Abstract
Advancements in next-generation sequencing (NGS) technologies are generating a vast amount of data. This exacerbates the current challenge of translating NGS data into actionable clinical interpretations. We have comprehensively combined germline and somatic nonsynonymous single-nucleotide variations (nsSNVs) that affect drug binding sites in order to investigate their prevalence. The integrated data thus generated in conjunction with exome or whole-genome sequencing can be used to identify patients who may not respond to a specific drug because of alterations in drug binding efficacy due to nsSNVs in the target protein's gene. To identify the nsSNVs that may affect drug binding, protein–drug complex structures were retrieved from Protein Data Bank (PDB) followed by identification of amino acids in the protein–drug binding sites using an occluded surface method. Then, the germline and somatic mutations were mapped to these amino acids to identify which of these alter protein–drug binding sites. Using this method we identified 12 993 amino acid–drug binding sites across 253 unique proteins bound to 235 unique drugs. The integration of amino acid–drug binding sites data with both germline and somatic nsSNVs data sets revealed 3133 nsSNVs affecting amino acid–drug binding sites. In addition, a comprehensive drug target discovery was conducted based on protein structure similarity and conservation of amino acid–drug binding sites. Using this method, 81 paralogs were identified that could serve as alternative drug targets. In addition, non-human mammalian proteins bound to drugs were used to identify 142 homologs in humans that can potentially bind to drugs. In the current protein–drug pairs that contain somatic mutations within their binding site, we identified 85 proteins with significant differential gene expression changes associated with specific cancer types. Information on protein–drug binding predicted drug target proteins and prevalence of both somatic and germline nsSNVs that disrupt these binding sites can provide valuable knowledge for personalized medicine treatment. A web portal is available where nsSNVs from individual patient can be checked by scanning against DrugVar to determine whether any of the SNVs affect the binding of any drug in the database.
Introduction
With the development of massively parallel sequencing, also known as next-generation sequencing (NGS), a vast amount of NGS data are being generated with greater throughput and decreased cost compared with its predecessor technology, Sanger sequencing.1, 2, 3, 4 The identification of single-nucleotide variations (SNVs) is one of the most common tasks in NGS data analysis.5 Although most SNVs are found in the intergenic region, many can also be found at more crucial locations such as within protein coding regions. For example, missense SNVs (msSNVs) may play a more direct role in causing or exacerbating disease by changing the protein structure or by other mechanisms.6, 7 Pharmacogenetic and pharmacogenomic studies have shown that SNVs can affect how a patient responds to administered drug treatments.8, 9, 10, 11 The most direct example is where msSNVs exists within the coding region of a gene coding for target protein and these SNVs alter the amino acid of binding site of the drug, resulting in changes in drug binding affinity and consequent therapeutic effect of the drug.12, 13, 14 In addition to msSNVs affecting the binding site, there are several other factors that can cause differences in drug responses including gene expression, drug metabolism and environmental factors.15 In this study we focus on the effects of msSNVs that affect protein–drug binding sites.
The Protein Data Bank (PDB),16 a three-dimensional structure database, contains structure data of proteins complexed with small molecules such as substrates, cofactors, inhibitors and drugs and is widely used in drug discovery research.17, 18, 19 Secondary databases, such as DGIdb, CREDO20 and FireDB,21 use data from PDB and provide value-added information through further analysis. The above databases do not provide comprehensive somatic mutation or polymorphism mapping; neither do they provide protein–drug interaction-centric information. Research has shown that individual's genetic makeup can contribute to differential drug response.8, 22, 23, 24 PharmGKB mines information from this type of research publications,25 and at the time of writing this paper it contained over 5000 variant annotations in more than 900 proteins related to over 600 drugs.
This paper describes identification and integration of amino acid–drug binding sites from PDB and nonsynonymous single-nucleotide variations (nsSNVs) compiled from various sources to create a comprehensive data set called DrugVar. This data set can be used to scan exome or whole-genome sequencing data from patients to see whether a patient has a missense mutation that affects any protein–drug binding site, and also how prevalent this SNV is in tumors or whether they exist as polymorphism in the population.
Materials and methods
Amino acid–drug binding data set
Amino acid–drug complex structure data was obtained from PDB database.26 The Anatomical Therapeutic Chemical Classification (ATC), a hierarchical representation of drugs, was used for identifying cancer and noncancer-related drugs.26 Data were manually curated in order to separate drugs from other small molecules. PDB amino acid sequence positions were mapped to UniProtKB accessions27 followed by pairwise alignment. For each atom of the amino acid residues in the drug binding pocket, occluded surface (buried) area by the drug in its binding pocket was computed using the program OS28 and a list of drug-interacting residues was generated based on ranking the occlusion percentage. All PDB IDs associated with the same protein and all protein–drug pairs were considered.
nsSNV data set and data integration
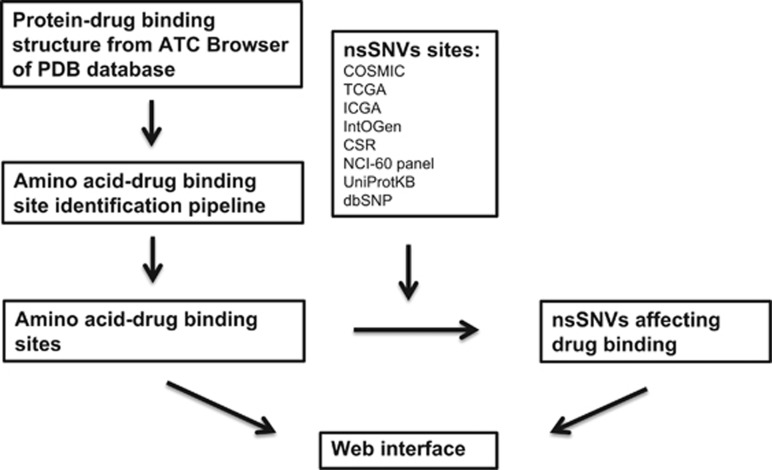
A comprehensive non-redundant data set of both germline and somatic nonsynonymous nsSNVs was generated using data from The Cancer Genome Atlas (TCGA; http://cancergenome.nih.gov/), International Cancer Genome Consortium (ICGC),29 IntOGen,30 CSR,31 cancer cell lines from NCI-60 panel,32 dbSNP33 and UniProtKB/Swiss-Prot.34 The integration was performed using methods described earlier.35 The final data set contains 1 705 286 somatic nsSNVs and 1 132 832 germline nsSNVs. Drug IDs were retrieved from DrugBank and literature to allow linking to detailed pharmacology and pharmaceutical knowledge and drug target information.36, 37, 38 Figure 1 shows the workflow used to generate the protein–drug interaction data set.
Figure 1.
Workflow for mapping nonsynonymous single-nucleotide variations (nsSNVs) on protein–drug binding sites. ATC, anatomical therapeutic chemical classification; PDB, protein data bank.
Analysis
A statistical significance analysis was conducted to investigate the distribution of binding sites between amino acids and drugs. Significance of observed versus expected of amino acids in drug binding sites was calculated using the Binomial statistic described by Mi and Thomas39 and applied in our previous studies.40 Protein Functional Class Enrichment analysis was performed using the PANTHER Classification System.41
Identification of homologs as alternative drug targets
For paralog identification, each protein in the data set was used to BLAST42 against the human proteome to identify paralogs43, 44 that might also bind the drug. The binding sites in the paralogs were checked using pairwise alignment to ensure they are conserved. Homology modeling of these paralogs was carried out using MODELLER.45 These homology models of paralogs were energy minimized using the AMBER force field in Molecular Operating Environmen (MOE) (Version 2014.06) package developed by the Chemical Computing Group, Montreal, Canada (www.chemcomp.com). For additional homolog identification, all proteins of non-human mammalian origin–drug complex were retrieved from the PDB database. Similar workflow as mentioned above was applied on these data for identification of human homologs that may serve as drug targets.
Structural and mutational modeling
Visualization of the amino acids in the binding site for the amino acid–drug complexes was carried out using UCSF CHIMERA.46 To understand the effect of mutations on a drug binding to a protein target, the X-ray structure of carbonic anhydrase 2 (CA2) bound to the drug Lacosamide (PDB ID: 3IEO)47 was chosen. The wild-type amino acid–drug and the mutated amino acid–drug complexes were energy minimized using the AMBER force field in MOE and binding energies computed.
Neighboring mutations and gene expression analysis
Even if there are no amino acid variations in protein–drug binding sites, the variations existing in a binding site's neighboring region may structurally block the protein–drug binding. Therefore, for each protein–drug binding site, mutations within its upstream and downstream region were identified. The total number of variations within the expanded protein–drug binding region was calculated. In addition, as the gene expression level is another factor that may influence drug efficacy, cancer-related gene expression data were retrieved from BioXpress.48 It analyzes and normalizes raw count data of paired sample (tumor and normal) from TCGA using DEseq R package (http://bioconductor.org/packages/release/bioc/html/DESeq.html), a method that is regarded as one of the most robust RNA-seq normalization methods.49 If a protein–drug pair contained one or more amino acid variations in its binding site and if the variations were somatic mutations associated with a specific cancer type, the corresponding tumor versus healthy tissue differential gene expression was evaluated to rank promising validation targets.
Results and discussion
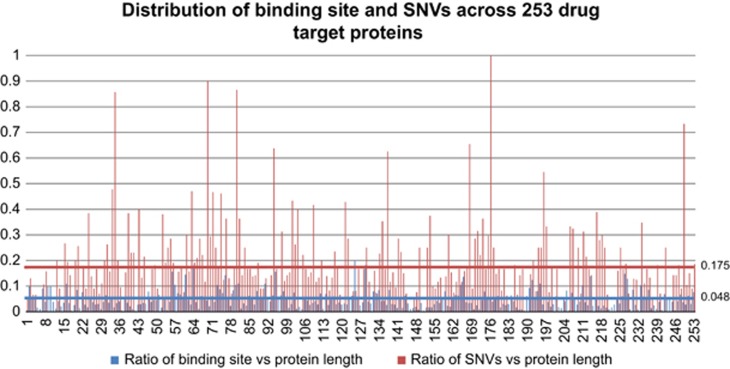
Out of 1 12 722 structures available in the PDB database, 20 385 PDB structures are associated with 1712 nonstandard polymeric components. After further filtering of crystallization stabilizers and proteins of non-human origin, a total number of 827 PDB structures were identified with drug bound to them. These PDB structures were mapped to 253 UniProtKB proteins binding to 235 unique drugs with 12 993 binding sites (Supplementary Table S1). Out of 253 proteins in the amino acid–drug binding data set, 210 proteins had 1618 drug binding site altering variations (Supplementary Tables S2 and S3). Figure 2 shows the distribution of binding sites and binding sites affected by SNVs across 253 drug target proteins. For each drug target protein, the blue bar indicates the ratio between number of drug binding sites and target protein length, whereas the red bar shows the ratio between number of binding site affecting SNVs and binding sites. In 253 identified drug target proteins, on average 4.8% of the amino acids are identified as drug binding sites and 14.8% of these drug binding sites have variation mapped to them. The number of binding sites for each protein is largely influenced by the number of ‘binding pockets' it contains and the structure of ‘binding pocket'. In all, 1162 binding sites are affected because of germline variations and 456 binding sites are affected by somatic mutations. The number of identified drugs and their target proteins is relatively small compared with the total number of FDA (Food and Drug Administration)-approved drugs (1602) because of the limitations of X-ray crystallography technology. With the advent of large-scale structural genomics projects, we expect to see increased coverage in the coming years. In addition, binding sites based on electrospray ionization mass spectrometry50 and mutation studies51 can be used to model additional sites.
Figure 2.
Distribution of binding sites and binding sites affecting single-nucleotide variations (SNVs) across 253 drug target proteins. The blue bar indicates the ratio between number of drug binding sites and target protein length, whereas the red bar shows the ratio between number of binding sites affecting SNVs and binding sites.
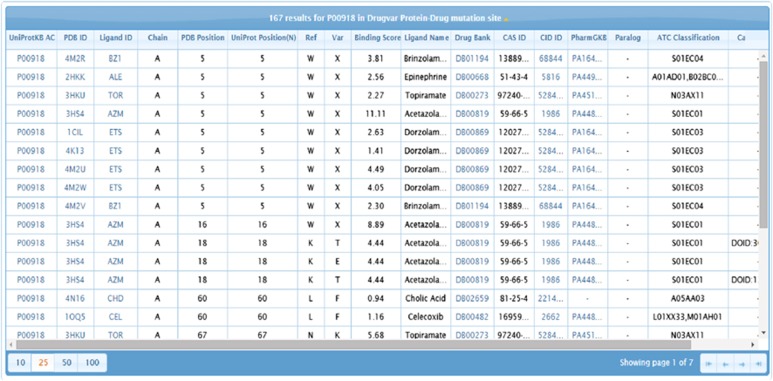
It is interesting to note that 83% of the identified drug target proteins contain mutations on their binding sites. Out of the large amount of identified mutations, 0.12% of germline mutation and 0.035% of somatic mutations are mapped to drug binding sites in our study. Amino acid–drug binding sites and nsSNV sites are available from the DrugVar portal and a snapshot of the web interface is shown in Figure 3. All drugs were classified into 14 groups based on ATC (Table 1).
Figure 3.
DrugVar website browser interface. Users can perform searches using Protein Data Bank (PDB) IDs, gene names, UniProtKB accessions and drug names or identifiers.
Table 1. Statistical summary of data set based on the anatomical therapeutic chemical (ATC) classification system.
| ATC classification | Number of protein | Number of drug |
|---|---|---|
| Cancer drugs | ||
| Antineoplastic agents | 36 | 25 |
| Noncancer drugs | ||
| Alimentary tract and metabolism | 56 | 36 |
| Blood and blood-forming organs | 28 | 9 |
| Cardiovascular system | 43 | 33 |
| Dermatologicals | 23 | 15 |
| Genitourinary system and sex hormones | 28 | 17 |
| Systemic hormonal preparations, excluding sex hormones and insulins | 8 | 4 |
| Antiinfectives for systemic use | 11 | 13 |
| Musculoskeletal system | 18 | 22 |
| Nervous system | 26 | 26 |
| Antiparasitic products, insecticides and repellents | 8 | 9 |
| Respiratory system | 13 | 5 |
| Sensory organs | 9 | 7 |
| Antidotea | 28 | 4 |
Antidote is a subclass of ‘Various' (ATC Code: V).
Antineoplastic drug binding sites and somatic mutation impact
Out of 12 993 drug binding sites, 1408 protein binding sites were found to be associated with 25 antineoplastic drugs (inhibiting or preventing the growth and spread of tumors or malignant cells; ATC Code: L01). Some of them act by disrupting the metabolism of DNA synthesis components. For example, folic acid analogs, such as methotrexate and raltitrexed, inhibit key enzymes that are necessary for synthesis of thymidylate, an essential component of DNA.52, 53 Similarly, purine analogs, such as cladribine, and pyrimidine analogs, such as gemcitabine, inhibit cancer cell growth by disrupting DNA synthesis, whereas nilotinib binds to the catalytic site of BCR-Abl, a protein kinase that only appears in cancer cells of chronic myelogenous leukemia patients, and inhibits the tumor growth.
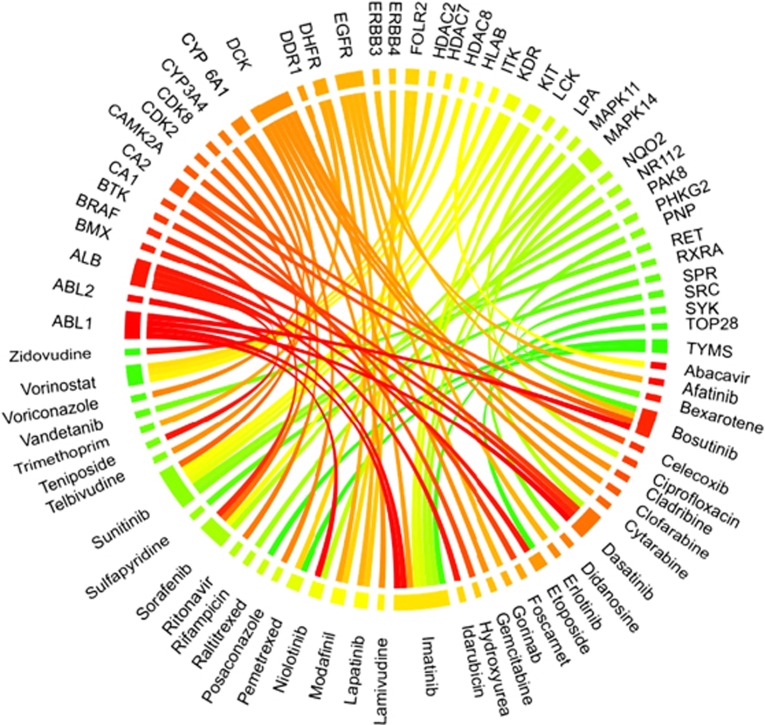
We further examined 25 unique antineoplastic agent drugs and 35 unique target proteins. Figure 4 shows a Circos plot54 representing the protein–drug binding relationship. Imatinib, a tyrosine kinase inhibitor used in the treatment of multiple cancers has been shown through solved three-dimensional structures to bind to as many as eight proteins (gene symbols: ABL1, ABL2, DDR1, KIT, LCK, MAPK14, NQO2 and SYK).55, 56, 57, 58, 59, 60 As one of the most well-known targeted therapy drugs for cancer treatment, imatinib inhibits the BCR-Abl kinase to cut off the energy source for cancer proliferation, thereby inhibiting the tumor growth.55, 61 It is also known that imatinib can also inhibit mast/stem cell growth factor receptor Kit (KIT)56 to stop tumor growth. However, the effect of imatinib on other proteins in Figure 4 still remains largely unexplored even though structures are available. Sunitinib can bind to six proteins (CDK2, KDR, KIT, PHKG2, ITK and MAPK14). It inhibits cellular signaling by targeting tyrosine kinase including platelet-derived growth factor receptor-β, vascular endothelial growth factor receptor 1, KIT and vascular endothelial growth factor receptor 2.62,63,64,65 However, further studies are needed for the physiological outcome of its binding to rest of the proteins.
Figure 4.
Circos plot representing the binding connections between 25 antineoplastic agents and their target proteins. Proteins are presented with gene names. Ribbon colors are assigned for visualization purposes and the ribbon width indicates the number of target proteins.
For the antineoplastic drug target proteins, 608 out of 3133 variations were found on binding sites between 25 antineoplastic drugs and their 36 target proteins. Out of these, 178 are somatic mutations and 429 are germline variations. Gefitinib is primarily used in treatment of non-small-cell lung cancer.66 In the DrugVar data set, six lung cancer-associated somatic mutations exist within binding sites between epidermal growth factor receptor (EGFR) and gefitinib on amino acid positions 744, 766, 790, 792, 844 and 855. In addition, a total number of seven germline mutations exist within binding sites between EGFR epidermal growth factor receptor and gefitinib on amino acid positions 719, 726, 743, 788, 796, 800 and 854. It is possible that these mutations on EGFR may weaken binding efficacy of gefitinib to its target protein in lung cancer patients.
Drugs that bind to multiple proteins and proteins with multiple msSNVs
It is reasonable to assume that the more proteins a drug binds to, the more diverse its effects might be. Imatinib has eight target proteins (ABL1, LCK, KIT, NQO2, ABL2, SYK, DDR1 and MAPK14) in the DrugVar database. Out of them, 7 proteins have a total of 58 amino acid mutations in their binding sites with imatinib. As one of the primary target proteins, KIT is inhibited by imatinib67 and contains 11 possible variations in its imatinib binding sites. These mutations may affect the drug binding affinity and make imatinib less effective under regular dose intake. Tyrosine-protein kinase ABL1 (ABL1) is another imatinib targeted protein and contains 29 variations on binding sites. Out of 29 mutations, 19 are germline mutations and 8 are somatic mutations associated with diffuse large B-cell lymphoma. We report here many such amino acid variations in drug binding sites of proteins. The biological outcomes of these variations have not yet been elucidated. Such examples include epithelial discoidin domain-containing receptor 1 (DDR1) that is suggested to be a pharmacologically available target for cancer treatment,60, 68 tyrosine-protein kinase Lck (LCK), tyrosine-protein kinase SYK (SYK), mitogen-activated protein kinase 14 (MAPK14) and abelson tyrosine-protein kinase 2 (ABL2).
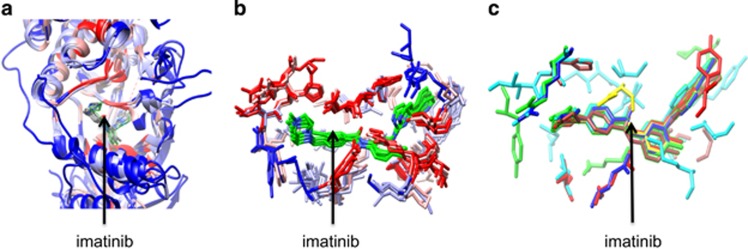
To better understand the binding of the drugs, the binding pocket of one of the drugs was further analyzed. Figure 5a shows the superposition of X-ray crystal structures of imatinib binding to its eight target proteins. The drug binds in the active site cleft of the kinase catalytic domain that is represented by ribbon structures. Figure 5b shows the side chains identified as binding site of amino acids with imatinib. They are marked by a green color in the pocket of target proteins. The structural differences could help to modify imatinib to improve the selectivity for a given protein target. Figure 5c shows the amino acids that are reported as mutations for each target protein in the amino acid–drug binding sites. The drug is located in the active site cleft of the kinases, each of which is shown in different color for clarity.
Figure 5.
Structural view of protein–drug interactions. (a) Superposition (c-alpha atoms) of imatinib binding to eight target protein X-ray structures (ABL1, LCK, KIT, NQO2, ABL2, SYK, DDR1 and MAPK14). The superimposed protein structures are colored. The blue to red color represents low to high conservation. The ligand is shown bound to protein pockets. (b) Imatinib binding to the same target proteins as shown in (a). Only the side chains of binding sites are shown. (c) Imatinib binding to its target proteins. The side chains of proteins are imatinib binding sites that are mutated.
Our study also emphasizes the importance of proteins that bind to specific drugs while still containing a large number of mutations on their binding sites. EGFR is a well-known protein target for antineoplastic drugs.69 Some drugs, such as lapatinib and erlotinib,70, 71 have been designed specifically as antagonists of EGFR to prevent its activation that is required for tumor proliferation.71, 72 We identified 25 mutations within EGFR binding sites with lapatinib, suggesting that lapatinib might vary in effectiveness based on the genomic profile of the patient.
Non-antineoplastic drug binding sites and germline mutation impact
There are 11 124 amino acid–drug binding sites associated with 200 non-antineoplastic drugs. These 200 non-antineoplastic drugs can be further categorized into 13 subclasses. Table 1 shows the number of proteins and drugs associated with each group categorized by ATC Classification System. Out of a total of 3133 amino acid variations, 2704 were found on 200 non-antineoplastic drug target proteins.
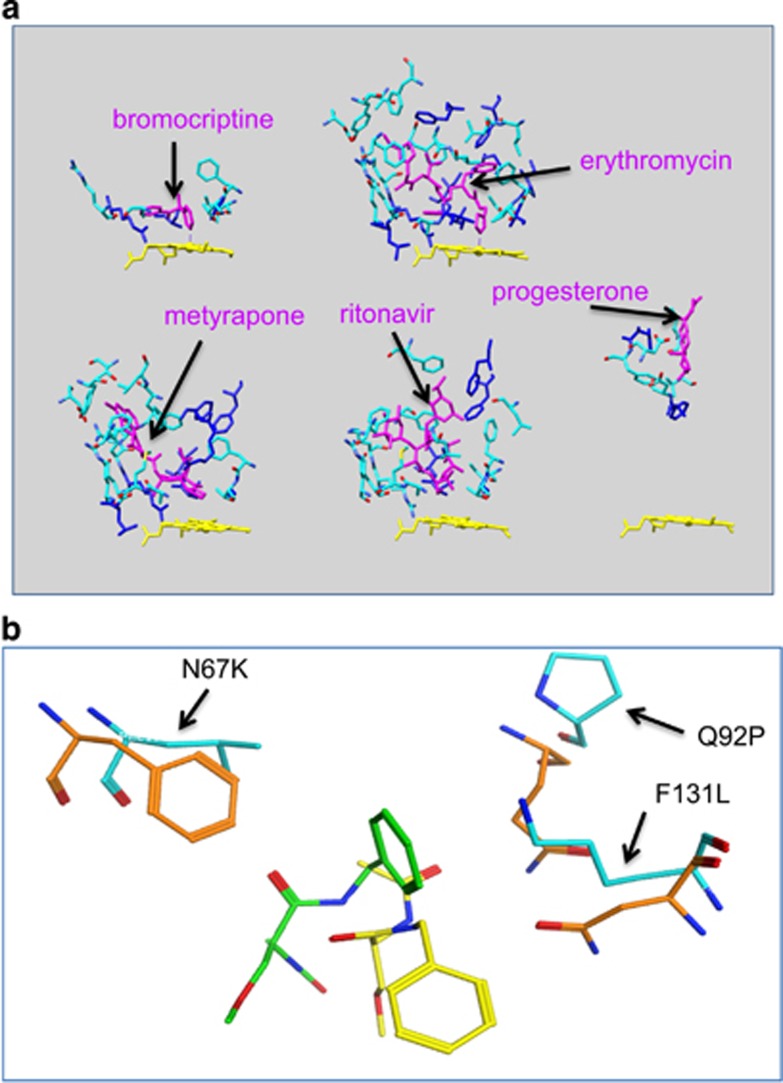
There are seven proteins in this data set that have at least seven drugs that can bind to them. The proteins are androgen receptor (AR), deoxycytidine kinase (DCK), aldo-keto reductase family 1 member C3 (AKR1C3), aldo-keto reductase family 1 member C2 (AKR1C2), transthyretin (TTR), CA2 and serum albumin (ALB). Most of them play important roles in drug transportation and in metabolic pathways. For example, ALB, a major plasma protein that functions as drug transporter, has 18 drug–protein complex structures available in PDB and contains 97 amino acid variation sites within the binding sites for these drugs. Similarly, TTR, another major drug transporter, also has 11 drug–protein complex structures available in PDB and contains 39 amino acid variations. As a drug that binds to the plasma protein will remain in circulation and only serve as a reservoir before it is unbound from plasma protein, the binding affinity between the drug and plasma protein directly influences the biological half-life of the drug. In addition, many proteins that are involved in drug metabolic pathways can bind to multiple drugs and several structures are available. Cytochrome P450 3A4 (CYP3A4), the major enzyme that modifies and detoxifies drugs by oxidation reaction in the liver, has 5 drug-bound protein structures in PDB and contains 28 variations within drug binding sites. Figure 6a shows the binding of different drugs (bromocriptine, erythromycin, metyrapone, ritonavir, progesterone) and CYP3A4. The drugs are shown in magenta color in the protein pocket bound to the amino acid residues that are shown in cyan and blue color. The blue colors mean the amino acids are altered because of nsSNV. The heme of CYP3A4 is marked in yellow. Panels on the top of Figure 6a show that bromocriptine and erythromycin chemically bind to heme, whereas panels on the bottom show that metyrapone, ritonavir and progesterone occupy the pocket without directly binding to heme. AKR1C2 and AKR1C3, the major enzymes to detoxify drugs by conjugation reaction, have 10 and 8 target proteins available in PDB and contain 15 and 20 mutations within protein binding sites, respectively. For these key proteins, we believe that the identified mutations on the drug binding sites are one of the determinants for drug binding affinity and, consequently, the drug efficacy and toxicity. As an example, Figure 6b shows the superposition of energy minimized structures for the wild-type (WT) CA2 bound to lacosamide (PDB: 3IEO) and the mutated models (N67K, Q92P and F131L) bound to the same drug. Only the WT (orange color) and mutated residues (cyan color) with the bound drugs (yellow: WT and green color: mutant) are shown. Because of the mutations, the drug has to shift from the WT location to another location in the binding pocket that causes an unfavorable binding energy by 4 kcal compared with the WT binding. Using the mutational profile, it is possible to understand the difference between a drug bound to WT and the mutant protein. Therefore, the identification of their mutational profile contributes to their pharmacogenetic and pharmacogenomic characterizations.
Figure 6.
Structural representation of protein–drug binding sites. (a) Cytochrome P450 3A4 bound to bromocriptine, erythromycin, metyrapone, ritonavir and progesterone, respectively. The drugs are shown in magenta color in the protein pocket bound to amino acid residues that are shown in cyan except the mutated amino acids marked in blue. The yellow color is the heme of Cytochrome P450 3A4 (CYP3A4). (b) Superposition of energy minimized structures for the wild-type carbonic anhydrase 2 (CA2) bound to lacosamide (PDB: 3IEO) and the mutated models (N67K, Q92P and F131L) bound to the same drug. PDB, protein data bank.
In terms of orthlogs of non-antineoplastic drug target proteins, 133 orthologs have been identified with 43 drugs involved.
Identification of paralogs and additional homologs for alternative drug target discovery
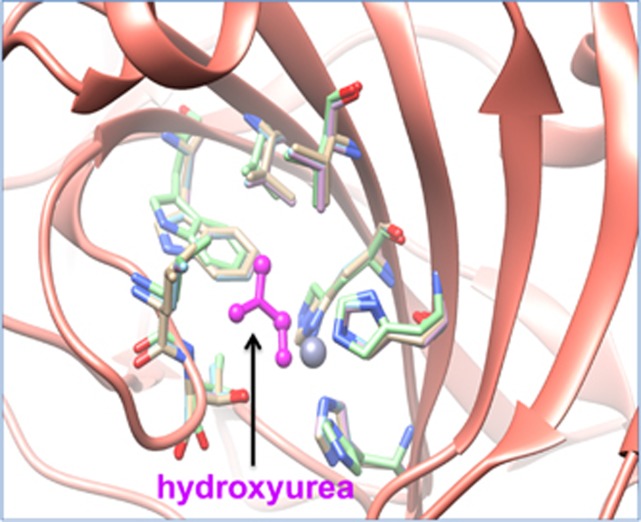
Within the 36 proteins that bind to 25 antineoplastic drugs, 5 proteins have at least one paralog that have the full binding site conserved. Carbonic anhydrase 5A (CA5A), carbonic anhydrase 5B (CA5B), carbonic anhydrase 7 (CA7) and carbonic anhydrase 13 (CA13) were identified as four paralogs of CA2 that is a target protein of hydroxyurea, an antineoplastic agent. Figure 7 shows a model of CA2 and its paralogs CA13 and CA7 binding to hydroxyurea (magenta color). The three-dimensional structure shows the consistent binding between side chains of CA2, CA13, CA7 and hydroxyurea and suggests the potential binding affinity between CA5B, CA5A and hydroxyurea. In the DrugBank database, the only listed target for hydroxyurea is ribonucleodise-diphosphate reductase large subunit (RRM1).73 In PharmaGKB, the only available target protein of hydroxyurea is mitogen-activated protein kinase kinase kinase 5 (MAP3K5). Therefore, the data we provide here are complimentary to what is available on DrugBank, PharmaGKB and other resources.
Figure 7.
Superposition of X-ray crystal structures of carbonic anhydrase 2 (CA2) and its paralogs (CA13, CA7) bound to hydroxyurea. The ribbon structure of CA2 and its paralogs is shown in pink color. The hydroxyurea in the protein pocket is shown in magenta color bound to amino acid residues that are conserved across CA2 and its paralogs.
In terms of 221 non-antineoplastic drug target proteins, we identified at least one paralog for 42 of them that have the drug binding site conserved. CA2 is also a target for non-antineoplastic drugs. It is part of the CA family and exists along with several of its family members ubiquitously across various tissues.74 It is a well-known target for diuretic and glaucoma drugs, such as ethoxzolamide, acetazolamide and methazolamide.75 However, its existence in vascular tissue also causes hypertension as a side effect by a number of drugs. Our study shows that CA2 has paralogs CA13, CA5A, CA5B and CA7 that have conserved drug binding sites. Search for druggable paralogs may lead to structurally modified alternative drugs that target its more tissue-selective paralogs and show similar pharmacological effect but less side effect. Roflumilast, another example, shows anti-inflammatory and anti-modulatory effects in the pulmonary system by selectively inhibiting phosphodiesterase-4.76 Amino acid–drug binding data retrieved from PDB show the binding between cAMP-specific 3',5'-cyclic phosphodiesterase 4D (PDE4D) and Roflumilast. Our paralogs survey shows that paralogs PDE4A, PDE4B and PDE4C are potential alternative target proteins.
For additional homolog discovery, a total number of 142 human homologs of non-human mammalian proteins were identified as potential drug targets (Supplementary Table S4). Among them, 19 are targeted by antineoplastic drugs and the rest of 123 human homologs are targeted by non-antineoplastic drugs.
Protein–drug binding affinity ranking
To further estimate the SNVs that may structurally affect protein–drug binding, we mapped msSNVs affecting within two and five amino acids upstream and downstream region of all the binding sites for each protein–drug pair. The results of 407 protein–drug pairs are shown in Supplementary Table S5. Generally, the number of binding site affecting SNVs increases proportionally as the binding region extends from two amino acids to five amino acids. Table 2 shows the top 10 protein–drug pairs and their somatic mutations and germline mutations within 2 and 5 amino acids upstream and downstream of binding sites. For germline mutation in Table 2, EGFR and tyrosine-protein kinase ABL1 appear to be two cancer drug target proteins with large number of somatic mutations within the drug binding region. The high variability of these two proteins shows potential risk in therapeutics when they are targeted for cancer treatment.
Table 2. Top 10 protein–drug pairs with highest number of somatic mutations and germline mutations within two and five amino acids upstream and downstream of binding sites.
|
Somatic mutations |
Germline mutations |
||||||
|---|---|---|---|---|---|---|---|
| UniProtKB Ac | Drug name | 2-Score | 5-Score | UniProtKB Ac | Drug name | 2-Score | 5-Score |
| P00533 | Lapatinib | 122 | 191 | P69905 | L-Methionine | 35 | 62 |
| P00533 | Erlotinib | 104 | 171 | P19652 | Disopyramide | 14 | 16 |
| P00533 | Gefitinib | 97 | 153 | P10635 | Thioridazine | 14 | 18 |
| P15056 | Sorafenib | 67 | 108 | P00519 | Nilotinib | 11 | 15 |
| P00533 | Afatinib | 37 | 62 | P00519 | Imatinib | 11 | 15 |
| P00519 | Nilotinib | 33 | 49 | P08263 | Ethacrynic acid | 11 | 13 |
| P00519 | Imatinib | 32 | 49 | P00519 | Dasatinib | 10 | 22 |
| P00519 | Dasatinib | 28 | 44 | P02768 | Levothyroxine | 10 | 24 |
| P02768 | Levothyroxine | 24 | 44 | P60022 | Glycine | 10 | 20 |
| P00519 | Bosutinib | 23 | 44 | P00533 | Lapatinib | 9 | 13 |
In addition to SNVs, gene expression changes in specific cancer types can also affect therapeutic effect.15 In the current protein–drug pairs that contain somatic mutations within their binding site, we identified 85 differential gene expression changes associated with specific cancer types (see Supplementary Table S6).48 Table 3 shows differential gene expression profiles for antineoplastic drug targets in specific cancer types. For example, KIT is significantly underexpressed in lung squamous cell carcinoma and colon adenocarcinoma. When sunitinib is used with normal dose in targeted therapy in cancer treatment, the therapeutic effect could be diminished because of the lower expression of target protein and somatic mutations within the binding sites between sunitinib and its target protein.
Table 3. Differential gene expression profiles for antineoplastic drug targets in specific cancer types.
| UniProtKB | Fold change | Adjusted | ||||||
|---|---|---|---|---|---|---|---|---|
| Ac | Position | Ref | Var | Drug name | P-value | Expression | Cancer type | |
| P02768 | 170 | H | Y | Teniposide | 9.78 | 1.4E−09 | Up | DOID:3907/lung squamous cell carcinoma |
| P10721 | 603 | V | D | Sunitinib | −5.8 | 1.2E−03 | Down | DOID:3907/lung squamous cell carcinoma |
| P10721 | 603 | V | D | Sunitinib | −5.8 | 1.2E−03 | Down | DOID:3907/ lung squamous cell carcinoma |
| P10721 | 623 | K | N | Sunitinib | −4.66 | 1.0E−03 | Down | DOID:234/colon adenocarcinoma |
| P10721 | 623 | K | N | Sunitinib | −4.66 | 1.0E−03 | Down | DOID:234/ colon adenocarcinoma |
| Q08881 | 499 | S | F | Sunitinib | 4.1 | 3.1E−05 | Up | DOID:3459 / breast carcinoma |
| P02768 | 170 | H | Y | Etoposide | 9.78 | 1.4E−09 | Up | DOID:3907/lung squamous cell carcinoma |
| Q15303 | 799 | M | L | Lapatinib | −10.52 | 1.4E−11 | Down | DOID:4467/renal clear cell carcinoma |
| P14207 | 119 | R | H | Pemetrexed | −4.06 | 5.8E−05 | Down | DOID:234/colon adenocarcinoma |
| P14207 | 119 | R | H | Pemetrexed | −4.06 | 5.8E−05 | Down | DOID:234/ colon adenocarcinoma |
| P00918 | 96 | H | Q | Hydroxyurea | −7.54 | 3.4E−05 | Down | DOID:234/colon adenocarcinoma |
| P10721 | 603 | V | D | Imatinib | −5.8 | 1.2E−03 | Down | DOID:3907/lung squamous cell carcinoma |
| P10721 | 623 | K | N | Imatinib | −4.66 | 1.0E−03 | Down | DOID:234/colon adenocarcinoma |
| P07949 | 756 | A | V | Vandetanib | −8.25 | 8.4E−04 | Down | DOID:234/colon adenocarcinoma |
Adjusted P-value is calculated using procedure of Benjamini and Hochberg.
DrugVar access
DrugVar (http://hive.biochemistry.gwu.edu/tools/drugvar) scan allows users to upload a comma/tab-delimited annotation files such as those produced by ANNOVAR77 variation annotation pipeline and see whether any of the variations can alter any of the drug binding sites. The interface also supports both protein-based (UniProtKB accession, PDB ID) and drug-based (DrugBank ID, CAS ID, CID ID) searches.
There are many more drug–protein interactions than what is available in structural databases such as PDB. It is expected that over time, because of structural genomics initiatives, more structural data will be available. We will update DrugVar at least once every year to capture all such new data.
Acknowledgments
This project is partially funded by NCI/EDRN contract no. 156620 to RM. We want to thank K Smith, S Goldweber and E Thompson for useful comments.
Footnotes
Supplementary Information accompanies the paper on The Pharmacogenomics Journal website (http://www.nature.com/tpj)
The authors declare no conflict of interest.
Supplementary Material
References
- Venter JC, Levy S, Stockwell T, Remington K, Halpern A. Massive parallelism, randomness and genomic advances. Nat Genet 2003; 33: 219–227. [DOI] [PubMed] [Google Scholar]
- Zhang J, Chiodini R, Badr A, Zhang G. The impact of next-generation sequencing on genomics. J Genet Genomics 2011; 38: 95–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gullapalli RR, Lyons-Weiler M, Petrosko P, Dhir R, Becich MJ, LaFramboise WA. Clinical integration of next-generation sequencing technology. Clin Lab Med 2012; 32: 585–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahassi, el M, Stambrook PJ. Next-generation sequencing technologies: breaking the sound barrier of human genetics. Mutagenesis 2014; 29: 303–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlopoulos GA, Oulas A, Iacucci E, Sifrim A, Moreau Y, Schneider R et al. Unraveling genomic variation from next generation sequencing data. BioData Min 2013; 6: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res 2002; 30: 3894–3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakken S, Alseth I, Rognes T. Computational prediction of the effects of non-synonymous single nucleotide polymorphisms in human DNA repair genes. Neuroscience 2007; 145: 1273–1279. [DOI] [PubMed] [Google Scholar]
- Giacomini KM, Brett CM, Altman RB, Benowitz NL, Dolan ME, Flockhart DA et al. The pharmacogenetics research network: from SNP discovery to clinical drug response. Clin Pharm Ther 2007; 81: 328–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans WE, Relling MV. Pharmacogenomics: translating functional genomics into rational therapeutics. Science 1999; 286: 487–491. [DOI] [PubMed] [Google Scholar]
- McLeod HL, Yu J. Cancer pharmacogenomics: SNPs, chips, and the individual patient. Cancer Invest 2003; 21: 630–640. [DOI] [PubMed] [Google Scholar]
- Shastry BS. SNPs and haplotypes: genetic markers for disease and drug response (review). Int J Mol Med 2003; 11: 379–382. [PubMed] [Google Scholar]
- Geisler T, Schaeffeler E, Gawaz M, Schwab M. Genetic variation of platelet function and pharmacology: an update of current knowledge. Thromb Haemost 2013; 110: 876–887. [DOI] [PubMed] [Google Scholar]
- Brandl EJ, Chowdhury NI, Tiwari AK, Lett TA, Meltzer HY, Kennedy JL et al. Genetic variation in CYP3A43 is associated with response to antipsychotic medication. J Neural Transm 2015; 122: 29–34. [DOI] [PubMed] [Google Scholar]
- Sun HY, Ji FQ, Fu LY, Wang ZY, Zhang HY. Structural and energetic analyses of SNPs in drug targets and implications for drug therapy. J Chem Inf Model 2013; 53: 3343–3351. [DOI] [PubMed] [Google Scholar]
- Ma Q, Lu AY. Pharmacogenetics pharmacogenomics, and individualized medicine. Pharmacol Rev 2011; 63: 437–459. [DOI] [PubMed] [Google Scholar]
- Berman HM, Battistuz T, Bhat TN, Bluhm WF, Bourne PE, Burkhardt K et al. The Protein Data Bank. Acta Crystallogr D Biol Crystallogr 2002; 58: 899–907. [DOI] [PubMed] [Google Scholar]
- Higueruelo AP, Schreyer A, Bickerton GR, Pitt WR, Groom CR, Blundell TL. Atomic interactions and profile of small molecules disrupting protein-protein interfaces: the TIMBAL database. Chem Biol Drug Des 2009; 74: 457–467. [DOI] [PubMed] [Google Scholar]
- Winter A, Higueruelo AP, Marsh M, Sigurdardottir A, Pitt WR, Blundell TL. Biophysical and computational fragment-based approaches to targeting protein-protein interactions: applications in structure-guided drug discovery. Q Rev Biophys 2012; 45: 383–426. [DOI] [PubMed] [Google Scholar]
- Idrees S, Ashfaq UA. Discovery and design of cyclic peptides as dengue virus inhibitors through structure-based molecular docking. Asian Pac J Trop Med 2014; 7: 513–516. [DOI] [PubMed] [Google Scholar]
- Schreyer AM, Blundell TL. CREDO: a structural interactomics database for drug discovery. Database 2013; 2013: bat049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez G, Valencia A, Tress M. FireDB—a database of functionally important residues from proteins of known structure. Nucleic Acids Res 2007; 35: D219–D223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalow W, Tang BK, Endrenyi L. Hypothesis: comparisons of inter- and intra-individual variations can substitute for twin studies in drug research. Pharmacogenetics 1998; 8: 283–289. [DOI] [PubMed] [Google Scholar]
- Mango R, Vecchione L, Raso B, Borgiani P, Brunetti E, Mehta JL et al. Pharmacogenomics in cardiovascular disease: the role of single nucleotide polymorphisms in improving drug therapy. Expert Opin Pharmacother 2005; 6: 2565–2576. [DOI] [PubMed] [Google Scholar]
- Wang Z, Wang J, Tantoso E, Wang B, Tai AY, Ooi LL et al. Signatures of recent positive selection at the ATP-binding cassette drug transporter superfamily gene loci. Hum Mol Genet 2007; 16: 1367–1380. [DOI] [PubMed] [Google Scholar]
- Thorn CF, Klein TE, Altman RB. Pharmacogenomics and bioinformatics: PharmGKB. Pharmacogenomics 2010; 11: 501–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose PW, Prlic A, Bi C, Bluhm WF, Christie CH, Dutta S et al. The RCSB Protein Data Bank: views of structural biology for basic and applied research and education. Nucleic Acids Res 2015; 43: D345–D356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, McGarvey PB, Suzek BE, Mazumder R, Zhang J, Chen Y et al. A comprehensive protein-centric ID mapping service for molecular data integration. Bioinformatics 2011; 27: 1190–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattabiraman N, Ward KB, Fleming PJ. Occluded molecular surface: analysis of protein packing. J Mol Recognit 1995; 8: 334–344. [DOI] [PubMed] [Google Scholar]
- Hudson TJ, Anderson W, Artez A, Barker AD, Bell C, Bernabe RR et al. International network of cancer genome projects. Nature 2010; 464: 993–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Perez A, Perez-Llamas C, Deu-Pons J, Tamborero D, Schroeder MP, Jene-Sanz A et al. IntOGen-mutations identifies cancer drivers across tumor types. Nat Methods 2013; 10: 1081–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole C, Krampis K, Karagiannis K, Almeida JS, Faison WJ, Motwani M et al. Non-synonymous variations in cancer and their effects on the human proteome: workflow for NGS data biocuration and proteome-wide analysis of TCGA data. BMC Bioinformatics 2014; 15: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abaan OD, Polley EC, Davis SR, Zhu YJ, Bilke S, Walker RL et al. The exomes of the NCI-60 panel: a genomic resource for cancer biology and systems pharmacology. Cancer Res 2013; 73: 4372–4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res 2001; 29: 308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt_Consortium. UniProt: a hub for protein information. Nucleic Acids Res 2015; 43: D204–D212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TJ, Shamsaddini A, Pan Y, Smith K, Crichton DJ, Simonyan V et al. A framework for organizing cancer-related variations from existing databases, publications and NGS data using a High-performance Integrated Virtual Environment (HIVE). Database 2014; 2014: bau022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law V, Knox C, Djoumbou Y, Jewison T, Guo AC, Liu Y et al. DrugBank 4.0: shedding new light on drug metabolism. Nucleic Acids Res 2014; 42: D1091–D1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox C, Law V, Jewison T, Liu P, Ly S, Frolkis A et al. DrugBank 3.0: a comprehensive resource for 'omics' research on drugs. Nucleic Acids Res 2011; 39: D1035–D1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whirl-Carrillo M, McDonagh EM, Hebert JM, Gong L, Sangkuhl K, Thorn CF et al. Pharmacogenomics knowledge for personalized medicine. Clin Pharmacol Ther 2012; 92: 414–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Thomas P. PANTHER pathway: an ontology-based pathway database coupled with data analysis tools. Methods Mol Biol 2009; 563: 123–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Karagiannis K, Zhang H, Dingerdissen H, Shamsaddini A, Wan Q et al. Human germline and pan-cancer variomes and their distinct functional profiles. Nucleic Acids Res 2014; 42: 11570–11588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res 2003; 13: 2129–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 1997; 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumder R, Vasudevan S. Structure-guided comparative analysis of proteins: principles, tools, and applications for predicting function. PLoS Comput Biol 2008; 4: e1000151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumder R, Vasudevan S, Nikolskaya AN. Protein functional annotation by homology. Methods Mol Biol 2008; 484: 465–490. [DOI] [PubMed] [Google Scholar]
- Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY et al. Comparative protein structure modeling using Modeller. Curr Protoc Bioinformatics 2006. Chapter 5: Unit 5.6. [DOI] [PMC free article] [PubMed]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 2004; 25: 1605–1612. [DOI] [PubMed] [Google Scholar]
- Temperini C, Innocenti A, Scozzafava A, Parkkila S, Supuran CT. The coumarin-binding site in carbonic anhydrase accommodates structurally diverse inhibitors: the antiepileptic lacosamide as an example and lead molecule for novel classes of carbonic anhydrase inhibitors. J Med Chem 2010; 53: 850–854. [DOI] [PubMed] [Google Scholar]
- Wan Q, Dingerdissen H, Fan Y, Gulzar N, Pan Y, Wu TJ et al. BioXpress: an integrated RNA-seq-derived gene expression database for pan-cancer analysis. Database 2015; 2015: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol 2010; 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkestock K, Edlund PO, Roeraade J. Electrospray ionization mass spectrometry as a tool for determination of drug binding sites to human serum albumin by noncovalent interaction. Rapid Commun Mass Spectrom 2005; 19: 1637–1643. [DOI] [PubMed] [Google Scholar]
- Safa AR. Identification and characterization of the binding sites of P-glycoprotein for multidrug resistance-related drugs and modulators. Curr Med Chem Anticancer Agents 2004; 4: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uga H, Kuramori C, Ohta A, Tsuboi Y, Tanaka H, Hatakeyama M et al. A new mechanism of methotrexate action revealed by target screening with affinity beads. Mol Pharmacol 2006; 70: 1832–1839. [DOI] [PubMed] [Google Scholar]
- Kindler HL. Pemetrexed in pancreatic cancer. Semin Oncol 2002; 29: 49–53. [DOI] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D et al. Circos: an information aesthetic for comparative genomics. Genome Res 2009; 19: 1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadal E, Olavarria E. Imatinib mesylate (Gleevec/Glivec) a molecular-targeted therapy for chronic myeloid leukaemia and other malignancies. Int J Clin Pract 2004; 58: 511–516. [DOI] [PubMed] [Google Scholar]
- Lee JL, Kim JY, Ryu MH, Kang HJ, Chang HM, Kim TW et al. Response to imatinib in KIT- and PDGFRA-wild type gastrointestinal stromal associated with neurofibromatosis type 1. Dig Dis Sci 2006; 51: 1043–1046. [DOI] [PubMed] [Google Scholar]
- de Groot JW, Plaza Menacho I, Schepers H, Drenth-Diephuis LJ, Osinga J, Plukker JT et al. Cellular effects of imatinib on medullary thyroid cancer cells harboring multiple endocrine neoplasia Type 2A and 2B associated RET mutations. Surgery 2006; 139: 806–814. [DOI] [PubMed] [Google Scholar]
- Delbaldo C. Pharmacokinetic-pharmacodynamics relationships of imatinib (Glivec). Therapie 2007; 62: 87–90. [DOI] [PubMed] [Google Scholar]
- Dewar AL, Farrugia AN, Condina MR, Bik To L, Hughes TP, Vernon-Roberts B et al. Imatinib as a potential antiresorptive therapy for bone disease. Blood 2006; 107: 4334–4337. [DOI] [PubMed] [Google Scholar]
- Xu L, Tong R, Cochran DM, Jain RK. Blocking platelet-derived growth factor-D/platelet-derived growth factor receptor beta signaling inhibits human renal cell carcinoma progression in an orthotopic mouse model. Cancer Res 2005; 65: 5711–5719. [DOI] [PubMed] [Google Scholar]
- Waller CF. Imatinib mesylate. Recent Results Cancer Res 2010; 184: 3–20. [DOI] [PubMed] [Google Scholar]
- Mendel DB, Laird AD, Xin X, Louie SG, Christensen JG, Li G et al. In vivo antitumor activity of SU11248, a novel tyrosine kinase inhibitor targeting vascular endothelial growth factor and platelet-derived growth factor receptors: determination of a pharmacokinetic/pharmacodynamic relationship. Clin Cancer Res 2003; 9: 327–337. [PubMed] [Google Scholar]
- O'Farrell AM, Foran JM, Fiedler W, Serve H, Paquette RL, Cooper MA et al. An innovative phase I clinical study demonstrates inhibition of FLT3 phosphorylation by SU11248 in acute myeloid leukemia patients. Clin Cancer Res 2003; 9: 5465–5476. [PubMed] [Google Scholar]
- Pietras K, Hanahan D. A multitargeted, metronomic, and maximum-tolerated dose "chemo-switch" regimen is antiangiogenic, producing objective responses and survival benefit in a mouse model of cancer. J Clin Oncol 2005; 23: 939–952. [DOI] [PubMed] [Google Scholar]
- Amino N, Ideyama Y, Yamano M, Kuromitsu S, Tajinda K, Samizu K et al. YM-359445, an orally bioavailable vascular endothelial growth factor receptor-2 tyrosine kinase inhibitor, has highly potent antitumor activity against established tumors. Clin Cancer Res 2006; 12: 1630–1638. [DOI] [PubMed] [Google Scholar]
- Ciardiello F, Caputo R, Bianco R, Damiano V, Pomatico G, De Placido S et al. Antitumor effect and potentiation of cytotoxic drugs activity in human cancer cells by ZD-1839 (Iressa), an epidermal growth factor receptor-selective tyrosine kinase inhibitor. Clin Cancer Res 2000; 6: 2053–2063. [PubMed] [Google Scholar]
- De Giorgi U. KIT mutations and imatinib dose effects in patients with gastrointestinal stromal tumors. J Clin Oncol 2007; 25: 1146–1147. [DOI] [PubMed] [Google Scholar]
- Gotlib J, Berube C, Growney JD, Chen CC, George TI, Williams C et al. Activity of the tyrosine kinase inhibitor PKC412 in a patient with mast cell leukemia with the D816V KIT mutation. Blood 2005; 106: 2865–2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer CJ. Emerging role of epidermal growth factor receptor inhibition in therapy for advanced malignancy: focus on NSCLC. Int J Radiat Oncol Biol Phys 2004; 58: 991–1002. [DOI] [PubMed] [Google Scholar]
- Medina PJ, Goodin S. Lapatinib: a dual inhibitor of human epidermal growth factor receptor tyrosine kinases. Clin Ther 2008; 30: 1426–1447. [DOI] [PubMed] [Google Scholar]
- Tevaarwerk AJ, Kolesar JM. Lapatinib: a small-molecule inhibitor of epidermal growth factor receptor and human epidermal growth factor receptor-2 tyrosine kinases used in the treatment of breast cancer. Clin Ther 2009; 31: 2332–2348. [DOI] [PubMed] [Google Scholar]
- Bulgaru AM, Mani S, Goel S, Perez-Soler R. Erlotinib (Tarceva): a promising drug targeting epidermal growth factor receptor tyrosine kinase. Expert Rev Anticancer Ther 2003; 3: 269–279. [DOI] [PubMed] [Google Scholar]
- Jiang W, Xie J, Varano PT, Krebs C, Bollinger JM Jr. Two distinct mechanisms of inactivation of the class Ic ribonucleotide reductase from Chlamydia trachomatis by hydroxyurea: implications for the protein gating of intersubunit electron transfer. Biochemistry 2010; 49: 5340–5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supuran CT. Carbonic anhydrases as drug targets—an overview. Curr Top Med Chem 2007; 7: 825–833. [DOI] [PubMed] [Google Scholar]
- Mincione F, Scozzafava A, Supuran CT. The development of topically acting carbonic anhydrase inhibitors as antiglaucoma agents. Curr Pharm Des 2008; 14: 649–654. [DOI] [PubMed] [Google Scholar]
- Barone FC, Barton ME, White RF, Legos JJ, Kikkawa H, Shimamura M et al. Inhibition of phosphodiesterase type 4 decreases stress-induced defecation in rats and mice. Pharmacology 2008; 81: 11–17. [DOI] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010; 38: e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.