Human epidermal growth factor receptor 2 (HER2), also known as Neu, ErbB-2, CD340 or p185, plays a significant role in the development and progression of certain aggressive types of breast cancer. Recently, HER2 has become an important biomarker and therapeutic target for approximately 30% of breast cancer patients. HER2 testing is performed in breast cancer patients to assess their prognosis and to determine their suitability for trastuzumab therapy. A novel HER2 aptamer (HB5) has recently been developed using systematic evolution of ligands by exponential enrichment technology (SELEX). However, the application of HB5 in clinical diagnosis has not been extensively studied. Here, we tried to use HB5 instead of a specific anti-HER2 mAb in HER2 testing. Samples from 214 breast cancer cases were stained with HB5 for immunohistochemistry (IHC), and the results were compared with those obtained FDA-approved commercial HER2 testing kits. Stronger membrane staining is observed using HB5 than using the commercial anti-HER2 mAb, especially in equivocal samples. More importantly, the HB5 staining signal can be further enhanced by using rolling circle amplification (RCA). In addition, HB5 also showed relatively strong binding to HER2-positive breast cancer cells (SK-BR-3) and weak binding to HER2-negative cells (MDA-MB-231). Collectively, a newly developed DNA aptamer (HB5) was found to be capable of specifically binding to HER2-positive breast cancer cells. Therefore, we believe that HB5 may have application potentials in the clinical diagnosis of HER2 status in cancers.
Worldwide, breast cancer accounts for 22.9% of all cancers in women. Almost 1.4 million women were diagnosed with breast cancer in 2008, and approximately 459,000 deaths were recorded (13.7% of cancer deaths in women).1 The prognosis and survival rates for breast cancer vary greatly depending on the cancer type. According to the St. Gallen International Breast Cancer Conference in 2011,2 breast cancer can be divided into four molecular subtypes: (i) Luminal A: ER and/or PR positive and Ki67 low (<14%), and HER2 negative; (ii) Luminal B: ER and/or PR positive and Ki67 high (>14%) or HER2 positive; (iii) HER2 overexpression: ER and PR absent, HER2 overexpressed or amplified; and (iv) Basal-like: triple negative for ER, PR and HER2. The amplification or overexpression of HER2 occurs in approximately 15–30% of breast cancers and is strongly associated with increased disease recurrence and a poor prognosis.3,4 The overexpression of HER2 is also known to occur in ovarian and stomach cancers and in aggressive forms of uterine cancer.
HER2 is a transmembrane protein that in humans is encoded by the ERBB2 gene, located at the long arm of human chromosome 17 (17q21). The ErbB family comprises four plasma membrane-bound receptor tyrosine kinases, including Erb-1 (EGFR), Erb-2 (HER2/neu), Erb-3 and ErbB-4. All four contain an extracellular ligand-binding domain, a transmembrane domain and an intracellular domain. HER2 comprises an extracellular region of four domains (I/L1, II/CR1, III/L2 and IV/CR2). Several methods are available to detect the HER2 status of breast cancer, including IHC and in situ hybridization (ISH).5,6 IHC using a specific anti-HER2 mAb is essential for identifying patients who can benefit from HER2-targeted therapy.
In 1990, two labs independently developed the technique of selection: the Gold lab, using the term SELEX for their process of selecting RNA ligands against T4 DNA polymerase;7 and the Szostak lab, coining the term in vitro selection for selecting RNA ligands against various organic dyes. SELEX products were named ‘aptamers,' which are single-stranded RNA or DNA oligonucleotides that bind with high affinity to specific targets such as proteins, small molecules, nucleic acids and cells.
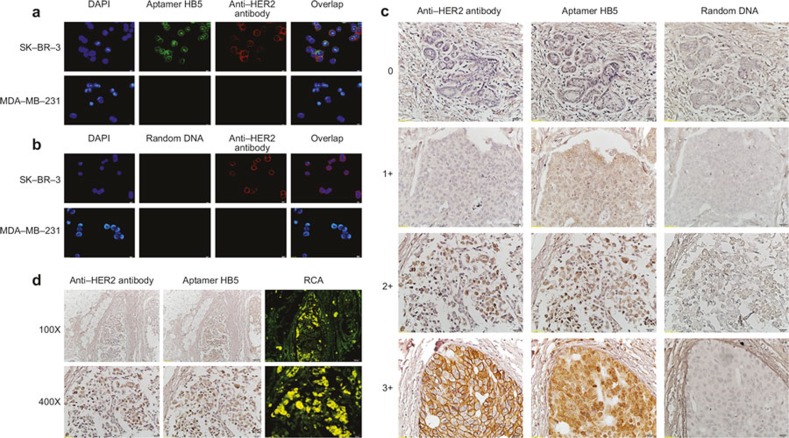
Recently, a new HB5 was developed using SELEX. The selected aptamer was an 86-nucleotide DNA molecule that bound to an epitope peptide of HER2 with a Kd of 18.9 nM. The aptamer also specifically bound to the extracellular domain of HER2 protein with a Kd of 316 nM.8 Thus, we wondered if HB5 could be used in IHC to identify the HER2 status of breast cancer. We first identified the binding specificity of the aptamer HB5 using confocal microscopy compared with the specific anti-HER2 antibody. We found that the aptamer HB5 exhibited a relatively strong binding to HER2-positive breast cancer cells (SK-BR-3) but minimal binding to HER2-negative cells (MDA-MB-231; Figure 1A). In addition, random DNA showed no binding preference for all of the cell lines tested (Figure 1B). These results indicated that the aptamer HB5 could specifically bind to HER2-positive breast cancer cells, possibly by recognizing HER2 in these cells. More importantly, the aptamer HB5 shared the same binding sites on the membrane and in the nucleus with the specific anti-HER2 antibody. We also found the same scoring criteria as found using the FDA-approved commercial kits (Figure 1C). Stronger staining of HER2-positive tumor cells was observed using the aptamer HB5 compared with the specific anti-HER2 antibody. Thus, it is appropriate to use aptamer IHC testing as the sole methodology to evaluate the HER2 status, especially in equivocal cases. Furthermore, the HB5 staining signal can be further enhanced by RCA, which is a newly developed technique involving a process of unidirectional nucleic acid replication that can rapidly synthesize multiple copies of circular molecules of DNA or RNA9,10 (Figure 1D).
Figure 1.
Figure 1. Evaluation of HER2 in breast cancer with the aptamer HB5. (a) Binding profiles of the aptamer HB5 to HER2-positive (SKBR3) or HER2-negative (MDA-MB-231) breast cancer cells. (b) The binding profiles of random DNA to breast cancer cells. (c) Examples of IHC staining for HER2 in invasive ductal carcinoma with a commercial anti-HER2 antibody or the aptamer HB5. (d) The detection of HER2 expression in situ based on the RCA.
Herein, because of the need to evaluate the HER2 status in situ, we propose a novel IHC method using a newly developed DNA aptamer (HB5). The aptamer HB5 was shown to be capable of specifically binding to HER2 protein and HER2-positive breast cancer cells, with minimal binding to HER2-negative cells. An improved IHC method was then established using the aptamer HB5 to evaluate the expression profile of HER2 in 214 samples. This method was shown to be more effective and specific than the FDA-approved commercial kits. In addition, the aptamer staining signal can be further enhanced by using RCA. We believe that these new developments in IHC testing using aptamers may have the potential for application in future clinicopathological diagnoses.
References
- Youlden DR, Cramb SM, Dunn NA, Muller JM, Pyke CM, Baade PD. The descriptive epidemiology of female breast cancer: an international comparison of screening, incidence, survival and mortality. Cancer Epidemiol 2012; 36: 237–248. [DOI] [PubMed] [Google Scholar]
- Goldhirsch A, Wood WC, Coates AS, Gelber RD, Thurlimann B, Senn HJ et al. Strategies for subtypes—dealing with the diversity of breast cancer: highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann Oncol 2011; 22: 1736–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke T, Reeves J, Lanigan A, Stanton P. HER2 as a prognostic and predictive marker for breast cancer. Ann Oncol 2001; 12: S23–S28. [DOI] [PubMed] [Google Scholar]
- Holbro T, Civenni G, Hynes NE. The ErbB receptors and their role in cancer progression. Exp Cell Res 2003; 284: 99–110. [DOI] [PubMed] [Google Scholar]
- Hoang MP, Sahin AA, Ordonez NG, Sneige N. HER-2/neu gene amplification compared with HER-2/neu protein overexpression and interobserver reproducibility in invasive breast carcinoma. Am J Clin Pathol 2000; 113: 852–859. [DOI] [PubMed] [Google Scholar]
- Lebeau A, Deimling D, Kaltz C, Sendelhofert A, Iff A, Luthardt B et al. Her-2/neu analysis in archival tissue samples of human breast cancer: comparison of immunohistochemistry and fluorescence in situ hybridization. J Clin Oncol 2001; 19: 354–363. [DOI] [PubMed] [Google Scholar]
- Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990; 249: 505–510. [DOI] [PubMed] [Google Scholar]
- Konry T, Hayman RB, Walt DR. Microspherebased rolling circle amplification microarray for the detection of DNA and proteins in a single assay. Anal Chem 2009; 81: 5777–5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Duan JH, Song YM, Ma J, Wang FD, Lu X et al. Novel HER2 aptamer selectively delivers cytotoxic drug to HER2-positive breast cancer cells in vitro. J Transl Med 2012; 10: 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baner J, Nilsson M, Mendel-Hartvig M, Landegren U. Signal amplification of padlock probes by rolling circle replication. Nucleic Acids Res 1998; 26: 5073–5078. [DOI] [PMC free article] [PubMed] [Google Scholar]