Abstract
Currently, antiviral drugs that target specific viral protein functions are available for the treatment of influenza; however, concern regarding the emergence of drug-resistant viruses is warranted, as is the urgent need for new antiviral targets, including non-viral targets, such as host cellular factors. Viruses rely on host cellular functions to replicate, and therefore a thorough understanding of the roles of virus-host interactions during influenza virus replication is essential to develop novel anti-influenza drugs that target the host factors involved in virus replication. Here, we review recent studies that used several approaches to identify host factors involved in influenza virus replication. These studies have permitted the construction of an interactome map of virus-host interactions in the influenza virus life cycle, clarifying the entire life cycle of this virus and accelerating the development of new antiviral drugs with a low propensity for the development of resistance.
Introduction
Influenza viruses cause annual epidemics and recurring pandemics, such as the Spanish influenza in 1918–1919, the Asian influenza in 1957, the Hong Kong influenza in 1968, and the swine-origin H1N1 2009 pandemic influenza in 2009; these outbreaks have a huge impact on public health and the global economy [1]. In addition, recent sporadic human infections with H5N1 and H7N9 avian influenza viruses have raised concerns regarding the pandemic potential of these viruses [2-5]. Currently, two classes of FDA-approved compounds are available for prophylaxis and treatment of influenza virus infection: M2 ion channel inhibitors (amantadine, rimantadine) and neuraminidase (NA) inhibitors (oseltamivir, zanamivir) [6,7]. In addition, several new antiviral compounds have been approved for human use in some countries, including two NA inhibitors (peramivir and laninamivir) [8-11], and a viral polymerase inhibitor [favipiravir (T-705)] [12-14]. However, a major problem concerning anti-influenza drugs that target viral proteins is the frequent emergence of drug-resistant viruses. Indeed, most human H3N2 and H1N1, including pandemic 2009 H1N1, influenza viruses are now resistant to amantadine/rimantadine [15,16], and oseltamivir-resistant H1N1 viruses have emerged in many countries frequently [17]. Therefore, concern regarding the emergence of drug-resistant viruses is clearly warranted, as is the urgent need for new antiviral targets, including non-viral targets, to treat influenza virus infection while minimizing the emergence of drug resistance.
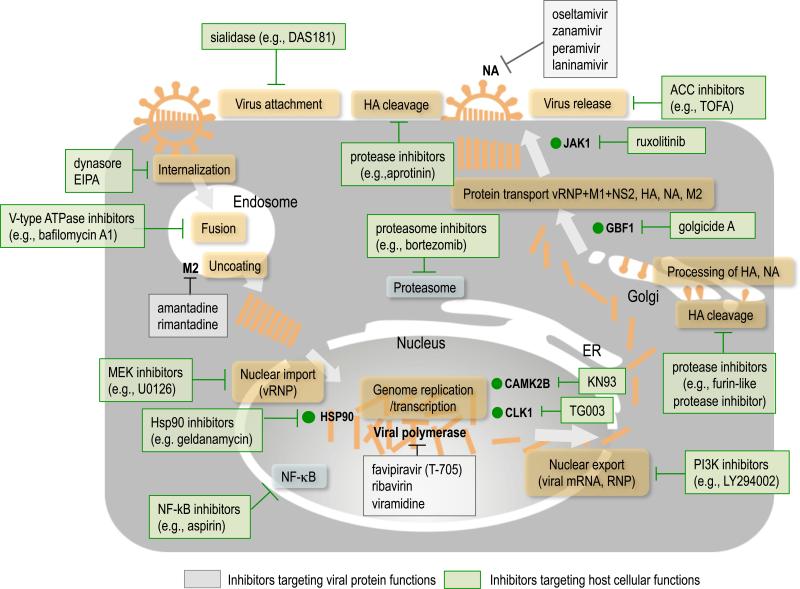
Host factors have been in the spotlight recently as potential antiviral targets to overcome the problems described above. Emergence of resistance could be less frequent when host factors are targeted compared with directly targeting viral proteins. Recently, several drugs targeting host factors identified to be important for virus replication have been developed. For example, the antiretroviral drug maraviroc, which was approved by the FDA in 2007, targets C-C chemokine receptor type 5 (CCR5), a co-receptor for human immunodeficiency virus type 1 (HIV-1), preventing HIV-1 entry [18]. Another example is alisporivir, a cyclophilin inhibitor, which was under clinical evaluation for the treatment of chronic hepatitis C [19,20]; in 2012, however, the FDA halted the clinical trials due to the possible side effect of pancreatitis. Alisporivir inhibits the function of cyclophilin A, which has an essential role in hepatitis C virus (HCV) replication and virus production [21]. In the case of influenza, the sialidase DAS181 (Fludase), which is currently in clinical trials, targets influenza virus receptors, sialic acids, and prevents viral attachment via removal of the sialic acids from the epithelial cells of the respiratory tract (Figure 1) [22-27]. Several other compounds, including protease inhibitors, block cellular proteases, which mediate the HA cleavage that is required for membrane fusion between the viral envelope and the endosomes [28-34], the MEK inhibitor U0126, NF-κB inhibitors such as acetylsalicylic acid (known as aspirin), and agonists of sphingosine-1-phosphate receptors AAL-R, all of which target host cellular functions involved in influenza virus replication, have been studied to explore their potential as new antiviral drugs against influenza (Figure 1) [26,35-41]. In addition, combination therapy with a protease inhibitor and conventional anti-influenza drugs has been recently tested in vitro [42].
Figure 1.
A schematic diagram of compounds that inhibit influenza virus replication and their known or putative points of action. There are two types of compound: one targets viral protein functions and the other targets host cellular functions. Examples of the former include M2 ion channel inhibitors (e.g., amantadine, rimantadine), NA inhibitors (e.g., oseltamivir, zanamivir, peramivir, and laninamivir) and viral polymerase inhibitors (e.g., favipiravir, rivavirin, and viramidine). Examples of the latter include sialidase (e.g., DAS181) [22-27], dynamin inhibitors (e.g., dynarose) [81], micropinocytosis inhibitors (e.g., EIPA) [81], MEK (MAPK/ERK kinase) inhibitor (e.g., U0126) [35,36], V-type ATPase (vacuolar-type H+ -ATPase) inhibitors (e.g., bafilomycin A) [82], protease inhibitors (e.g., aprotinin [28], which inhibits proteases that cleave cell surface HA proteins that have a single arginine at their cleavage site, and furin-like protease inhibitors, including peptidemimetics derived from decanoylated basic tetrapeptides, such as decRVKR chloromethylketone, which inhibit the HA cleavage of highly pathogenic H5 and H7 viruses [32-34] in the trans-Golgi, resulting in the inhibition of membrane fusion between the viral envelope and the endosomes), Hsp90 inhibitors (e.g., geldamycin) [83], NF-κB inhibitors (e.g., aspirin) [37], PI3K (phosphatidylinositol 3-kinase) inhibitors (e.g., LY294002) [84], ACC (acetyl-CoA carboxylase-α) inhibitors (e.g., TOFA) [85], and proteasome inhibitors (e.g., bortezomib) [86]. Compounds targeting host factors that have been shown to be involved in influenza virus replication in recent RNAi-based screens are also shown (i.e., KN93, TG003, ruxolitinib, and golgicide A) [43,47,71].
The advent of the possibility to target host factors involved in virus replication places us at the threshold of a new era of antiviral drug development. However, to explore targets for antiviral therapy, a systematic understanding of the mechanisms of influenza virus replication is essential. Recent efforts to identify host factors involved in influenza virus replication have provided the missing pieces of the virus-host interaction map of the influenza virus life cycle, helping us to move toward the development of host-targeting antiviral drugs. Here, we review the recent progress in the identification of host factors involved in influenza virus replication and the latest efforts to develop antiviral drugs that target host factors.
Strategies to identify host factors involved in influenza virus replication
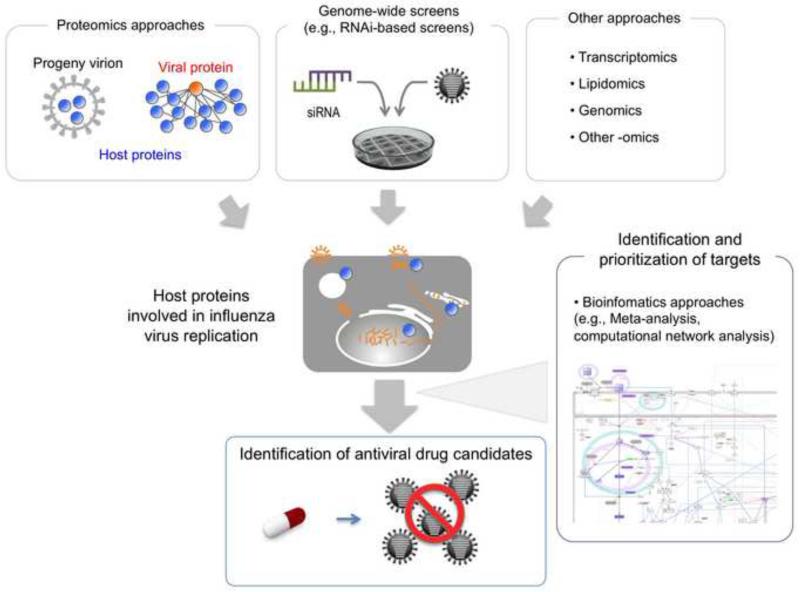
Recent technological advances have made it possible for researchers to take various approaches to identifying host factors involved in the life cycles of several viruses, including HIV-1, HCV, Dengue virus, and West Nile virus, as well as influenza virus [43-54]. For influenza virus, several strategies have been applied to the search for host factors involved in viral replication, as described below (Figure 2).
Figure 2.
Overview of the strategies used to explore antiviral drugs targeting host factors involved in influenza virus replication. Various approaches, such as genome-wide screens, proteomics, transcriptomics, and lipidomics have been used to identify host factors involved in the influenza virus life cycle. More detailed experimental studies and bioinformatics analyses will help identify and prioritize host factors with potential as therapeutic targets.
Genome-wide screens
The yeast single-gene deletion library consists of almost 5,000 single-gene deletion strains and covers about 80% of the yeast genome. This library has been used extensively in the yeast community for genomic studies. Naito et al. [55] used it to identify host genes involved in influenza virus replication. They used Saccharomyces cerevisiae to established a system that supported influenza viral genome replication and transcription and then screened a sub-library that lacked 354 genes encoding putative nucleic acid-binding and related functional proteins. Several host factors that affect influenza viral RNA synthesis, including Tat stimulatory factor 1 (Tat-SF1), were identified in this study[55].
Genome-wide RNAi-based screens also represent an excellent approach to searches for host factors involved in virus replication. In the absence of RNAi-based screening systems in mammalian cells, Hao et al. [48] used Drosophila RNAi technology to screen for host factors involved in influenza virus replication. They tested an RNAi library against more than 13,000 genes (approximately 90% of the Drosophila genome) and identified 110 Drosophila genes that, when depleted, affected influenza virus replication in Drosophila cells. Of those 110 candidates, further analysis validated roles for three host proteins––ATP6V0D1 (an ATPase), COX6A1 (a cytochrome C oxidase subunit), and NXF1 (a nuclear RNA export factor)–– in influenza virus replication in mammalian cells.
When RNAi-based screening systems in mammalian cells were subsequently established, the roles of mammalian host proteins in influenza virus replication could be comprehensively analyzed. Three key genome-wide RNAi-based screen studies were published in 2009 and 2010 [43,46,47]. Brass et al. [46] identified 133 host factors potentially involved in the influenza life cycle steps of virus entry, uncoating, vRNP nuclear import, genome transcription, and viral protein translation by examining the effects of siRNAs targeting over 17,000 human genes on influenza virus replication in human osteosarcoma. Konig et al. [43] performed a similar study using the human lung cell line A549 and identified 295 host factors that may also be involved in these early to middle steps of the influenza life cycle. In the third key genome-wide RNAi-based screen study, Karlas et al. [47] looked at the entire influenza virus life cycle. By measuring virus titers in the culture supernatant of siRNA-treated, virus-infected cells they identified 287 host factors important for influenza virus replication [47].
Sui et al. [56] applied ‘Random Homozygous Gene Perturbation’ (RHGP) to identify host targets that are required for influenza virus infection. RHGP uses a lentiviral-based genetic element that integrates into the genome leading to abrogation or enhancement of gene expression and an altered phenotype. The authors generated an RHGP library in mammalian cells, which they then infected with influenza virus. When they isolated the cell clones that survived and sequenced their genomes, they were able to identify 110 host genes that were associated with host cell resistance to influenza virus infection.
Proteomics approaches
Yeast two-hybrid analyses and proteomics approaches have been successfully used to identify host molecules that interact with the influenza viral proteins (reviewed in [57]), and identified several host proteins involved in viral genome replication and transcription [58-65], as have NS1 protein-interacting host factors involved in the immune response to infection [66]. This system also identified 87 human proteins as interaction partners of ten influenza viral proteins and 109 human proteins as interaction partners of three viral polymerase subunits [49,67], making it possible to built interactome maps consisting of influenza viral-host protein interactions.
By using a mass spectrometry approach, Mayer et al. [68] identified 41 host proteins that interact with the influenza viral ribonucleoprotein complex. Of these 41 proteins, they demonstrated a potential role for nucleoplasmin in viral RNA synthesis. Shaw et al. [69] and Hutchinson et al. [70] also used a mass spectrometry approach to identify host cellular proteins that are incorporated into progeny virion particles. They found that influenza virions contain abundant host proteins that make a substantial contribution to the influenza virion architecture. Shaw et al. [69] identified 36 host proteins incorporated in virions, including cytoskeletal proteins, annexins, glycolytic enzymes, and tetraspanin. Hutchinson et al. [70] found that the host proteins that are part of influenza virions produced in virus-infected mammalian cells largely overlapped with those produced in virus-infected chicken eggs, suggesting that these common host proteins are important in the formation of influenza virion particles.
Combining proteomics and RNAi-based screen approaches to construct the interactome map of viral-host protein interactions in influenza virus replication
Shapira et al. [49] used a combination of proteomics and functional genomics to construct an interactome map of the viral-host protein interactions that occur during influenza virus replication. They used a yeast two-hybrid screen to identify host proteins that interact with viral proteins and then built a physical map of these interactions. They also used transcriptional profiling to determine whether the host cell genes were differentially expressed in primary human bronchial epithelial cells upon influenza virus infection (or exposure to influenza viral RNA). By using this approach, they identified 1,745 host factors with potential involvement in influenza virus replication. When they tested siRNAs targeted to these 1,745 human genes, they found 616 host factors with involvement in influenza virus replication.
Watanabe et al. [71] used mass spectrometry and RNAi-based screening approaches to elucidate the physical and functional host-viral interactions during influenza virus replication. They first identified 1,292 host proteins that co-immunoprecipitated with influenza viral proteins that were transiently expressed in human cells. Then, they validated the roles of 324 of these host factors in influenza virus replication by testing the effects of siRNAs targeted to the 1,292 influenza viral host interaction partners on influenza virus production. Further extensive analyses of the 91 top-ranked host factors allowed them to define the steps of the viral life cycle that were affected and to generate an interactome map of the virus-host protein interactions that are involved in the influenza virus replication cycle.
Exploring antiviral drugs designed to target host factors involved in influenza virus replication
The studies described above have produced a vast amount of data on the host factors that are involved in influenza virus replication. The next step is to select host factors that may be suitable targets for antiviral drugs. Small compounds that inhibit the cellular functions of particular host proteins have been used to validate the requirement of a specific host protein for virus replication (Figure 1). Konig et al. [43] tested the compound KN93, which is a selective inhibitor of CAMK2B (calcium/calmodulin-dependent kinase IIβ) that was shown to be involved in the regulation of viral RNA transcription in their genome-wide screen, and demonstrated that the compound reduced virus titers in influenza virus-infected cells. Karlas et al. [47] showed that treatment with the compound TG003, a small molecule inhibitor of CDC-like kinase 1 (CLK1) identified as a host candidate involved in influenza virus replication in their screen, led to reduced virus production in human cells probably by affecting the splicing of viral RNAs. Brass et al. [46] found that the interferon-inducible transmembrane proteins IFITM1, 2, and 3 hampered the early steps of influenza virus replication in vitro, and also suggested that expression of IFITM3 defends host against influenza virus infection in vivo [72]. Watanabe et al. [71] tested 11 compounds that inhibit the cellular functions of several host factors identified in their screen. Of these compounds they found that ruxolitinib and gogicide A (which are inhibitors of JAK1 and the Golgi-specific brefeldin A-resistant guanine nucleotide exchange factor GBF1, respectively) markedly decreased virus titers in human cells infected with influenza virus without affecting cell viability.
In addition to approaches targeting host cellular proteins, Morita et al. [73] assessed the possible involvement of lipid products in influenza virus replication. By conducting mediator lipidomics and a bioactive lipid screen, they discovered that the DHA-derived protectin D1 isomer (10S, 17S-dihydroxydocosahexaenoic acid) markedly hampers influenza virus replication in vitro by inhibiting the nuclear export of viral mRNA. More importantly, they demonstrated that intravenous administration of protectin D1 protected mice from influenza virus infection, suggesting that protectin D1 has potential as a new therapeutic tool in the treatment and prevention of influenza.
Besides the experimental studies described above, bioinformatics analysis could be an attractive approach to identifying and prioritizing host factors with potential as therapeutic targets (Figure 2). Chassey et al. [74] conducted a meta-analysis using the datasets from the six independent genome-wide screens described above [43,46-49,56] and made a list of 925 host factors important for influenza virus replication (note that this list differs from lists obtained by others [75-77] due to the use of a different data evaluation strategy). By retrieving information from the DrugBank database (http://www.drugbank.ca/), Chassey and colleagues searched for molecules that interacted with these 925 host factors [74]. They found 298 molecules that potentially interact with 100 host factors. Importantly, of the 298 molecules, 204 are already approved by FDA, and therefore, could be highly prioritized candidates for influenza therapy [74].
Matsuoka et al. [78] attempted to apply another bioinfomatics approach to predict and prioritize host factors that could be therapeutic targets by using a comprehensive map of the influenza virus life cycle called ‘FluMap’ (accessible at http://www.influenza-x.org/flumap/) (Figure 2). FluMap can be used to visualize the entire influenza virus life cycle and was constructed on the basis of information obtained from the literature and publicly available pathway databases. The authors applied a computational network analysis, called a controllability analysis [79,80], to FluMap to identify host molecules that disregulated the virus replication cycle when their functions were disrupted, resulting in inhibition of virus replication. By using the controllability analysis, Matsuoka and colleagues identified 112 critical host molecules [78]. Further in silico analysis led them to prioritize several important molecules, such as the nuclear pore complex, Akt, PKC, and the Ran/GTPase complex, which are essential and highly connected within the virus-host network [78]. Accordingly, these molecules can be prioritized as potential new therapeutic targets.
Concluding remarks
Given the high rates of emergence of influenza virus strains that are resistant to the currently available anti-influenza drugs that target viral protein functions, host cellular functions involved in influenza virus replication are attractive targets for antiviral therapy. The host cellular systems are formed by highly complicated and sophisticated networks that consist of interactions between numerous cellular components; viruses hijack these host cellular system, leading to the re-construction of different networks to meet their need to replicate. Recent progress in our understanding of the virus-host interactions during influenza virus replication and of the human interactome network can accelerate the construction of a precise map of the virus-host interactome network during influenza virus replication, leading to a systematic understanding of the mechanisms involved in the influenza virus life cycle. Combining detailed functional approaches (e.g., in vitro and in vivo analyses) and extensive in silico approaches with such a comprehensive map of influenza virus replication could help to identify and prioritize novel host factors with potential as therapeutic targets.
HIGHLIGHTS.
Host factors are attractive targets for antiviral therapy.
Recent screens identified host factors involved in influenza virus replication.
Virus-host interactome screens are powerful tools to identify therapeutic targets.
Acknowledgements
We thank Yukiko Matsuoka for helpful discussions. We also thank Dr. Susan Watson for editing the manuscript. This work was supported by National Institute of Allergy and Infectious Diseases Public Health Service research grants (RO1 AI080598 and R56 AI099275); the Japan Initiative for Global Research Network on Infectious Diseases from the Ministry of Education, Culture, Sports, Science and Technology, Japan; by grants-in-aid from the Ministry of Health, Labour and Welfare, Japan; by ERATO; by grants from the Strategic Basic Research Program of the Japan Science and Technology Agency; and by the Advanced Research & Development Programs for Medical Innovation from the Japan Agency for Medical Research and Development (AMED).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Wright PF, Neumann G, Kawaoka Y. In: Orthomyxoviruses. Fields Virology. 6 th edition Knipe PMH DM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE, editors. Lippincott Williams & Wilkins; Philadelphia: 2013. pp. 1186–1243. [Google Scholar]
- 2.Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, Chen J, Jie Z, Qiu H, Xu K, et al. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med. 2013;368:1888–1897. doi: 10.1056/NEJMoa1304459. [DOI] [PubMed] [Google Scholar]
- 3.Li Q, Zhou L, Zhou M, Chen Z, Li F, Wu H, Xiang N, Chen E, Tang F, Wang D, et al. Epidemiology of human infections with avian influenza A(H7N9) virus in China. N Engl J Med. 2014;370:520–532. doi: 10.1056/NEJMoa1304617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Webster RG, Govorkova EA. H5N1 influenza--continuing evolution and spread. N Engl J Med. 2006;355:2174–2177. doi: 10.1056/NEJMp068205. [DOI] [PubMed] [Google Scholar]
- 5.Yen HL, Webster RG. Pandemic influenza as a current threat. Curr Top Microbiol Immunol. 2009;333:3–24. doi: 10.1007/978-3-540-92165-3_1. [DOI] [PubMed] [Google Scholar]
- 6.Davies WL, Grunert RR, Haff RF, McGahen JW, Neumayer EM, Paulshock M, Watts JC, Wood TR, Hermann EC, Hoffmann CE. Antiviral Activity of 1-Adamantanamine (Amantadine). Science. 1964;144:862–863. doi: 10.1126/science.144.3620.862. [DOI] [PubMed] [Google Scholar]
- 7.Hayden FG. Perspectives on antiviral use during pandemic influenza. Philos Trans R Soc Lond B Biol Sci. 2001;356:1877–1884. doi: 10.1098/rstb.2001.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kamali A, Holodniy M. Influenza treatment and prophylaxis with neuraminidase inhibitors: a review. Infect Drug Resist. 2013;6:187–198. doi: 10.2147/IDR.S36601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kiso M, Kubo S, Ozawa M, Le QM, Nidom CA, Yamashita M, Kawaoka Y. Efficacy of the new neuraminidase inhibitor CS-8958 against H5N1 influenza viruses. PLoS Pathog. 2010;6:e1000786. doi: 10.1371/journal.ppat.1000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Birnkrant D, Cox E. The Emergency Use Authorization of peramivir for treatment of 2009 H1N1 influenza. N Engl J Med. 2009;361:2204–2207. doi: 10.1056/NEJMp0910479. [DOI] [PubMed] [Google Scholar]
- 11.Hernandez JE, Adiga R, Armstrong R, Bazan J, Bonilla H, Bradley J, Dretler R, Ison MG, Mangino JE, Maroushek S, et al. Clinical experience in adults and children treated with intravenous peramivir for 2009 influenza A (H1N1) under an Emergency IND program in the United States. Clin Infect Dis. 2011;52:695–706. doi: 10.1093/cid/cir001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Furuta Y, Takahashi K, Kuno-Maekawa M, Sangawa H, Uehara S, Kozaki K, Nomura N, Egawa H, Shiraki K. Mechanism of action of T-705 against influenza virus. Antimicrob Agents Chemother. 2005;49:981–986. doi: 10.1128/AAC.49.3.981-986.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kiso M, Takahashi K, Sakai-Tagawa Y, Shinya K, Sakabe S, Le QM, Ozawa M, Furuta Y, Kawaoka Y. T-705 (favipiravir) activity against lethal H5N1 influenza A viruses. Proc Natl Acad Sci U S A. 2010;107:882–887. doi: 10.1073/pnas.0909603107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sleeman K, Mishin VP, Deyde VM, Furuta Y, Klimov AI, Gubareva LV. In vitro antiviral activity of favipiravir (T-705) against drug-resistant influenza and 2009 A(H1N1) viruses. Antimicrob Agents Chemother. 2010;54:2517–2524. doi: 10.1128/AAC.01739-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bright RA, Medina MJ, Xu X, Perez-Oronoz G, Wallis TR, Davis XM, Povinelli L, Cox NJ, Klimov AI. Incidence of adamantane resistance among influenza A (H3N2) viruses isolated worldwide from 1994 to 2005: a cause for concern. Lancet. 2005;366:1175–1181. doi: 10.1016/S0140-6736(05)67338-2. [DOI] [PubMed] [Google Scholar]
- 16.Bright RA, Shay DK, Shu B, Cox NJ, Klimov AI. Adamantane resistance among influenza A viruses isolated early during the 2005-2006 influenza season in the United States. JAMA. 2006;295:891–894. doi: 10.1001/jama.295.8.joc60020. [DOI] [PubMed] [Google Scholar]
- 17.Nicoll A, Ciancio B, Kramarz P. Observed oseltamivir resistance in seasonal influenza viruses in Europe interpretation and potential implications. Euro Surveill. 2008;13 doi: 10.2807/ese.13.05.08025-en. [DOI] [PubMed] [Google Scholar]
- 18.MacArthur RD, Novak RM. Reviews of anti-infective agents: maraviroc: the first of a new class of antiretroviral agents. Clin Infect Dis. 2008;47:236–241. doi: 10.1086/589289. [DOI] [PubMed] [Google Scholar]
- 19.Watashi K, Shimotohno K. Chemical genetics approach to hepatitis C virus replication: cyclophilin as a target for anti-hepatitis C virus strategy. Rev Med Virol. 2007;17:245–252. doi: 10.1002/rmv.534. [DOI] [PubMed] [Google Scholar]
- 20.Gallay PA, Lin K. Profile of alisporivir and its potential in the treatment of hepatitis C. Drug Des Devel Ther. 2013;7:105–115. doi: 10.2147/DDDT.S30946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaul A, Stauffer S, Berger C, Pertel T, Schmitt J, Kallis S, Zayas M, Lohmann V, Luban J, Bartenschlager R. Essential role of cyclophilin A for hepatitis C virus replication and virus production and possible link to polyprotein cleavage kinetics. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan RW, Chan MC, Wong AC, Karamanska R, Dell A, Haslam SM, Sihoe AD, Chui WH, Triana-Baltzer G, Li Q, et al. DAS181 inhibits H5N1 influenza virus infection of human lung tissues. Antimicrob Agents Chemother. 2009;53:3935–3941. doi: 10.1128/AAC.00389-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Triana-Baltzer GB, Gubareva LV, Klimov AI, Wurtman DF, Moss RB, Hedlund M, Larson JL, Belshe RB, Fang F. Inhibition of neuraminidase inhibitor-resistant influenza virus by DAS181, a novel sialidase fusion protein. PLoS One. 2009;4:e7838. doi: 10.1371/journal.pone.0007838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Triana-Baltzer GB, Gubareva LV, Nicholls JM, Pearce MB, Mishin VP, Belser JA, Chen LM, Chan RW, Chan MC, Hedlund M, et al. Novel pandemic influenza A(H1N1) viruses are potently inhibited by DAS181, a sialidase fusion protein. PLoS One. 2009;4:e7788. doi: 10.1371/journal.pone.0007788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edinger TO, Pohl MO, Stertz S. Entry of influenza A virus: host factors and antiviral targets. J Gen Virol. 2014;95:263–277. doi: 10.1099/vir.0.059477-0. [DOI] [PubMed] [Google Scholar]
- 26.Lee SM, Yen HL. Targeting the host or the virus: current and novel concepts for antiviral approaches against influenza virus infection. Antiviral Res. 2012;96:391–404. doi: 10.1016/j.antiviral.2012.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malakhov MP, Aschenbrenner LM, Smee DF, Wandersee MK, Sidwell RW, Gubareva LV, Mishin VP, Hayden FG, Kim DH, Ing A, et al. Sialidase fusion protein as a novel broad-spectrum inhibitor of influenza virus infection. Antimicrob Agents Chemother. 2006;50:1470–1479. doi: 10.1128/AAC.50.4.1470-1479.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhirnov OP, Klenk HD, Wright PF. Aprotinin and similar protease inhibitors as drugs against influenza. Antiviral Res. 2011;92:27–36. doi: 10.1016/j.antiviral.2011.07.014. [DOI] [PubMed] [Google Scholar]
- 29.Zhirnov OP, Ovcharenko AV, Bukrinskaya AG. Suppression of influenza virus replication in infected mice by protease inhibitors. J Gen Virol. 1984;65(Pt 1):191–196. doi: 10.1099/0022-1317-65-1-191. [DOI] [PubMed] [Google Scholar]
- 30.Bottcher-Friebertshauser E, Freuer C, Sielaff F, Schmidt S, Eickmann M, Uhlendorff J, Steinmetzer T, Klenk HD, Garten W. Cleavage of influenza virus hemagglutinin by airway proteases TMPRSS2 and HAT differs in subcellular localization and susceptibility to protease inhibitors. J Virol. 2010;84:5605–5614. doi: 10.1128/JVI.00140-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sielaff F, Bottcher-Friebertshauser E, Meyer D, Saupe SM, Volk IM, Garten W, Steinmetzer T. Development of substrate analogue inhibitors for the human airway trypsin-like protease HAT. Bioorg Med Chem Lett. 2011;21:4860–4864. doi: 10.1016/j.bmcl.2011.06.033. [DOI] [PubMed] [Google Scholar]
- 32.Gagnon H, Beauchemin S, Kwiatkowska A, Couture F, D'Anjou F, Levesque C, Dufour F, Desbiens AR, Vaillancourt R, Bernard S, et al. Optimization of furin inhibitors to protect against the activation of influenza hemagglutinin H5 and Shiga toxin. J Med Chem. 2014;57:29–41. doi: 10.1021/jm400633d. [DOI] [PubMed] [Google Scholar]
- 33.Garten W, Hallenberger S, Ortmann D, Schafer W, Vey M, Angliker H, Shaw E, Klenk HD. Processing of viral glycoproteins by the subtilisin-like endoprotease furin and its inhibition by specific peptidylchloroalkylketones. Biochimie. 1994;76:217–225. doi: 10.1016/0300-9084(94)90149-x. [DOI] [PubMed] [Google Scholar]
- 34.Stieneke-Grober A, Vey M, Angliker H, Shaw E, Thomas G, Roberts C, Klenk HD, Garten W. Influenza virus hemagglutinin with multibasic cleavage site is activated by furin, a subtilisin-like endoprotease. EMBO J. 1992;11:2407–2414. doi: 10.1002/j.1460-2075.1992.tb05305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pleschka S, Wolff T, Ehrhardt C, Hobom G, Planz O, Rapp UR, Ludwig S. Influenza virus propagation is impaired by inhibition of the Raf/MEK/ERK signalling cascade. Nat Cell Biol. 2001;3:301–305. doi: 10.1038/35060098. [DOI] [PubMed] [Google Scholar]
- 36.Droebner K, Pleschka S, Ludwig S, Planz O. Antiviral activity of the MEK-inhibitor U0126 against pandemic H1N1v and highly pathogenic avian influenza virus in vitro and in vivo. Antiviral Res. 2011;92:195–203. doi: 10.1016/j.antiviral.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 37.Mazur I, Wurzer WJ, Ehrhardt C, Pleschka S, Puthavathana P, Silberzahn T, Wolff T, Planz O, Ludwig S. Acetylsalicylic acid (ASA) blocks influenza virus propagation via its NF-kappaB-inhibiting activity. Cell Microbiol. 2007;9:1683–1694. doi: 10.1111/j.1462-5822.2007.00902.x. [DOI] [PubMed] [Google Scholar]
- 38.Eyers S, Weatherall M, Shirtcliffe P, Perrin K, Beasley R. The effect on mortality of antipyretics in the treatment of influenza infection: systematic review and meta-analysis. J R Soc Med. 2010;103:403–411. doi: 10.1258/jrsm.2010.090441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marsolais D, Hahm B, Walsh KB, Edelmann KH, McGavern D, Hatta Y, Kawaoka Y, Rosen H, Oldstone MB. A critical role for the sphingosine analog AAL-R in dampening the cytokine response during influenza virus infection. Proc Natl Acad Sci U S A. 2009;106:1560–1565. doi: 10.1073/pnas.0812689106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walsh KB, Teijaro JR, Wilker PR, Jatzek A, Fremgen DM, Das SC, Watanabe T, Hatta M, Shinya K, Suresh M, et al. Suppression of cytokine storm with a sphingosine analog provides protection against pathogenic influenza virus. Proc Natl Acad Sci U S A. 2011;108:12018–12023. doi: 10.1073/pnas.1107024108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muller KH, Kakkola L, Nagaraj AS, Cheltsov AV, Anastasina M, Kainov DE. Emerging cellular targets for influenza antiviral agents. Trends Pharmacol Sci. 2012;33:89–99. doi: 10.1016/j.tips.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 42.Lu Y, Hardes K, Dahms SO, Bottcher-Friebertshauser E, Steinmetzer T, Than ME, Klenk HD, Garten W. Peptidomimetic furin inhibitor MI-701 in combination with oseltamivir and ribavirin efficiently blocks propagation of highly pathogenic avian influenza viruses and delays high level oseltamivir resistance in MDCK cells. Antiviral Res. 2015;120:89–100. doi: 10.1016/j.antiviral.2015.05.006. [DOI] [PubMed] [Google Scholar]
- 43••.Konig R, Stertz S, Zhou Y, Inoue A, Hoffmann HH, Bhattacharyya S, Alamares JG, Tscherne DM, Ortigoza MB, Liang Y, et al. Human host factors required for influenza virus replication. Nature. 2010;463:813–817. doi: 10.1038/nature08699. [One of the important genome-wide RNAi-based screening studies in mammalian cells to identify host factors involved in influenza virus replication.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Konig R, Zhou Y, Elleder D, Diamond TL, Bonamy GM, Irelan JT, Chiang CY, Tu BP, De Jesus PD, Lilley CE, et al. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell. 2008;135:49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, Lieberman J, Elledge SJ. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- 46••.Brass AL, Huang IC, Benita Y, John SP, Krishnan MN, Feeley EM, Ryan BJ, Weyer JL, van der Weyden L, Fikrig E, et al. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell. 2009;139:1243–1254. doi: 10.1016/j.cell.2009.12.017. [One of the important genome-wide RNAi-based screening studies in mammalian cells to identify host factors involved in influenza virus replication.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47••.Karlas A, Machuy N, Shin Y, Pleissner KP, Artarini A, Heuer D, Becker D, Khalil H, Ogilvie LA, Hess S, et al. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature. 2010;463:818–822. doi: 10.1038/nature08760. [One of the important genome-wide RNAi-based screening studies in mammalian cells to identify host factors involved in influenza virus replication.] [DOI] [PubMed] [Google Scholar]
- 48••.Hao L, Sakurai A, Watanabe T, Sorensen E, Nidom CA, Newton MA, Ahlquist P, Kawaoka Y. Drosophila RNAi screen identifies host genes important for influenza virus replication. Nature. 2008;454:890–893. doi: 10.1038/nature07151. [The first reported genome-wide RNAi-based screen to identify host factors important for influenza virus replication. This screen was conducted in Drosophila cells due to the lack of available mammalian RNAi technology at the time.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49••.Shapira SD, Gat-Viks I, Shum BO, Dricot A, de Grace MM, Wu L, Gupta PB, Hao T, Silver SJ, Root DE, et al. A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell. 2009;139:1255–1267. doi: 10.1016/j.cell.2009.12.018. [This study utilized a combination of transcriptional profiling, a yeast-two hybrid screen, and an RNAi-based screen to identify host factors involved in influenza virus replication.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cherry S, Doukas T, Armknecht S, Whelan S, Wang H, Sarnow P, Perrimon N. Genome-wide RNAi screen reveals a specific sensitivity of IRES-containing RNA viruses to host translation inhibition. Genes Dev. 2005;19:445–452. doi: 10.1101/gad.1267905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goff SP. Knockdown screens to knockout HIV-1. Cell. 2008;135:417–420. doi: 10.1016/j.cell.2008.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krishnan MN, Ng A, Sukumaran B, Gilfoy FD, Uchil PD, Sultana H, Brass AL, Adametz R, Tsui M, Qian F, et al. RNA interference screen for human genes associated with West Nile virus infection. Nature. 2008;455:242–245. doi: 10.1038/nature07207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Q, Brass AL, Ng A, Hu Z, Xavier RJ, Liang TJ, Elledge SJ. A genome-wide genetic screen for host factors required for hepatitis C virus propagation. Proc Natl Acad Sci U S A. 2009;106:16410–16415. doi: 10.1073/pnas.0907439106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou H, Xu M, Huang Q, Gates AT, Zhang XD, Castle JC, Stec E, Ferrer M, Strulovici B, Hazuda DJ, et al. Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe. 2008;4:495–504. doi: 10.1016/j.chom.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 55•.Naito T, Kiyasu Y, Sugiyama K, Kimura A, Nakano R, Matsukage A, Nagata K. An influenza virus replicon system in yeast identified Tat-SF1 as a stimulatory host factor for viral RNA synthesis. Proc Natl Acad Sci U S A. 2007;104:18235–18240. doi: 10.1073/pnas.0705856104. [By using the yeast single-gene deletion library, the authors identified host factors involved in influenza viral genome replication/transcription.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56•.Sui B, Bamba D, Weng K, Ung H, Chang S, Van Dyke J, Goldblatt M, Duan R, Kinch MS, Li WB. The use of Random Homozygous Gene Perturbation to identify novel host-oriented targets for influenza. Virology. 2009;387:473–481. doi: 10.1016/j.virol.2009.02.046. [By using 'Random Homozygous Gene Pertubation' technology, the authors conducted a genome-wide screen and identified host targets important for influenza virus replication.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nagata K, Kawaguchi A, Naito T. Host factors for replication and transcription of the influenza virus genome. Rev Med Virol. 2008;18:247–260. doi: 10.1002/rmv.575. [DOI] [PubMed] [Google Scholar]
- 58.Engelhardt OG, Smith M, Fodor E. Association of the influenza A virus RNA-dependent RNA polymerase with cellular RNA polymerase II. J Virol. 2005;79:5812–5818. doi: 10.1128/JVI.79.9.5812-5818.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Deng T, Engelhardt OG, Thomas B, Akoulitchev AV, Brownlee GG, Fodor E. Role of ran binding protein 5 in nuclear import and assembly of the influenza virus RNA polymerase complex. J Virol. 2006;80:11911–11919. doi: 10.1128/JVI.01565-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huarte M, Sanz-Ezquerro JJ, Roncal F, Ortin J, Nieto A. PA subunit from influenza virus polymerase complex interacts with a cellular protein with homology to a family of transcriptional activators. J Virol. 2001;75:8597–8604. doi: 10.1128/JVI.75.18.8597-8604.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jorba N, Juarez S, Torreira E, Gastaminza P, Zamarreno N, Albar JP, Ortin J. Analysis of the interaction of influenza virus polymerase complex with human cell factors. Proteomics. 2008;8:2077–2088. doi: 10.1002/pmic.200700508. [DOI] [PubMed] [Google Scholar]
- 62.Kawaguchi A, Nagata K. De novo replication of the influenza virus RNA genome is regulated by DNA replicative helicase, MCM. EMBO J. 2007;26:4566–4575. doi: 10.1038/sj.emboj.7601881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Momose F, Basler CF, O'Neill RE, Iwamatsu A, Palese P, Nagata K. Cellular splicing factor RAF-2p48/NPI-5/BAT1/UAP56 interacts with the influenza virus nucleoprotein and enhances viral RNA synthesis. J Virol. 2001;75:1899–1908. doi: 10.1128/JVI.75.4.1899-1908.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Momose F, Naito T, Yano K, Sugimoto S, Morikawa Y, Nagata K. Identification of Hsp90 as a stimulatory host factor involved in influenza virus RNA synthesis. J Biol Chem. 2002;277:45306–45314. doi: 10.1074/jbc.M206822200. [DOI] [PubMed] [Google Scholar]
- 65.Resa-Infante P, Jorba N, Zamarreno N, Fernandez Y, Juarez S, Ortin J. The host-dependent interaction of alpha-importins with influenza PB2 polymerase subunit is required for virus RNA replication. PLoS One. 2008;3:e3904. doi: 10.1371/journal.pone.0003904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hale BG, Randall RE, Ortin J, Jackson D. The multifunctional NS1 protein of influenza A viruses. J Gen Virol. 2008;89:2359–2376. doi: 10.1099/vir.0.2008/004606-0. [DOI] [PubMed] [Google Scholar]
- 67.Tafforeau L, Chantier T, Pradezynski F, Pellet J, Mangeot PE, Vidalain PO, Andre P, Rabourdin-Combe C, Lotteau V. Generation and comprehensive analysis of an influenza virus polymerase cellular interaction network. J Virol. 2011;85:13010–13018. doi: 10.1128/JVI.02651-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mayer D, Molawi K, Martinez-Sobrido L, Ghanem A, Thomas S, Baginsky S, Grossmann J, Garcia-Sastre A, Schwemmle M. Identification of cellular interaction partners of the influenza virus ribonucleoprotein complex and polymerase complex using proteomic-based approaches. J Proteome Res. 2007;6:672–682. doi: 10.1021/pr060432u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69•.Shaw ML, Stone KL, Colangelo CM, Gulcicek EE, Palese P. Cellular proteins in influenza virus particles. PLoS Pathog. 2008;4:e1000085. doi: 10.1371/journal.ppat.1000085. [The authors used a mass spectrometry approach to identify host cellular proteins that are incorporated into progeny virion particles.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70•.Hutchinson EC, Charles PD, Hester SS, Thomas B, Trudgian D, Martinez-Alonso M, Fodor E. Conserved and host-specific features of influenza virion architecture. Nat Commun. 2014;5:4816. doi: 10.1038/ncomms5816. [The authors used a mass spectrometry approach to identify host cellular proteins that are incorporated into progeny virion particles.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71••.Watanabe T, Kawakami E, Shoemaker JE, Lopes TJ, Matsuoka Y, Tomita Y, Kozuka-Hata H, Gorai T, Kuwahara T, Takeda E, et al. Influenza virus-host interactome screen as a platform for antiviral drug development. Cell Host Microbe. 2014;16:795–805. doi: 10.1016/j.chom.2014.11.002. [This study used mass spectrometry and RNAi-based screening approaches to elucidate physical and functional host-viral interactions during influenza virus replication.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Everitt AR, Clare S, Pertel T, John SP, Wash RS, Smith SE, Chin CR, Feeley EM, Sims JS, Adams DJ, et al. IFITM3 restricts the morbidity and mortality associated with influenza. Nature. 2012;484:519–523. doi: 10.1038/nature10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73•.Morita M, Kuba K, Ichikawa A, Nakayama M, Katahira J, Iwamoto R, Watanebe T, Sakabe S, Daidoji T, Nakamura S, et al. The lipid mediator protectin D1 inhibits influenza virus replication and improves severe influenza. Cell. 2013;153:112–125. doi: 10.1016/j.cell.2013.02.027. [By using mediator lipidomics and conducting a bioactive lipid screen, the authors found that protectin D decreases influenza virus production in vitro and in vivo, indicating the involvement of lipid products in influenza virus replication.] [DOI] [PubMed] [Google Scholar]
- 74•.de Chassey B, Meyniel-Schicklin L, Aublin-Gex A, Andre P, Lotteau V. Genetic screens for the control of influenza virus replication: from meta-analysis to drug discovery. Mol Biosyst. 2012;8:1297–1303. doi: 10.1039/c2mb05416g. [The authors performed a meta-analysis of datasets from several genome-wide screens that identified host factors important for influenza virus replication. They then searched for molecules that interact with the identified host factors by retrieving information from the DrugBank database.] [DOI] [PubMed] [Google Scholar]
- 75.Watanabe T, Watanabe S, Kawaoka Y. Cellular networks involved in the influenza virus life cycle. Cell Host Microbe. 2010;7:427–439. doi: 10.1016/j.chom.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shaw ML. The host interactome of influenza virus presents new potential targets for antiviral drugs. Rev Med Virol. 2011;21:358–369. doi: 10.1002/rmv.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stertz S, Shaw ML. Uncovering the global host cell requirements for influenza virus replication via RNAi screening. Microbes Infect. 2011;13:516–525. doi: 10.1016/j.micinf.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78••.Matsuoka Y, Matsumae H, Katoh M, Eisfeld AJ, Neumann G, Hase T, Ghosh S, Shoemaker JE, Lopes TJ, Watanabe T, et al. A comprehensive map of the influenza A virus replication cycle. BMC Syst Biol. 2013;7:97. doi: 10.1186/1752-0509-7-97. [The authors constructed a comprehensive map of influenza virus replication (called 'FluMap') by using information obtained from the literature and publicly available databases. They applied a computational network analysis to FluMap to explore host targets for antiviral therapy.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu YY, Slotine JJ, Barabasi AL. Controllability of complex networks. Nature. 2011;473:167–173. doi: 10.1038/nature10011. [DOI] [PubMed] [Google Scholar]
- 80.Luni C, Shoemaker JE, Sanft KR, Petzold LR, Doyle FJ., 3rd Confidence from uncertainty--a multi-target drug screening method from robust control theory. BMC Syst Biol. 2010;4:161. doi: 10.1186/1752-0509-4-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.de Vries E, Tscherne DM, Wienholts MJ, Cobos-Jimenez V, Scholte F, Garcia-Sastre A, Rottier PJ, de Haan CA. Dissection of the influenza A virus endocytic routes reveals macropinocytosis as an alternative entry pathway. PLoS Pathog. 2011;7:e1001329. doi: 10.1371/journal.ppat.1001329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Muller KH, Kainov DE, El Bakkouri K, Saelens X, De Brabander JK, Kittel C, Samm E, Muller CP. The proton translocation domain of cellular vacuolar ATPase provides a target for the treatment of influenza A virus infections. Br J Pharmacol. 2011;164:344–357. doi: 10.1111/j.1476-5381.2011.01346.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chase G, Deng T, Fodor E, Leung BW, Mayer D, Schwemmle M, Brownlee G. Hsp90 inhibitors reduce influenza virus replication in cell culture. Virology. 2008;377:431–439. doi: 10.1016/j.virol.2008.04.040. [DOI] [PubMed] [Google Scholar]
- 84.Shin YK, Liu Q, Tikoo SK, Babiuk LA, Zhou Y. Effect of the phosphatidylinositol 3-kinase/Akt pathway on influenza A virus propagation. J Gen Virol. 2007;88:942–950. doi: 10.1099/vir.0.82483-0. [DOI] [PubMed] [Google Scholar]
- 85.Munger J, Bennett BD, Parikh A, Feng XJ, McArdle J, Rabitz HA, Shenk T, Rabinowitz JD. Systems-level metabolic flux profiling identifies fatty acid synthesis as a target for antiviral therapy. Nat Biotechnol. 2008;26:1179–1186. doi: 10.1038/nbt.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Widjaja I, de Vries E, Tscherne DM, Garcia-Sastre A, Rottier PJ, de Haan CA. Inhibition of the ubiquitin-proteasome system affects influenza A virus infection at a postfusion step. J Virol. 2010;84:9625–9631. doi: 10.1128/JVI.01048-10. [DOI] [PMC free article] [PubMed] [Google Scholar]