Abstract
Variability in drug responsivity has prompted the development of Personalized Medicine, which has shown great promise in utilizing genotypic information to develop safer and more effective drug regimens for patients. Similarly, individual variability in learning outcomes has puzzled researchers who seek to create optimal learning environments for students. “Personalized Learning” seeks to identify genetic, neural and behavioral predictors of individual differences in learning and aims to use predictors to help create optimal teaching paradigms. Evidence for Personalized Learning can be observed by connecting research in pharmacogenomics, cognitive genetics and behavioral experiments across domains of learning, which provides a framework for conducting empirical studies from the laboratory to the classroom and holds promise for addressing learning effectiveness in the individual learners. Evidence can also be seen in the subdomain of speech learning, thus providing initial support for the applicability of Personalized Learning to language.
Keywords: individual differences, language learning, neurogenetics, personalized learning
Introduction
Research in educational sciences and related clinical disciplines has strived to identify the most efficacious educational and interventional programs. Results from numerous high-quality empirical studies provided converging evidence for many areas of best educational practices, ranging from early childhood cognitive development to mathematics, sciences and literacy. In asking what factors and interventions predict and produce the best learning outcomes, high-quality research in educational science often focuses on learners as a group. More recently, research has begun to examine the impact that individual differences and learning-centered factors may have on responsivity to a given intervention.
This review discusses Personalized Learning, a translational line of inquiry that stands in parallel with pharmacogenomics and Personalized Medicine (Wang et al., 2011). The concept of using genetic information to improve patient outcomes has already been established by pharmacogenomic research. This concept has great potential to be extended to developing personalized educational practices as well as personalized treatment of behavioral disorders, communication disorders and learning disabilities (see also Gabrieli, Ghosh, & Whitfield-Gabrieli, 2015). Recent advances in genomics have expanded beyond understanding of the molecular genetics of cellular functions and diseases (McCormack et al., 2011; Pare et al., 2010) directly relevant to drug therapies and have begun to also shed light onto the genetic basis of higher order human functions, including executive functions and memory (Papassotiropoulos and de Quervain, 2011) as well as domain-specific behaviors, such as motor learning (Adkins et al., 2006). Exploring how genetic, neural and behavioral predictors can be used to customize learning paradigms across various modalities of learning would lay the foundation for optimizing both learning and behavioral treatment to the individual, a shift away from learning paradigms that focus on cognitive training at the group level. After briefly reviewing Personalized Medicine, the framework for Personalized Learning will be discussed followed by an example of personalized language learning in the subdomain of speech learning and general considerations for implementations.
Personalized Medicine
Recognizing individual variability in drug responsivity and safety, researchers and clinicians in fields of medicine have begun to seek ways to tailor medical treatments to patients on an individual level. Often termed Individualized Medicine, Personalized Medicine and more recently Precision Medicine (Collins & Vamus, 2015), this new area of intensive research has resulted in a number of treatment strategies that have shown great promise in improving patient outcomes (Ma and Lu, 2011; Wang et al., 2011). Facilitated by recent developments and reduced costs in human genomics, researchers have demonstrated that in the pharmacological treatment of certain diseases, genetic variation contributes to differences in patients' responses to medication (Mallal et al., 2008). A well-studied example of genetic polymorphisms affecting treatment response in Personalized Medicine is warfarin, an anticoagulant whose responsivity is associated with variants of CYP2C9, a group of genes that encode enzymes that are responsible for the metabolic clearance of warfarin, and VKORC1, which encodes for an enzyme involved in recycling vitamin K-, a cofactor necessary for the formation of various clotting factors. These discoveries have led to FDA approval of clinical tests for genetic variants and labeling changes on medication that provide considerations of CYP2C9 and VKORC1 polymorphisms in deciding on a dosing regimen (Schwarz et al., 2008). Other pharmacological therapies that have shown similar successes in treatment responses include abacavir (Mallal et al., 2008), gefitinib (Lynch et al., 2004), clopidogrel (Mega et al., 2010; Pare et al., 2010), carbamazepine (McCormack et al., 2011; Phillips and Mallal, 2011), and hepatitis C treatments (Thomas et al., 2009). These successes in Personalized Medicine can potentially be extended to domains of learning.
Personalized Learning: The Framework
Personalized Learning depends on three conditions. First, individual differences in learning need to be identified, demonstrating that not every person learns optimally under the same training paradigm. Second, genotypic, endogenotypic (neural) and/or behavioral (e.g., cognitive-perceptual) factors that are predictive of individual differences in learning should be determined. Third, these predictors should be used to place learners into the most optimal training conditions, individualized to their specific learning needs.
Individual Differences in Learning
Research on cognition and across domains of learning has shown individual degrees of success in learning vary, even under the same learning paradigms. Widespread individual differences have been found in learning achievement across mathematics, science and reading literacy (Halberda et al., 2008; Martin & Mullis, 2013), from the learning of facts to the acquisition of skill (Ackerman, 2007), in explicit forms as well as implicit forms of learning (Kaufman et al., 2010). Individual differences exist in how fast children normally acquire their native language (Bates et al., 1995) and in native language attainment (Street & Dabrowska, 2010). Compared to first language acquisition, individual differences are even more pervasive in second language acquisition (Birdsong, 2004). In clinical populations, treatment outcomes have also demonstrated variability. For example, children who are severely or profoundly deaf and have received cochlear implants have shown extensive variability in speech-and-language outcomes (Peterson et al., 2010).
It is worth mentioning that the identification of individual differences in learning outcomes is not the sole focus of Personalized Learning. It is important to consider what is being learned and how learning occurs. In this regard, individual differences in learning outcomes per se may be less useful for the goals of Personalized Learning than individual differences in the determinants (i.e., predictors) of learning (see next section). This is because the existence of individual differences in learning outcomes suggests that not everyone learns optimally under certain conditions, but these differences alone give little information as to why some learners do not succeed. Individual differences in predictors of learning are important because these differences can further elucidate the learning conditions that may impede or facilitate learning. Such differences can ultimately be used to improve learning. Important questions arise: To what extent can individual differences in learning be predicted? Are there available objective predictors that do not require extensive testing of individual performance? Can predictors be used to modify approaches to individual learning?
Predictors of Learning
In Personalized Medicine, genetic polymorphisms (genetic variations across individuals) have been shown to be predictive of patient responsivity to certain pharmacological treatments (Pare et al., 2010). These genomic advances have also extended beyond life-threatening diseases that have clear molecular origins. In learning and higher-level functions, numerous studies have emerged that demonstrate the predictive ability of individual genetic differences on learning and cognition. Individual differences in episodic memory have been found to be associated with a cluster of genes related to the glutamate system (de Quervain and Papassotiropoulos, 2006; Papassotiropoulos and de Quervain, 2011). Individual differences in various aspects of procedural learning have been tied to polymorphisms of genes in the dopaminergic D2 receptor and striatal systems (Frank et al., 2009; Klein et al., 2007). Individual differences in working memory have also been linked to polymorphisms of dopaminergic D1 receptor genes (Egan et al., 2001; Rybakowski et al., 2005), such that homozygous carriers of the G allele of DRD1 seemed to show worse performance. Not suprisingly, some of these genes have recently been found to be associated with cognitive and psychiatric disorders (Bilder et al., 2011). As cognitive functions are strongly linked across domains of learning (Klahr et al., 2011), these cognition-related genes are likely to be predictive of aspects of learning although future studies are required to examine their actual predictive power.
Despite numerous cases of success (e.g., Franke et al., 2008; Gateva et al., 2009; Soronen et al., 2010), it is important to point out the enormous challenges in finding genetic predictors. For example, many previously identified associations based on genome-wide association studies (GWAS) have failed to replicate (Campa et al., 2012; Cousin et al., 2011; Molendijk et al., 2012). Even the best examples of replications only explain a small proportion of phenotypic variance (Queitsch et al., 2012). In a recent study, Harlaar et al. (2014) used a genome-wide association approach to identify genetic variants associated with individual differences in receptive language in a sample of 2329 children. No associations were found that survived the conventional statistical significance threshold correcting for multiple comparisons; it was suggested that larger sample sizes and newer sequencing methods are required for future studies.
As finding genetic predictors will continue to be challenging, Personalized Learning also aims to incorporate other predictors outside of genetic markers for successful learning. Among non-genetic markers, having been shown to have predictive powers across diverse areas of learning are general cognitive factors such as psychometric intelligence (Neisser et al., 1996), executive functioning (Bull et al., 2008) and working memory (Alloway & Alloway, 2010). For example, fluid intelligence measured at age 11 could predict academic achievement at age 16 across an extensive list of school subjects, from mathematics and sciences (physics, chemistry, biology) to arts and humanities that included native and foreign languages (Deary et al., 2007). Based on past behavioral studies, in addition to executive functioning and working memory, candidate markers of language learning may include measures of ability to learn specific associations between stimuli (Ellis, 2008) and also declarative memory (Ullman, 2005), implicit learning of sequential regularities (Kaufman et al., 2010) and also procedural memory (Ullman, 2005), and perceptual sensitivity such as pitch contour perception (Wong & Perrachione, 2007). Electrophysiological and neuroimaging studies further suggest that event-related negative response seen in younger infants can serve as a predictor of later language development (Kooijman et al., 2013) while volumes in the left Heschl's gyrus and white matter connectivity around Broca's area can serve as predictors of phonetic learning and grammar learning, respectively (Golestani, 2014).
The goal of Personalized Learning is not to find predictors to identify “good learners” regardless of the type of learning, but rather to predict learning outcomes under a particular learning paradigm. Personalized Learning crucially requires finding predictors of learning across different training paradigms within the same domain of learning and across different domains of learning. The same predictor (e.g., working memory) may predict better or worse learning, depending on the type of learning in question. For example, although higher working memory has been linked to better reading abilities (Carpenter and Daneman, 1980) and second language learning (Slevc & Miyake, 2006), individuals with higher working memory were found to require more trials to learn information-integration category structures compared to individuals with lower working memory; thus, higher working memory may sometimes result in less efficient learning (Decaro et al., 2008). Furthermore, connections between working memory and language learning may be more robust under explicit training contexts than under implicit ones (Tagarelli et al., 2011). Under the latter conditions, individuals with lower spans may perform similarly to individuals with higher spans (see also Unsworth & Engle, 2005). Thus, higher working memory or other cognitive and biological predictors does not always result in better learning. Finding predictors across learning paradigms and domains is crucial because it provides converging evidence for understanding which learning conditions best benefiting which learners and how.
Using Predictors for Optimizing Learning
The third and perhaps most crucial component of Personalized Learning is the ability to use predictors to place learners into the most optimal learning paradigm. A crucial assumption in this idea is that different kinds of training and processing strategies can lead to the same learning and behavioral outcome. Research studies across domains of learning and processing support this assumption. For example, although different ERP neural components were found in the processing of grammatical structures by native and high-proficiency non-native speakers, both groups achieve the same level of accuracy in grammaticality judgment (Mueller et al., 2007). Similarly, both explicit and implicit language training could result in similar levels of proficiency, though the two types of training rely on different neural processes (e.g. Morgan-Short et al., 2012). More recently, Cohen and Schneidman (2013) found that changing teaching methods in visual learning could result in similarly high levels of learning. What is being highlighted in Personalized Learning is that different training could be optimally effective across individual learners.
This third component of Personalized Learning is also a component which directly shapes future educational practices and provides opportunities for future research. While ample evidence supports the first two components of Personalized Learning, direct empirical evidence for this component is relatively scarce. Series of recent studies in motor learning seem to provide partial support especially in the context of identifying viable genetic predictors. In the first of a series of experiments examining the brain-derived neurotrophic factor (BDNF) gene, subjects who were homozygous for the val allele (val/val) were found to have higher motor-evoked potentials and more expanded motor maps (see Figure 1A-B) (Kleim et al., 2006) following fine-motor exercises relative to those subjects who were heterozygous for val (val/met) or homozygous for the met allele (met/met). This first establishes BDNF as a genetic predictor of fine-motor learning. Subsequently, sustained motor learning was found to help the met-group (val/met and met/met subjects) achieve the same level of cortical plasticity as the val/val group after 5 and 12 days of training (see Figure 1C) (McHughen et al., 2011). In other words BDNF polymorphisms were found to be associated with responsivity to the dosage of motor training and associated cortical plasticity. It is important to note, however, that these studies were not designed to directly test whether predictors of motor learning could be used to place learners into the most optimal learning situation. Furthermore, these studies focused on cortical plasticity as the outcome measure, rather than behavioral motor performance. Thus, support for Personalized Learning is not complete. However, they speak to the possibility that identification of a BDNF polymorphism before training could potentially help determine whether a stronger or weaker dosage of training should be prescribed.
Figure 1. BDNF val66met polymorphism is associated with responsivity to dosage of fine motor learning.
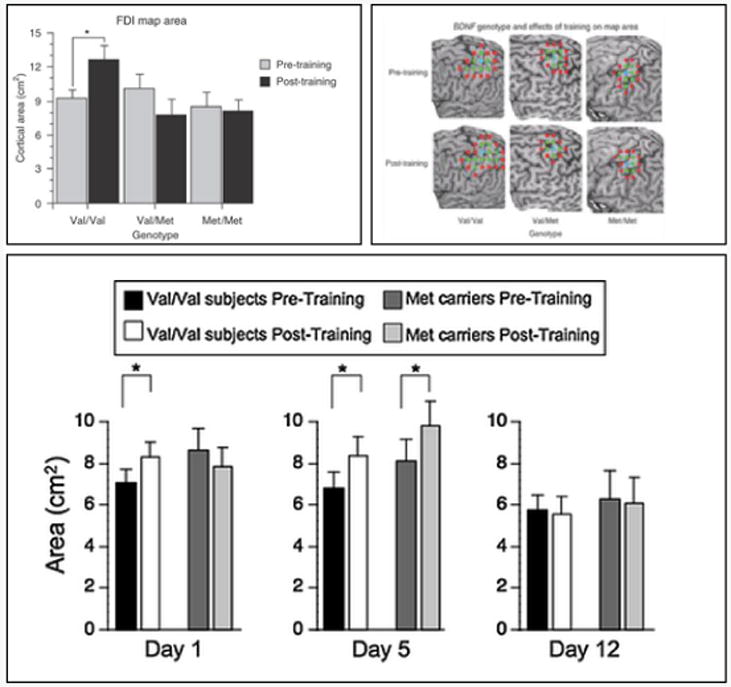
(A) In one study, subjects performed exercises to improve movements of the first dorsal interosseous muscle (FDI) (Kleim et al., 2006). Cortical plasticity in the form of cortical motor map expansion was measured using transcranial magnetic stimulation. Subjects were trained for 30 minutes. Only subjects with the val/val genotype showed significant cortical map expansion: average data (*p < .05) and (B) cortical maps from representative subjects from each genotype group are depicted. (C) In a subsequent study performed by an independent laboratory, new subjects participated in a multi-day training program (McHughen et al., 2011). As in the initial study (Kleim et al., 2006), only the val/val group showed cortical changes after the first day of training. Most importantly, genotypic effects disappeared after 5 and 12 days of training (*p < .05).
In addition to the tremendous challenge in identifying genetic predictors, we must also acknowledge that it would be even more difficult to attempt to use genetic predictors for optimizing learning. Nevertheless, the motor learning evidence cited above provides an illustration of how genetic information could be useful. As discussed, predictors of learning can extend beyond genetic markers. Evidence from speech learning summarized below provides a further illustration for the feasibility of Personalized Learning in language.
Personalized Language Learning: An Example
The majority of world's languages mark word meanings based on pitch patterns of the speakers' voice (Yip, 2002). For example, in Mandarin Chinese, a tone language, the syllable /ma/ can have four different lexical meanings depending on the pitch of the syllable. Research from our group has focused on the acquisition of nonnative tones in adulthood. In a series of training studies, we have found marked individual differences in tone perception and acquisition (see Figure 2A). Individual differences in learning can be successfully predicted by a set of behavioral, neural, and possibly genetic markers (see Figure 2B-C). Preliminary research suggests that markers can be used to place learners into training paradigms that support personalized learning (see Figure 2D).
Figure 2. Applying Personalized Learning to Language.

(A) Individual differences in tone learning success: In our studies, we taught native English-speaking adults to incorporate pitch in lexically meaningful contexts. We found a range of learning success in these adults (Deng et al., in press). (B) Genetic association between tone perception and load of ASPM-G allele (Wong et al., 2013). (C) Neural markers of tone learning: Brain activation revealed by successful versus less successful learners in pre-training contrast (Wong et al., 2007; upper panel). HG volume of successful versus less successful learners in the left and right hemispheres (Wong et al., 2008; lower panel, **p < 0.007 and *p < 0.05). (D) Using predictors for optimizing tone learning: High variability training significantly enhanced learning for better pitch perceivers (HAL), whereas poorer pitcher perceivers (LAL) were significantly impaired by increased stimulus variability (Perrachione et al., 2011; left panel, ordinate values have been arcsine transformed). Lexical pitch-pattern training given before lexical training improved learning more than lexical training alone, and more so for poorer pitch perceivers (Ingvalson et al., 2013; right panel, arcsine-transformed accuracy, the dashed line indicates perfect identification performance, error bars represent SEM).
Individual differences in the acquisition of nonnative tones
In our studies, we taught native English-speaking adults to incorporate pitch in lexically meaningful contexts. For example, the syllable [peʃ] spoken with a high-level pitch pattern means ‘glass’ while the same syllable spoken with a rising pitch pattern means ‘pencil’ (Wong & Perrachione, 2007). We classified these adults into two groups, successful learners and less successful learners, based on whether they achieved a 95% accuracy learning criterion. As can be seen in Figure 2A, we found a range of learning success in these adults (see also Asaridou et al., 2016).
Predictors of tone learning success
To identify predictors of tone learning success, we focused on pretraining variables that distinguished successful from less successful learners. In several behavioral studies, we found that successful learners were better at identifying pitch patterns in a pitch contour perception test (Wong & Perrachione, 2007), and they attended more to pitch directional cues than pitch height whereas less successful learners showed less marked difference in the relative weighting of these cues (Chandrasekaran, Sampath, & Wong, 2010). Furthermore, successful learners showed greater activation in bilateral auditory cortex (Wong, Perrachione, & Parrish, 2007) and had greater volume in the left Heschl's Gyrus (Wong, Warrier, Penhune, Roy, Sadehh, & Parrish, & Zatorre, 2008). When we considered the three predictors, pitch pattern perception, bilateral auditory cortex activation, and volume of left Heschl's Gyrus, all measured before training, we found about 61% of the variance in learning explained (see Figure 2C, Wong et al., 2008). Removal of each of these predictors from the regression model significantly impacted the amount of variance explained, suggesting the importance of each. Our work suggests that other markers of tone learning include large-scale functional brain connectivity (Sheppard, Wang, & Wong, 2012), spontaneous brain activity during resting state (Deng et al., 2016), and musical training (Wong, 2007), perhaps partially due to the fact that musicians showed more faithful pitch tracking as early as the auditory brainstem (Wong, Skoe, Russo, Dees, & Kraus, 2007).
Using predictors for optimizing tone learning
The above findings suggest that pretraining perceptual aptitude is a predictor of tone learning success. In an initial attempt at finding optimal training for individual learners, we compared the efficacy of two speech training paradigms for learners with stronger and weaker perceptual aptitude (see Figure 2D) (Perrachione et al., 2011). Prior to training, we divided listeners into two groups, better and poorer pitch perceivers, based on their performance on the pitch contour perception test. Listeners from both groups were equally assigned to a training paradigm emphasizing trial-by-trial stimulus variability or to low-variability training, in either a single-talker or blocked-talker paradigm. The listeners were trained to asymptotic performance. Better pitch perceivers were found to better respond to high variability training, consistent with earlier work in this paradigm. However, poorer pitch perceivers were impaired by high variability. These listeners showed the greatest benefit when variability was limited. These results suggest that auditory perception ability could be used as a marker for placing learners into learning paradigms that differ in trial-by-trial stimulus variability.
In another study, we used the same predictor (pretraining sensitivity to pitch patterns) but explored a different way to improve learning for poorer pitch perceivers (Ingvalson, Barr, & Wong, 2013). Prior to lexical pitch-to-word learning, both better and poorer perceivers received pitch-pattern training that emphasized differences among tone contrasts. Results showed that such training resulted in better learning than lexical training alone, primarily in the poorer perceivers. Thus, using predictors can help us find the right training paradigms especially for poorer learners. Although these studies examined non-genetic markers, it is interesting to note that in a recent study we found in an independent group of subjects an association between the abnormal spindle-like microcephaly-associated (ASPM) gene and pitch pattern perception, making ASPM a potential genetic marker candidate for speech learning involving pitch (see Figure 2B; Wong et al., 2012).
Considerations for Implementation
The examples cited above in motor learning and speech learning provide some evidence for Personalized Learning as a concept but extensive testing in both laboratory and authentic learning settings remains to be completed. There are a number of challenges that need to be overcome before Personalized Learning can be implemented in practice.
Finding Predictors across Multiple Sources and Time Scales
Personalized Learning requires identification of predictors that are both efficient and objective measures. While earlier studies on individual differences of learning have identified cognitive predictors, other objective measurements such as genetic testing remain to be explored. As discussed above, several genetic markers have been found for different types of learning (Klein et al., 2007) and cognitive abilities (Egan et al., 2001), with support from multiple replication studies (Jocham et al., 2009). Clearly, one single-nucleotide polymorphism (SNP) alone cannot be perfectly mapped onto behaviors. Rather, the emphasis here is on identifying genotypic information from multiple sources along with other factors (e.g., neural and behavioral) for predicting learning success across a number of domains. Because the focus of Personalized Medicine is responsivity to molecular compounds, linking molecular genetics to drug treatments has quickly demonstrated its feasibility. However, the genetic basis of complex traits, including cognition and learning, is widely recognized as multifaceted. Nevertheless, recent advances in the genetic basis of higher-order behaviors show great promise in helping to identify genetic pathways and environmental factors that shape human cognition and learning. The development of newer technologies (e.g., whole genome sequencing) (Lupski et al., 2010) and analytic techniques (Manolio et al., 2009) will hopefully assist in finding the best predictors for Personalized Learning as they can likely increase the sensitivity and specifcity of the investigation.
In finding predictors, we must keep in mind the dynamics of learning. For example, the acquisition of cognitive skill has been characterized as proceeding through series of transitions, from declarative to procedural stages (Anderson, 1982) and from controlled processing to automatic processing (Shiffrin & Schneider, 1977). Language development involves vocabulary building (Marchman & Bates, 1994) and simple phrase learning (Lieven et al., 2003) which provide the basis for the emergence of grammar. As learning proceeds through phases, predictors of learning may change. For example, because the initial phase of skill acquisition poses high demand on general cognitive ability (e.g., required for understanding the task), general intelligence can be predictive of individual differences in skill acquisition during this phase; as cognitive demand decreases in subsequent stages the predictive power of general cognitive ability can decline with task practice (Ackerman, 1988). Thus, it is important to attend to the dynamics of learning, and individual differences in this dynamics, in evaluating predictors for any aspects of learning.
In considering the use of predictors, a distinction needs to be made between predictiveness and usefulness. Predictors that are high on predictiveness are not necessarily most useful for the goals of Personalized Learning1. General intelligence is an excellent predictor of learning achievement across multiple domains. However, as a complex measure of thinking ability it is not immediately obvious how this marker can be used for optimizing language learning. A more specific variable such as perceptual aptitude for speech may have poor predictive power beyond language but nevertheless can be useful for optimizing language learning, as shown above.
Frameworks for Optimizing Learning
Personalized Learning calls for optimization of learning for the individual learners based on predictors. Theoretical frameworks for guiding such an optimization process are needed. The declarative/procedural model (Ullman, 2004, 2005) is one potential framework for research on first and second language acquisition across normal development and disorders including Specific Language Impairment (Ullman & Pierpont, 2005). This model posits two distinct neurocognitive components supporting language learning, a declarative memory component (well-suited for vocabulary acquisition) and a procedural memory component (better suited for grammar acquisition). Recent development suggests that declarative memory can compensate for aspects of grammatical function when procedural memory is compromised (Ullman & Pullnam, 2015). Thus, according to the model, declarative/procedural memory profiles can serve as predictors for language learning and these profiles can be used for optimizing language learning across individuals. For example, it is expected that individuals with functioning declarative memory and weak or otherwise compromised procedural memory would best benefit from grammar training paradigms that target the declarative component.
Computational models of learning and cognition could provide other avenues for guiding optimization. One notable example is the working memory-augmented model of reinforcement learning (Collins & Frank, 2012; Frank et al., 2009) which provides a unifying neurogenetic framework connecting aspects of motivation, learning and cognition. The model posits a reinforcement learning system for slow incremental learning from positive and negative feedback and a working memory (WM) component for maintaining encoded information in memory – the WM component has a faster learning rate but is subject to decay and capacity limitation. The model also makes specific predictions on the contribution of the components over the course of learning: Due to fast learning rate WM can predict outcomes better during initial acquisition but the incremental accumulation of reinforcement values would be predictive of outcomes over time. Extensive computational and empirical work has linked individual differences in WM to prefrontal dopamine function (associated, for example, with the COMT gene) and individual differences in reinforcement learning to striatal dopaminergic function – genetic polymorphisms associated with striatal D1 and D2 function, such as polymorphisms of the DARPP-32 gene and the DRD2 gene, have been found predictive of learning from positive and negative outcomes, respectively (e.g., Frank et al., 2007, Frank et al., 2009).
These findings especially the identification of different neurogenetic components for learning from positive and negative feedback have important implications. In research on language learning, connectionist models have emphasized the role of (implicit) negative feedback (Rohde & Plaut, 1999); recent modeling work further attributed age-related differences in language learning to differential sensitivity to positive and negative evidence across age groups (whether the evidence is consistent or inconsistent with the learner's internal hypothesis, Rische & Komarova, 2016). In a preliminary study, we have found an association between the dopamine receptor D2 gene (DRD2) and learning of an artificial grammar modeled after Shimakonde, a Bantu language spoken in Mozambique (Wong, Ettlinger, & Zheng, 2013). DRD2-TAQ-IA polymorphism (rs1800497) is associated with dopamine receptor D2 distribution and dopamine impact in the human striatum, such that A1 allele carriers show reduced D2 receptor binding relative to carriers who are homozygous for the A2 allele. We found that learners who were homozygous for the A2 allele were better at learning the artificial grammar. These learners also had higher striatal responses relative to those who have at least one A1 allele. Connecting our findings to the reinforcement learning framework above, one potential venue for optimizing grammar learning is to build training paradigms that take into account the relative weightings of positive and negative feedback for individuals with DRD2 (and DARPP-32) polymorphisms.
With the appropriate theoretical framework for redesigning training paradigms, prospective studies can be conducted that assess how predictors can be used to place learners in the most effective learning environments.
Ecological Validity and Cost Effectiveness
As the vast majority of the studies cited are from laboratory-based learning, the ecological validity of the concept is called into question. While linking basic genetic information to actual authentic learning might seem far-fetched, one study has already demonstrated the increasing need of appropriate educational reading programs for children who are genetically predisposed to having reading problems (Taylor et al., 2010). Taylor et al. (2010) examined oral reading fluency in 280 monozygotic (MZ) and 526 dizygotic (DZ) twin pairs in the Florida Twin Project on Reading. They found an interesting interaction between estimates of genetic variance and teaching quality in predicting reading achievement such that when teacher quality is higher, the genetic variance associated with reading increases. Thus, measurably, high quality teaching is needed for learners to achieve their full genetic potential at least as far as reading in authentic learning environments is concerned.
Future studies should consider not only the applicability of Personalized Learning in authentic learning settings but also feasibility (e.g., costs). Although tailoring training to the individual learners may seem costly at first, sub-dividing learners into classrooms using different pedagogical methods that best suit their learning rather than duplicating the same methods across multiple classrooms could cost the same amount of funding. Furthermore, providing the most optimal training environment early on prevents the need for any additional teaching in the future to fill in gaps where initial training may have been lacking, and may ultimately prove to be more cost effective.
In essence, Personalized Learning overlaps with several other conceptual ideas including the concept of “learning style” (e.g., visual vs. auditory learners; Kolb, 1985) and Differentiated Instruction (Hall, 2002). The concept of learning style has received a lot of interest in the education field; however, a review found that high-quality evidence for learning styles is virtually non-existent (Pashler et al., 2008), primarily because the research reviewed failed to find a learner-by-instruction interaction. Differentiated Instruction is an approach to teaching that aims to modify teaching and learning activities to address the needs of individual students. A descriptive review (Tomlinson et al., 2003) found favorable evidence while also pointing out that key evidence was missing – including which of various models of teaching and learning best suit individual students and the relative effect of differentiating instruction across learning profiles. Personalized Learning distinguishes from these approaches in highlighting the importance of evaluating predictors across learning paradigms and domains. In this respect, Personalized Learning is more similar to “aptitude-treatment interaction research” (DeKeyser, 2012), which focuses on interactions between individual variables (such as aptitude and age) and linguistic as well as treatment variables (such as inductive or deductive teaching), although this research has been limited to investigating behavioral processes and outcomes of second language learning.
Ethical Considerations
Most critically, as in all research and practice concerning genetics and human performance, ethical considerations are paramount. The question addressed by Personalized Learning is not one that concerns who can or cannot learn, but one that seeks to determine how each and every individual can be provided with an optimal learning environment. It is worth emphasizing again that better cognitive abilities alone do not guarantee successful learning in all learning situations (Beilock and Carr, 2005; Decaro et al., 2008), so it is important to ascertain how training can be tailored to individuals. Perhaps, the most ethical and cost-effective way of teaching is one that does not simply assume that everyone learns the same way.
To achieve optimal learning for each learner, effective teaching could include a combination of common classroom learning where all learners acquire a set of core skills, smaller tutorial sessions for subgroups of learners with different learning needs, and an e-learning platform for personalizing reviewing materials (Lindsey et al., 2014). Within the same classroom teachers who recognize learning differences could also structure learning activities to allow different degrees and types of lecture delivery, feedback, and questions to engage the diversity of learners in the classroom. Importantly, because the long-term retention of knowledge and skills requires practice and review (Soderstrom & Bjork, 2015), effective teaching and learning must go beyond the initial classroom experience. Lindsey et al. (2014) used a Bayesian inference model to determine review materials for individual students over one semester of Spanish as a foreign language. They found that personalized review resulted in better student retention (by 16.5%) over typical (massed) practice. Rather than being restricted to in-class arrangements, Personalized Learning can be implemented in activities outside the common classroom which nevertheless lie at the heart of promoting long-term learning and retention. Thus, we are not necessarily advocating a complete “segregation” of learners based on predictors of learning, but rather a form of inclusive learning that also recognizes the possibility that individual learners indeed learn differently and can benefit from different forms of learning support.
Conclusions
Practitioners and researchers in educational sciences and behavioral health have observed individual differences in learning and treatment outcomes. The goal of this review is to bring forth representative efforts that demonstrate the feasibility of developing educational and treatment solutions that could enable each learner to learn optimally. Central to Personalized Learning, we argue that it is critical for us to attend to individual differences across domains of learning, to identify efficient and objective predictors of individual differences, and to use predictors to help design training paradigms that enable more effective learning at the individual level. This forward-looking framework is undoubtedly filled with challenges, especially in identification of genetic predictors and using such predictors to optimize learning. Implementation in authentic learning situations will require more empirical laboratory studies followed by studies conducted in the field. These studies will complement the numerous high-quality studies that have identified effective pedagogy and treatment at the group level.
By grounding our framework in biological (neural and genetic) and cognitive predictors and by using the appropriate experimental methodology to demonstrate learner-by-instruction interaction under clear and uniform outcome measures, we hope to propel a new era of high-quality educational and health science research under Personalized Learning. In the end, we hope that it will prove to allow for better and more efficient learning for all learners. This framework not only can be applied to classroom learning, but also to behavioral health which often requires substantial rehabilitation of learning components such as in speech-language, occupational, and physical therapies.
Highlights.
Individual differences exist across diverse areas including language learning
A new framework, Personalized Learning, is proposed for addressing such differences
Evidence connecting cognitive genetics, neural and behavioral research is reviewed
Implications for language learning are discussed
Acknowledgments
We are grateful to Michael Ullman and two anonymous reviewers for constructive feedback. We also thank Tyler Perrachione, Ken Paller, John Sheppard, Erin Ingvalson, and Mark Antoniou for their insightful comments. This work is supported by grants from the National Institutes of Health (R01DC008333), National Science Foundation (BCS-1125144) and the Dr. Stanley Ho Medical Development Foundation.
Footnotes
We thank Michael Ullman for raising this point.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Patrick C. M. Wong, Dept of Linguistics & Modern Languages and Brain and Mind Institute, The Chinese University of Hong Kong
Loan Vuong, Dept of Linguistics & Modern Languages and Brain and Mind Institute, The Chinese University of Hong Kong.
Kevin Liu, Feinberg School of Medicine, Northwestern University.
References
- Adkins DL, Boychuk J, Remple MS, Kleim JA. Motor training induces experience-specific patterns of plasticity across motor cortex and spinal cord. J Appl Physiol. 2006;101:1776–1782. doi: 10.1152/japplphysiol.00515.2006. [DOI] [PubMed] [Google Scholar]
- Ackerman PL. Determinants of individual differences during skill acquisition: Cognitive abilities and information processing. Journal of Experimental Psychology General. 1988;117(3):288–318. [Google Scholar]
- Ackerman PL. New Developments in Understanding Skilled Performance. Current Directions in Psychological Science. 2007;16(5):235–239. [Google Scholar]
- Alexander L, Martray C. The development of an abbreviated version of the Mathematics Anxiety Rating Scale. Meas Eval Couns Dev. 1989:143–150. [Google Scholar]
- Alloway TP, Alloway RG. Investigating the predictive roles of working memory and IQ in academic attainment. Journal of Experimental Child Psychology. 2010;106(1):20–29. doi: 10.1016/j.jecp.2009.11.003. [DOI] [PubMed] [Google Scholar]
- Anderson JR. Acquisition of cognitive skill. Psychological Review. 1982;89(4):369–406. [Google Scholar]
- Asaridou SS, Takashima A, Dediu D, Hagoort P, McQueen JM. Repetition suppression in the Left Inferior Frontal Gyrus predicts tone learning performance. Cerebral Cortex. 2016;26:2728–2742. doi: 10.1093/cercor/bhv126. [DOI] [PubMed] [Google Scholar]
- Ashcraft MH, Kirk EP. The relationships among working memory, math anxiety, and performance. J Exp Psychol Gen. 2001;130:224–237. doi: 10.1037//0096-3445.130.2.224. [DOI] [PubMed] [Google Scholar]
- Bates E, Dale P, Thal D. Individual differences and their implications for theories of language development. In: Fletcher P, MacWhinney B, editors. The handbook of child language. Oxford: Basil Blackwell; 1995. pp. 96–151. [Google Scholar]
- Beilock SL, Carr TH. When high-powered people fail: working memory and “choking under pressure” in math. Psychological science. 2005;16:101–105. doi: 10.1111/j.0956-7976.2005.00789.x. [DOI] [PubMed] [Google Scholar]
- Berg EA. A simple objective technique for measuring flexibility in thinking. J Gen Psychol. 1948;39:15–22. doi: 10.1080/00221309.1948.9918159. [DOI] [PubMed] [Google Scholar]
- Bilder RM, Howe A, Novak N, Sabb FW, Parker DS. The genetics of cognitive impairment in schizophrenia: a phenomic perspective. Trends Cogn Sci. 2011;15:428–435. doi: 10.1016/j.tics.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birdsong D. Second language acquisition and ultimate attainment. In: Davies A, Elder C, editors. The handbook of applied linguistics. Malden, MA: Blackwell; 2004. pp. 82–105. [Google Scholar]
- Bull R, Espy KA, Wiebe SA. Short-term memory, working memory, and executive functioning in preschoolers: Longitudinal predictors of mathematical achievement at age 7 years. Developmental Neuropsychology. 2008;33(3):205–228. doi: 10.1080/87565640801982312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunge SA, Klingberg T, Jacobsen RB, Gabrieli JD. A resource model of the neural basis of executive working memory. Proc Natl Acad Sci U S A. 2000;97:3573–3578. doi: 10.1073/pnas.050583797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campa D, Rizzato C, Bauer AS, Werner J, Capurso G, Costello E, Talar-Wojnarowska R, Jamroziak K, Pezzilli R, Gazouli M, Khaw KT, Key TJ, Bambi F, Mohelnikova-Duchonova B, Heller A, Landi S, Vodickova L, Theodoropoulos G, Bugert P, Vodicka P, Hoheisel J, Delle Fave G, Neoptolemos J, Soucek P, Buchler MW, Giese NA, Canzian F. Lack of replication of seven pancreatic cancer susceptibility loci identified in two Asian populations. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2012 doi: 10.1158/1055-9965.EPI-12-1182. [DOI] [PubMed] [Google Scholar]
- Carpenter PA, Daneman M. Individual Differences in Working Memory and Reading. Journal of Verbal Learning and Verbal Behavior. 1980;19:450–466. [Google Scholar]
- Chandrasekaran B, Sampath PD, Wong PCM. Individual variability in cue-weighting and lexical tone learning. The Journal of the Acoustical Society of America. 2010;128(1):456–565. doi: 10.1121/1.3445785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen Y, Schneidman E. High-order feature-based mixture models of classification learning predict individual learning curves and enable personalized teaching. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:684–689. doi: 10.1073/pnas.1211606110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins AGE, Frank MJ. How much of reinforcement learning is working memory, not reinforcement learning? A behavioral, computational, and neurogenetic analysis. European Journal of Neuroscience. 2012;35(7):1024–1035. doi: 10.1111/j.1460-9568.2011.07980.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins FS, Varmus H. A new initiative on precision medicine. New England Journal of Medicine. 2015;372(9):793–795. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousin E, Mace S, Rocher C, Dib C, Muzard G, Hannequin D, Pradier L, Deleuze JF, Genin E, Brice A, Campion D. No replication of genetic association between candidate polymorphisms and Alzheimer's disease. Neurobiology of aging. 2011;32:1443–1451. doi: 10.1016/j.neurobiolaging.2009.09.004. [DOI] [PubMed] [Google Scholar]
- Deary IJ, Strand S, Smith P, Fernandes C. Intelligence and educational achievement. Intelligence. 2007;35(1):13–21. [Google Scholar]
- Deng Z, Chandrasekaran B, Wang S, Wong PCM. Resting-state low-frequency fluctuations reflect individual differences in spoken language learning. Cortex. 2016;76:63–78. doi: 10.1016/j.cortex.2015.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Quervain DJ, Papassotiropoulos A. Identification of a genetic cluster influencing memory performance and hippocampal activity in humans. Proc Natl Acad Sci U S A. 2006;103:4270–4274. doi: 10.1073/pnas.0510212103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaro MS, Thomas RD, Beilock SL. Individual differences in category learning: sometimes less working memory capacity is better than more. Cognition. 2008;107:284–294. doi: 10.1016/j.cognition.2007.07.001. [DOI] [PubMed] [Google Scholar]
- Dekeyser R. Interactions Between Individual Differences, Treatments, and Structures in SLA. Language Learning. 2012;62:189–200. [Google Scholar]
- Egan MF, Goldberg TE, Kolachana BS, Callicott JH, Mazzanti CM, Straub RE, Goldman D, Weinberger DR. Effect of COMT Val108/158 Met genotype on frontal lobe function and risk for schizophrenia. Proc Natl Acad Sci U S A. 2001;98:6917–6922. doi: 10.1073/pnas.111134598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis NC. Usage-based and form-focused language acquisition: The associative learning of constructions, learned attention, and the limited L2 endstate. In: Robinson P, Ellis N, editors. Handbook of Cognitive Linguistics and Second Language Acquisition. London: Routledge; 2008. pp. 372–405. [Google Scholar]
- Evans TM, Kochalka J, Ngoon TJ, Wu SS, Qin S, Battista C, Menon V. Brain structural integrity and intrinsic functional connectivity forecast 6 year longitudinal growth in children's numerical abilities. Journal of Neuroscience. 2015;35(33):11743–11750. doi: 10.1523/JNEUROSCI.0216-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust MW, Ashcraft MH, Fleck DE. Mathematics anxiety effects in simple and complex addition. Mathemat Cogn. 1996:25–62. [Google Scholar]
- Frank MJ, Doll BB, Oas-Terpstra J, Moreno F. Prefrontal and striatal dopaminergic genes predict individual differences in exploration and exploitation. Nat Neurosci. 2009;12:1062–1068. doi: 10.1038/nn.2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank MJ, Moustafa AA, Haughey HM, Curran T, Hutchison KE. Genetic triple dissociation reveals multiple roles for dopamine in reinforcement learning. Proceedings of the National Academy of Sciences. 2007;104(41):16311–16316. doi: 10.1073/pnas.0706111104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke A, Balschun T, Karlsen TH, Hedderich J, May S, Lu T, Schuldt D, Nikolaus S, Rosenstiel P, Krawczak M, Schreiber S. Replication of signals from recent studies of Crohn's disease identifies previously unknown disease loci for ulcerative colitis. Nature genetics. 2008;40:713–715. doi: 10.1038/ng.148. [DOI] [PubMed] [Google Scholar]
- Gabrieli JDE, Ghosh SS, Whitfield-Gabrieli S. Prediction as a humanitarian and pragmatic contribution from human cognitive neuroscience. Neuron. 2015;85(1):11–26. doi: 10.1016/j.neuron.2014.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gateva V, Sandling JK, Hom G, Taylor KE, Chung SA, Sun X, Ortmann W, Kosoy R, Ferreira RC, Nordmark G, Gunnarsson I, Svenungsson E, Padyukov L, Sturfelt G, Jonsen A, Bengtsson AA, Rantapaa-Dahlqvist S, Baechler EC, Brown EE, Alarcon GS, Edberg JC, Ramsey-Goldman R, McGwin G, Jr, Reveille JD, Vila LM, Kimberly RP, Manzi S, Petri MA, Lee A, Gregersen PK, Seldin MF, Ronnblom L, Criswell LA, Syvanen AC, Behrens TW, Graham RR. A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nature genetics. 2009;41:1228–1233. doi: 10.1038/ng.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golestani N. Brain structural correlates of individual differences at low-to high-levels of the language processing hierarchy: A review of new approaches to imaging research. International Journal of Bilingualism. 2014;18(1):6–34. [Google Scholar]
- Halberda J, Mazzocco MM, Feigenson L. Individual differences in non-verbal number acuity correlate with maths achievement. Nature. 2008;455:665–668. doi: 10.1038/nature07246. [DOI] [PubMed] [Google Scholar]
- Hall T. Differentiated instruction. Wakefield, MA: National Center on Accessing the General Curriculum; 2002. Available: http://www.cast.org/publications/ncac/ncac_diffinstruc.html. [Google Scholar]
- Harlaar N, Meaburn EL, Hayiou-Thomas ME, Davis OSP, Docherty S, Hanscombe KB, et al. Genome-Wide Association Study of Receptive Language Ability of 12-Year-Olds. Journal of Speech, Language, and Hearing Research. 2014;57:96–105. doi: 10.1044/1092-4388(2013/12-0303). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingvalson EM, Barr AM, Wong PCM. Poorer Phonetic Perceivers Show Greater Benefit in Phonetic-Phonological Speech Learning. Journal of Speech, Language, and Hearing Research. 2013;56:1045–50. doi: 10.1044/1092-4388(2012/12-0024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jocham G, Klein TA, Neumann J, von Cramon DY, Reuter M, Ullsperger M. Dopamine DRD2 polymorphism alters reversal learning and associated neural activity. J Neurosci. 2009;29:3695–3704. doi: 10.1523/JNEUROSCI.5195-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman SB, Deyoung CG, Gray JR, Brown J, Mackintosh N. Implicit learning as an ability. Cognition. 2010;116(3):321–340. doi: 10.1016/j.cognition.2010.05.011. [DOI] [PubMed] [Google Scholar]
- King J, Just MA. Individual Differences in Syntactic Processing: The Role of Working Memory. J Mem Lang. 1991;30:580–602. [Google Scholar]
- Klahr D, Zimmerman C, Jirout J. Educational interventions to advance children's scientific thinking. Science. 2011;333:971–975. doi: 10.1126/science.1204528. [DOI] [PubMed] [Google Scholar]
- Kleim JA, Chan S, Pringle E, Schallert K, Procaccio V, Jimenez R, Cramer SC. BDNF val66met polymorphism is associated with modified experience-dependent plasticity in human motor cortex. Nat Neurosci. 2006;9:735–737. doi: 10.1038/nn1699. [DOI] [PubMed] [Google Scholar]
- Klein TA, Neumann J, Reuter M, Hennig J, von Cramon DY, Ullsperger M. Genetically determined differences in learning from errors. Science. 2007;318:1642–1645. doi: 10.1126/science.1145044. [DOI] [PubMed] [Google Scholar]
- Kolb DA. Learning Style Inventory. Hay Group; Boston, MA: 1985. [Google Scholar]
- Kooijman V, Junge C, Johnson EK, Hagoort P, Cutler A. Predictive brain signals of linguistic development. Frontiers in Psychology. 2013;4:25. doi: 10.3389/fpsyg.2013.00025. Published online 08 Feburary, 2015: http://doi.org/10.3389/fpsyg.2013.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeFevre J, Greenham SL, Waheed N. The development of procedural and conceptual knowledge in computational estimation. Cognition Instruct. 1993;11:95–132. [Google Scholar]
- Lieven E, Behrens H, Speares J, Tomasello M. Early syntactic creativity: a usage-based approach. Journal of Child Language. 2003;30(2):333–370. [PubMed] [Google Scholar]
- Lindsey RV, Shroyer JD, Pashler H, Mozer MC. Improving students' long-term knowledge retention through personalized review. Psychological Science. 2014;25(3):639–647. doi: 10.1177/0956797613504302. [DOI] [PubMed] [Google Scholar]
- Lupski JR, Reid JG, Gonzaga-Jauregui C, Rio Deiros D, Chen DC, Nazareth L, Bainbridge M, Dinh H, Jing C, Wheeler DA, McGuire AL, Zhang F, Stankiewicz P, Halperin JJ, Yang C, Gehman C, Guo D, Irikat RK, Tom W, Fantin NJ, Muzny DM, Gibbs RA. Whole-genome sequencing in a patient with Charcot-Marie-Tooth neuropathy. The New England Journal of Medicine. 2010;362:1181–1191. doi: 10.1056/NEJMoa0908094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, Louis DN, Christiani DC, Settleman J, Haber DA. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- Ma Q, Lu AY. Pharmacogenetics, pharmacogenomics, and individualized medicine. Pharmacol Rev. 2011;63:437–459. doi: 10.1124/pr.110.003533. [DOI] [PubMed] [Google Scholar]
- Mallal S, Phillips E, Carosi G, Molina JM, Workman C, Tomazic J, Jagel-Guedes E, Rugina S, Kozyrev O, Cid JF, Hay P, Nolan D, Hughes S, Hughes A, Ryan S, Fitch N, Thorborn D, Benbow A, Team PS. HLA-B*5701 screening for hypersensitivity to abacavir. N Engl J Med. 2008;358:568–579. doi: 10.1056/NEJMoa0706135. [DOI] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TF, McCarroll SA, Visscher PM. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchman VA, Bates E. Continuity in lexical and morphological development: A test of the critical mass hypothesis. Journal of Child Language. 1994;21:339–366. doi: 10.1017/s0305000900009302. [DOI] [PubMed] [Google Scholar]
- Martin MO, Mullis IVS, editors. TIMMS and PIRLS 2011: Relationships among reading, mathematics, and science achievement at the fourth grade—Implications for early learning. Chestnut Hill, MA: TIMSS & PIRLS International Study Center, Boston College; 2013. [Google Scholar]
- McCormack M, Alfirevic A, Bourgeois S, Farrell JJ, Kasperaviciute D, Carrington M, Sills GJ, Marson T, Jia X, de Bakker PI, Chinthapalli K, Molokhia M, Johnson MR, O'Connor GD, Chaila E, Alhusaini S, Shianna KV, Radtke RA, Heinzen EL, Walley N, Pandolfo M, Pichler W, Park BK, Depondt C, Sisodiya SM, Goldstein DB, Deloukas P, Delanty N, Cavalleri GL, Pirmohamed M. HLA-A*3101 and carbamazepine-induced hypersensitivity reactions in Europeans. N Engl J Med. 2011;364:1134–1143. doi: 10.1056/NEJMoa1013297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHughen SA, Pearson-Fuhrhop K, Ngo VK, Cramer SC. Intense training overcomes effects of the val66met BDNF polymorphism on short-term plasticity. Exp Brain Res. 2011;213:415–422. doi: 10.1007/s00221-011-2791-z. [DOI] [PubMed] [Google Scholar]
- McNab F, Varrone A, Farde L, Jucaite A, Bystritsky P, Forssberg H, Klingberg T. Changes in cortical dopamine D1 receptor binding associated with cognitive training. Science. 2009;323:800–802. doi: 10.1126/science.1166102. [DOI] [PubMed] [Google Scholar]
- Mega JL, Simon T, Collet JP, Anderson JL, Antman EM, Bliden K, Cannon CP, Danchin N, Giusti B, Gurbel P, Horne BD, Hulot JS, Kastrati A, Montalescot G, Neumann FJ, Shen L, Sibbing D, Steg PG, Trenk D, Wiviott SD, Sabatine MS. Reduced-function CYP2C19 genotype and risk of adverse clinical outcomes among patients treated with clopidogrel predominantly for PCI: a meta-analysis. JAMA. 2010;304:1821–1830. doi: 10.1001/jama.2010.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molendijk ML, Bus BA, Spinhoven P, Kaimatzoglou A, Oude Voshaar RC, Penninx BW, van IMH, Elzinga BM. A systematic review and meta-analysis on the association between BDNF val(66)met and hippocampal volume--a genuine effect or a winners curse? American journal of medical genetics. Part B, Neuropsychiatric genetics : the official publication of the International Society of Psychiatric Genetics. 2012;159B:731–740. doi: 10.1002/ajmg.b.32078. [DOI] [PubMed] [Google Scholar]
- Monchi O, Petrides M, Petre V, Worsley K, Dagher A. Wisconsin Card Sorting revisited: distinct neural circuits participating in different stages of the task identified by event-related functional magnetic resonance imaging. J Neurosci. 2001;21:7733–7741. doi: 10.1523/JNEUROSCI.21-19-07733.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan-Short K, Steinhauer K, Sanz C, Ullman MT. Explicit and implicit second language training differentially affect the achievement of native-like brain activation patterns. Journal of cognitive neuroscience. 2012;24:933–947. doi: 10.1162/jocn_a_00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller JL, Hirotani M, Friederici AD. ERP evidence for different strategies in the processing of case markers in native speakers and non-native learners. BMC neuroscience. 2007;8:18. doi: 10.1186/1471-2202-8-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neisser U, Boodoo G, Bouchard TJ., Jr Intelligence: knowns and unknowns. American Psychologist. 1996;51:77–101. [Google Scholar]
- Papassotiropoulos A, de Quervain DJ. Genetics of human episodic memory: dealing with complexity. Trends Cogn Sci. 2011;15:381–387. doi: 10.1016/j.tics.2011.07.005. [DOI] [PubMed] [Google Scholar]
- Pare G, Mehta SR, Yusuf S, Anand SS, Connolly SJ, Hirsh J, Simonsen K, Bhatt DL, Fox KA, Eikelboom JW. Effects of CYP2C19 genotype on outcomes of clopidogrel treatment. N Engl J Med. 2010;363:1704–1714. doi: 10.1056/NEJMoa1008410. [DOI] [PubMed] [Google Scholar]
- Pashler H, Mcdaniel M, Rohrer D, Bjork R. Learning styles: concepts and evidence. Psychol Sci. 2008;9:105–119. doi: 10.1111/j.1539-6053.2009.01038.x. [DOI] [PubMed] [Google Scholar]
- Perrachione TK, Lee J, Ha LY, Wong PC. Learning a novel phonological contrast depends on interactions between individual differences and training paradigm design. J Acoust Soc Am. 2011;130:461–472. doi: 10.1121/1.3593366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson NR, Pisoni DB, Miyamoto RT. Cochlear implants and spoken language processing abilities: review and assessment of the literature. Restor Neurol Neurosci. 2010;28:237–250. doi: 10.3233/RNN-2010-0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips EJ, Mallal SA. HLA-B*1502 screening and toxic effects of carbamazepine. N Engl J Med. 2011;365:672. doi: 10.1056/NEJMc1105467. [DOI] [PubMed] [Google Scholar]
- Queitsch C, Carlson KD, Girirajan S. Lessons from model organisms: phenotypic robustness and missing heritability in complex disease. PLoS genetics. 2012;8:e1003041. doi: 10.1371/journal.pgen.1003041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rische JL, Komarova NL. Regularization of languages by adults and children: A mathematical framework. Cognitive Psychology. 2016;84:1–30. doi: 10.1016/j.cogpsych.2015.10.001. [DOI] [PubMed] [Google Scholar]
- Rohde DL, Plaut DC. Language acquisition in the absence of explicit negative evidence: How important is starting small? Cognition. 1999;72:67–109. doi: 10.1016/s0010-0277(99)00031-1. [DOI] [PubMed] [Google Scholar]
- Rybakowski JK, Borkowska A, Czerski PM, Kapelski P, Dmitrzak-Weglarz M, Hauser J. An association study of dopamine receptors polymorphisms and the Wisconsin Card Sorting Test in schizophrenia. J Neural Transm. 2005;112:1575–1582. doi: 10.1007/s00702-005-0292-6. [DOI] [PubMed] [Google Scholar]
- Schneider W, Shiffrin RM. Controlled and automatic human information processing: I. Detection, search, and attention. Psychological Review. 1977;84:1–66. [Google Scholar]
- Schwarz UI, Ritchie MD, Bradford Y, Li C, Dudek SM, Frye-Anderson A, Kim RB, Roden DM, Stein CM. Genetic determinants of response to warfarin during initial anticoagulation. N Engl J Med. 2008;358:999–1008. doi: 10.1056/NEJMoa0708078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard JP, Wang JP, Wong P. Large-scale cortical network properties predict future sound-to-word learning success. Journal of Cognitive Neuroscience. 2012;24:1087–1103. doi: 10.1162/jocn_a_00210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiffrin RM, Schneider W. Controlled and automatic human information processing: II. Perceptual learning, automatic attending, and a general theory. Psychological Review. 1977;84:127–190. [Google Scholar]
- Slevc LR, Miyake A. Individual differences in second-language proficiency: does musical ability matter? Psychological science. 2006;17:675–681. doi: 10.1111/j.1467-9280.2006.01765.x. [DOI] [PubMed] [Google Scholar]
- Soderstrom NC, Bjork RA. Learning Versus Performance: An Integrative Review. Perspectives on Psychological Science. 2015;10(2):176–199. doi: 10.1177/1745691615569000. [DOI] [PubMed] [Google Scholar]
- Soronen P, Ollila HM, Antila M, Silander K, Palo OM, Kieseppa T, Lonnqvist J, Peltonen L, Tuulio-Henriksson A, Partonen T, Paunio T. Replication of GWAS of bipolar disorder: association of SNPs near CDH7 with bipolar disorder and visual processing. Molecular psychiatry. 2010;15:4–6. doi: 10.1038/mp.2009.86. [DOI] [PubMed] [Google Scholar]
- Street JA, Dabrowska ED. More individual differences in language attainment: How much do adult native speakers of English know about passives and quantifiers? Lingua. 2010;120(8):2080–2094. [Google Scholar]
- Tagarelli KM, Borges Mota M, Rebuschat P. The role of working memory in implicit and explicit language learning. In: Carlson L, Holscher C, Holscher T, editors. Proceedings of the 33rd Annual Conference of the Cognitive Science Society. Austin, TX: Cognitive Science Society; 2011. pp. 2061–2066. [Google Scholar]
- Taylor J, Roehrig AD, Soden Hensler B, Connor CM, Schatschneider C. Teacher quality moderates the genetic effects on early reading. Science. 2010;328:512–514. doi: 10.1126/science.1186149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas DL, Thio CL, Martin MP, Qi Y, Ge D, O'Huigin C, Kidd J, Kidd K, Khakoo SI, Alexander G, Goedert JJ, Kirk GD, Donfield SM, Rosen HR, Tobler LH, Busch MP, McHutchison JG, Goldstein DB, Carrington M. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461:798–801. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson CA, Brighton C, Hertberg H, Callahan CM, Moon TR, Brimijoin K, et al. Differentiating Instruction in Response to Student Readiness, Interest, and Learning Profile in Academically Diverse Classrooms: A Review of Literature. Journal for the Education of the Gifted. 2003;27(2-3):119–145. [Google Scholar]
- Ullman MT. Contributions of memory circuits to language: the declarative/procedural model. Cognition. 2004;92(1-2):231–270. doi: 10.1016/j.cognition.2003.10.008. [DOI] [PubMed] [Google Scholar]
- Ullman MT. A cognitive neuroscience perspective on second language acquisition: The declarative/procedural model. Mind and Context in Adult Second Language Acquisition. 2005:141–178. [Google Scholar]
- Ullman MT, Pierpont EI. Specific language impairment is not specific to language: The procedural deficit hypothesis. Cortex. 2005;41(3):399–433. doi: 10.1016/s0010-9452(08)70276-4. [DOI] [PubMed] [Google Scholar]
- Ullman MT, Pullman MY. A compensatory role for declarative memory in neurodevelopmental disorders. Neuroscience and Biobehavioral Reviews. 2015;51:205–222. doi: 10.1016/j.neubiorev.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unsworth N, Engle R. Individual differences in working memory capacity and learning: Evidence from the serial reaction time task. Memory and Cognition. 2005;33(2):213–220. doi: 10.3758/bf03195310. [DOI] [PubMed] [Google Scholar]
- Vijayraghavan S, Wang M, Birnbaum SG, Williams GV, Arnsten AF. Inverted-U dopamine D1 receptor actions on prefrontal neurons engaged in working memory. Nat Neurosci. 2007;10:376–384. doi: 10.1038/nn1846. [DOI] [PubMed] [Google Scholar]
- Wang L, McLeod HL, Weinshilboum RM. Genomics and drug response. N Engl J Med. 2011;364:1144–1153. doi: 10.1056/NEJMra1010600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams GV, Goldman-Rakic PS. Modulation of memory fields by dopamine D1 receptors in prefrontal cortex. Nature. 1995;376:572–575. doi: 10.1038/376572a0. [DOI] [PubMed] [Google Scholar]
- Wong PC, Chandrasekaran B, Zheng J. The derived allele of ASPM is associated with lexical tone perception. PloS one. 2012;7:e34243. doi: 10.1371/journal.pone.0034243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong PCM, Ettlinger M, Zheng J. Linguistic Grammar Learning and DRD2-TAQ-IA Polymorphism. PLoS ONE. 2013;8(5):e64983. doi: 10.1371/journal.pone.0064983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong PCM, Perrachione TK, Parrish TB. Neural characteristics of successful and less successful speech and word learning in adults. Human Brain Mapping. 2007;28:995–1006. doi: 10.1002/hbm.20330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong PCM, Skoe E, Russo NM, Dees T, Kraus N. Musical experience shapes human brainstem encoding of linguistic pitch patterns. Nature Neuroscience. 2007;10:420–422. doi: 10.1038/nn1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong PCM, Warrier CM, Penhune VB, Roy AK, Sadehh A, Parrish TB, Zatorre RJ. Volume of left Heschl's Gyrus and linguistic pitch learning. Cereb. Cortex. 2008;18:828–836. doi: 10.1093/cercor/bhm115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip M. Tone. Cambridge, MA: Cambridge University Press; 2002. [Google Scholar]


