Abstract
We describe two modifications that parallelize and reorganize caching in the well-known Greedy Equivalence Search (GES) algorithm for discovering directed acyclic graphs on random variables from sample values. We apply one of these modifications, the Fast Greedy Search (FGS) assuming faithfulness, to an i.i.d. sample of 1,000 units to recover with high precision and good recall an average degree 2 directed acyclic graph (DAG) with one million Gaussian variables. We describe a modification of the algorithm to rapidly find the Markov Blanket of any variable in a high dimensional system. Using 51,000 voxels that parcellate an entire human cortex, we apply the FGS algorithm to Blood Oxygenation Level Dependent (BOLD) time series obtained from resting state fMRI.
Introduction
High-dimensional data sets afford the possibility of extracting causal information about complex systems from samples. Causal information is now commonly represented by directed graphs associated with a family of probability distributions. Fast, accurate recovery is needed for such systems.
An acyclic directed graphical (DAG) model < G(V, E), P> consists of a directed acyclic graph, G, whose vertices v are random variables with directed edges, e, and a probability distribution, P, satisfying the Markov factorization for all assignments of values to variables in V having positive support:
| (1) |
where PA(a(v)) denotes a value assignment to each member of the set of variables with edges directed into v (the parents of v). The Markov Equivalence Class (MEC) of a DAG G is the set of all DAGs G having the same adjacencies as G and the same “v-structures”—substructures x → y ← z, where x and y are not adjacent in G.
Such graphs can be used simply as devices for computing conditional probabilities. When, however, a causal interpretation is appropriate, search for DAG models offers insight into mechanisms and the effects of potential interventions, finds alternatives to a priori hypotheses, and in appropriate domains suggests experiments. Under a causal interpretation of G, a directed edge x → y represents the proposition that there exist values of all variables in V\{x, y} such that if these variables were (hypothetically) fixed at those values (not: conditioned on those values), some exogenous variation of x would be associated with variation in y.
A causal interpretation is inappropriate for Pearson correlations of time series, because the correlations are symmetric and do not identify causal pathways leading from one variable to another; correlation searches will return not only an undirected edge (adjacency) between the intermediate variables in a causal chain, but also adjacencies for the transitive closure of a chain of causal connections. Causal interpretations are usually not correct for Markov Random Field (MRF) models, including those obtained for high dimensions by various penalized inverse covariance methods such as GLASSO and LASSO [1] [2], in part because like correlation graphs MRF graphs are undirected, and in the large sample limit specify an undirected edge between two variables that are conditionally independent but dependent when a third variable is conditioned on. This pattern of dependencies is characteristic of circumstances in which two conditionally independent variables are jointly causes of a third. It should be of interest therefore whether DAG search procedures can be speeded up to very high dimensions, with what accuracies. Ideally, one should be able to analyze tens of thousands of variables on a laptop in a work session, and be able to analyze problems with a million variables or more on a supercomputer overnight. Such a capability would be useful, for example, in cognitive neuroscience and in learning DAGs from high dimensional biomedical datasets.
Strategies
Various algorithmic strategies have been developed for searching data for DAG models. One strategy, incorporated in GES, iteratively adds edges starting with an empty graph according to maximal increases in a score, generally the BIC score [3] for Gaussian variables, and the BDeu score [4] for multinomial variables, although many other scores are possible, including modifications of the Fisher Z score [5] for conditional independence tests. The algorithms return a MEC of models, with the guarantee that if the distribution P is in the MEC for some DAG over the sample variables then asymptotically in probability the models returned have the highest posterior probability. There is no guarantee that on finite samples the model or models returned are those with the highest posterior probability, which is known to be an NP-hard problem [6].
The GES algorithm, as previously implemented, has trouble with large variable sets. Smith et al. gave up running GES on low dimensional problems (e.g., 50 variables and 200 datapoints) on the grounds that too much time was required for the multiple runs their simulations required [7].
Another area of interest for scaling up MEC search has been the search for Markov Blankets of variables, the minimal set of variables, not including a target variable t, conditional on which all other recorded variables are independent of t [9] [10] [11]. This has an obvious advantage for scalability; if the nodes surrounding a target variable in a large data set can be identified, the number of relations among nodes that need to be assessed is reduced. Since the literature has not suggested a way to estimate Markov Blankets using a GES-style algorithm, we provide one here. It is scalable for sparse graphs, and yields estimates of graphical structure about the target that are simple restrictions of the MEC of the graphical causal structure generating the data.
Fast Greedy Search
The elements of a GES search and relevant proofs, but no precise algorithm, were given by Chickering [12]; the basic idea is given by Meek [13]. Free public implementations of GES are available in the pcalg package in R and the Tetrad software suite. GES searches iteratively through the MECs of one edge additions to the current MEC, beginning with the totally disconnected graph on the recorded variables. At each stage of the forward phase all alternative one-edge additions are scored; the edge with the best score is added and the resulting MEC formed, until no more improvements in the score can be made by a single edge addition; in the backward stage, the procedure is repeated but this time removing edges, starting with the MEC of the forward stage, until no more improvements in the score can be made by single edge deletions. For multinomial distributions, the algorithm has two intrinsic tuning parameters: a “structure prior” that penalizes for the number of parents of a variable in a graph, and a “sample prior” required for the Dirichlet prior distribution. For Gaussian distributions, the algorithm has one tuning parameter, the complexity penalty in the BIC score. The original BIC score has a penalty proportional to the number of variables, but without changing asymptotic convergence to the MEC of DAGs with the maximum posterior probability, this penalty can be increased or decreased (to a positive fraction) to control false positive or false negative adjacencies. Increasing the penalty forces sparser graphs on finite samples.
It is important to note that the proof of correctness of GES assumes causal sufficiency—that is, it assumes that a common cause of any pair of variables in the set of variables over which the algorithm reasons is in the set of variables. That is, if the algorithm is run over a set of variables X, and x and y are in X, and x ← L → y for some variable L, then L is in X as well. Thus, GES assumes that there are no unmeasured common causes. If there are unmeasured common causes, then GES will systematically introduce extra edges. GES also assumes that there are no feedback loops that would have to be represented by a finite cyclic graph.
The FGS procedure uses a similar strategy with some important differences. First, in accord with (1), the scores are decomposable by graphical structure, so that the score for an entire graph may be the sum of scores for fragments of it. The updating of the score for a potential edge addition is done by caching scores of potential edge additions from previous steps, and where a new edge addition will not (for graphical reasons) alter the score of a fragment of the graph, uses the cached score, saving time in graph scoring. This requires an increase in memory, since the local scores for the relevant fragments must be stored.
Second, each step of the FGS procedure may be parallelized. For most of the running time of the algorithm, a parallelization can be given for which all processors are used, even on a large machine, radically improving the running time of the algorithm.
Third, if the BIC score is used, the penalty for this score can be increased, forcing estimation of a sparser graph. The sparser the graph, the faster the search returns but at the risk of false negative adjacencies.
Fourth, one can make the assumption that an edge x → y, where x and y are uncorrelated, is not added to the graph at any step in the forward search. This is a limited version of the so-called “faithfulness” assumption [5] and allows a search procedure to be speeded up considerably at the expense of allowing graphs to violate the Markov factorization, (1), for the conditional dependence and independence relations estimated from the sample. This weak faithfulness assumption can be included in FGS, or not. For low dimensional problems where speed is not an issue, there is no compelling reason to assume this kind of one-edge faithfulness, but in high dimensions the search can be much faster if the assumption is made. There are two situations in which assuming one-edge faithfulness leads to incorrect graphs. The first is perfectly canceling paths. Consider a model A → B → C → D, with A → D, where the two paths exactly cancel. FGS without the weak faithfulness assumption will (asymptotically) identify the correct model; with that assumption the A → D edge will go missing. In this regard, FGS with the assumption behaves like any algorithm whose convergence proof assumes faithfulness, such as the PC algorithm [5], although only with regard to single-edge violations of faithfulness. Faithfulness conditions for perfectly canceling paths have been shown to hold measure 1 for continuous measures over the parameter spaces of either discrete or continuous joint probability distributions on variable values, but in estimates of joint distributions from finite samples faithfulness can be violated for much larger sets of parameter values [5]. We will use this one-edge faithfulness assumption in what follows.
FGS is implemented, with publicly accessible code, in the development branch of https://github.com/cmu-phil/tetrad. Pseudo-code is given in the supplement to this paper.
The continuous BIC score for the difference between a graph with a set of parents Z of y, Z → y, and a graph with an additional parent x of Y, Z U {x} → y, is BIC(Z ∪ {x}, y) –BIC (Z, y), where for an arbitrary set X of variables, BIC(X, y) = 2 L − c k ln n, with L the log likelihood of the linear model X → y with Gaussian residual (equal to −n/2 ln s2 + C), k = |X| + 1, n is the sample size, s2 is the variance of the residual of y regressed on X, and c is a constant (“penalty discount”) used to increase or decrease the penalty of the score. Chickering [12] shows this score is positive if it is not the case that (x_||_y | Z) and negative if x _||_ y | Z.
The discrete BDeu score is as given by Chickering [12]. We use differences of these scores as for the continuous case above. We use a modified structure prior, as described in Section 4.
The correctness of the implementation from d-separation [14] can be tested using Chickering’s “locally consistent scoring criterion” theorem. Using d-separation, any DAG can be consistently scored by +1 if it is not the case that dsep(x, y | Z) and −1 if dsep(x, y | Z). This score may be used to run FGS directly from a DAG using d-separation facts alone and can be used to check the correctness of the implementation of the algorithm without using data and the difference scores. Running FGS using the graph score should recover the MEC of G. We have tested the implementation in this way on more than 100,000 random DAGs with 10 nodes and 10 or 20 edges without an error.
Multiple Samples with FGS
Appending data sets can lead to associations among variables that are independent in each data set. For that reason, the IMaGES algorithm [15] runs each step of GES separately on each data set, and updates the inferred MEC by adding (or in the backwards phase, deleting) the edge whose total score summed over the scores from the several data sets is best. The same strategy can be used with FGS. The scoring over the separate data sets can of course be parallelized to reduce runtime almost in proportion to the number of data sets, but we have not yet done so in our published software.
Simulations with BIC and BDeu
Our simulations generated data from directed acyclic graphs parameterized with linear relations and independent, identically distributed Gaussian noise, or with categorical, three valued variables, parameterized as multinomial. For continuous distributions, all coefficients were sampled uniformly from (−1.5, −.5) ∪ (.5, 1.5). For categorical variables, for each cell of the conditional probability table, random values were drawn uniformly from (0, 1) and rows were normalized. All simulations were conducted using Pittsburgh Supercomputing Center (PSC) Blacklight computer. The number of processors (hardware threads) used varied from 2 (for 1000 variables) to 120 (for one million variables). All simulations used samples of size 1000. Where repeated simulations were done for the same dimension, a new graph, parameterization, and sample were used for each trial. The BIC penalty was multiplied by 4 in all searches with continuous data. The BDeu score used is as by Chickering [12], except that we used the following structure prior (for each vertex in a possible DAG):
| (2) |
where v is the number of variables in a DAG, p is the number of parents of that vertex and e is a prior weight, by default equal to 1. In (2), whether a node Xi has a node Xj as a parent is modeled as a Bernoulli trial with probability pj of Xj being a parent of Xi and (1 – pj) of Xj not being a parent of Xi. In simulations we have found this structure prior yields more accurate estimates of the DAG than the structure prior suggested by Chickering (2002). We used a sample prior of 1.
For continuous data, we have run FGS with the penalized BIC score defined above for (1) DAGs with 1000 nodes and 1000 or 2000 edges, for 100 repetitions, (2) DAGs with 30,000 nodes and 30,000 or 60,000 edges, for 10 repetitions, and (3) a DAG with 1,000,000 nodes and 1,000,000 edges, in all cases assuming the weak faithfulness condition noted above. For discrete data, we have run FGS with the BDeu only for circumstances (1) and (2) above. Random graphs were generated by constructing a list of variables and then adding forward edges by random iteration, avoiding cycles. The node degree distribution returned by the search from samples from random graphs with 30,000 nodes and 30,000 or 60,000 edges generated by our method is shown in Figure 1. Continuous results are tabulated in Table 1; discrete results are tabulated in Table 2. Where M1 is the true MEC of the DAG and M2 is the estimated MEC returned by FGS, precision for adjacencies was calculate as TP/(TP + FP); recall as TP/(TP + FN), where TP is the number of adjacencies shared by the true (M1) and estimated (M2) MECs of the DAG„ FP is the number of adjacencies in M2 but not in M1, and FN is the number of adjacencies in M1 but not in M2.
Fig 1.
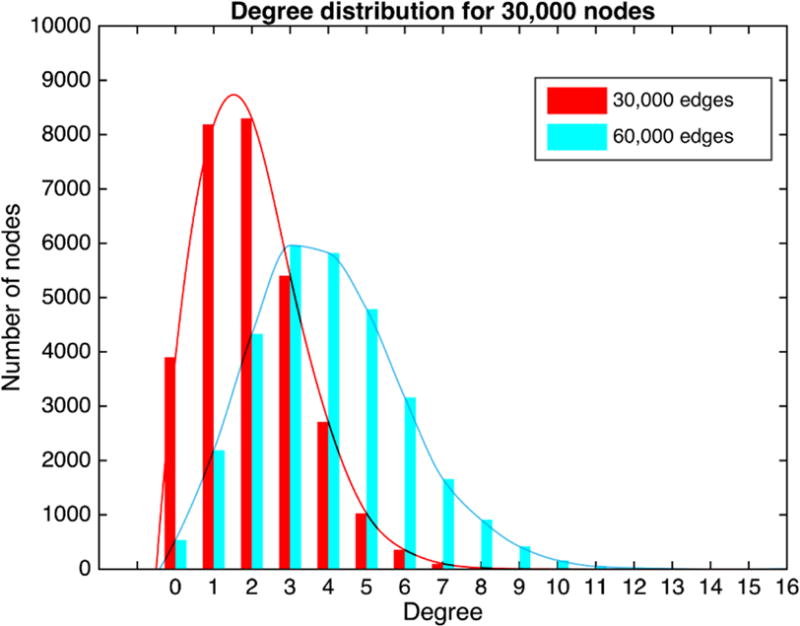
Node degree distribution for simulated graphs with 30,000 nodes and 30,000 or 60,000 edges
Table 1.
Accuracy and time results for continuous data.
| # Nodes | # Edges | # Rep | Adj Prec | Adj Rec | Arr Prec | Arr Rec | # Processors | Elapsed |
|---|---|---|---|---|---|---|---|---|
| 1000 | 1000 | 100 | 98.92% | 94.77% | 98.92% | 90.05% | 2 | 1.2 s |
| 1000 | 2000 | 100 | 98.43% | 88.04% | 96.27% | 85.74% | 4 | 8.5 s |
| 30,000 | 30,000 | 10 | 99.77% | 94.60% | 99.04% | 89.97% | 120 | 53.5 s |
| 30,000 | 60,000 | 10 | 99.81% | 86.72% | 99.23% | 84.47% | 120 | 3.4 m |
| 1,000,000 | 1,000,000 | 1 | 93.90% | 94.83% | 83.11% | 90.57% | 120 | 11.0 h |
Table 2.
Accuracy and time results for discrete data.
| # Nodes | # Edges | # Rep | Adj Prec | Adj Rec | Arr Prec | Arr Rec | # Processors | Elapsed |
|---|---|---|---|---|---|---|---|---|
| 1000 | 1000 | 100 | 99.65% | 74.98% | 89.52% | 43.79% | 2 | 2.1 s |
| 1000 | 2000 | 100 | 48.48% | 82.70% | 82.70% | 28.41% | 4 | 2.2 s |
| 30,000 | 30,000 | 10 | 99.96% | 72.18% | 92.46% | 37.97% | 120 | 2.6 m |
| 30,000 | 60,000 | 10 | 99.97% | 45.85% | 86.39% | 23.89% | 120 | 3.2 m |
Arrowhead precision and recall were calculated in a similar way. An arrowhead was taken to be in M1 and M2 for each variable A and B such that A → B in both M1 and M2, and an arrowhead was taken to be in one MEC but not the other for each variable A and B such that A → B in one but A ← B in the other, or A—B in the other, or A and B are not adjacent in the other.
In all cases, we generate a DAG G randomly with the given number of nodes and edges, parameterize it as a linear, Gaussian structural equation model, run FGS as described using the penalized BIC score with BIC penalty multiplied by 4, and calculate average precision and recall for adjacencies and arrowheads with respect to the MEC of G. We record average running time. Times are shown in hours (h), minutes (m), or seconds (s).
In all cases, we generated a DAG G randomly with the given number of nodes and edges, parameterize it as a multinomial model with 3 categories for each variable, ran FGS as described using the penalized BDeu score with sample prior 1 and structure prior 1, and calculated average precision and recall for adjacencies and arrowheads with respect to the MEC of G. We record average running time.
For time comparisons (accuracies were the same), searches were also run on a 3.1 GHz MacBook Pro laptop computer with 2 cores and 4 hardware threads, for 1000 and 30,000 variable continuous problems. Runtime for 1000 nodes with 1000 edges was 3.6 s. 30,000 variable problems with 30,000 edges required 5.6 minutes. On the same machine, 1000 variable categorical search with 2000 edges required 3.9 seconds, and 30,000 variables with 60,000 edges required (because of a 16 GB memory constraint) 40.3 minutes.
The results in Tables 1 and 2 use the one-edge faithfulness assumption, above. Using the implementation described in the appendix, this assumption does not need to be made if time is not of the essence,. The running time is, however, considerably longer, as shown in Table 3 for the case of 30,000 node graphs. Accuracy is not appreciably better.
Table 3.
Accuracy and time results for the 30,000 node cases, for continuous data, not assuming one-edge faithfulness
| # Nodes | # Edges | # Rep | Adj Prec | Adj Rec | Arr Prec | Arr Rec | # Processors | Elapsed |
|---|---|---|---|---|---|---|---|---|
| 30,000 | 30,000 | 1 | 99.75% | 94.55% | 98.88% | 89.91% | 120 | 54.8 s |
| 30,000 | 60,000 | 1 | 99.83% | 87.74% | 99.32% | 85.63% | 120 | 8.9 m |
Markov Blanket search
A number of algorithms have been proposed to calculate the Markov Blanket of an arbitrary target variable t without calculating the graphical structure over the nodes. Other algorithms attempt to identify the graphical structure over these nodes (that is, the parents of t, the children of t, and the parents of children of t). (An excellent overview of the field as of 2010 is given by Aliferis et. al [10].) A simple restriction of FGS, FGS-MB, finds the Markov Blanket of any specified variable and estimates the graph over those variables including all orientations that FGS would estimate for edges in that graph.
For the Markov blanket search, one calculates a set A of adjacencies x–y about t as follows. First, given a score I(a, b, C), one finds the set of variables x such that I(x, t, ∅) > 0 and adds x–t to A for each x found. Then for each such variable x found, one finds the set of variables y such that I(y, x, ∅) > 0 and adds y–x to A for each such y found. The general form of this calculation is familiar for Markov blanket searches, though in this case it is carried out using a score difference function. One then simply runs the rest of FGS, restricting adjacencies used in the search to the adjacencies in A in the first pass and then marrying parents as necessary and re-running the backward search to get the remaining unshielded colliders, inferring additional orientations as necessary. The resulting graph may then simply be trimmed to the variables in A and returned.
That this procedure returns the same result as running FGS over the entire set of variables and then trimming it to the variables in A is implied by the role of common causes in the search, assuming one-edge faithfulness. A consists of all variables d-connected to the target or d-connected to a variable d-connected to the target. But any common cause of any two variables in this the Markov blanket of t is immediately in this set and so will be taken account of by FGS-MB. Trimming the search over A to the nodes that are adjacents of t or spouses of t will then produce the same result (and the correct Markov blanket) as running FGS over all of the (causally sufficient) variables and then trimming that graph in the same way to the Markov blanket of t.
For example, consider how FGS-MB is able to orient a single parent of t. Let {x, y}->w->r->t be the generating DAG. The step above will form a clique over x, y, w, r, and t (except for the edge x—y). It will then orient x->w<-y, either directly or by shielding in the forward step and then removing the shield and orienting the collider in the backward step. This collider orientation will then be propagated to t by the Meek rules, yielding graph {x, y}->w->r->t. This graph will then be trimmed to r->t, the correct restriction of the FGS output to the Markov blanket of t as judged by FGS.
As with constraint-based Markov blanket procedures, FGS-MB can be modified to search arbitrarily far from the target and can trim the graph in different ways, either to the Markov blanket, or the adjacents and adjacents of adjacents, or more simply, just to the parents and children of the target. One only needs to construct an appropriate set of adjacencies over which FGS should search and then run the rest of the steps of the FGS algorithm restricted to those adjacencies. Even for several targets taken together, the running time of FGS-MB over the appropriate adjacency set will generally be much less than the running time of the full FGS procedure over all variables and can usually be carried out on a laptop even for very large datasets.
To illustrate that FGS-MB can run on large data sets on a laptop, Tables 4 and 5 show running times and average counts for FGS-MB for random targets selected from random models with 1000 nodes and 1000 or 2000 edges, 30,000 nodes with 30,000 or 60,000 edges, for continuous and discrete simulations, in the same style as for Tables 1 and 2. For the continuous case, a simulation with 1,000,000 nodes and 1,000,000 edges is included as well. Because the number of variables in the Markov Blanket varies considerably, accuracies are given in average absolute numbers of edges rather than in percentages.
Table 4.
Average accuracy and time results for FGS-MB run on continuous data in simulation.
| # Nodes | # Edges | # Rep | # Correct Adj | # Correct Arr | # FP Adj | # FN Adj | Elapsed |
|---|---|---|---|---|---|---|---|
| 1000 | 1000 | 100 | 3.1 | 3.1 | 0.0 | 0.2 | 99 ms |
| 1000 | 2000 | 100 | 7.5 | 7.1 | 0.5 | 2.4 | 3.7 s |
| 30,000 | 30,000 | 10 | 5.3 | 4.7 | 0.0 | 0.3 | 418 ms |
| 30,000 | 60,000 | 10 | 5.4 | 5.4 | 0.0 | 1.8 | 13.2 s |
| 1,000,000 | 1,000,000 | 1 | 2.0 | 2.0 | 0.0 | 1.0 | 42.9 s |
Table 5.
Accuracy and time results for FGS-MB run on discrete data in simulation.
| # Nodes | # Edges | # Rep | # Correct Adj | # Correct Arr | # FP Adj | # FN Adj | Elapsed |
|---|---|---|---|---|---|---|---|
| 1000 | 1000 | 100 | 3.1 | 3.1 | 0.0 | 0.2 | 99 ms |
| 1000 | 2000 | 100 | 3.0 | 2.3 | 0.1 | 6.3 | 65 ms |
| 30,000 | 30,000 | 10 | 2.2 | 2.0 | 0.0 | 0.6 | 1.5 s |
| 30,000 | 60,000 | 10 | 5.3 | 4.3 | 0.0 | 6.8 | 3.2 m |
For each row of the table, a single DAG was randomly generated and parameterized as a linear, Gaussian structural equation model as described in the text. Then random nodes in the model were selected and their Markov blanket MECs assessed using the FGS-MB algorithm, using the penalized BIC score with BIC penalty multiplied by 4; these were then compared to the restriction of the true MEC of the generating DAG, restricted to the Markov blanket variables. Average number of correct adjacencies (# Correct Adj), correctly oriented edges (# Correct Arr), number of false positive adjacencies (# FP Adj) and number of false negative adjacencies (# FN Adj) are reported. Times are shown in milliseconds (ms) and seconds (s). The average number of mis-directed edges for each row of Table 4 is given by the difference of the Correct Adjacency value and the Correct Arrowhead values.
For each row of the table, a single DAG was generated and parameterized as a Bayes Net with multinomial distributed categorical variables each taking 3 possible values. Conditional probabilities were randomly selected from a uniform [0, 1] distribution for each cell and the rows in the data table normalized. Then random nodes in the model were selected and their Markov blanket MECs assessed using the FGS-MB algorithm, using the penalized BIC score with BIC penalty multiplied by 4; these were then compared to the restriction of the true MEC of the generating DAG, restricted to the Markov blanket variables. Average number of correct adjacencies (# Correct Adj), correctly oriented edges (# Correct Arr), number of false positive adjacencies (# FP Adj) and number of false negative adjacencies (# FN Adj) are reported. Altogether for each row in Table 5, 100 nodes were used to calculate the averages shown. Times are shown in milliseconds (ms) and seconds (s).
A Voxel Level Causal Model of the Resting State Human Cortex
To provide an empirical illustration of the algorithm, we applied FGS to all of the cortical voxels in a resting state fMRI scan (approximately 51,000 voxels). Details of measurements and data preparation are given in the supplement. Because of the density of cortical connections, we applied the algorithm at penalties 40, 20, 10, and 4. Lower penalty analyses were not computationally feasible. All runs were done on the Pittsburgh Supercomputing Center (PSC) Bridges computer, using 24 nodes. Runtime for the penalty 4 FGS search was 14 hours.
Tables 6 and 7 show the percentage of adjacencies and directed edges retained between each penalty run and the absolute numbers of adjacencies and directed edges returned at each penalty. Note that the great majority of adjacencies are directed.
Table 6.
Adjacencies retained at decreasing BIC penalty from resting state FGS searches over all cortical voxels.
| Penalty | # Adjacencies | % in 40 | % in 20 | % in 10 | % in 4 |
|---|---|---|---|---|---|
| 40 | 6,013 | 1.00 | 0.99 | 0.96 | 0.93 |
| 20 | 16,722 | 1.00 | 0.95 | 0.91 | |
| 10 | 40,982 | 1.00 | 0.90 | ||
| 4 | 127,533 | 1.00 |
Table 7.
Directed edges retained at decreasing BIC penalty from resting state FGS searches over all cortical voxels.
| Penalty | # Directed Edges | % in 40 | % in 20 | % in 10 | % in 4 |
|---|---|---|---|---|---|
| 40 | 5,850 | 1.00 | 0.99 | 0.96 | 0.93 |
| 20 | 16,453 | 1.00 | 0.95 | 0.91 | |
| 10 | 39,999 | 1.00 | 0.89 | ||
| 4 | 127,281 | 1.00 |
For the penalty 4 run, the distribution of path lengths (in voxel units) between voxels (not counting zero lengths) is almost Gaussian (Fig 2). The distribution of total degrees is inverse exponential as expected (Fig 3). Fig 4 illustrates the parents and children of a single voxel.
Fig 2.
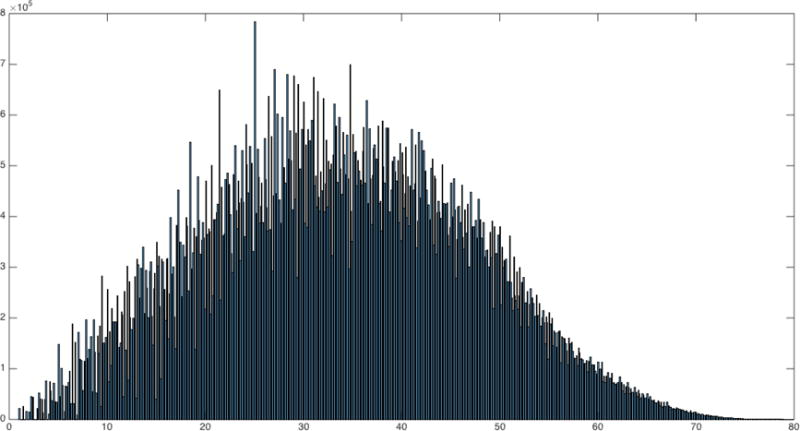
Distribution of path distances (in voxel units) in penalty 4 run
Fig 3.
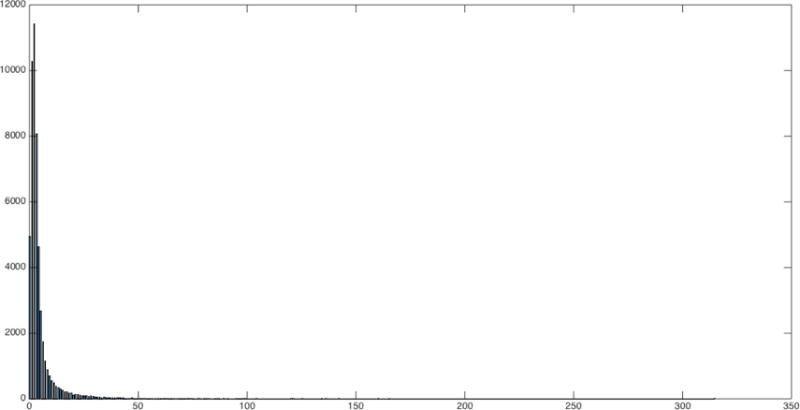
Histogram of total degrees in penalty 4 run
Fig 4.
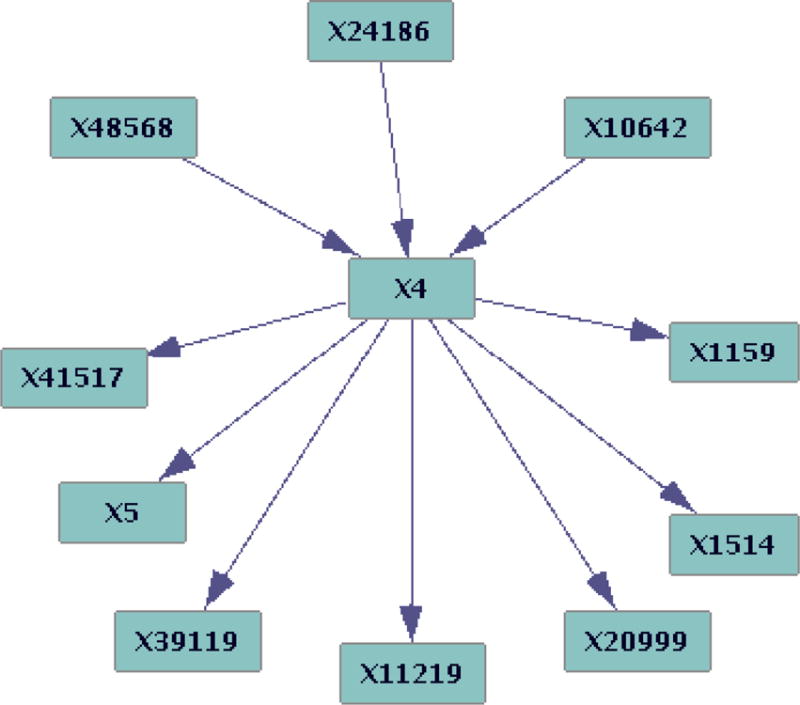
Parents and children of a voxel
Discussion
For the sparse models depicted in Tables 1 and 2, for FGS, we see high precisions throughout for both adjacencies and arrowheads, although for the million node case with continuous models precision suffers, and for the discrete models direction of edge precision suffers for the denser models. For discrete models, sample sizes are on the low side for the conditional probability tables we have estimated, and it is not surprising that recall is comparably low. If we had not assumed the weak faithfulness condition, run times would have been considerably longer; for small models, recall would have been higher, though for large models our experience suggests they would have been approximately the same.
Tables 3 and 4, for FGS-MB, show excellent and fast recovery of Markov blankets for the sparse continuous case, less recall for the denser continuous case, and less recall than that for the discrete cases. The runtime for FGS-MB, for single targets, is a fraction of the runtime for FGS for all variables on a large data set—for the continuous, sparse, million node case, it is reduced from 11 hours to 42.9 seconds for single runs of those algorithms. If only the structure around a target node is needed, or structure along a path, it makes more sense to use FGS-MB or some simple variant of it than to estimate the entire graph and extract the portion of it that is needed. If a great deal of the structure is needed, it may make sense to estimate the entire graph.
Biological systems tend to be scale-free, implying that there are nodes of very high degree. The complexity of the search increases with the number of parents, and recall accuracy decreases. FGS deals better with systems, as in scale-free structures, in which high-degree nodes are parents (or “hubs”) rather than children of their adjacent nodes. If prior knowledge limits the number of parents of a variable, FGS can deal with high-degree nodes by limiting the number of edges oriented as causes of a given node.
These simulations do not address cases in which both categorical and continuous variables occur in datasets. Sedgewick et al. have proposed in such cases to first estimate an undirected graph using a pseudo-likelihood and then pruning and directing edges [16]. It seems worth investigating whether such a mixed strategy proves more accurate than discretizing all variables and using FGS; it would be better still to have a suitable likelihood function for DAG models with mixed variable types.
The fMRI application only illustrates possibilities. In practice, fMRI measurements, even in the same subject, may shift voxel locations by millimeters. Voxel identification across scans is therefore unreliable, which at the voxel level of resolution means that quite different DAGs will be recovered from different scans. Usually in fMRI analyses, clusters of several hundred voxels (regions of interest) are formed based on anatomy or correlations using any one of a great many clustering algorithms, and connections are estimated from correlations of the average BOLD signals from each cluster. FGS at the voxel level offers the prospect of using causal connections among voxels to build supervoxel regions whose estimated effective connections are stable across scans, a strategy we are currently investigating.
Neural signaling connections in the brain have both feedback and unrecorded common causes that are not captured by any present methods at high dimensions, although low dimensional algorithms have been developed for these problems [5]. It seems important to investigate the possibility of scaling up and/or modifying these algorithms for high dimensional problems, in part through parallelization, and investigating their accuracies.
Supplementary Material
Acknowledgments
We thank Gregory Cooper for helpful advice and Russell Poldrack for data.
References
- 1.Hastie T, Tibshirani R, Friedman J, Franklin J. The elements of statistical learning: data mining, inference and prediction. The Mathematical Intelligencer. 2005;27(2):83–85. [Google Scholar]
- 2.Friedman J, Hastie T, Tibshirani R. Sparse inverse covariance estimation with the graphical lasso. Biostatistics. 2008;9(3):432–441. doi: 10.1093/biostatistics/kxm045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schwarz G. Estimating the dimension of a model. Ann Stat. 1978;6(2):461–464. [Google Scholar]
- 4.Chickering DM. Optimal structure identification with greedy search. J Mach Learn Res. 2003;3:507–554. [Google Scholar]
- 5.Spirtes P, Glymour CN, Scheines R. Causation, prediction, and search. 2nd. Vol. 2000 MIT press; 2000. [Google Scholar]
- 6.Chickering DM, Heckerman D, Meek C. Large-sample learning of Bayesian networks is NP-hard. J Mach Learn Res. 2004;5:1287–1330. [Google Scholar]
- 7.Smith SM, Miller KL, Salimi-Khorshidi G, Webster M, Beckmann CF, Nichols TE, et al. Network modelling methods for FMRI. Neuroimage. 2011;54(2):875–891. doi: 10.1016/j.neuroimage.2010.08.063. [DOI] [PubMed] [Google Scholar]
- 8.Ramsey J, Sanchez-Romero R, Glymour C. Non-Gaussian methods and high-pass filters in the estimation of effective connections. Neuroimage. 2013;84:986–1006. doi: 10.1016/j.neuroimage.2013.09.062. [DOI] [PubMed] [Google Scholar]
- 9.Aliferis CF, Tsamardinos I, Statnikov A. HITON: a novel Markov Blanket algorithm for optimal variable selection. AMIA Annu Symp Proc. 2003;2003:21. [PMC free article] [PubMed] [Google Scholar]
- 10.Aliferis CF, Statnikov A, Tsamardinos I, Mani S, Koutsoukos XD. Local causal and markov blanket induction for causal discovery and feature selection for classification part i: Algorithms and empirical evaluation. J Mach Learn Res. 2010;11:171–234. [Google Scholar]
- 11.Aliferis CF, Statnikov A, Tsamardinos I, Mani S, Koutsoukos XD. Local causal and markov blanket induction for causal discovery and feature selection for classification part ii: Analysis and extensions. J Mach Learn Res. 2010;11:235–284. [Google Scholar]
- 12.Chickering DM. Learning Equivalence Classes of Bayesian-Network Structures. J Mach Learn Res. 2002;2:445–498. [Google Scholar]
- 13.Meek C. Causal inference and causal explanation with background knowledge. Proceedings of the Eleventh Conference on Uncertainty in Artificial Intelligence. 1995;1995:403–410. [Google Scholar]
- 14.Pearl J. Probabilistic Reasoning in Intelligent Systems: Networks of Plausible Inference. 1st. San Francisco: Morgan Kaufmann Publishers Inc; 1988. [Google Scholar]
- 15.Ramsey JD, Hanson SJ, Hanson C, Halchenko YO, Poldrack RA, Glymour C. Six problems for causal inference from fMRI. Neuroimage. 2010;49(2):1545–1558. doi: 10.1016/j.neuroimage.2009.08.065. [DOI] [PubMed] [Google Scholar]
- 16.Sedgewick AJ, Shi I, Donovan RM, Benos PV. Learning mixed graphical models with separate sparsity parameters and stability-based model selection. BMC Bioinformatics. 2016;17 doi: 10.1186/s12859-016-1039-0. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsamardinos I, Aliferis CF, Statnikov AR, Statnikov E. Algorithms for Large Scale Markov Blanket Discovery. Proc Int Fla Al Res Soc Conf. 2003 May;:2. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


