Abstract
Foodborne outbreaks associated with fresh fruits and vegetables are on the rise worldwide. Biofilm formation is one of the important traits of pathogens making them strongly attached to substrates as well as express virulence phenotypes. Present study investigates the biofilm forming ability of E. coli and Salmonella sp. isolated from fresh fruits and vegetables. A total of 53 strains, including 35 E. coli and 18 Salmonella sp. isolated from different fruit and vegetable samples were taken into account for the study. Initial screening for biofilm formation was done using Congo Red agar plate test. Results revealed that 22.8% E. coli and 22.2% Salmonella sp. were potential biofilm formers. However, the MTP (Micro-Titre Plate) assay suggested more isolates of both E. coli and Salmonella sp. were moderate to strong biofilm producers. Agar plate diffusion assay with Agrobacterium tumefaciens NTL-4 showed the production of quorum signaling molecules (AHLs) by three isolates of E. coli and one Salmonella sp. Two E. coli isolates showed a significant amount of EPS production indicating higher biofilm forming potential. The Presence of LUX R homologue gene (sdiA) in two of the Salmonella isolates were confirmed by PCR which demonstrated their potential pathogenicity. Results of the work underline the biofilm forming and potentially virulent capacities of isolates from the surface of fruits and vegetables.
Keywords: Fresh fruits and vegetables, E. coli, Salmonella sp., Biofilm formation, Quorum sensing
Introduction
Biofilms are aggregates of microorganisms which adhere to each other as well as to surfaces and remain embedded in a self-produced extracellular polymeric substance matrix. Biofilm formation is triggered as a result of many internal and external factors. They can be formed by single or multiple bacterial species, as a single layer or three-dimensional structures. Percival et al. (2000) showed that bacteria in biofilms are capable of adapting to changing environmental conditions in a rapid way at both phenotypic and genotypic levels. Biofilm formation generally occurs under diverse conditions in both natural and man-made environments. They consist of aggregates of microbial cells along with the exopolysaccharide component, making it a heterogenous environment (Sutherland 2001).
Biofilm formation is a serious concern in food industry, as pathogenic microorganisms are capable of attaching and growing on food surfaces and surfaces of other processing equipments. Earlier investigations have shown that the presence of pathogens with ability to form biofilms is one of the leading causes of food contamination and precipitation of disease. Islam et al. (2004a, b) reported soil contamination with S. enterica which affected crops such as radish, lettuce, carrot, parsley, and persisted for more than 6 months. Similar reports were documented by Barak and Liang (2008) who showed that a wide range of agricultural crops including tomatoes are affected by S. enterica. Teplitski et al. (2009) have also reported the highly persistent nature of Salmonella in plant environments. They can, therefore act as vehicles of contamination of these pathogens to humans (Berger et al. 2010). Further, their complete destruction or removal in such cases is difficult. Fresh produce including fresh fruits and are on the rise in markets and the cases of foodborne illnesses resulting from their consumption have also been increasing in the past few decades (Berger et al. 2010; Byrne et al. 2014; Kozak et al. 2013), making the microbiologists develop strategies to reduce the occurrence of foodborne outbreaks worldwide (Lynch et al. 2009). Quorum sensing has been found to be a key factor involved in biofilm formation by foodborne pathogens. Smith et al. (2004) reported that quorum sensing is a regulated system in which bacterial cells interact with each other such that it benefits an entire biofilm population. Though conventional sanitation techniques have been employed for cleaning and processing of fresh produce, it doesn’t completely remove the risk of microbiological contamination. Recently, Denis et al. (2016) analysed the prevalence and trends of bacterial contamination in fresh fruits and vegetables sold in Canadian markets and found various pathogenic organisms with notable seasonal variations in their incidence although the prevalence was considerably low in general. However, the scenario is likely to be different in India. The present study, therefore, is aimed at determining the biofilm formation ability of bacteria isolated from fresh produce, including different fruits and vegetables. E. coli and Salmonella spp., the two prominent members of Enterobacteriaceae family, known for their involvement in foodborne outbreaks have been taken into account for the study. It would help in determining the potential of these microbes in causing foodborne illness. Further research in understanding the nature of biofilm formation and regulation can lead to minimize the occurrence of these microorganisms on fresh produce and help in controlling the incidence of foodborne disease outbreaks. Thus, the objectives of the present study were proposed as follows:
To investigate the biofilm forming ability in the isolates from fresh fruits and vegetables by microbiological and molecular methods.
To assess the motility behavior and quorum sensing in the isolates along with their biofilm forming ability.
Materials and methods
Congo red agar plate test
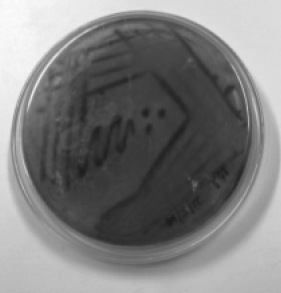
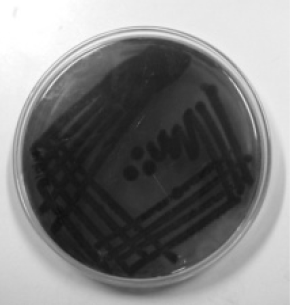
A total of 53 strains including 35 E. coli and 18 Salmonella sp. were isolated from different fruits and vegetables consumed raw by the enrichment method. All the isolates were stored in nutrient agar slants and screened for biofilm formation ability using Congo Red Agar (CRA) plates. The CRA medium (per litre) contains 0.8 g of Congo red, 36 g of Sucrose and 11 g of Brain Heart Infusion agar. Test cultures were streaked onto CRA agar plates and incubated for 24–48 h at 30 °C. Biofilm forming strains were observed as dark black colonies while non-formers were observed as red colonies on the CRA plates. They were categorized into strong, moderate and weak biofilm formers according to the visible colour of the colonies.
Morphological identification of biofilms
For Scanning electron microscopy (SEM), sample preparation included swabbing of the test isolates on fruits surfaces and drying at room temperature until complete dry. An overnight culture of E. coli and Salmonella sp. was swabbed onto the surface of uniformly cut pieces of Guava, Apple, and Cucumber. The samples were then analysed by using a SEM (Hitachi S-3000H, Japan).
Test tube method
E. coli and Salmonella isolates were inoculated in test tubes containing Tryptone Soybean broth and incubated at 37 °C for 24 h, following which the tubes were washed with PBS and crystal violet solution. The tubes were then dried for about 10–15 min, and observed for biofilm formation on sides and bottom of the tube (Annous et al. 2005). The results were noted according to the results of the control strains, E. coli (MTCC 727) and Salmonella typhimurium (ATCC 14028). A visible purple film lining the walls and bottom of the tubes indicated positive result for biofilm formation. The amount of biofilm formed was scored as weak, moderate and strong depending on the intensity of the visible film.
Swimming and swarming assay
For swimming assay, the media consisted of 1% tryptone, 0.5% NaCl and 0.3% agar and overnight cultures of the test bacteria were point inoculated at the center of the medium. Similarly, in the case of swarming, test bacteria were point inoculated at the center of the medium consisting of 1% peptone, 0.5% NaCl, 0.5% agar and 0.5% of filter sterilized d-glucose. The plates were then incubated at 37 °C in an upright position for 16 h to observe the motility pattern.
Microtitre plate assay
In a microtitre plate, 100 µl of the isolates including 8 E. coli and 4 Salmonella sp. were incubated for 18 h at 37 °C. The content of the wells were removed after incubation and washed with 0.2 ml of phosphate buffer saline (PBS pH 7.2) to remove free-floating planktonic bacteria. The wells were then stained with 20 µl of crystal violet. Excess stain was rinsed off by thorough washing with distilled water, and 100 µl of decolouriser was added. Optical density (OD) of stained adherent bacteria was determined with an Eon micro-ELISA auto reader at a wavelength of 580 nm.
Screening for AHL production
12 isolates including 8 E. coli and 4 Salmonella sp. were screened for quorum sensing by using the reporter strain Agrobacterium tumefaciens NTL-4. The reporter strain was swabbed onto the agar medium containing X-Gal (5-bromo-4chloro-3-indolyl-β-d-galactopyranoside) at a concentration of 5 mg/100 ml of media followed by spot inoculation of the test isolates and incubation at 30 °C. Development of greenish to blue colour around colony indicated the production of quorum signalling molecule AHLs.
Exopolysaccharide (EPS) production
Test isolates were grown in LB medium at 37 °C. Following overnight incubation for about 16 h, the cells were harvested during late log phase by centrifuging at 8000 rpm, 30 min at 20 °C. The supernatant was filtered after which three volumes of ice cold 100% ethanol was added and stored at −20 °C overnight for precipitation of exopolysaccharide (EPS). The precipitated EPS was collected by centrifugation at 7000 rpm for 30 min at 5 °C. The pellet was then dissolved in 1 ml of MilliQ water and the EPS was quantified by phenol sulphuric acid method (Dubois et al. 1956).
Detection of biofilm operon
The representative 8 E. coli and 4 Salmonella isolates were analyzed for the presence of Ica (intercellular adhesion) locus by using PCR with the icaA gene specific primers, and LUX R homologue sdiA primers (Table 1). The PCR reaction was performed with a total volume of 50 µl using the TaqMan Universal PCR Master Mix. The template DNA (4 µl) was mixed with 25 µl Mastermix along with the forward and reverse primer set (5 µl each) and made up to a volume of 50 µl with MilliQ water.
Table 1.
PCR primers and cycling conditions
| S. No | Primer | Primer sequence | Cycling conditions |
|---|---|---|---|
| 1. | icaA | 5′-TCTCTTGCAGGAGCAATCAA-3′ | 94 °C, 30 s |
| 5′-TCAGGCACTAACATCCAGCA-3′ | 55.5 °C, 30 s 72 °C, 30 s |
||
| 2. | sdi A | 5′-AATATCGCTTCGTACCAC-3′ | 94 °C, 30 s 52 °C, 40 s |
| 5′-GTAGGTAAACGAGGAGCAG-3′ | 72 °C, 7 min |
Results and discussion
Screening for biofilm formation
The CRA plate test was performed for the screening of biofilm production. Phenotypic production of slime by all the isolates were analyzed and interpreted according to the standard colour variation from pink/red to black. Black colonies represented biofilm formers while red represented non-biofilm formers. Results indicated that 48.5% of E. coli and 50% of Salmonella sp. were strong biofilm producers while the rest were found to be moderate and weak biofilm producers (Table 2). Silva et al. (2002) reported biofilm production in only 25% of strains from clinical specimens of newborns in a neonatal intensive care unit, while the proportion was significantly higher in other reports (Cafiso et al. 2004) in which 83% of Staphylococci were positive on CRA plates. Though CRA plate test is fast and easy to perform among the phenotypic methods, its sensitivity, specificity, and accuracy are low when compared to other qualitative and quantitative methods (Fitzpatrick et al. 2005).
Table 2.
Screening of biofilm formers
| Biofilm formation | CRA plate test | E. coli | Salmonella spp. | Tube test | E. coli | Salmonella spp. |
|---|---|---|---|---|---|---|
| Weak |
|
13 | 6 |

|
8 | 2 |
| Moderate |
|
5 | 3 |

|
19 | 12 |
| Strong |
|
17 | 9 |

|
8 | 4 |
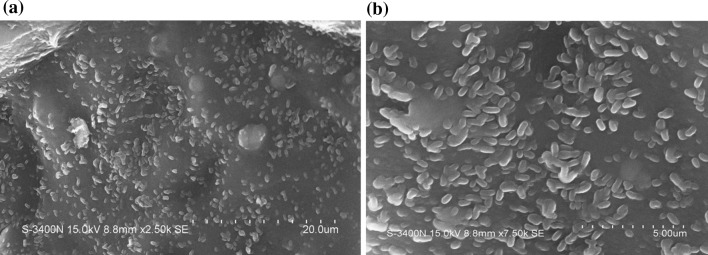
For morphological confirmation, the isolates were subjected to Scanning electron microscopy (SEM) analysis. (Annous et al. 2005) and Iturriaga et al. (2007) have reported that many bacteria are capable of forming distinct biofilms on surfaces of fruits and vegetables, including E. coli, Salmonella, Listeria, Campylobacter and Shigella. The SEM micrographs revealed biofilms in the form of an extensive aggregation of microbial cells on the surfaces of fruits such as apple and guava (Fig. 1). Earlier reports have shown that isolates of Salmonella sp. are better adhered and attached to tomato compared with non-biofilm-producers (Cevallos–Cevallos et al. 2012).
Fig. 1.
SEM micrograph showing cells of E. coli (a) and Salmonella spp. (b) aggregating in the form of biofilms on the surfaces of Apple and Guava respectively
The test tube method for detection of biofilm production suggested that a significantly lower percentage of isolates as strong biofilm producers when compared to CRA plate test results, indicating the specificity of the method. Results indicated that 22.8% E. coli and 22.2% of Salmonella sp. were strong biofilm formers. Hassan et al. (2011) reported biofilm formation in clinical isolates by test tube method and detected 49% isolates as biofilm producers and 51% as non-biofilm producers. For further analysis, only the strong biofilm formers including 8 E. coli and 4 Salmonella sp. were taken into account.
Verstraeten et al. (2008) has demonstrated that biofilm formation and pathogenesis compulsorily requires the bacteria to exhibit their motility. All the four Salmonella spp. isolates screened showed featureless swimming pattern on the agar media, while only six E. coli showed the same pattern and two isolates didn’t show swimming pattern at all. The surface swarming ability of the isolates seemed towards an increase in colony diameter ranging from 1 to 4.5 cm after 24 h of incubation. Positive control strains of E. coli and Salmonella sp. were also tested for their motility, which showed colony diameters of 4 and 6.5 cm, respectively. However, the swarming motility of the tested strains in this study was found to be weak compared to earlier reports Kalaichelvam et al. (2014) and Kim and Surette (2005).
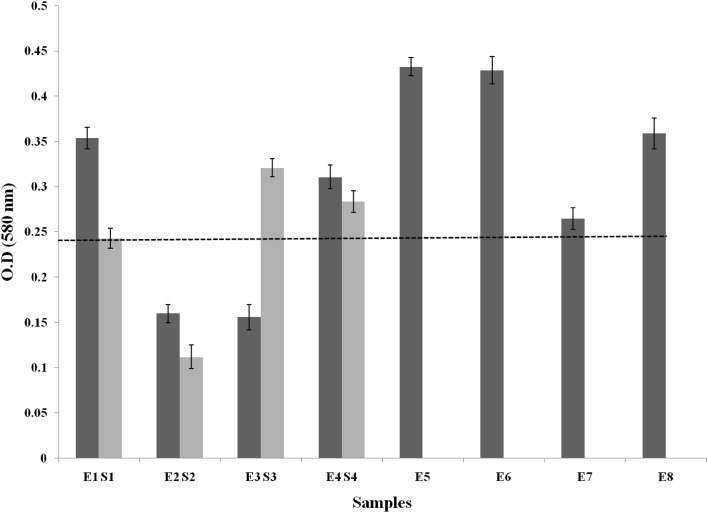
Results from the microtitre plate (MTP) assay suggested that 75% each of E. coli and Salmonella isolates belonged to the category of strong biofilm formers (with OD values > 0.24) while the rest were moderate and weak biofilm formers (with OD values between 0.12–0.24 and <0.12 respectively) (Fig. 2). Previous reports on MTP assay by Mathur et al. (2006) indicated 53.9% of clinical isolates of Staphylococci to be biofilm producers and 46% as non-biofilm producers. MTP assay yielded reliable results with higher accuracy when compared to qualitative tests such as CRA plate and test tube assay, which has been supported earlier (Deighton and Balkau 1990).
Fig. 2.
MTP Assay of E. coli(E) and Salmonella sp.(S) Black dotted line indicates the critical limit (0.24) for being strong biofilm formers
Screening for quorum signalling molecules (AHL)
Results of the agar plate diffusion assay indicated that four E. coli and one Salmonella sp. isolates were found to produce quorum sensing molecules (AHL), indicated by the production of bluish green colour around the colony on X-gal agar media (Fig. 3). When active forms of AHLs are produced, beta-galactosidase enzymatically cleaves X-Gal, resulting in the formation of 5, 5′ dibromo-4, 4′-dichloro-indigo, which is an insoluble blue precipitate.
Fig. 3.
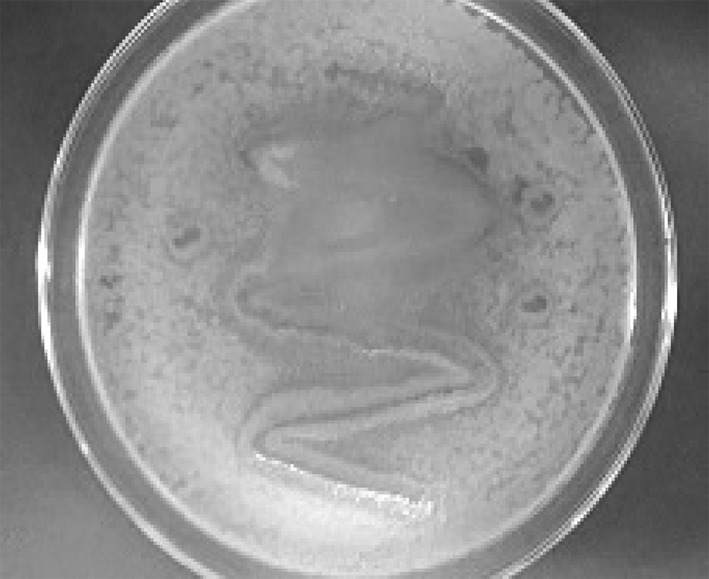
Screening for quorum sensing in the isolate E3. Pigmentation around the colony indicates the production of AHLs by the E. coli isolate
EPS production
Flemming et al. 2000 has reported that biofilms comprise 50–90% exopolysaccharide component. The test isolates were subjected to EPS extraction and quantification according to Dubois et al. 1956. It was observed that two E. coli strains (E1 and E3) produced a significantly higher amount of EPS with 507 and 275 mg/ml, respectively. The EPS yield among the rest of the isolates was ranging from 35 to 100 mg/ml, which was found to be much higher compared to previous reports by Ami et al. 2012, in which Weissella sp. isolated from fermented food produced 0.5 mg/ml of EPS.
Amplification of biofilm specific gene by PCR
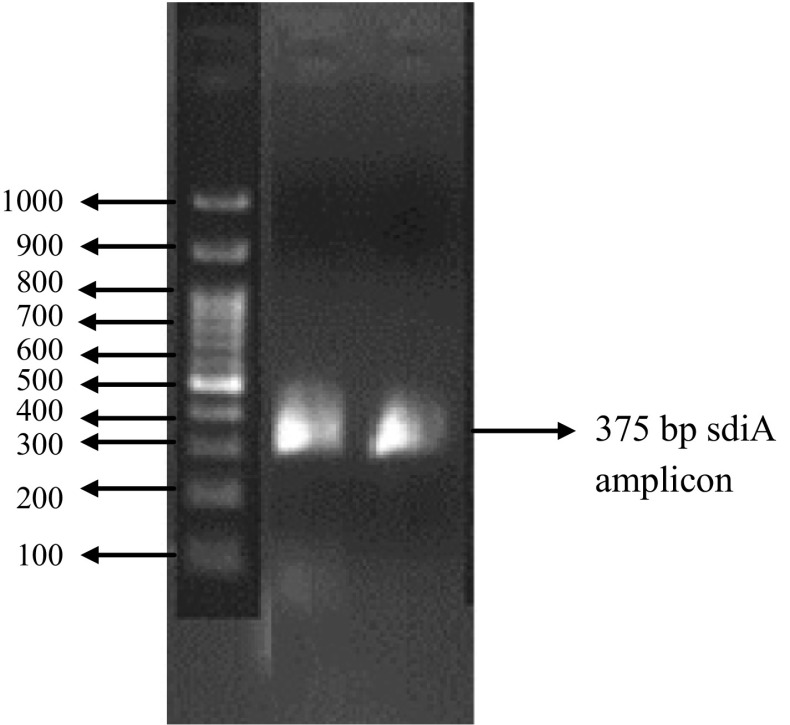
The E. coli and Salmonella isolates were analysed for the presence of Ica A gene and Sdi A gene, respectively. Two isolates of Salmonella sp. (S1 and S3) showed the presence of LUX R homologue sdiA (375 bp), there by indicating their potential in virulence and biofilm formation (Fig. 4). However, none of the E. coli isolates amplified Ica A operon, which is specific for E. coli. Lack of IcaA gene in any of the E. coli isolates indicate that their biofilm forming potential is not because of the presence of the particular gene and maybe due to other genes which are not taken into account in the study or can even be attributed to other genetic and environmental factors.
Fig. 4.
Amplicon size of 375 bp corresponding to the LUX R homologue sdiA observed in the Salmonella strains S1 and S3
Conclusion
The present study has examined biofilm forming potential of E. coli and Salmonella sp. isolated from fresh fruits and vegetables. The results clearly demonstrate that the microbial flora associated with the fresh fruits and vegetables are capable of biofilm formation, which is essential for virulence of the pathogenic strains. MTP assay and PCR yielded the most reliable results when compared to the phenotypic methods. Presence of the sdiA gene and screening for AHL production firmly established the biofilm forming potential of the isolates. Thus, the findings of the present study provide an insight into the pathogenicity and potential threat associated with microbial flora of fresh fruits and vegetables. It can therefore be concluded that systematic and hygienic handling of fruits and vegetables is essential for ensuring safe products and measures needs to be devised to maintain the standards of food safety.
Acknowledgements
Authors are grateful to the DST-PURSE Programme, DST, New Delhi, for providing the funding for the research work and Pondicherry University for providing the essential laboratory facilities to carry out the work.
References
- Ami P, Lindstrom C, Arti P, Prajapati JB, Olle H. Probiotic properties of exopolysaccharide producing lactic acid bacteria isolated from vegetables and traditional Indian fermented foods. Int J Ferment Foods. 2012;1:77–86. [Google Scholar]
- Annous BA, Solomon EB, Cooke PH, Burke Biofilm formation by Salmonella sp. on cantaloupe melons. J Food Saf. 2005;25:276–287. doi: 10.1111/j.1745-4565.2005.00024.x. [DOI] [Google Scholar]
- Barak JD, Liang AS. Role of soil, crop debris, and a plant pathogen in Salmonella enterica contamination of tomato plants. PLoS ONE. 2008;3:e1657. doi: 10.1371/journal.pone.0001657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger CN, Sodha SV, Shaw RK, Griffin PM, Pink D, Hand P. Fresh fruit and vegetables as vehicles for the transmission of human pathogens. Environ Microbiol. 2010;12:2385–2397. doi: 10.1111/j.1462-2920.2010.02297.x. [DOI] [PubMed] [Google Scholar]
- Byrne L, Fisher I, Peters T, Mather A, Thomson N, Rosner B, Bernard H, McKeown P, Cormican M, Cowden J, Aiyedun V, Lane C. A multi-country outbreak of Salmonella Newport gastroenteritis in Europe associated with watermelon from Brazil, confirmed by whole genome sequencing: October 2011 to January 2012. Euro Surveill. 2014;19:6–13. doi: 10.2807/1560-7917.ES2014.19.31.20866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cafiso V, Bertuccio T, Santagati M, Campanile F, Amicosante G, Perilli MG. Presence of the ica operon in clinical isolates of Staphylococcus epidermidis and its role in biofilm production. Clin Microbiol Infect. 2004;10:1081–1088. doi: 10.1111/j.1469-0691.2004.01024.x. [DOI] [PubMed] [Google Scholar]
- Cevallos-Cevallos JM, Gu G, Danyluk MD, Van Bruggen AH. Adhesion and splash dispersal of Salmonella enterica Typhimurium on tomato leaflets: effects of rdar morphotype and trichome density. Int J Food Microbiol. 2012;160:58–64. doi: 10.1016/j.ijfoodmicro.2012.09.021. [DOI] [PubMed] [Google Scholar]
- Deighton MA, Balkau B. Adherence measured by microtiter assay as a virulence marker for Staphylococcus epidermidis infections. J Clin Microbiol. 1990;28:2442–2447. doi: 10.1128/jcm.28.11.2442-2447.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis N, Zhang H, Leroux A, Trudel R, Bietlot H. Prevalence and trends of bacterial contamination in fresh fruits and vegetables sold at retail in Canada. Food Control. 2016;67:225–234. doi: 10.1016/j.foodcont.2016.02.047. [DOI] [Google Scholar]
- Dubois MK, Gilles A, Hamilton K, Rebers PA, Smith F. Calorimetric method for determination of sugars and related substances. Anal Chem. 1956;28:350–356. doi: 10.1021/ac60111a017. [DOI] [Google Scholar]
- Fitzpatrick F, Humphreys H, O’Gara JP. Evidence for ica ADBC-independent biofilm development mechanism in methicillin-resistant Staphylococcus aureus clinical isolates. J Clin Microbiol. 2005;43:1973–1976. doi: 10.1128/JCM.43.4.1973-1976.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming HC, Wingender JG, Mayer C. Physico-chemical properties of biofilms. In: Evans LV, editor. Biofilms: recent advances in their study and control. Amsterdam: Harwood Academic Publishers; 2000. pp. 19–34. [Google Scholar]
- Hassan A, Usman J, Kaleem F, Omair M, Khalid A, Iqbal M. Evaluation of different detection methods of biofilm formation in the clinical isolates. Braz J Infect Dis. 2011;15:305–311. doi: 10.1016/S1413-8670(11)70197-0. [DOI] [PubMed] [Google Scholar]
- Islam M, Morgan J, Doyle MP, Phatak SC, Millner P, Jiang X. Fate of Salmonella enterica serovar Typhimurium on carrots and radishes grown in fields treated with contaminated manure composts or irrigation water. Appl Environ Microbiol. 2004;70:2497–2502. doi: 10.1128/AEM.70.4.2497-2502.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam M, Morgan J, Doyle MP, Phatak SC, Millner P, Jiang X. Persistence of Salmonella enterica serovar Typhimurium on lettuce and parsley and in soils on which they were grown in fields treated with contaminated manure composts or irrigation water. Foodborne Pathog Dis. 2004;1:27–35. doi: 10.1089/153531404772914437. [DOI] [PubMed] [Google Scholar]
- Iturriaga MH, Tamplin ML, Escartin EF. Colonization of tomatoes by Salmonella Montevideo is affected by relative humidity and storage temperature. J Food Prot. 2007;70:30–34. doi: 10.4315/0362-028X-70.1.30. [DOI] [PubMed] [Google Scholar]
- KalaiChelvam K, Chai LC, Thong KW. Variations in motility and biofilm formation of Salmonella enterica serovar Typhi. Gut pathog. 2014;6:2. doi: 10.1186/1757-4749-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Surette GM. Prevalence of surface swarming behavior in Salmonella. J Bacteriol. 2005;187:6580–6583. doi: 10.1128/JB.187.18.6580-6583.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak GK, MacDonald D, Landry L, Farber JM. Foodborne outbreaks in Canada linked to produce: 2001 through 2009. J Food Prot. 2013;76:173–183. doi: 10.4315/0362-028X.JFP-12-126. [DOI] [PubMed] [Google Scholar]
- Lynch MF, Tauxe RV, Hedberg CW. The growing burden of foodborne outbreaks due to contaminated fresh produce: risks and opportunities. Epidemiol Infect. 2009;137:307–315. doi: 10.1017/S0950268808001969. [DOI] [PubMed] [Google Scholar]
- Mathur T, Singhal S, Khan S, Upadhyay DJ, Fatma T, Rattan A. Detection of biofilm formation among the clinical isolates of Staphylococci: an evaluation of three different screening methods. Indian J of Med Microbiol. 2006;24:25–29. doi: 10.4103/0255-0857.19890. [DOI] [PubMed] [Google Scholar]
- Percival SL, Walker J, Hunter P. Microbiological aspects of biofilms and drinking water. New York: CRC Press; 2000. [Google Scholar]
- Silva De, Silva GDI, Kantzanou M, Justice A, Massey RC, Wilkinson AR. The ica operon and biofilm production in coagulase-negative staphylococci associated with carriage and disease in a neonatal intensive care unit. J Clin Microbiol. 2002;40:382–388. doi: 10.1128/JCM.40.02.382-388.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JL, Fratamico PM, Novak JS. Quorum sensing: a primer for food microbiologists. J Food Prot. 2004;67:1053–1070. doi: 10.4315/0362-028X-67.5.1053. [DOI] [PubMed] [Google Scholar]
- Sutherland IW. The biofilm matrix: an immobilized but dynamic microbial environment. Trends Microbiol. 2001;9:222–227. doi: 10.1016/S0966-842X(01)02012-1. [DOI] [PubMed] [Google Scholar]
- Teplitski M, Barak JD, Schneider KR. Human enteric pathogens in produce: un-answered ecological questions with direct implications for food safety. Curr Opin Biotechnol. 2009;20:166–171. doi: 10.1016/j.copbio.2009.03.002. [DOI] [PubMed] [Google Scholar]
- Verstraeten N, Braeken K, Debkumari B, Fauvart M, Fransaer J, Vermant J, Michiels J. Living on a surface: swarming and biofilm formation. Trends Microbiol. 2008;16:496–506. doi: 10.1016/j.tim.2008.07.004. [DOI] [PubMed] [Google Scholar]