Abstract
Background
Atrial fibrillation (AF) is common and has a substantial genetic basis. Identification of individuals at greatest AF risk could minimize the incidence of cardioembolic stroke.
Methods
To determine whether genetic data can stratify risk for development of AF, we examined associations between AF genetic risk scores and incident AF in five prospective studies comprising 18,919 individuals of European ancestry. We examined associations between AF genetic risk scores and ischemic stroke in a separate study of 509 ischemic stroke cases (202 cardioembolic [40%]) and 3,028 controls. Scores were based on 11 to 719 common variants (≥5%) associated with AF at P-values ranging from <1×10−3 to <1×10−8 in a prior independent genetic association study.
Results
Incident AF occurred in 1,032 (5.5%) individuals. AF genetic risk scores were associated with new-onset AF after adjusting for clinical risk factors. The pooled hazard ratio for incident AF for the highest versus lowest quartile of genetic risk scores ranged from 1.28 (719 variants; 95%CI, 1.13–1.46; P=1.5×10−4) to 1.67 (25 variants; 95%CI, 1.47–1.90; P=9.3×10−15). Discrimination of combined clinical and genetic risk scores varied across studies and scores (maximum C statistic, 0.629–0.811; maximum ΔC statistic from clinical score alone, 0.009–0.017). AF genetic risk was associated with stroke in age- and sex-adjusted models. For example, individuals in the highest quartile of a 127-variant score had a 2.49-fold increased odds of cardioembolic stroke, versus those in the lowest quartile (95%CI, 1.39–4.58; P=2.7×10−3). The effect persisted after excluding individuals (n=70) with known AF (odds ratio, 2.25; 95%CI, 1.20–4.40; P=0.01).
Conclusions
Comprehensive AF genetic risk scores were associated with incident AF beyond clinical AF risk factors, with magnitudes of risk comparable to other clinical risk factors, though offered small improvements in discrimination. AF genetic risk was also associated with cardioembolic stroke in age- and sex-adjusted analyses. Efforts to determine whether AF genetic risk may improve identification of subclinical AF or distinguish stroke mechanisms are warranted.
Keywords: atrial fibrillation, stroke, genetic, risk, prediction
Atrial fibrillation (AF) is a heritable1 and common arrhythmia associated with substantial morbidity and economic costs.2 Approximately one in five ischemic strokes are attributable to cardioembolic events from AF.3 Strokes due to AF are associated with more disability and mortality than strokes from other etiologies.4 Since many strokes caused by AF are preventable with effective anticoagulation,5 and because AF may be undetected in some individuals, there is a critical need to identify those at greatest risk for the arrhythmia.
In recent years, risk models for AF prediction have been developed based on clinical and demographic variables.6–9 We and others have identified common genetic variants associated with AF,10–17 and some of these have been associated with incident AF18 and ischemic stroke19 after adjustment for clinical risk factors. Yet it remains unclear whether a comprehensive AF genetic risk score can facilitate identification of individuals at greatest risk for AF or cardioembolic stroke, since such individuals might benefit from stroke prevention efforts.
We therefore sought to determine whether comprehensive AF genetic risk scores are associated with incident AF beyond clinical risk factors, and might facilitate identification of individuals at greatest risk for the arrhythmia. In addition, we sought to examine whether AF genetic risk is associated with ischemic stroke, and in particular, cardioembolic stroke.
METHODS
Participants
We examined the association between AF genetic risk and incident AF in five prospective studies. Briefly, these studies were the Malmö Diet and Cancer Study (MDCS),20 the Multi-Ethnic Study of Atherosclerosis (MESA),21 the Prevention of Renal and Vascular Endstage Disease (PREVEND) study,22 the PROspective Study of Pravastatin in the Elderly at Risk (PROSPER),23 and the Vanderbilt University de-identified DNA biobank (BioVU).24 We also examined the association between AF genetic risk and stroke in the Massachusetts General Hospital Genes Associated with Stroke Risk and Outcomes Study (MGH-GASROS), a hospital-based case-control study of acute ischemic stroke patients (enrolled between July 2000 and 2011) and referent individuals from the Myocardial Infarction Genetics Consortium (without a history of myocardial infarction).25,26 All stroke cases in MGH-GASROS underwent etiologic stroke subtyping in a uniform fashion, according to the Trial of Org 10172 in Acute Stroke Treatment (TOAST) criteria.27 Descriptions of each study are provided in the online supplement, including details on clinical risk factor and outcome ascertainment, genotyping, and imputation. For all analyses, samples were restricted to individuals of self-reported European ancestry. Each study was approved by its Institutional Review Board, and participants provided written informed consent.
AF genetic risk
To estimate genetic risk using a minimal set of single nucleotide polymorphisms (SNPs), we selected uncorrelated SNPs by pruning28 2.2 million HapMap variants included in a prior independent meta-analysis of genome-wide association studies for AF from the AFGen consortium (6,707 individuals with and 53,436 without AF).15 We considered all SNPs that had allele frequencies ≥ 5% and were nominally associated with AF (P<1×10−3). We then selected the most significantly associated SNP within a given 250 kilobase locus that was not in linkage disequilibrium with another more significantly associated SNP at that locus (r2<0.1). In total, 719 uncorrelated SNPs were selected for construction of genetic risk scores (Supplemental Table 1).
For each individual, we calculated AF genetic risk scores by summing the dosage of each AF risk allele (ranging from 0 to 2) weighted by the natural logarithm of the relative risk for each SNP. Weights were determined in our earlier, independent meta-analysis.15 Thus, a genetic risk score for an individual is a single linear predictor variable. Since the optimum number of risk alleles that should be used for genetic risk scores has not been fixed, we constructed seven different scores for each individual based on the strength of association between each SNP and AF in the earlier analysis.15 We selected the seven different significance thresholds a priori: P<1×10−3, <1×10−4, <1×10−5, <1×10−6, <1×10−7, <5×10−8, and <1×10−8. Liberal inclusion of SNPs was motivated by observations that uncorrelated SNPs demonstrating less significant associations with a trait may still explain a substantial proportion of the heritability of the trait.29–32
Statistical analysis
Within each prospective study, we used proportional hazards regression to examine associations between the different AF genetic risk scores and incident AF over a 5-year time horizon. For all incident AF analyses, person-time in each cohort began at DNA collection or baseline enrollment. Individuals were treated as censored at the time of death or loss to follow-up. Models were adjusted for variables included in a previously validated composite risk score for 5-year AF risk prediction (CHARGE-AF risk score).9 The composite CHARGE-AF risk score included age, height, weight, systolic and diastolic blood pressures, smoking status, antihypertensive medication use, diabetes status, heart failure status, myocardial infarction status, electrocardiographic evidence of left ventricular hypertrophy, and PR interval. Electrocardiographic variables that were not available were omitted from the scores on a study-by-study basis (left ventricular hypertrophy was unavailable in MDCS, MESA, PREVEND, and PROSPER; PR interval was unavailable in MDCS, PREVEND, PROSPER, and BioVU). Race was not included in the models since we restricted our sample to individuals of European ancestry. Proportional hazards assumptions were verified with multiplicative interaction terms between covariates and the natural logarithm of follow-up time.
For each model, we calculated goodness-of-fit statistics using Akaike’s Information Criterion, a penalized likelihood metric in which lower values indicate better fit.33 We also assessed discrimination using the C statistic for time-to-event data.34 Calibration of the prediction models was assessed using the Hosmer-Lemeshow statistic modified for survival analysis.35
In exploratory analyses we combined model parameters from each study by use of an inverse variance random-effects meta-analysis approach, and calculated heterogeneity using the I2 statistic.36 We utilized a random-effects approach owing to inherent differences in study design (see supplemental methods for details). We then multiplied the summary score beta coefficient by the difference between the 12.5th and 87.5th percentiles of AF genetic risk scores from a common reference population (Supplemental Table 2). The resulting values estimate the relative risk comparing individuals in the highest and lowest quartiles across each study and score, in a standard fashion. The common reference population used was a pooled sample of 12,801 individuals from the Framingham Heart Study (n=2,551),37 the Atherosclerosis Risk in Communities Study (n=7,278),38 and the Cardiovascular Health Study (n=2,972)39 with genome-wide genotyping data.15
We then examined whether AF genetic risk was associated with AF, ischemic stroke, and cardioembolic stroke in MGH-GASROS using multivariable logistic regression. Since several of the identified pruned AF SNPs were not available in the MGH-GASROS sample, we utilized proxy SNPs on the basis of linkage disequilibrium when available (Supplemental Table 1). The number of SNPs in some genetic risk scores differed slightly based on inability to identify proxies. Models were adjusted for age and sex only, because extended clinical information was not available in the referent participants. Since AF was ascertained only in stroke cases, we assumed that AF was not present among referents for analyses of AF (an assumption that would be expected to bias the results toward a null association between genetic risk and AF due to the potential for misclassified individuals who have AF among the referent sample). We then examined associations between AF genetic risk and ischemic stroke, as well as the association with the TOAST cardioembolic stroke classification (a subset of ischemic stroke). We utilized the same referent sample set for analyses of ischemic and cardioembolic stroke. Because AF may occur as a subclinical condition, we examined in exploratory analyses whether AF genetic risk scores were associated with stroke in individuals without known AF, again assuming that referent subjects did not have AF.
None of the studies in our analysis of incident AF were used in any aspect of the derivation of genetic risk or the CHARGE-AF scores. The a priori significance threshold for all analyses was P<0.05 using two-sided tests. Meta-analyses were conducted using the rmeta40 package in R.41 Other software utilized for analyses is described in the supplement.
RESULTS
AF genetic risk scores and incident AF
Among 18,919 individuals across all studies in our analyses of incident AF, the mean age ranged from 58–75 years, and the proportion of women ranged from 47–52%. During the 5-year follow-up window, 1,032 (5.5%) individuals developed incident AF (Table 1). AF genetic risk scores were associated with incident AF after accounting for clinical risk factors (Supplemental Figure 1 and Supplemental Table 3). Heterogeneity of effect estimates was modest between studies. Generally, the models with the best fit included scores with between 25 and 129 SNPs, as indicated by the AIC (Supplemental Table 3).
Table 1.
Characteristics of participants included in analyses of incident atrial fibrillation.
| MDCS | MESA | PREVEND | PROSPER* | BioVU | |
|---|---|---|---|---|---|
| No. total | 8,226 | 2,451 | 1,624 | 5,212 | 1,388 |
| No. incident AF | 190 | 76 | 34 | 503 | 229 |
| Age, years | 59±7 | 63±10 | 58±8 | 75±3 | 60±11 |
| Women | 4,275 (52) | 1,321 (52) | 770 (47) | 2,716 (52) | 678 (49) |
| Height, cm | 169±9 | 169±10 | 172±9 | 165±9 | 171±11 |
| Weight, kg | 75±14 | 79±16 | 80±14 | 73±13 | 86±22 |
| Systolic blood pressure, mmHg | 145±20 | 124±20 | 135±21 | 155±22 | 131±20 |
| Diastolic blood pressure, mmHg | 87±10 | 75±10 | 77±10 | 84±11 | 75±30 |
| History of smoking | 2,513 (31) | 1,401 (55) | 671 (41) | 1,388 (27) | 619 (45) |
| Antihypertensive medication | 1,799 (22) | 840 (33) | 362 (22) | 3,854 (74) | 1,339 (96) |
| History of diabetes | 542 (7) | 151 (6) | 98 (6) | 540 (10) | 359 (26) |
| History of heart failure | 39 (0.5) | 52 (2) | 4 (0.2) | NA | 161 (12) |
| History of myocardial infarction | 487 (9) | 63 (3) | 71 (4) | 697 (13) | 284 (20) |
Data presented as mean ± standard deviation, or No. (%)
Maximum follow-up in PROSPER was 4 years.
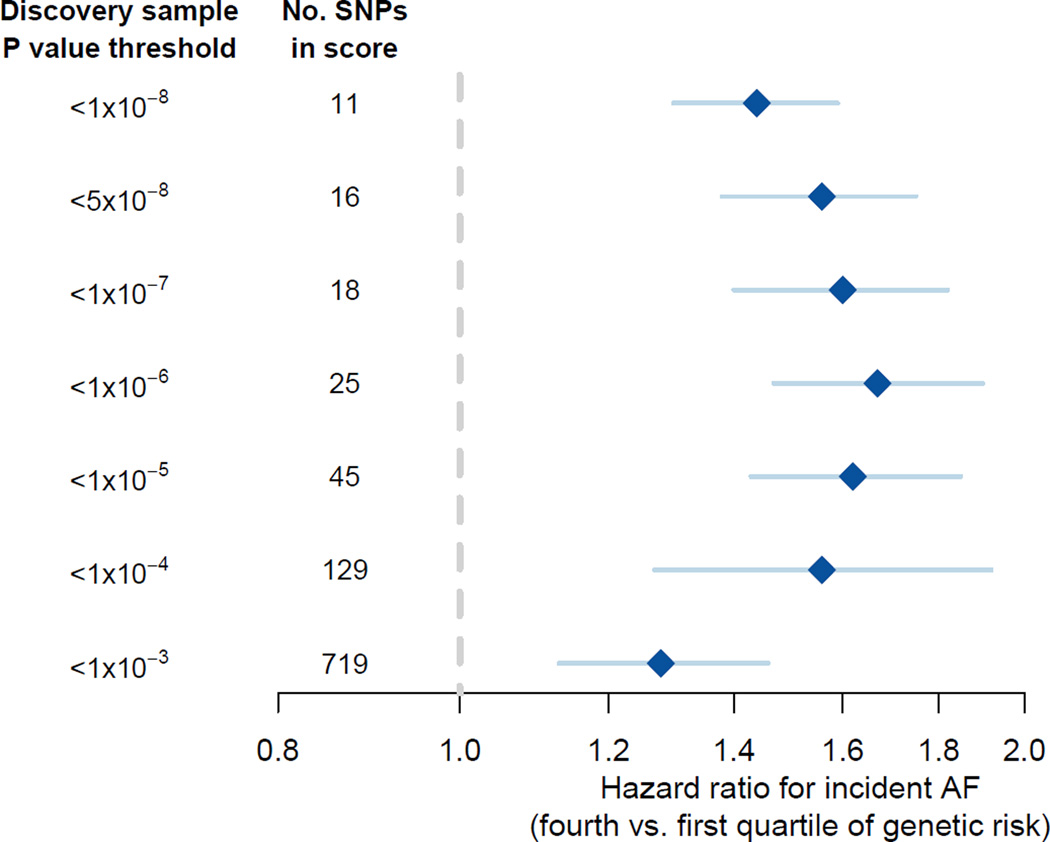
For each of the seven groups of genetic risk scores, we estimated hazard ratios comparing individuals in the highest quartile of each genetic risk score with those in the lowest quartile. Across the genetic risk scores, those in the highest quartile had a 1.28-fold (719 SNPs; 95% CI, 1.13–1.46; P=1.5×10−4) to 1.67-fold (25 SNPs; 95%CI, 1.47–1.90; P=9.3×10−15) increased hazard for AF (Figure 1). C statistics for the clinical risk factor model without AF genetic risk scores ranged from 0.615 to 0.802 across cohorts (Supplemental Table 3). Adding AF genetic risk scores to the clinical risk factor model resulted in a maximum change in the C-statistic of between 0.009 and 0.017 across all cohorts and scores. The maximum change of up to 0.065 in PROSPER may have been driven by the small sample size and was considered an outlier. To illustrate the impact of clinical and genetic risk on incident AF detection, we plotted the cumulative incidence of AF stratified by dichotomized clinical risk, as well by both clinical and genetic risk together, for one representative study (MDCS) in Supplemental Figure 2.
Figure 1.
Pooled 5-year relative hazard of incident atrial fibrillation among individuals in the highest quartile of AF genetic risk relative to those in the lowest quartile.
SNPs included in scores were derived using different thresholds of association between each SNP and atrial fibrillation in an earlier, independent study.15
AF genetic risk scores and ischemic stroke
We examined the association between AF genetic risk scores and stroke among 509 independent individuals with stroke from MGH-GASROS and 3,028 controls (Table 2). Among the stroke cases, 202 (40%) were classified as having had a cardioembolic stroke by TOAST criteria. In total, 87 (17%) individuals with ischemic stroke had documented AF.
Table 2.
Characteristics of participants of European ancestry included in analyses of ischemic stroke from MGH-GASROS and referents.
| Cases | Referents | |
|---|---|---|
| N | 509 | 3,028 |
| Age, years | 66.9 ± 14.4 | 42.3 ± 7.8 |
| Women | 214 (24.2) | 732 (42.0) |
| Atrial fibrillation | 87 (17) | – |
Data presented as mean ± standard deviation, or No. (%)
Stroke etiologic subtype: cardioembolic (n=202, 39%), large artery (n=114, 22%), small vessel / lacunar (n=62, 12%), other (n=124, 24%), undetermined (n=7, 1%).
P for comparison of age and sex between cases and controls <0.001.
In MGH-GASROS, modest associations between AF genetic risk scores and AF, ischemic stroke (all subtypes), and the subset of cases with cardioembolic stroke were observed using continuous genetic risk scores (Supplemental Table 4). The most significantly associated score with AF, as judged by the score with the smallest P-value, occurred with a score constructed from 127 SNPs, corresponding to SNPs with P values <1×10−4 for associations with AF in the prior independent AFGen analysis.15 Individuals in the highest quartile of the 127-SNP genetic risk score had a 3.13-fold (95%CI, 1.47–7.21; P=0.005) increased odds of AF relative to those in the lowest quartile.
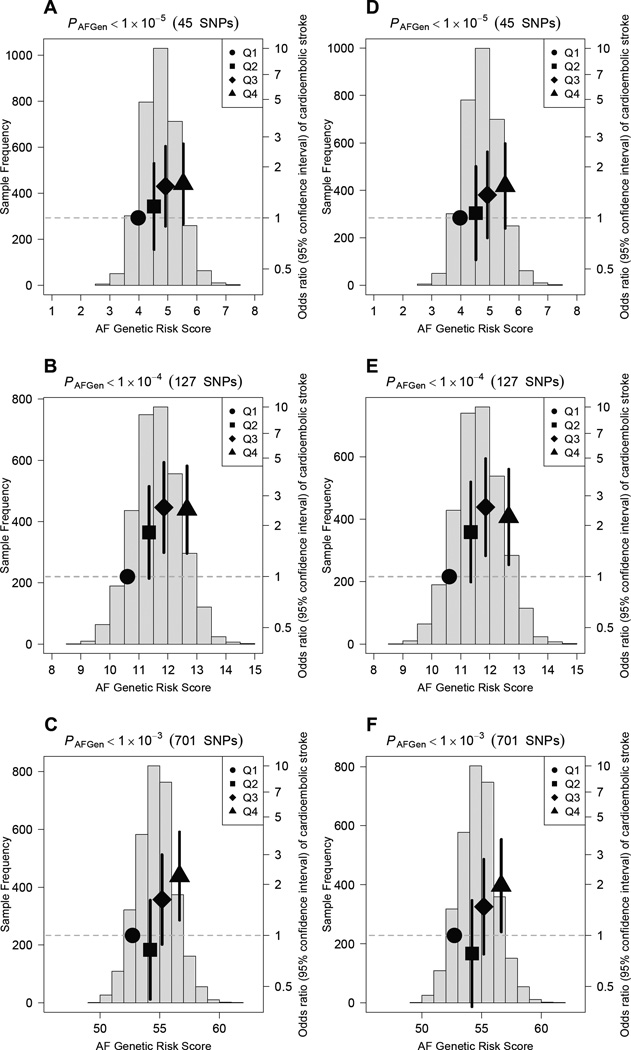
In the analysis of ischemic stroke cases and referent individuals, AF genetic risk scores were also modestly associated with both ischemic stroke (all subtypes) and cardioembolic stroke (Supplemental Table 3). Those in the highest quartile of the 127-SNP genetic risk score had a 1.73-fold (95%CI, 1.15–2.61; P=9.0×10−3) increased odds of ischemic stroke, and a 2.49-fold (95%CI, 1.39–4.58; P=2.7×10−3) increased odds of cardioembolic stroke (after excluding other stroke subtypes, Figure 2). After omitting the 87 stroke cases with known AF (70 of whom had cardioembolic strokes), the associations between AF genetic risk and both ischemic and cardioembolic stroke remained but were slightly attenuated (Supplemental Table 5). Specifically, the relative odds of ischemic stroke comparing those in the highest with those in the lowest quartile of a 127-SNP AF genetic risk score were 1.55 (95%CI, 1.03–2.36; P=0.04) for ischemic stroke, and 2.25 (95%CI, 1.20–4.40; P=0.01) for cardioembolic stroke (Figure 2).
Figure 2.
Risk of cardioembolic stroke in MGH-GASROS according to atrial fibrillation genetic risk.
Odds ratios for cardioembolic stroke in relation to atrial fibrillation genetic risk scores among cardioembolic stroke cases and 3,028 controls. Blue histograms show distributions of genetic risk scores among cases and controls. Black dots indicate odds ratios for stroke for each quartile of genetic risk scores (bars indicate 95% confidence intervals). For panels A–C, genetic risk scores were based on 45 (A), 127 (B), and 701 (C) SNPs among 202 cardioembolic stroke cases (including 70 with known AF) and controls. For panels D-F, genetic risk scores were based 45 (D), 127 (E), and 701 (F) SNPs among 152 cardioembolic stroke cases (none with known AF) and controls. SNP totals may not equal those used in the incident atrial fibrillation analysis since some SNPs were unavailable in MGH-GASROS, in which case proxies were used when available (Supplemental Table 1).
DISCUSSION
In our analysis of nearly 19,000 individuals of European ancestry, scores reflecting the burden of AF risk alleles were associated with 5-year risks of new-onset AF, after adjusting for clinical risk factors. Individuals in the highest quartile of the genetic scores had up to a 67% higher risk of new-onset AF than those in the lowest quartile, although incremental discrimination beyond clinical risk factors was small regardless of the number of SNPs included in the genetic risk score. In an independent sample, individuals in the highest quartile of a score comprised of 127 AF-associated genetic markers had roughly two-fold higher odds of cardioembolic stroke, compared with those in the lowest quartile after adjustment for age and sex. Associations between AF genetic risk scores and cardioembolic stroke persisted after excluding individuals with known AF.
Our findings support and extend prior observations that AF genetic risk is associated with both AF and stroke. We previously observed an association between familial AF and incident AF in the Framingham Heart Study, beyond associations for clinical risk factors.1 Subsequently, we observed an approximately 4 to 5-fold gradient in risk between those in the highest versus lowest tails of a 12-SNP AF genetic risk score (based on nine loci) in case-referent and cohort studies.16 The Women’s Genome Health Study reported an association between an AF genetic risk score based on 12 SNPs and occurrence of incident AF,18 although the AF-associated SNPs used in the analysis were identified in a previous discovery study using the same study sample. Earlier work also described associations between the top AF-associated variants on chromosomes 4q25 and 16q22 with ischemic (and in particular, cardioembolic) stroke.13,26,42–44 Recently, we and others reported a 2-fold increased hazard of AF and a 1.23-fold increased hazard of ischemic stroke for individuals in the highest versus lowest quintiles of scores based on a 12-SNP genetic risk model during an average follow-up of 14 years in the MDCS, subjects of which were included in the present analysis of incident AF.19 Thus, by using well-characterized independent study samples, our current findings extend prior reports that AF genetic risk is associated with incident AF, as well as ischemic stroke.
Our observations have three major implications. First, our finding that AF genetic risk is associated with incident AF beyond the effects observed for accepted clinical risk factors highlights the ability of common genetic variation to capture complementary information. Indeed, the 28%-67% increased risk of AF among individuals in the highest versus the lowest quartile of genetic risk is comparable to the magnitude of risk conferred by traditional clinical risk factors for AF.9 Nevertheless, even by including a large number of genetic variants and assessing associations with incident AF in large cohorts, the magnitudes of risk associated with genetic risk improved discrimination minimally beyond clinical factors. Such findings underscore the challenges of improving clinical prediction models even when including highly associated predictors.45
Second, our observations, coupled with prior findings that AF genetic risk may be preferentially associated with cardioembolic stroke,13,42,43 raise the possibility that AF genetic risk may serve as a signature for strokes caused by thromboembolism due to AF. Our observation that AF genetic risk was associated with an increased risk of cardioembolic stroke even after excluding individuals with known AF is consistent with the hypothesis that AF genetic risk may be a clinically relevant marker for subclinical, or previously undiagnosed, AF. Although AF genetic risk has a limited impact beyond knowledge of clinical risk factors on AF prediction over a 5-year time horizon, it is possible that such genetic profiling may provide insights into stroke mechanisms and therefore screening and treatment options for secondary prevention. Future analyses are warranted to determine if AF genetic risk discriminates effectively between different stroke subtypes, to assess whether AF genetic risk can identify cryptogenic stroke patients at elevated risk for recurrent stroke due to AF, and whether estimating AF risk can enhance secondary stroke prevention efforts.
Third, our observation that genetic risk scores constructed from liberally selected SNPs were nevertheless associated with AF and AF-related stroke emphasizes the polygenic nature of AF. Therefore, true AF susceptibility variants are likely to exist even though they may not meet the stringent genome-wide significance criteria currently utilized. Future genetic discovery efforts in larger samples with better power are warranted to identify additional AF susceptibility signals. Indeed, since publication of the most recent AFGen meta-analysis,15 additional bona fide subthreshold AF signals have been identified, and some appear to be associated with stroke.17 It remains to be determined whether future assessment of AF genetic risk based on associations derived from larger samples will enhance specificity of prediction models.
Our study should be interpreted in the context of the study design. First, all participants were of European descent, and therefore our findings may not be generalizable to individuals of other ancestral groups. Second, the genetic risk models were linear in nature with a single predictor variable, and did not account for potential non-additive genetic effects, interactions between genetic variants, or interactions between genetic variants and environmental factors. Additional modeling methods, including penalized regression or other techniques, may yield more precise genetic risk models. Third, other important determinants of AF risk were not available in our study, including plasma biomarkers such as brain natriuretic peptide.46 Similarly, in analyses of ischemic stroke, clinical covariates beyond age and sex were unavailable, so we could not evaluate whether the genetic risk score adds appreciably to prediction afforded by the CHA2DS2-VASc score47 or individual stroke risk factors. Future studies are warranted to determine whether genetic risk adds additional information to other clinical and biomarker factors related to AF and stroke. Fourth, our genetic risk models were comprised of common SNPs genotyped in the HapMap reference populations,48 many of which are likely tag-SNPs and serve as proxies for true causal variation. Through the use of larger sample sizes and newer techniques to comprehensively assess genomic variation, such as whole genome sequencing, we anticipate better power to identify causal variants underlying AF in the future. Inclusion of causal variants in genetic risk scores may improve the specificity of the models. Fifth, the genetic predictors of prevalent stroke may not be identical to those of incident stroke due to potential survival biases. Therefore, the clinical utility of AF genetic risk factors for identifying individuals at risk for incident stroke merits future study.
Conclusions
We observed that comprehensive AF genetic risk scores were associated with incident AF, exceeding effects of clinical risk factors, in individuals of European ancestry. We further observed that AF genetic risk is associated with both ischemic and cardioembolic stroke after adjustment for age and sex, even among individuals with cardioembolic stroke without established AF. Our findings underscore the polygenic nature of AF and the independent value of genetic information beyond clinical risk factors for the identification of individuals at risk for AF. However, although genetic risk scores are highly associated with AF, genetic information currently affords small improvements in discrimination of AF risk, and therefore does not yet need to be incorporated into routine clinical decision-making. Future clinical trials are necessary to rigorously assess whether AF genetic risk is an effective clinical marker of cardioembolic stroke etiology, and can identify individuals with subclinical AF.
Supplementary Material
CLINICAL PERSPECTIVE.
What is new?
Studies have identified several genetic loci associated with AF, yet it is unclear whether genetic profiling can identify individuals at greatest risk for AF or cardioembolic stroke.
Using genome-wide data from an independent large-scale analysis, we tested comprehensive AF genetic risk scores for association with new-onset AF in five prospective studies, and with stroke in a separate stroke case-control sample.
Genetic risk scores were associated with AF beyond established clinical risk factors, but improved prediction minimally.
AF genetic risk was strongly associated with cardioembolic stroke, suggesting that elevated AF genetic risk might serve as a surrogate for thromboembolism from AF.
What are the clinical implications?
Our findings underscore the complementary information provided by both clinical and genetic factors.
However, since genetic information currently affords small improvements in discrimination of AF risk, widespread use of genetic risk profiling does not need to be incorporated into routine clinical decision-making at this time.
Our findings raise the possibility that AF genetic risk may serve as a signature for strokes caused by thromboembolism from AF.
Future studies are warranted to determine whether AF genetic risk can distinguish stroke etiologic mechanisms, or identify individuals with strokes that have subclinical AF.
Acknowledgments
Sources of funding: Please refer to supplemental material for detailed funding support. The sponsors did not have any input into the study design or conduct; data collection, management, analysis, or interpretation; nor did they influence the preparation, review, or approval of the manuscript.
Disclosures: Dr. Ellinor is a principal investigator on a grant from Bayer HealthCare to the Broad Institute.
References
- 1.Lubitz SA, Yin X, Fontes JD, Magnani JW, Rienstra M, Pai M, Villalon ML, Vasan RS, Pencina MJ, Levy D, Larson MG, Ellinor PT, Benjamin EJ. Association between familial atrial fibrillation and risk of new-onset atrial fibrillation. JAMA. 2010;304:2263–2269. doi: 10.1001/jama.2010.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Writing Group M, Mozaffarian D, Benjamin EJ, Go AS, Arnett DK, Blaha MJ, Cushman M, Das SR, de Ferranti S, Despres JP, Fullerton HJ, Howard VJ, Huffman MD, Isasi CR, Jimenez MC, Judd SE, Kissela BM, Lichtman JH, Lisabeth LD, Liu S, Mackey RH, Magid DJ, McGuire DK, Mohler ER, 3rd, Moy CS, Muntner P, Mussolino ME, Nasir K, Neumar RW, Nichol G, Palaniappan L, Pandey DK, Reeves MJ, Rodriguez CJ, Rosamond W, Sorlie PD, Stein J, Towfighi A, Turan TN, Virani SS, Woo D, Yeh RW, Turner MB American Heart Association Statistics C, Stroke Statistics S. Heart Disease and Stroke Statistics-2016 Update: A Report From the American Heart Association. Circulation. 2016;133:e38–e60. doi: 10.1161/CIR.0000000000000350. [DOI] [PubMed] [Google Scholar]
- 3.Marini C, De Santis F, Sacco S, Russo T, Olivieri L, Totaro R, Carolei A. Contribution of atrial fibrillation to incidence and outcome of ischemic stroke: results from a population-based study. Stroke. 2005;36:1115–1119. doi: 10.1161/01.STR.0000166053.83476.4a. [DOI] [PubMed] [Google Scholar]
- 4.Lamassa M, Di Carlo A, Pracucci G, Basile AM, Trefoloni G, Vanni P, Spolveri S, Baruffi MC, Landini G, Ghetti A, Wolfe CD, Inzitari D. Characteristics, outcome, and care of stroke associated with atrial fibrillation in Europe: data from a multicenter multinational hospital-based registry (The European Community Stroke Project) Stroke. 2001;32:392–398. doi: 10.1161/01.str.32.2.392. [DOI] [PubMed] [Google Scholar]
- 5.Hart RG, Pearce LA, Aguilar MI. Meta-analysis: antithrombotic therapy to prevent stroke in patients who have nonvalvular atrial fibrillation. Ann Intern Med. 2007;146:857–867. doi: 10.7326/0003-4819-146-12-200706190-00007. [DOI] [PubMed] [Google Scholar]
- 6.Schnabel RB, Sullivan LM, Levy D, Pencina MJ, Massaro JM, D’Agostino RB, Sr. Newton-Cheh C, Yamamoto JF, Magnani JW, Tadros TM, Kannel WB, Wang TJ, Ellinor PT, Wolf PA, Vasan RS, Benjamin EJ. Development of a risk score for atrial fibrillation (Framingham Heart Study): a community-based cohort study. Lancet. 2009;373:739–745. doi: 10.1016/S0140-6736(09)60443-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schnabel RB, Aspelund T, Li G, Sullivan LM, Suchy-Dicey A, Harris TB, Pencina MJ, D’Agostino RB, Sr. Levy D, Kannel WB, Wang TJ, Kronmal RA, Wolf PA, Burke GL, Launer LJ, Vasan RS, Psaty BM, Benjamin EJ, Gudnason V, Heckbert SR. Validation of an atrial fibrillation risk algorithm in whites and African Americans. Archives of internal medicine. 2010;170:1909–1917. doi: 10.1001/archinternmed.2010.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chamberlain AM, Agarwal SK, Folsom AR, Soliman EZ, Chambless LE, Crow R, Ambrose M, Alonso A. A clinical risk score for atrial fibrillation in a biracial prospective cohort (from the Atherosclerosis Risk in Communities [ARIC] study) The American journal of cardiology. 2011;107:85–91. doi: 10.1016/j.amjcard.2010.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alonso A, Krijthe BP, Aspelund T, Stepas KA, Pencina MJ, Moser CB, Sinner MF, Sotoodehnia N, Fontes JD, Janssens AC, Kronmal RA, Magnani JW, Witteman JC, Chamberlain AM, Lubitz SA, Schnabel RB, Agarwal SK, McManus DD, Ellinor PT, Larson MG, Burke GL, Launer LJ, Hofman A, Levy D, Gottdiener JS, Kaab S, Couper D, Harris TB, Soliman EZ, Stricker BH, Gudnason V, Heckbert SR, Benjamin EJ. Simple Risk Model Predicts Incidence of Atrial Fibrillation in a Racially and Geographically Diverse Population: the CHARGE-AF Consortium. J Am Heart Assoc. 2013;2:e000102. doi: 10.1161/JAHA.112.000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gudbjartsson DF, Arnar DO, Helgadottir A, Gretarsdottir S, Holm H, Sigurdsson A, Jonasdottir A, Baker A, Thorleifsson G, Kristjansson K, Palsson A, Blondal T, Sulem P, Backman VM, Hardarson GA, Palsdottir E, Helgason A, Sigurjonsdottir R, Sverrisson JT, Kostulas K, Ng MC, Baum L, So WY, Wong KS, Chan JC, Furie KL, Greenberg SM, Sale M, Kelly P, MacRae CA, Smith EE, Rosand J, Hillert J, Ma RC, Ellinor PT, Thorgeirsson G, Gulcher JR, Kong A, Thorsteinsdottir U, Stefansson K. Variants conferring risk of atrial fibrillation on chromosome 4q25. Nature. 2007;448:353–357. doi: 10.1038/nature06007. [DOI] [PubMed] [Google Scholar]
- 11.Benjamin EJ, Rice KM, Arking DE, Pfeufer A, van Noord C, Smith AV, Schnabel RB, Bis JC, Boerwinkle E, Sinner MF, Dehghan A, Lubitz SA, D’Agostino RB, Sr. Lumley T, Ehret GB, Heeringa J, Aspelund T, Newton-Cheh C, Larson MG, Marciante KD, Soliman EZ, Rivadeneira F, Wang TJ, Eiriksdottir G, Levy D, Psaty BM, Li M, Chamberlain AM, Hofman A, Vasan RS, Harris TB, Rotter JI, Kao WH, Agarwal SK, Stricker BH, Wang K, Launer LJ, Smith NL, Chakravarti A, Uitterlinden AG, Wolf PA, Sotoodehnia N, Kottgen A, van Duijn CM, Meitinger T, Mueller M, Perz S, Steinbeck G, Wichmann HE, Lunetta KL, Heckbert SR, Gudnason V, Alonso A, Kaab S, Ellinor PT, Witteman JC. Variants in ZFHX3 are associated with atrial fibrillation in individuals of European ancestry. Nat Genet. 2009;41:879–881. doi: 10.1038/ng.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellinor PT, Lunetta KLNLG, Pfeufer A, Alonso A, Chung MK, Sinner MF, de Bakker PI, Mueller M, Lubitz SA, Fox E, Darbar D, Smith NL, Smith JD, Schnabel R, Soliman EZ, Rice K, Van Wagoner DR, Beckmann BM, van Noord C, Wang K, Ehret GB, Rotter JI, Hazen S, Steinbeck G, Makino S, Nelis M, Milan DJ, Perz S, Esko T, Kottgen A, Moebus S, Newton-Cheh C, Li M, Mohlenkamp S, Wang TJ, Kao WH, Vasan RS, Nothen MM, MacRae CA, Levy D, Boerwinkle E, Metspalu A, Topol EJ, Chakravarti A, Psaty BM, Roden D, T M, Wichmann HE, Witteman JC, Barnard J, Arking DE, Benjamin EJ, Heckbert SR, Kaab S. Common Variants in KCNN3 are Associated with Lone Atrial Fibrillation. Nat Genet. 2010;42:240–244. doi: 10.1038/ng.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gudbjartsson DF, Holm H, Gretarsdottir S, Thorleifsson G, Walters GB, Thorgeirsson G, Gulcher J, Mathiesen EB, Njolstad I, Nyrnes A, Wilsgaard T, Hald EM, Hveem K, Stoltenberg C, Kucera G, Stubblefield T, Carter S, Roden D, Ng MC, Baum L, So WY, Wong KS, Chan JC, Gieger C, Wichmann HE, Gschwendtner A, Dichgans M, Kuhlenbaumer G, Berger K, Ringelstein EB, Bevan S, Markus HS, Kostulas K, Hillert J, Sveinbjornsdottir S, Valdimarsson EM, Lochen ML, Ma RC, Darbar D, Kong A, Arnar DO, Thorsteinsdottir U, Stefansson K. A sequence variant in ZFHX3 on 16q22 associates with atrial fibrillation and ischemic stroke. Nat Genet. 2009;41:876–878. doi: 10.1038/ng.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schnabel RB, Kerr KF, Lubitz SA, Alkylbekova EL, Marcus GM, Sinner MF, Magnani JW, Wolf PA, Deo R, Lloyd-Jones DM, Lunetta KL, Mehra R, Levy D, Fox ER, Arking DE, Mosley TH, Muller-Nurasyid M, Young TR, Wichmann HE, Seshadri S, Farlow DN, Rotter JI, Soliman EZ, Glazer NL, Wilson JG, Breteler MM, Sotoodehnia N, Newton-Cheh C, Kaab S, Ellinor PT, Alonso A, Benjamin EJ, Heckbert SR. Large-scale candidate gene analysis in whites and African Americans identifies IL6R polymorphism in relation to atrial fibrillation: the National Heart, Lung, and Blood Institute’s Candidate Gene Association Resource (CARe) project. Circulation. Cardiovascular genetics. 2011;4:557–564. doi: 10.1161/CIRCGENETICS.110.959197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ellinor PT, Lunetta KL, Albert CM, Glazer NL, Ritchie MD, Smith AV, Arking DE, Muller-Nurasyid M, Krijthe BP, Lubitz SA, Bis JC, Chung MK, Dorr M, Ozaki K, Roberts JD, Smith JG, Pfeufer A, Sinner MF, Lohman K, Ding J, Smith NL, Smith JD, Rienstra M, Rice KM, Van Wagoner DR, Magnani JW, Wakili R, Clauss S, Rotter JI, Steinbeck G, Launer LJ, Davies RW, Borkovich M, Harris TB, Lin H, Volker U, Volzke H, Milan DJ, Hofman A, Boerwinkle E, Chen LY, Soliman EZ, Voight BF, Li G, Chakravarti A, Kubo M, Tedrow UB, Rose LM, Ridker PM, Conen D, Tsunoda T, Furukawa T, Sotoodehnia N, Xu S, Kamatani N, Levy D, Nakamura Y, Parvez B, Mahida S, Furie KL, Rosand J, Muhammad R, Psaty BM, Meitinger T, Perz S, Wichmann HE, Witteman JC, Kao WH, Kathiresan S, Roden DM, Uitterlinden AG, Rivadeneira F, McKnight B, Sjogren M, Newman AB, Liu Y, Gollob MH, Melander O, Tanaka T, Stricker BH, Felix SB, Alonso A, Darbar D, Barnard J, Chasman DI, Heckbert SR, Benjamin EJ, Gudnason V, Kaab S. Meta-analysis identifies six new susceptibility loci for atrial fibrillation. Nat Genet. 2012;44:670–675. doi: 10.1038/ng.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lubitz SA, Lunetta KL, Lin H, Arking DE, Trompet S, Li G, Krijthe BP, Chasman DI, Barnard J, Kleber ME, Dorr M, Ozaki K, Smith AV, Muller-Nurasyid M, Walter S, Agarwal SK, Bis JC, Brody JA, Chen LY, Everett BM, Ford I, Franco OH, Harris TB, Hofman A, Kaab S, Mahida S, Kathiresan S, Kubo M, Launer LJ, Macfarlane PW, Magnani JW, McKnight B, McManus DD, Peters A, Psaty BM, Rose LM, Rotter JI, Silbernagel G, Smith JD, Sotoodehnia N, Stott DJ, Taylor KD, Tomaschitz A, Tsunoda T, Uitterlinden AG, Van Wagoner DR, Volker U, Volzke H, Murabito JM, Sinner MF, Gudnason V, Felix SB, Marz W, Chung M, Albert CM, Stricker BH, Tanaka T, Heckbert SR, Jukema JW, Alonso A, Benjamin EJ, Ellinor PT. Novel genetic markers associate with atrial fibrillation risk in Europeans and Japanese. J Am Coll Cardiol. 2014;63:1200–1210. doi: 10.1016/j.jacc.2013.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sinner MF, Tucker NR, Lunetta KL, Ozaki K, Smith JG, Trompet S, Bis JC, Lin H, Chung MK, Nielsen JB, Lubitz SA, Krijthe BP, Magnani JW, Ye J, Gollob MH, Tsunoda T, Muller-Nurasyid M, Lichtner P, Peters A, Dolmatova E, Kubo M, Smith JD, Psaty BM, Smith NL, Jukema JW, Chasman DI, Albert CM, Ebana Y, Furukawa T, MacFarlane P, Harris TB, Darbar D, Dorr M, Holst AG, Svendsen JH, Hofman A, Uitterlinden A, Gudnason V, Isobe M, Malik R, Dichgans M, Rosand J, Van Wagoner DR, Consortium M, Consortium AF, Benjamin EJ, Milan DJ, Melander O, Heckbert S, Ford I, Liu Y, Barnard J, Olesen MS, Stricker BH, Tanaka T, Kaab S, Ellinor PT. Integrating Genetic, Transcriptional, and Functional Analyses to Identify Five Novel Genes for Atrial Fibrillation. Circulation. 2014 doi: 10.1161/CIRCULATIONAHA.114.009892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Everett BM, Cook NR, Conen D, Chasman DI, Ridker PM, Albert CM. Novel genetic markers improve measures of atrial fibrillation risk prediction. Eur Heart J. 2013;34:2243–2251. doi: 10.1093/eurheartj/eht033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tada H, Shiffman D, Smith JG, Sjogren M, Lubitz SA, Ellinor PT, Louie JZ, Catanese JJ, Engstrom G, Devlin JJ, Kathiresan S, Melander O. Twelve-single nucleotide polymorphism genetic risk score identifies individuals at increased risk for future atrial fibrillation and stroke. Stroke. 2014;45:2856–2862. doi: 10.1161/STROKEAHA.114.006072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith JG, Platonov PG, Hedblad B, Engstrom G, Melander O. Atrial fibrillation in the Malmo Diet and Cancer study: a study of occurrence, risk factors and diagnostic validity. European journal of epidemiology. 2010;25:95–102. doi: 10.1007/s10654-009-9404-1. [DOI] [PubMed] [Google Scholar]
- 21.Bild DE, Bluemke DA, Burke GL, Detrano R, Diez Roux AV, Folsom AR, Greenland P, Jacob DR, Jr, Kronmal R, Liu K, Nelson JC, O’Leary D, Saad MF, Shea S, Szklo M, Tracy RP. Multi-Ethnic Study of Atherosclerosis: objectives and design. Am J Epidemiol. 2002;156:871–881. doi: 10.1093/aje/kwf113. [DOI] [PubMed] [Google Scholar]
- 22.Vermond RA, Geelhoed B, Verweij N, Tieleman RG, Van der Harst P, Hillege HL, Van Gilst WH, Van Gelder IC, Rienstra M. Incidence of Atrial Fibrillation and Relationship With Cardiovascular Events, Heart Failure, and Mortality: A Community-Based Study From the Netherlands. J Am Coll Cardiol. 2015;66:1000–1007. doi: 10.1016/j.jacc.2015.06.1314. [DOI] [PubMed] [Google Scholar]
- 23.Shepherd J, Blauw GJ, Murphy MB, Bollen EL, Buckley BM, Cobbe SM, Ford I, Gaw A, Hyland M, Jukema JW, Kamper AM, Macfarlane PW, Meinders AE, Norrie J, Packard CJ, Perry IJ, Stott DJ, Sweeney BJ, Twomey C, Westendorp RG. Pravastatin in elderly individuals at risk of vascular disease (PROSPER): a randomised controlled trial. Lancet. 2002;360:1623–1630. doi: 10.1016/s0140-6736(02)11600-x. [DOI] [PubMed] [Google Scholar]
- 24.Roden DM, Pulley JM, Basford MA, Bernard GR, Clayton EW, Balser JR, Masys DR. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther. 2008;84:362–369. doi: 10.1038/clpt.2008.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anderson CD, Biffi A, Rahman R, Ross OA, Jagiella JM, Kissela B, Cole JW, Cortellini L, Rost NS, Cheng YC, Greenberg SM, de Bakker PI, Brown RD, Jr, Brott TG, Mitchell BD, Broderick JP, Worrall BB, Furie KL, Kittner SJ, Woo D, Slowik A, Meschia JF, Saxena R, Rosand J International Stroke Genetics C. Common mitochondrial sequence variants in ischemic stroke. Annals of neurology. 2011;69:471–480. doi: 10.1002/ana.22108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.International Stroke Genetics C, Wellcome Trust Case Control C. Bellenguez C, Bevan S, Gschwendtner A, Spencer CC, Burgess AI, Pirinen M, Jackson CA, Traylor M, Strange A, Su Z, Band G, Syme PD, Malik R, Pera J, Norrving B, Lemmens R, Freeman C, Schanz R, James T, Poole D, Murphy L, Segal H, Cortellini L, Cheng YC, Woo D, Nalls MA, Muller-Myhsok B, Meisinger C, Seedorf U, Ross-Adams H, Boonen S, Wloch-Kopec D, Valant V, Slark J, Furie K, Delavaran H, Langford C, Deloukas P, Edkins S, Hunt S, Gray E, Dronov S, Peltonen L, Gretarsdottir S, Thorleifsson G, Thorsteinsdottir U, Stefansson K, Boncoraglio GB, Parati EA, Attia J, Holliday E, Levi C, Franzosi MG, Goel A, Helgadottir A, Blackwell JM, Bramon E, Brown MA, Casas JP, Corvin A, Duncanson A, Jankowski J, Mathew CG, Palmer CN, Plomin R, Rautanen A, Sawcer SJ, Trembath RC, Viswanathan AC, Wood NW, Worrall BB, Kittner SJ, Mitchell BD, Kissela B, Meschia JF, Thijs V, Lindgren A, Macleod MJ, Slowik A, Walters M, Rosand J, Sharma P, Farrall M, Sudlow CL, Rothwell PM, Dichgans M, Donnelly P, Markus HS. Genome-wide association study identifies a variant in HDAC9 associated with large vessel ischemic stroke. Nat Genet. 2012;44:328–333. doi: 10.1038/ng.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adams HP, Jr, Bendixen BH, Kappelle LJ, Biller J, Love BB, Gordon DL, Marsh EE., 3rd Classification of subtype of acute ischemic stroke. Definitions for use in a multicenter clinical trial TOAST. Trial of Org 10172 in Acute Stroke Treatment. Stroke. 1993;24:35–41. doi: 10.1161/01.str.24.1.35. [DOI] [PubMed] [Google Scholar]
- 28.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Evans DM, Visscher PM, Wray NR. Harnessing the information contained within genome-wide association studies to improve individual prediction of complex disease risk. Human molecular genetics. 2009;18:3525–3531. doi: 10.1093/hmg/ddp295. [DOI] [PubMed] [Google Scholar]
- 30.Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, Sullivan PF, Sklar P. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lango Allen H, Estrada K, Lettre G, Berndt SI, Weedon MN, Rivadeneira F, Willer CJ, Jackson AU, Vedantam S, Raychaudhuri S, Ferreira T, Wood AR, Weyant RJ, Segre AV, Speliotes EK, Wheeler E, Soranzo N, Park JH, Yang J, Gudbjartsson D, Heard-Costa NL, Randall JC, Qi L, Vernon Smith A, Magi R, Pastinen T, Liang L, Heid IM, Luan J, Thorleifsson G, Winkler TW, Goddard ME, Sin Lo K, Palmer C, Workalemahu T, Aulchenko YS, Johansson A, Zillikens MC, Feitosa MF, Esko T, Johnson T, Ketkar S, Kraft P, Mangino M, Prokopenko I, Absher D, Albrecht E, Ernst F, Glazer NL, Hayward C, Hottenga JJ, Jacobs KB, Knowles JW, Kutalik Z, Monda KL, Polasek O, Preuss M, Rayner NW, Robertson NR, Steinthorsdottir V, Tyrer JP, Voight BF, Wiklund F, Xu J, Zhao JH, Nyholt DR, Pellikka N, Perola M, Perry JR, Surakka I, Tammesoo ML, Altmaier EL, Amin N, Aspelund T, Bhangale T, Boucher G, Chasman DI, Chen C, Coin L, Cooper MN, Dixon AL, Gibson Q, Grundberg E, Hao K, Juhani Junttila M, Kaplan LM, Kettunen J, Konig IR, Kwan T, Lawrence RW, Levinson DF, Lorentzon M, McKnight B, Morris AP, Muller M, Suh Ngwa J, Purcell S, Rafelt S, Salem RM, Salvi E, Sanna S, Shi J, Sovio U, Thompson JR, Turchin MC, Vandenput L, Verlaan DJ, Vitart V, White CC, Ziegler A, Almgren P, Balmforth AJ, Campbell H, Citterio L, De Grandi A, Dominiczak A, Duan J, Elliott P, Elosua R, Eriksson JG, Freimer NB, Geus EJ, Glorioso N, Haiqing S, Hartikainen AL, Havulinna AS, Hicks AA, Hui J, Igl W, Illig T, Jula A, Kajantie E, Kilpelainen TO, Koiranen M, Kolcic I, Koskinen S, Kovacs P, Laitinen J, Liu J, Lokki ML, Marusic A, Maschio A, Meitinger T, Mulas A, Pare G, Parker AN, Peden JF, Petersmann A, Pichler I, Pietilainen KH, Pouta A, Ridderstrale M, Rotter JI, Sambrook JG, Sanders AR, Schmidt CO, Sinisalo J, Smit JH, Stringham HM, Bragi Walters G, Widen E, Wild SH, Willemsen G, Zagato L, Zgaga L, Zitting P, Alavere H, Farrall M, McArdle WL, Nelis M, Peters MJ, Ripatti S, van Meurs JB, Aben KK, Ardlie KG, Beckmann JS, Beilby JP, Bergman RN, Bergmann S, Collins FS, Cusi D, den Heijer M, Eiriksdottir G, Gejman PV, Hall AS, Hamsten A, Huikuri HV, Iribarren C, Kahonen M, Kaprio J, Kathiresan S, Kiemeney L, Kocher T, Launer LJ, Lehtimaki T, Melander O, Mosley TH, Jr, Musk AW, Nieminen MS, O’Donnell CJ, Ohlsson C, Oostra B, Palmer LJ, Raitakari O, Ridker PM, Rioux JD, Rissanen A, Rivolta C, Schunkert H, Shuldiner AR, Siscovick DS, Stumvoll M, Tonjes A, Tuomilehto J, van Ommen GJ, Viikari J, Heath AC, Martin NG, Montgomery GW, Province MA, Kayser M, Arnold AM, Atwood LD, Boerwinkle E, Chanock SJ, Deloukas P, Gieger C, Gronberg H, Hall P, Hattersley AT, Hengstenberg C, Hoffman W, Lathrop GM, Salomaa V, Schreiber S, Uda M, Waterworth D, Wright AF, Assimes TL, Barroso I, Hofman A, Mohlke KL, Boomsma DI, Caulfield MJ, Cupples LA, Erdmann J, Fox CS, Gudnason V, Gyllensten U, Harris TB, Hayes RB, Jarvelin MR, Mooser V, Munroe PB, Ouwehand WH, Penninx BW, Pramstaller PP, Quertermous T, Rudan I, Samani NJ, Spector TD, Volzke H, Watkins H, Wilson JF, Groop LC, Haritunians T, Hu FB, Kaplan RC, Metspalu A, North KE, Schlessinger D, Wareham NJ, Hunter DJ, O’Connell JR, Strachan DP, Wichmann HE, Borecki IB, van Duijn CM, Schadt EE, Thorsteinsdottir U, Peltonen L, Uitterlinden AG, Visscher PM, Chatterjee N, Loos RJ, Boehnke M, McCarthy MI, Ingelsson E, Lindgren CM, Abecasis GR, Stefansson K, Frayling TM, Hirschhorn JN. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ehret GB, Lamparter D, Hoggart CJ, Whittaker JC, Beckmann JS, Kutalik Z. A Multi-SNP Locus-Association Method Reveals a Substantial Fraction of the Missing Heritability. American journal of human genetics. 2012;91:863–871. doi: 10.1016/j.ajhg.2012.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Akaike H. A New Look at the Statistical Identification Model. IEEE Trans Automat Contr. 1974;19:716–723. [Google Scholar]
- 34.Pencina MJ, D’Agostino RB. Overall C as a measure of discrimination in survival analysis: model specific population value and confidence interval estimation. Stat Med. 2004;23:2109–2123. doi: 10.1002/sim.1802. [DOI] [PubMed] [Google Scholar]
- 35.D’Agostino R, Nam B. Handbook of Statistics. Amsterdam: Elsevier; 2004. Evaluation of the performance of survival analysis models: discrimantion and calibration measures; pp. 1–25. [Google Scholar]
- 36.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. Bmj. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dawber TR, Meadors GF, Moore FE., Jr Epidemiological approaches to heart disease: the Framingham Study. Am J Public Health Nations Health. 1951;41:279–281. doi: 10.2105/ajph.41.3.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.The ARICInvestigators. The Atherosclerosis Risk in Communities (ARIC) study: design and objectives. Am J Epidemiol. 1989;129:687–702. [PubMed] [Google Scholar]
- 39.Fried LP, Borhani NO, Enright P, Furberg CD, Gardin JM, Kronmal RA, Kuller LH, Manolio TA, Mittelmark MB, Newman A, O’Leary DH, Psaty BM, Rautaharju P, Tracy RP, Weiler PG. The Cardiovascular Health Study: design and rationale. Ann Epidemiol. 1991;1:263–276. doi: 10.1016/1047-2797(91)90005-w. [DOI] [PubMed] [Google Scholar]
- 40.Lumley T. rmeta: Meta-analysis. R package version 2.16. 2012 https://CRAN.R-project.org/package=rmeta.
- 41.Team RC. R: A language and environment for statistical computing. 2014 http://www.R-project.org.
- 42.Gretarsdottir S, Thorleifsson G, Manolescu A, Styrkarsdottir U, Helgadottir A, Gschwendtner A, Kostulas K, Kuhlenbaumer G, Bevan S, Jonsdottir T, Bjarnason H, Saemundsdottir J, Palsson S, Arnar DO, Holm H, Thorgeirsson G, Valdimarsson EM, Sveinbjornsdottir S, Gieger C, Berger K, Wichmann HE, Hillert J, Markus H, Gulcher JR, Ringelstein EB, Kong A, Dichgans M, Gudbjartsson DF, Thorsteinsdottir U, Stefansson K. Risk variants for atrial fibrillation on chromosome 4q25 associate with ischemic stroke. Ann Neurol. 2008;64:402–409. doi: 10.1002/ana.21480. [DOI] [PubMed] [Google Scholar]
- 43.Lemmens R, Buysschaert I, Geelen V, Fernandez-Cadenas I, Montaner J, Schmidt H, Schmidt R, Attia J, Maguire J, Levi C, Jood K, Blomstrand C, Jern C, Wnuk M, Slowik A, Lambrechts D, Thijs V. The association of the 4q25 susceptibility variant for atrial fibrillation with stroke is limited to stroke of cardioembolic etiology. Stroke; a journal of cerebral Circulation. 2010;41:1850–1857. doi: 10.1161/STROKEAHA.110.587980. [DOI] [PubMed] [Google Scholar]
- 44.Traylor M, Farrall M, Holliday EG, Sudlow C, Hopewell JC, Cheng YC, Fornage M, Ikram MA, Malik R, Bevan S, Thorsteinsdottir U, Nalls MA, Longstreth W, Wiggins KL, Yadav S, Parati EA, Destefano AL, Worrall BB, Kittner SJ, Khan MS, Reiner AP, Helgadottir A, Achterberg S, Fernandez-Cadenas I, Abboud S, Schmidt R, Walters M, Chen WM, Ringelstein EB, O’Donnell M, Ho WK, Pera J, Lemmens R, Norrving B, Higgins P, Benn M, Sale M, Kuhlenbaumer G, Doney AS, Vicente AM, Delavaran H, Algra A, Davies G, Oliveira SA, Palmer CN, Deary I, Schmidt H, Pandolfo M, Montaner J, Carty C, de Bakker PI, Kostulas K, Ferro JM, van Zuydam NR, Valdimarsson E, Nordestgaard BG, Lindgren A, Thijs V, Slowik A, Saleheen D, Pare G, Berger K, Thorleifsson G Australian Stroke Genetics Collaborative WTCCC; Hofman A, Mosley TH, Mitchell BD, Furie K, Clarke R, Levi C, Seshadri S, Gschwendtner A, Boncoraglio GB, Sharma P, Bis JC, Gretarsdottir S, Psaty BM, Rothwell PM, Rosand J, Meschia JF, Stefansson K, Dichgans M, Markus HS International Stroke Genetics C. Genetic risk factors for ischaemic stroke and its subtypes (the METASTROKE collaboration): a meta-analysis of genome-wide association studies. Lancet Neurol. 2012;11:951–962. doi: 10.1016/S1474-4422(12)70234-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cook NR. Use and misuse of the receiver operating characteristic curve in risk prediction. Circulation. 2007;115:928–935. doi: 10.1161/CIRCULATIONAHA.106.672402. [DOI] [PubMed] [Google Scholar]
- 46.Sinner MF, Stepas KA, Moser CB, Krijthe BP, Aspelund T, Sotoodehnia N, Fontes JD, Janssens AC, Kronmal RA, Magnani JW, Witteman JC, Chamberlain AM, Lubitz SA, Schnabel RB, Vasan RS, Wang TJ, Agarwal SK, McManus DD, Franco OH, Yin X, Larson MG, Burke GL, Launer LJ, Hofman A, Levy D, Gottdiener JS, Kaab S, Couper D, Harris TB, Astor BC, Ballantyne CM, Hoogeveen RC, Arai AE, Soliman EZ, Ellinor PT, Stricker BH, Gudnason V, Heckbert SR, Pencina MJ, Benjamin EJ, Alonso A. B-type natriuretic peptide and C-reactive protein in the prediction of atrial fibrillation risk: the CHARGE-AF Consortium of community-based cohort studies. Europace. 2014;16:1426–1433. doi: 10.1093/europace/euu175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lip GY, Nieuwlaat R, Pisters R, Lane DA, Crijns HJ. Refining clinical risk stratification for predicting stroke and thromboembolism in atrial fibrillation using a novel risk factor-based approach: the euro heart survey on atrial fibrillation. Chest. 2010;137:263–272. doi: 10.1378/chest.09-1584. [DOI] [PubMed] [Google Scholar]
- 48.The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.