Abstract
Purpose
Antibiotic prophylaxis is critical to ophthalmology and other surgical specialties. We performed natural language processing (NLP) of 743,838 operative notes recorded for 315,246 surgeries to ascertain two variables needed to study the comparative effectiveness of antibiotic prophylaxis in cataract surgery. The first key variable was an exposure variable, intracameral antibiotic injection. The second was an intraoperative complication, posterior capsular rupture (PCR), that functioned as a potential confounder. To help other researchers use NLP in their settings, we describe our NLP protocol and lessons learned.
Methods
For each of the two variables, we used SAS Text Miner and other SAS text-processing modules with a training set of 10,000 (1.3%) operative notes to develop a lexicon. The lexica identified misspellings, abbreviations, and negations, and linked words into concepts (e.g., “antibiotic” linked with “injection”). We confirmed the NLP tools by iteratively obtaining random samples of 2,000 (0.3%) notes, with replacement.
Results
The NLP tools identified approximately 60,000 intracameral antibiotic injections and 3,500 cases of PCR. The positive and negative predictive values for intracameral antibiotic injection exceeded 99%. For the intraoperative complication, they exceeded 94%.
Conclusion
NLP was a valid and feasible method for obtaining critical variables needed for a research study of surgical safety. These NLP tools were intended for use in the study sample. Use with external datasets or future datasets in our own setting would require further testing.
Keywords: Surgical-site Infection, Prophylaxis, Practice Variation, Comparative Effectiveness Research, Electronic Health Record, Natural Language Processing
INTRODUCTION
During the past decade, implementation of the electronic health record (EHR) in healthcare delivery systems has created a valuable resource for research and quality improvement. However, much of the pertinent information, such as intraoperative exposures and complications, are in unstructured notes that are not readily retrievable. Natural language processing (NLP) is a computational method that extracts information from unstructured natural language, processes the information, and maps the information to structured (i.e., coded) variables.1 NLP has been used to ascertain acute renal failure, venous thromboembolism, sepsis, and myocardial infarction in Veteran’s Administration data; Kawasaki disease in a pediatric emergency department; and tumor progression from magnetic resonance imaging, among other applications.2–5 Notwithstanding these applications, the technology and methods used to develop NLP for epidemiologic research are only now emerging.
For a study of surgical safety, we used NLP to ascertain two intraoperative variables in cataract surgery: (1) the exposure variable, intracameral antibiotic injection, and (2) an intraoperative complication that functions as a potential confounder, posterior capsular rupture (PCR).6, 7 Intracameral antibiotic injection provides direct delivery of antibiotic into the anterior chamber (i.e., the fluid-filled space between the cornea and the lens) by the surgeon at the close of the operation.8–10 PCR is a breach in the continuity of the eye’s posterior chamber, where the lens is located. PCR is highly associated with the risk of surgical site infection.11
This report describes our protocol for developing the NLP tools and presents the lessons that we learned from this experience. We wrote this report to help other researchers develop and implement NLP protocols in their own settings using notes from their own EHRs. It is not our intention to recommend that the NLP tools we developed as described in this report be applied to EHR data in external settings without further testing.
METHODS
The study was approved by an Institutional Review Board. The parent study tested the question: To prevent surgical-site infection following cataract surgery, what is the effectiveness of intraoperative injection of intracameral antibiotic compared with postoperative topical administration? PCR was a potential confounder of the relationship. This report is focused on NLP development and testing.
Setting and Population
The research setting is important in developing an NLP tool, because the design of the tool will reflect the organization of care and clinical workflows, as well as the particular way each provider writes their notes. The parent study was set during 2005–2012, when cataract surgery was performed at 38 surgical centers across Kaiser Permanente Northern and Southern California. As detailed in our earlier report, the study included 182 surgeons who performed 315,246 cataract surgeries on 204,515 patients in a hospital, outpatient surgical center or ophthalmology department surgical center.6. 7 The health plan uses an Epic®-based EHR to store detailed medical information. Over time, as Epic® has been upgraded and surgeons gained skills with Epic®, documentation practices have evolved. For example, documentation of the surgical episode has shifted from use of notes with fewer structured fields to use of systems designed specifically for operating room workflows. Various kinds of notes are stored together in the EHR, including long operative notes, brief operative notes, procedure notes, progress notes, nursing notes, and clinic notes.
Overview to the NLP Protocol
NLP has features in common with predictive modeling12 but is not the same as predictive modeling. In predictive modeling, the goal is to build a model in one dataset that will perform well in future datasets from the same setting and in external settings. A general approach to predictive modeling is to subset the dataset into parts. A training set is used to learn about the data. An independent validation set is used to fine-tune the model. An independent test set is used to assess the likely performance of the model in a new dataset that will be created in the future in the same setting. Finally, external validation should be performed to assess the performance of the model in other settings, although this often does not happen.
Similarly, NLP tools can be developed with the goal of dissemination, but this is not essential. Rather, NLP tools can be developed with the goal of performing with the highest possible validity in only a single dataset. This was our goal. Therefore, for the parent study, we divided the dataset into a training set and a validation set. Because we did not intend to disseminate the tools, we did not use a test set (i.e., hold-out sample). Because we used multiple validation sets that were non-independent of each another (i.e., used sampling with replacement), we refer to these as “confirmation sets” instead of “validation sets”.
In developing the NLP tools for intracameral antibiotic injection and PCR, we used the note as the unit of observation, with the full dataset containing 743,838 notes. Our approach to NLP involved 5 steps (Figure 1). Step 1 was a broad exploration of surgical workflows and documentation. At the end of Step 1, we identified which notes to obtain and we created a lexicon for each of the two variables. In Step 2, we selected a random sample of 10,000 notes (1.3%) to create a training set. At the end of Step 2, we wrote the two NLP tools into computer programs. In Step 3, we applied each NLP tool to the full dataset. In Step 4, we fine-tuned the NLP tools by obtaining random samples of 2,000 notes, of which 1,000 were coded by the NLP tool as “exposed” to the variable (intracameral antibiotic injection or PCR) and 1,000 were coded as “unexposed” to the variable. This was done over several iterations, using sampling with replacement. In Step 5, we measured the performance (positive and negative predictive values) of the final NLP tools using a confirmation set. The following sections provide greater details about these 5 steps.
Figure 1.
NLP Flowchart
Step 1. Gain a Broad Understanding of Surgical Workflows and Documentation
We interviewed four senior surgeons, spanning four surgical centers, to obtain an historical understanding of clinical workflows and workflow changes, including changes in medical documentation. We also observed cataract surgery, including the way various members of the surgical team used the EHR’s front-end interface as it pertained to the cataract surgery clinical workflow, including preoperative, intraoperative, and postoperative tasks. We learned that over the course of the study period, four kinds of notes have been used to record cataract surgery: long operative notes, brief operative notes, procedure notes, and progress notes. We then determined that the 315,246 cataract surgeries were documented in 743,838 notes, with some surgeries being documented in more than one note. For example, a surgery might be documented in both a long operative note and a brief operative note.
We learned that most cataract surgeons, although not all, had created a template for their own use. A template is a standard report, written by the surgeon, which can be copied from one patient to the next but is edited for each individual patient. For example, a template might be edited to indicate the occurrence of PCR. In addition, we were told that many surgeons had created their own “smart phrases”, i.e., shortcuts in the EHR that enabled the surgeon to pull up an entire paragraph by typing a short line of characters. For example, typing the characters “.pcr” might bring up the sentence, “A break in the posterior capsular bag was noted.” As we listened to the surgeons, we noted the words, phrases, and concepts they used to describe intracameral antibiotic injection and PCR.
For each of the 182 surgeons included in the study, we read five operative reports, recorded over the period of their surgical practice, to gain broad insight into each surgeon’s charting practices and use of language. During this step, we noted that the operative reports were lengthy and detailed. We further noted that nearly every surgeon made one or two significant changes in their clinical workflow, use of EHR templates, and charting habits during the study period. We further noted that within the same surgeon, the operative note showed a least some variation from one patient to the next. This mitigated the concern that the surgeons relied too heavily on templates and did not revise them with each patient. We also learned that notes that exceeded 5,000 characters were cut at the 5,000th character, and that surgeries with operative notes exceeding 5,000 characters were stored in the EHR as two or three extracts. Thus, the 743,843 notes were stored in the EHR as 1,183,406 extracts of ≤5,000 characters each.
The programming code that comprises an NLP tool is built upon a lexicon (i.e., an inventory of the words and phrases used to talk about a concept). Using the information gleaned from our explorations, and working in close collaboration with surgeons, we developed two lexica, one for intracameral antibiotic injection and one for PCR, that included concepts, synonyms, abbreviations, misspellings, and negations. The lexicon for intracameral antibiotic injection included intracameral, cefuroxime, moxifloxacin, vigamox, vancomycin, and antibiotic. The lexicon for PCR included posterior capsular rupture, vitreous loss/prolapse (a sequelae of PCR), retained lens (a sequelae of PCR), and vitrectomy (a procedure used to remove vitreous and retained lens from the posterior capsule, where they do not belong).
Step 2. Develop the NLP Tools in the Training Set
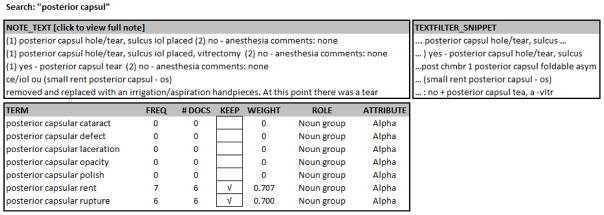
The training set included 10,000 (1.3%) randomly selected notes from the full set of 743,838 notes. Using the two lexica, we applied SAS Text Miner to identify synonyms, abbreviations, misspellings, and negations for each of the two NLP tools. Figure 2 shows Text Miner results for occurrences of the phrase “posterior capsul,” which was represented by “pc”, “pcap”, “post cap” and other variations. Text Miner identified 20 different spellings of the word “vitreous.” It also rolled words up to their root form (e.g., ruptured to rupture), and identified negations (e.g., previous, prior, history of, status post, no loss of vitreous, allergic to cefuroxime, etc.). Text Miner presents these occurrences in context, together with the frequency of their occurrence. The data scientist worked with a surgeon to judge whether each of the occurrences should be coded as PCR or not. To command SAS to include the occurrence of a text string in the NLP tool, the data scientist checked a box, which was built into the Text Miner tool.
Figure 2.
Text Miner
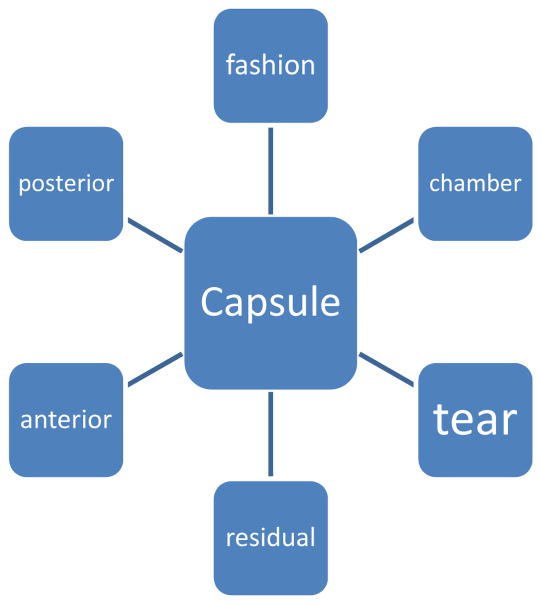
SAS Text Miner can also perform concept linking (Figure 3). Concept linking is the process of finding and displaying terms that are spatially associated with terms in the lexicon. For example, Text Miner linked “vitreous” with the modifier “loss,” i.e., “vitreous loss” and “loss of vitreous.” Likewise, Text Miner linked “capsule” with “break.” Concept linking is essential because a single concept (e.g., PCR) can include multiple terms that are spatially separated within a sentence, for example, “a tear of the posterior capsular bag was noted”.
Figure 3.
Concept Linking*
*Concept linking is the processing finding and displaying terms that are spatially associated with a term (e.g., “capsule”) that is part of the lexicon.
To further improve the NLP tool for intracameral antibiotic injection, we identified patients who did not have a record of receiving any prophylactic antibiotic (intraoperative intracameral injection or postoperative topical) and performed spot checks of these surgical cases. Similarly, for PCR, we identified patients of surgeons whose rates of PCR were at the extreme low or high end, and performed spot checks of these cases.
Step 3. Apply the NLP Tool to the Full Dataset
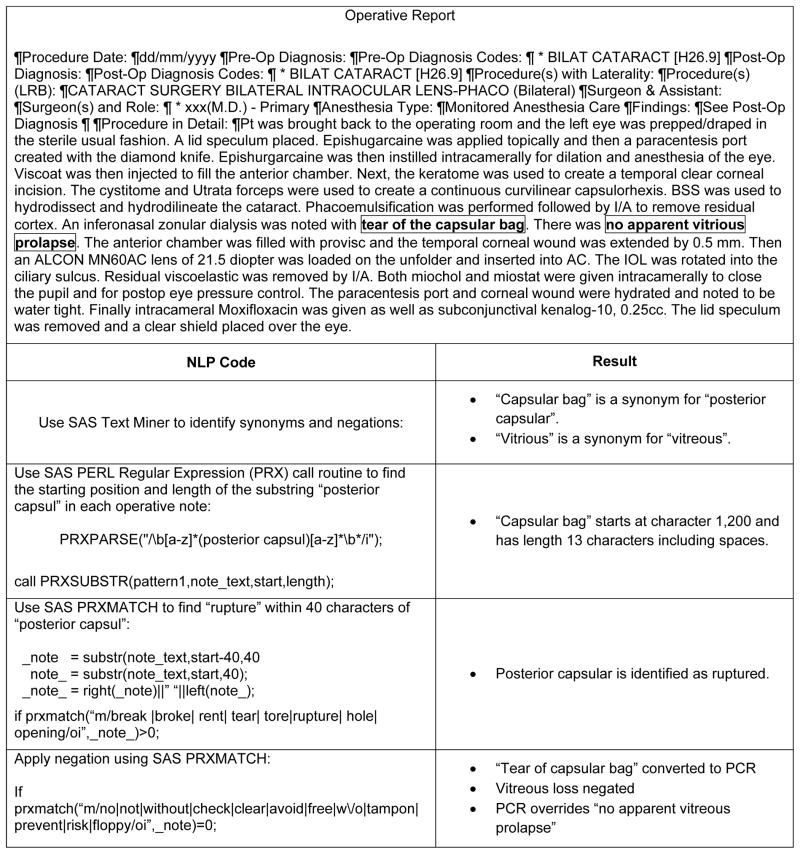
We used SAS PERL regular expressions (PRX)13, 14 to process the 743,838 notes for the 315,246 surgeries to obtain structured data for the two variables, intracameral antibiotic injection and PCR. “Regular expressions” is a set of ways to describe text. For example, text can be described as character or digit. A string of text can be described as having a length in terms of characters. One string of text can be identical to another string, but for one misspelled character. Text is organized into sentences, and within a sentence, terms might occur together, such as “posterior,” “capsular,” and “rupture,” that can be combined to form a concept. SAS PERL regular expressions is a highly versatile computer module, but it is technical and requires the expertise of a data analyst or data scientist. The computer processing time needed to run SAS PRX on the 743,838 notes was about 20 minutes for intracameral antibiotic injection and about 60 minutes for PCR. Figure 4 illustrates how SAS PRX extracted information from the note, processed the information, and mapped the information to the variable PCR.
Figure 4.
Development of NLP to Code Posterior Capsular Rupture (PCR) from an Operative Report
Step 4. Fine-tune the NLP Tools
Next, and separately for the two variables (intracameral antibiotic injection and PCR), we performed chart review of the operative report to identify opportunities to fine-tune the NLP tools. For this task, we randomly sampled 2,000 notes for each variable, of which 1,000 had been coded by the NLP tool as positive for the variable (intracameral antibiotic injection or PCR) and 1,000 had been coded as negative. These samples were selected without knowledge of the outcome variable, surgical site infection. To increase the efficiency of the chart review effort, we downloaded the operative reports into a word processing application (i.e., Microsoft Word®) and highlighted key terms as indicated in Figure 4. To further improve the NLP tools, we repeated this process two times for intracameral antibiotic injection and five times for PCR, until no clear opportunities for improvement remained. Random samples were obtained with replacement.
Step 5. Measure the Performance of the NLP Tools in the Confirmation Set
For each of the two variables, after we finished fine-tuning the NLP tool, we obtained a final confirmation set by again sampling notes from the full dataset (intracameral antibiotic injection, 1,000 coded tool as positive and 1,000 coded as negative; PCR, 400 coded as positive and 400 coded as negative). For PCR, we used fewer than 1,000 subjects for confirmation to reduce the level of effort. We then measured the final positive and negative predictive values using chart review as the gold standard. The chart reviewer had knowledge of how the NLP tool had classified the note (e.g., positive or negative for PCR) when reviewing the chart.
RESULTS
Using NLP, we identified 63,241 intracameral antibiotic injections (prevalence, 20.1%) and 3,551 cases of PCR (1.13%). The key concepts that identified intracameral antibiotic injection and PCR are shown in Table 1. The majority of intracameral injections were identified using the names of two antibiotics that are specific to intracameral injection, cefuroxime (56.6%) and moxifloxacin (33.4%). In addition, we identified 10% of intracameral injections through use of the concept “intracameral injection of antibiotic.” The majority of PCR cases were identified because they underwent vitrectomy, an intraoperative intervention used to address PCR.
Table 1.
Key concepts that identified intracameral injection and PCR
| Concept | N | Cases identified using the concept, % |
|---|---|---|
|
| ||
| Intracameral injection of antibiotics (N=63,241) | ||
| Cefuroxime | 35,781 | 57% |
| Moxifloxacin | 21,150 | 33% |
| Injection of antibiotic | 6,310 | 10% |
|
| ||
| PCR (N=3,551) | ||
| Vitrectomy | 2,046 | 58% |
| Posterior capsular rupture | 1,329 | 37% |
| Vitreous loss/vitreous prolapse | 134 | 4% |
| Retained lens | 42 | 1% |
Confirmation of the NLP tool for intracameral antibiotic gave positive and negative predictive values of 99.9% (95% CIs, 99.4–100%), respectively. Confirmation of the PCR tool gave positive and negative predictive values of 94.4% (95% CIs, 92.6–95.9%) and 99.9% (95% CI, 99.3–100.0%).
DISCUSSION
This report describes the protocol we used to develop and confirm two NLP tools for an observational comparative-effectiveness study of antibiotic prophylaxis in cataract surgery. The two variables were the exposure variable, intracameral antibiotic injection, and an intraoperative complication, PCR, that functions as a potential confounding factor. Surgical site infection is generally rare, so that large datasets are needed for research. Alternatively, we could have performed a matched case-control study with chart review. However, matched studies are less informative than cohort studies with respect to estimating incidence rates and attributable risks. In addition, cohort studies are more accessible than case-control studies to some readers.
The NLP tools we developed were tuned to the organizational structure, clinical workflows, EHR system, and dialects of the ophthalmologists who were included in our research study. Because we did not seek to create NLP tools that could be disseminated to other settings, this report focuses on our methodology, not the products of the methodology. NLP tools developed for retrospective research studies can be highly valid and reliable, but they are not necessarily generalizable. Nor is it essential that they be generalizable, because these tools can be developed and tested quite rapidly using rigorous research approaches.
The study included surgeons in Kaiser Permanente Northern and Southern California, which are two separate medical groups. We found that differences in clinical workflows between the Northern and Southern California groups resulted in striking differences in clinical workflows and charting for PCR requiring vitrectomy (58% of PCR cases). In one region, the same surgeon performed both the cataract surgery and the vitrectomy, so that both procedures were documented in the same operative report. In the other region, the two procedures were performed back-to-back by two separate surgeons and recorded in two separate operative notes. The operative note containing the vitrectomy had to be captured separately from the note containing the cataract surgery. This difference in clinical workflows would have reduced the performance of the NLP tool had we not learned about it. We believe it is unrealistic to expect clinical workflows, medical documentation, and use of language to be consistent across settings. Even within a setting, clinical workflows change, the EHR is frequently updated, and dialects evolve with personnel changes. For these reasons, we would not apply NLP tools developed for one study dataset to different study dataset without fine-tuning the tools and testing their performance characteristics. This would be our practice for studies set in the same healthcare system but at a different time points, and for studies set in an altogether different healthcare system. We strongly recommend that other researchers do the same, because NLP is feasible, and the improved face validity and test validity will offset the time needed to develop and optimize an NLP tool.
Although we are enthusiastic about the potential for NLP in studies using EHR data, we learned several important lessons while executing this work. First, by downloading and highlighting key terms, chart review can be accomplished very quickly. Nonetheless, review of 2,000 charts to confirm the NLP tool for intracameral antibiotic injection was excessive and unnecessary. It was for this reason that we reduced the confirmation set to 800 for PCR. Second, PCR is rare, and for confirmation of rare events, it may be most sensible to confirm every identified case rather than perform random sampling. With hindsight, we should have done this. And third, to increase validity, we ought to have blinded the chart reviewer to the NLP result. This could have been accomplished by mixing cases with and without intracameral antibiotic injection (or PCR) into a single list and randomizing the order of review. One might also argue that a test set, or hold-out sample, be partitioned from the overall dataset, so that the performance of the NLP tool in a hypothetical future dataset can be assessed. As we have already argued, we would perform fine-tuning and validation before using an established NLP tool in a future dataset.
NLP development was easier and more reliable for intracameral antibiotic injection than for PCR, because certain drug names (e.g., cefuroxime) are entirely specific to intracameral antibiotic injection. In contrast, the words “posterior”, “capsular”, and “rupture” have a wide variety of meanings in the context of cataract surgery. We particularly noticed that many different words were used to represent “rupture”, including “tear”, “break”, “rent”, and “hole”, among others. Furthermore, variations of the word “broke” were used in phrases unrelated to PCR, such as “broke open the package.” The subtle and various meanings of “posterior”, “capsule”, and “broke” and the variety of synonyms for “broke” limited the performance of the NLP tool for PCR.
The positive and negative predictive values of the NLP tools we developed (>94%) were outstanding. Again, it is important to remember that, by design, the tools were tuned to the study dataset. The NLP tool for PCR had excellent external validity. We observed the incidence of PCR to be similar to the rate reported in the British Health Services (1.13% compared to 1.92%).15 Furthermore, the association between PCR and risk of surgical site infection (OR 3.7)11 was nearly identical to that (OR 3.4) measured by a randomized controlled trial.9
We used SAS Text Miner in combination with SAS PERL regular expression (PRX) to develop the NLP tools because this approach was agile, flexible, and integrated with the SAS tools we use for nearly all our data processing and analysis. The present application was focused on highly specific medical concepts recorded exclusively into operative notes. In general, operative notes are concise and use few metaphors or literary devices. NLP tools developed for more nuanced concepts, such as “social support”, or using a wider diversity of documents might benefit from other NLP packages, including machine learning. Although our experience with these other NLP packages is limited, our impression is that they offer the researcher less control over NLP tool development, with decision rules being created inside a “black box”.
The use of NLP for medical research is an evolving topic. To advance the use of NLP for medical research and to increase the transparency of reporting, we recommend that investigators specify the goal of their work (single use, internally generalizable tool, externally generalizable tool); discuss the specificity of the concepts inherent in the variables being sought; provide detailed information about the setting, including a description of the clinicians, clinical workflows, EHR structure, and diversity of notes; state which NLP software was used (e.g., SAS vs. other specific commercial applications); state the size and partitioning of the dataset; and describe the approach to confirmation and validation.
In conclusion, in a study of 743,848 operative notes documenting cataract surgery, we found NLP to be valid and feasible for coding highly specific concepts, including antibiotic administration and an intraoperative complication. We are enthusiastic about the potential of NLP for broadening the scope and increasing the validity of the comparative effectiveness research, and recommend investment into further developing NLP methods for medical research. Rigorous reporting of NLP methods will advance the field. Additional research is needed to characterize the trade-offs between feasibility and validity when applying NLP tools across time frames and settings, to optimize NLP development and testing workflows, and to articulate best practices to strengthen the reporting of NLP tools.
Key Points.
Natural language processing (NLP) can be used to obtain information from notes in the electronic medical record.
SAS Text Miner in combination with SAS PERL regular expressions are practical tools for coding key variables (exposures, outcomes, potential confounders and effect modifiers) using NLP.
Integration of NLP tools and approaches can increase the informativeness and the validity of comparative effectiveness research.
Acknowledgments
Role of Study Sponsor: The National Eye Institute (R21EY022989) funded this research.
This research was supported by a grant from the National Eye Institute (R21EY022989). Drs. Shorstein and Herrinton were also funded by grants from the Kaiser Permanente Community Benefit program and from the Kaiser Permanente Garfield Fund.
Abbreviations
- EHR
electronic health record
- PPV
positive predictive value
- NPV
negative predictive value
- CI
confidence interval
- NLP
Natural Language Processing
Footnotes
Disclosures: In the past three years, Drs. Herrinton was funded by a research contract with MedImmune that was unrelated to the present project. The other authors have nothing to disclose.
References Cited
- 1.Boan S, Conway M, Phuong TM, Ohno-Machado L. Natural language processing in biomedicine: a unified system architecture overview. Methods Mol Biol. 2014;1168:275–94. doi: 10.1007/978-1-4939-0847-9_16. [DOI] [PubMed] [Google Scholar]
- 2.Hou JK, Imler TD, Imperiale TF. Current and future applications of natural language processing in the field of digestive disease. Clin Gastroenterol Hepatol. 2014;12:1257–61. doi: 10.1016/j.cgh.2014.05.013. [DOI] [PubMed] [Google Scholar]
- 3.Murff HJ, FitzHenry F, Matheny ME. Automated Identification of Postoperative Complications Within an Electronic Medical Record Using Natural Language Processing. JAMA. 2011;306(8):848–855. doi: 10.1001/jama.2011.1204. [DOI] [PubMed] [Google Scholar]
- 4.Doan S, Maehara CK, Chaparro JD, Lu S, et al. Building a Natural Language Processing Tool to Identify Patients with High Clinical Suspicion for Kawasaki Disease from Emergency Department Notes. Acad Emerg Med. 2016;23:628–367. doi: 10.1111/acem.12925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng LT, Zheng J, Savova JK, et al. Discerning Tumor Status from Unstructured MRI Reports—Completeness of Information in Existing Reports and Utility of Automated Natural Language Processing. J Digital Imaging. 2010;23:119–132. doi: 10.1007/s10278-009-9215-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shorstein NH, Liu L, Waxman MD, Herrinton LJ. Comparative Effectiveness of Three Prophylactic Strategies to Prevent Clinical Macular Edema after Phacoemulsification Surgery. Ophthalmol. 2015;122:2450–2456. doi: 10.1016/j.ophtha.2015.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herrinton LJ, Shorstein NH, Paschal JF, et al. Comparative Effectiveness of Antibiotic Prophylaxis in Cataract Surgery. Ophthalmol. 2016;123:287–294. doi: 10.1016/j.ophtha.2015.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donnenfeld ED, Comstock TL, Proksch JW. Human aqueous humor concentrations of besifloxacin, moxifloxacin, and gatifloxacin after topical ocular application. J Cataract Refract Surg. 2011;37:1082–9. doi: 10.1016/j.jcrs.2010.12.046. [DOI] [PubMed] [Google Scholar]
- 9.ESCRS Endophthalmitis Study Group. Prophylaxis of postoperative endophthalmitis following cataract surgery: results of the ESCRS multicenter study and identification of risk factors. J Cataract Refract Surg. 2007;33:978–988. doi: 10.1016/j.jcrs.2007.02.032. [DOI] [PubMed] [Google Scholar]
- 10.American Academy of Ophthalmology. Cataract in the Adult Eye; Preferred Practice Patterns. San Francisco, CA: American Academy of Ophthalmology; 2011. [Accessed May 25, 2016]. http://www.aao.org/preferred-practice-pattern/cataract-in-adult-eye-ppp--october-2011. [Google Scholar]
- 11.Herrinton LJ, Shorstein NH, Paschal JF, Liu L, Contreras R, Winthrop KL, Chang WJ, Melles RB, Fong DS. Comparative Effectiveness of Antibiotic Prophylaxis in Cataract Surgery. Ophthalmol. 2016;123:287–94. doi: 10.1016/j.ophtha.2015.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Steyerberg EW, Vergouwe Y. Towards better clinical prediction models: seven steps for development and an ABCD for validation. Eur Heart J. 2014;35:1925–31. doi: 10.1093/eurheartj/ehu207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. [Accessed November 4, 2016];Using Perl Regular Expressions in the DATA Step. http://support.sas.com/documentation/cdl/en/lrdict/64316/HTML/default/viewer.htm#a002291852.htm.
- 14. [Accessed November 4, 2016];Perl Regular Expressions Tip Sheet. https://support.sas.com/rnd/base/datastep/perl_regexp/regexp-tip-sheet.pdf.
- 15.Jaycock P, Johnston RL, Taylor H, Adams M, Tole DM, Galloway P, Canning C, Sparrow JM UK EPR user group. The Cataract National Dataset electronic multi-centre audit of 55,567 operations: updating benchmark standards of care in the United Kingdom and internationally. Eye (Lond) 2009;23:38–49. doi: 10.1038/sj.eye.6703015. [DOI] [PubMed] [Google Scholar]