR activation results in JAK/STAT3-mediated ZEB1 transcription, and ZEB1-dependent expression of CSC marker CD44. Consequently, a TME high in OSM creates more aggressive PDAC cells with greater tumorigenicity, increased invasive and metastatic potential, and resistance to gemcitabine, classified here as a CD24LOW/CD44HIpopulation. However, early PDAC stem cells were identified as CD24HI/CD44HI, yet we did not observe a CD24HI/CD44HI population following OSM exposure (10). Rather, CD24HI cells typically reduced CD24 expression first, then gained expression of CD44. This finding underscores the heterogeneity of CSC populations and the difficulties in identifying/targeting cellular plasticity, and points to the importance of using functional assays when defining PDAC stem biology. Moreover, OSMR is induced by OSM ligand, suggesting that a positive feedback loop is engaged to promote greater OSMR/JAK/STAT3 activation and maintain the induced mesenchymal/CSC cell state. This OSM/OSMR positive feedback loop provides a number of potential therapeutic targets, which if disrupted may prevent the emergence of aggressive mesenchymal/CSC in response OSM. In line with our observations, a recent study also demonstrated that OSM induces EMT in CFPAC1 pancreatic cancer cells, in addition to lung cancer cell lines. OSM induced mesenchymal properties and differential 3D growth and colony architecture that was regulated through a JAK-dependent mechanism. Our studies build upon these observations by demonstrating that the mesenchymal/CSC phenotype induced by OSM also promotes widespread metastasis and gemcitabine resistance(40).
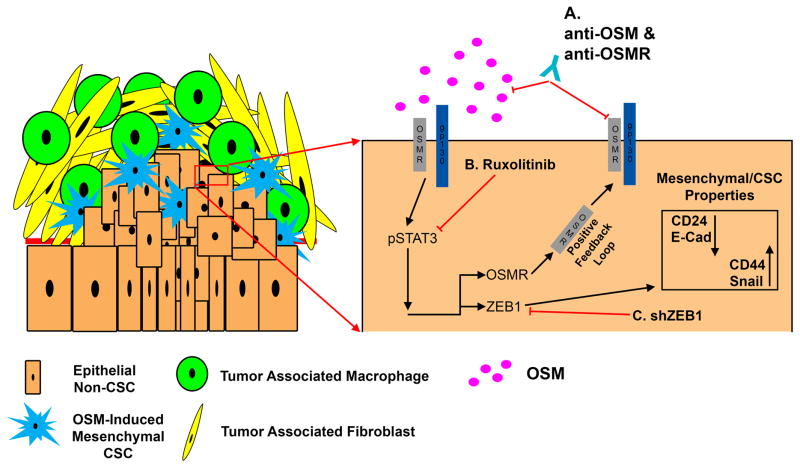
Figure 7. Schematic of OSM-induced mesenchymal/CSC plasticity.
Elevated OSM within the pancreatic tumor microenvironment induces STAT3 activation (Y705 phosphorylation), leading to transcriptional activation of ZEB1. As ZEB1 accumulates, E-Cadherin and CD24 surface expression are repressed, while Snail and CD44 surface expression are increased. The resulting EMT and acquisition of CSC properties decrease gemcitabine sensitivity, and increase tumor initiating capacity and metastasis. (A–C) Points where the aggressive phenotypes engaged by OSM can be inhibited (including OSM neutralizing or OSMR blocking antibodies, chemical inhibition of activated OSMR/JAK, or ZEB1 silencing).
Elevated levels of OSM in the tumor microenvironment (TME) have been associated with highly aggressive metastatic cancers, increased risk of tumor recurrence, and a poor prognosis in a variety of cancers (24, 41–44). OSM in the TME may originate from a host of immune cells as well as cancer-associated fibroblasts or adipose tissue (45–47). There is mounting evidence that the OSM in the TME contributes to tumor progression in many ways. First, macrophages secreting OSM are localized at the advancing, infiltrative margins of carcinomas, which may implicate OSM in tumor invasion (48). Second, highly aggressive basal-like and triple negative breast cancer subtypes express higher levels of OSMR which is associated with adverse clinical outcomes and increased expression of the CSC marker CD44 (47). Finally, DNA damaging chemotherapy induces additional OSM secretion from peritoneal and bone marrow-derived macrophages, potentially exacerbating the aggressive properties associated with mesenchymal and CSC properties following treatment with genotoxic therapies (49, 50).
Elevated serum levels of OSM are found in patients with PDAC, in line with the significant increase in OSM mRNA (19, 51). In PDAC patients, sustained OSM serum levels following Gemcitabine was considered a poor prognostic marker, consistent with our observation that OSM rendered PDAC cells highly resistant to first-line gemcitabine treatment. We attribute the ability of OSM to induce gemcitabine resistance with its ability to reprogram epithelial/non-CSC into mesenchymal/CSC. This observation is also in line with recent studies by Zheng et al. and Fischer et al., showing that the acquisition of mesenchymal properties led to therapeutic resistance in pancreatic and breast cancer cells (38, 52).
While the IL-6 family of cytokines can exhibit redundancy in their biological responses due to the shared use of the gp130 transmembrane receptor, in our studies, OSM has unique biological functions not recapitulated by IL-6 (53). Gp130/OSMR heterodimers have characteristics unique from other IL-6 family co-receptors that may account for the distinct signaling and biological effects of OSM relative to other IL-6 family members, such as enhanced JAK/STAT3 and MAPK activation. When coupled with the observation that OSM and OSMR are significantly elevated in PDAC, and OSMR is induced by OSM, the unique signaling emanating from the OSMR justifies a more thorough analysis of OSM and the signaling cascades described here.
Downstream of OSM/OSMR, JAK/STAT3 activation is increasingly gaining popularity in the field of cancer therapeutics, with ongoing clinical trials using the JAK inhibitor Ruxolitinib, including patients with advanced metastatic pancreatic cancer. Ruxolitinib, given as the second-line treatment along with capecitabine, improved overall survival in patients with systemic inflammation and those resistant to first-line therapies (54). STAT3-inhibiting drugs given in combination with additional targeted therapies is proving beneficial in oncogene-addicted cancer cells (such as those expressing the active KRAS mutant) that acquire resistance to MEK inhibitors, gemcitabine, and erlotinib by engaging a feedback activation of STAT3 (27). Furthermore, the recent development of a STAT3 inhibitor BBI-608 (55) as a CSC-selective agent highlights the importance of STAT3 to the CSC phenotype. We propose that suppressing OSM function (by neutralizing OSM in the TME or inhibiting OSMR activity) will prevent cancer cells from engaging a favored escape mechanism, which is to acquire and maintain mesenchymal/CSC properties (Figure 7A & B). Directly targeting OSM and OSMR has significant advantages, as both OSM and OSMR protein are expressed at lower levels in normal human tissues when compared to other components of the OSM/OSMR/JAK/STAT3 axis (Supplementary Figure S8). Elevated OSM expression occurs only in inflammatory, pathological conditions (such as cancer, arthritis, and inflammatory heart disease), which vastly improves the therapeutic window to target OSM/OSMR. Neutralizing or blocking antibodies, and decoy receptors that prevent OSM from engaging OSMR can ameliorate arthritis or prevent inflammatory heart failure in mouse models (56). Of note, blocking antibodies targeting the extracellular domain of OSMR reduced the OSM-mediated dedifferentiation of cardiomyocytes responsible for heart failure (57). We propose that OSMR targeting antibodies could be conjugated with cytotoxic agents, resulting in more selective targeting of tumor cells, similar to current approaches linking HER2 targeting antibodies with DM1 (58).
Finally, we found that suppression of OSMR/JAK/STAT3-mediated ZEB1 expression markedly reverted the mesenchymal/CSC phenotype, implicating ZEB1 as a crucial driver of OSM-induced CSC properties. In addition to defining ZEB1 as a key downstream effector of OSMR/JAK/STAT3 activation, these findings demonstrated the reversible nature of OSM induced mesenchymal/CSC plasticity. ZEB1 links EMT and CSC features in pancreatic cancer cells, by inhibiting the expression of the microRNA-200 family, which represses stem cell factors and induces epithelial differentiation. Moreover, ZEB1 and CD44s were shown to participate in a self-sustaining loop to maintain cancer stem-cell features in pancreatic cancer (59). The net result of OSMR/JAK/STAT3/ZEB1 pathway activation is a reversibly induced mesenchymal/CSC population encompassing aggressive cancer cell properties. By gaining a better understanding of TME cytokines, such as OSM, that induce mesenchymal/CSC plasticity, one may envision tailoring therapies directed at the source of the cytokine or the associated signaling so as to enhance efficacy of current therapies and ultimately improve patient survival.
Supplementary Material
Implications.
Therapeutic targeting the OSM/OSMR axis within the TME may prevent or reverse the aggressive mesenchymal and CSC phenotypes associated with poor outcomes in patients with PDAC.
Acknowledgments
Financial support: MWJ is supported by the US National Institutes of Health (R01CA138421) and the American Cancer Society (Research Scholar Award # RSG CCG-122517).
We gratefully acknowledge the research support provided by the Case Comprehensive Cancer Center (P30 CA43703; Flow cytometry, Imaging and Microscopy core, Athymic Animal and Xenograft core and Small Animal Imaging Research Center).
Footnotes
Conflict of interest disclosure: The authors disclose no potential conflicts of interest.
References
- 1.Howlader NNA, Krapcho M, Garshell J, Miller D, Altekruse SF, Kosary CL, Yu M, Ruhl J, Tatalovich Z, Mariotto A, Lewis DR, Chen HS, Feuer EJ, Cronin KA, editors. SEER Cancer Statistics Review, 1975–2012. National Cancer Institute; Bethesda, MD: Apr, 2015. http://seer.cancer.gov/csr/1975_2012/, based on November 2014 SEER data submission, posted to the SEER web site. [Google Scholar]
- 2.Rhim AD, Mirek ET, Aiello NM, Maitra A, Bailey JM, McAllister F, et al. EMT and dissemination precede pancreatic tumor formation. Cell. 2012;148(1–2):349–61. doi: 10.1016/j.cell.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schneider G, Siveke JT, Eckel F, Schmid RM. Pancreatic Cancer: Basic and Clinical Aspects. Gastroenterology. 2005;128(6):1606–25. doi: 10.1053/j.gastro.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 4.Batlle E, Sancho E, Franci C, Dominguez D, Monfar M, Baulida J, et al. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol. 2000;2(2):84–9. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- 5.Aigner K, Dampier B, Descovich L, Mikula M, Sultan A, Schreiber M, et al. The transcription factor ZEB1 (deltaEF1) promotes tumour cell dedifferentiation by repressing master regulators of epithelial polarity. Oncogene. 2007;26(49):6979–88. doi: 10.1038/sj.onc.1210508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119(6):1420–8. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133(4):704–15. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8(10):755–68. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 9.Liu G, Yuan X, Zeng Z, Tunici P, Ng H, Abdulkadir IR, et al. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol Cancer. 2006;5:67. doi: 10.1186/1476-4598-5-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, et al. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67(3):1030–7. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- 11.Hermann PC, Huber SL, Herrler T, Aicher A, Ellwart JW, Guba M, et al. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1(3):313–23. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 12.Junk DJ, Cipriano R, Bryson BL, Gilmore HL, Jackson MW. Tumor Microenvironmental Signaling Elicits Epithelial-Mesenchymal Plasticity through Cooperation with Transforming Genetic Events. Neoplasia. 2013;15(9):1100–9. doi: 10.1593/neo.131114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doherty MR, Smigiel JM, Junk DJ, Jackson MW. Cancer Stem Cell Plasticity Drives Therapeutic Resistance. Cancers (Basel) 2016;8(1) doi: 10.3390/cancers8010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sullivan NJ, Sasser AK, Axel AE, Vesuna F, Raman V, Ramirez N, et al. Interleukin-6 induces an epithelial-mesenchymal transition phenotype in human breast cancer cells. Oncogene. 2009;28(33):2940–7. doi: 10.1038/onc.2009.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Techasen A, Loilome W, Namwat N, Dokduang H, Jongthawin J, Yongvanit P. Cytokines released from activated human macrophages induce epithelial mesenchymal transition markers of cholangiocarcinoma cells. Asian Pac J Cancer Prev. 2012;13(Suppl):115–8. [PubMed] [Google Scholar]
- 16.Chang DZ, Ma Y, Ji B, Wang H, Deng D, Liu Y, et al. Mast cells in tumor microenvironment promotes the in vivo growth of pancreatic ductal adenocarcinoma. Clin Cancer Res. 2011;17(22):7015–23. doi: 10.1158/1078-0432.CCR-11-0607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whatcott CJ, Diep CH, Jiang P, Watanabe A, LoBello J, Sima C, et al. Desmoplasia in Primary Tumors and Metastatic Lesions of Pancreatic Cancer. Clin Cancer Res. 2015;21(15):3561–8. doi: 10.1158/1078-0432.CCR-14-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pandol S, Edderkaoui M, Gukovsky I, Lugea A, Gukovskaya A. Desmoplasia of pancreatic ductal adenocarcinoma. Clin Gastroenterol Hepatol. 2009;7(11 Suppl):S44–7. doi: 10.1016/j.cgh.2009.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Torres C, Perales S, Alejandre MJ, Iglesias J, Palomino RJ, Martin M, et al. Serum cytokine profile in patients with pancreatic cancer. Pancreas. 2014;43(7):1042–9. doi: 10.1097/MPA.0000000000000155. [DOI] [PubMed] [Google Scholar]
- 20.Benson DD, Meng X, Fullerton DA, Moore EE, Lee JH, Ao L, et al. Activation state of stromal inflammatory cells in murine metastatic pancreatic adenocarcinoma. Am J Physiol Regul Integr Comp Physiol. 2012;302(9):R1067–75. doi: 10.1152/ajpregu.00320.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Denley SM, Jamieson NB, McCall P, Oien KA, Morton JP, Carter CR, et al. Activation of the IL-6R/Jak/stat pathway is associated with a poor outcome in resected pancreatic ductal adenocarcinoma. J Gastrointest Surg. 2013;17(5):887–98. doi: 10.1007/s11605-013-2168-7. [DOI] [PubMed] [Google Scholar]
- 22.Corcoran RB, Contino G, Deshpande V, Tzatsos A, Conrad C, Benes CH, et al. STAT3 plays a critical role in KRAS-induced pancreatic tumorigenesis. Cancer Res. 2011;71(14):5020–9. doi: 10.1158/0008-5472.CAN-11-0908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richards CD. The enigmatic cytokine oncostatin m and roles in disease. ISRN Inflamm. 2013;2013:512103. doi: 10.1155/2013/512103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kan CE, Cipriano R, Jackson MW. c-MYC functions as a molecular switch to alter the response of human mammary epithelial cells to oncostatin M. Cancer Res. 2011;71(22):6930–9. doi: 10.1158/0008-5472.CAN-10-3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garbe JC, Bhattacharya S, Merchant B, Bassett E, Swisshelm K, Feiler HS, et al. Molecular distinctions between stasis and telomere attrition senescence barriers shown by long-term culture of normal human mammary epithelial cells. Cancer Res. 2009;69(19):7557–68. doi: 10.1158/0008-5472.CAN-09-0270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cipriano R, Miskimen KL, Bryson BL, Foy CR, Bartel CA, Jackson MW. FAM83B-mediated activation of PI3K/AKT and MAPK signaling cooperates to promote epithelial cell transformation and resistance to targeted therapies. Oncotarget. 2013;4(5):729–38. doi: 10.18632/oncotarget.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang B, Raviv A, Esposito D, Flanders KC, Daniel C, Nghiem BT, et al. A flexible reporter system for direct observation and isolation of cancer stem cells. Stem Cell Reports. 2015;4(1):155–69. doi: 10.1016/j.stemcr.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu H, Patel MR, Prescher JA, Patsialou A, Qian D, Lin J, et al. Cancer stem cells from human breast tumors are involved in spontaneous metastases in orthotopic mouse models. Proc Natl Acad Sci U S A. 2010;107(42):18115–20. doi: 10.1073/pnas.1006732107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishikawa M, Yoshida K, Yamashita Y, Ota J, Takada S, Kisanuki H, et al. Experimental trial for diagnosis of pancreatic ductal carcinoma based on gene expression profiles of pancreatic ductal cells. Cancer Science. 2005;96(7):387–93. doi: 10.1111/j.1349-7006.2005.00064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Badea LHV, Dima SO, Dumitrascu T, Popescu I. Combined gene expression analysis of whole-tissue and microdissected pancreatic ductal adenocarcinoma identifies genes specifically overexpressed in tumor epithelia. Hepatogastroenterology. 2008;55(88) [PubMed] [Google Scholar]
- 31.Logsdon CD, Simeone DM, Binkley C, Arumugam T, Greenson JK, Giordano TJ, et al. Molecular Profiling of Pancreatic Adenocarcinoma and Chronic Pancreatitis Identifies Multiple Genes Differentially Regulated in Pancreatic Cancer. Cancer Res. 2003;63(10):8. [PubMed] [Google Scholar]
- 32.Segara D. Expression of HOXB2, a Retinoic Acid Signaling Target in Pancreatic Cancer and Pancreatic Intraepithelial Neoplasia. Clinical Cancer Research. 2005;11(9):3587–96. doi: 10.1158/1078-0432.CCR-04-1813. [DOI] [PubMed] [Google Scholar]
- 33.Takaishi S, Okumura T, Tu S, Wang SS, Shibata W, Vigneshwaran R, et al. Identification of gastric cancer stem cells using the cell surface marker CD44. Stem Cells. 2009;27(5):1006–20. doi: 10.1002/stem.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Wei J, Wang H, Xue X, An Y, Tang D, et al. Epithelial mesenchymal transition correlates with CD24+CD44+ and CD133+ cells in pancreatic cancer. Oncol Rep. 2012;27(5):1599–605. doi: 10.3892/or.2012.1681. [DOI] [PubMed] [Google Scholar]
- 35.Lesina M, Kurkowski MU, Ludes K, Rose-John S, Treiber M, Kloppel G, et al. Stat3/Socs3 activation by IL-6 transsignaling promotes progression of pancreatic intraepithelial neoplasia and development of pancreatic cancer. Cancer Cell. 2011;19(4):456–69. doi: 10.1016/j.ccr.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 36.Shah AN, Summy JM, Zhang J, Park SI, Parikh NU, Gallick GE. Development and characterization of gemcitabine-resistant pancreatic tumor cells. Ann Surg Oncol. 2007;14(12):3629–37. doi: 10.1245/s10434-007-9583-5. [DOI] [PubMed] [Google Scholar]
- 37.Schober M, Jesenofsky R, Faissner R, Weidenauer C, Hagmann W, Michl P, et al. Desmoplasia and chemoresistance in pancreatic cancer. Cancers (Basel) 2014;6(4):2137–54. doi: 10.3390/cancers6042137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng X, Carstens JL, Kim J, Scheible M, Kaye J, Sugimoto H, et al. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature. 2015;527(7579):525–30. doi: 10.1038/nature16064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kleeff J, Beckhove P, Esposito I, Herzig S, Huber PE, Lohr JM, et al. Pancreatic cancer microenvironment. Int J Cancer. 2007;121(4):699–705. doi: 10.1002/ijc.22871. [DOI] [PubMed] [Google Scholar]
- 40.Argast GM, Mercado P, Mulford IJ, O’Connor M, Keane DM, Shaaban S, et al. Cooperative Signaling between Oncostatin M, Hepatocyte Growth Factor and Transforming Growth Factor-β Enhances Epithelial to Mesenchymal Transition in Lung and Pancreatic Tumor Models. Cells Tissues Organs. 2011;193(1–2):114–32. doi: 10.1159/000320179. [DOI] [PubMed] [Google Scholar]
- 41.Caffarel MM, Coleman N. Oncostatin M receptor is a novel therapeutic target in cervical squamous cell carcinoma. J Pathol. 2014;232(4):386–90. doi: 10.1002/path.4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.West NR, Murray JI, Watson PH. Oncostatin-M promotes phenotypic changes associated with mesenchymal and stem cell-like differentiation in breast cancer. Oncogene. 2014;33(12):1485–94. doi: 10.1038/onc.2013.105. [DOI] [PubMed] [Google Scholar]
- 43.Ryan RE, Martin B, Mellor L, Jacob RB, Tawara K, McDougal OM, et al. Oncostatin M binds to extracellular matrix in a bioactive conformation: implications for inflammation and metastasis. Cytokine. 2015;72(1):71–85. doi: 10.1016/j.cyto.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu M, Che Q, Liao Y, Wang H, Wang J, Chen Z, et al. Oncostatin M activates STAT3 to promote endometrial cancer invasion and angiogenesis. Oncol Rep. 2015;34(1):129–38. doi: 10.3892/or.2015.3951. [DOI] [PubMed] [Google Scholar]
- 45.Grenier A, Dehoux M, Boutten A, Arce-Vicioso M, Durand G, Gougerot-Pocidalo MA, et al. Oncostatin M production and regulation by human polymorphonuclear neutrophils. Blood. 1999;93(4):1413–21. [PubMed] [Google Scholar]
- 46.Suda T, Chida K, Todate A, Ide K, Asada K, Nakamura Y, et al. Oncostatin M production by human dendritic cells in response to bacterial products. Cytokine. 2002;17(6):335–40. doi: 10.1006/cyto.2002.1023. [DOI] [PubMed] [Google Scholar]
- 47.Levano KS, Jung EH, Kenny PA. Breast cancer subtypes express distinct receptor repertoires for tumor-associated macrophage derived cytokines. Biochem Biophys Res Commun. 2011;411(1):107–10. doi: 10.1016/j.bbrc.2011.06.102. [DOI] [PubMed] [Google Scholar]
- 48.Vlaicu P, Mertins P, Mayr T, Widschwendter P, Ataseven B, Högel B, et al. Monocytes/macrophages support mammary tumor invasivity by co-secreting lineage-specific EGFR ligands and a STAT3 activator. BMC Cancer. 2013;13(1) doi: 10.1186/1471-2407-13-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sodhi A, Shishodia S, Shrivastava A. Cisplatin-stimulated murine bone marrow-derived macrophages secrete oncostatin M. Immunol Cell Biol. 1997;75(5):492–6. doi: 10.1038/icb.1997.76. [DOI] [PubMed] [Google Scholar]
- 50.Singh RAK, Sodhi A. Cisplatin-treated macrophages produce oncostatin M: regulation by serine/threonine and protein tyrosine kinases/phosphatases and Ca2+/calmodulin. Immunology Letters. 1998;62(3):159–64. doi: 10.1016/s0165-2478(98)00040-6. [DOI] [PubMed] [Google Scholar]
- 51.Wingren C, Sandstrom A, Segersvard R, Carlsson A, Andersson R, Lohr M, et al. Identification of serum biomarker signatures associated with pancreatic cancer. Cancer Res. 2012;72(10):2481–90. doi: 10.1158/0008-5472.CAN-11-2883. [DOI] [PubMed] [Google Scholar]
- 52.Fischer KR, Durrans A, Lee S, Sheng J, Li F, Wong ST, et al. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature. 2015;527(7579):472–6. doi: 10.1038/nature15748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grant SL, Begley CG. The oncostatin M signalling pathway: reversing the neoplastic phenotype? Molecular Medicine Today. 1999;5(9):406–12. doi: 10.1016/s1357-4310(99)01540-3. [DOI] [PubMed] [Google Scholar]
- 54.Hurwitz HI, Uppal N, Wagner SA, Bendell JC, Beck JT, Wade SM, 3rd, et al. Randomized, Double-Blind, Phase II Study of Ruxolitinib or Placebo in Combination With Capecitabine in Patients With Metastatic Pancreatic Cancer for Whom Therapy With Gemcitabine Has Failed. J Clin Oncol. 2015;33(34):4039–47. doi: 10.1200/JCO.2015.61.4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li Y, Rogoff HA, Keates S, Gao Y, Murikipudi S, Mikule K, et al. Suppression of cancer relapse and metastasis by inhibiting cancer stemness. Proc Natl Acad Sci U S A. 2015;112(6):1839–44. doi: 10.1073/pnas.1424171112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Plater-Zyberk C, Buckton J, Thompson S, Spaull J, Zanders E, Papworth J, et al. Amelioration of arthritis in two murine models using antibodies to oncostatin M. Arthritis Rheum. 2001;44(11):2697–702. doi: 10.1002/1529-0131(200111)44:11<2697::aid-art450>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 57.Poling J, Gajawada P, Richter M, Lorchner H, Polyakova V, Kostin S, et al. Therapeutic targeting of the oncostatin M receptor-beta prevents inflammatory heart failure. Basic Res Cardiol. 2014;109(1):396. doi: 10.1007/s00395-013-0396-3. [DOI] [PubMed] [Google Scholar]
- 58.Lewis Phillips GD, Li G, Dugger DL, Crocker LM, Parsons KL, Mai E, et al. Targeting HER2-positive breast cancer with trastuzumab-DM1, an antibody-cytotoxic drug conjugate. Cancer Res. 2008;68(22):9280–90. doi: 10.1158/0008-5472.CAN-08-1776. [DOI] [PubMed] [Google Scholar]
- 59.Preca BT, Bajdak K, Mock K, Sundararajan V, Pfannstiel J, Maurer J, et al. A self-enforcing CD44s/ZEB1 feedback loop maintains EMT and stemness properties in cancer cells. Int J Cancer. 2015;137(11):2566–77. doi: 10.1002/ijc.29642. [DOI] [PubMed] [Google Scholar]
- 60.Pei H, Li L, Fridley BL, Jenkins GD, Kalari KR, Lingle W, et al. FKBP51 affects cancer cell response to chemotherapy by negatively regulating Akt. Cancer Cell. 2009;16(3):259–66. doi: 10.1016/j.ccr.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.