Abstract
Background: Non-small cell lung cancer (NSCLC) accounts for approximately 85% of clinical lung cancer cases. MicroRNA-93 (miR-93) is an oncomiR in many types of human cancer, exerting pivotal effects in the development and progression of malignancies, including NSCLC. However, the mechanism underlying miR-93 involvement in NSCLC is unknown. Our purpose was to reveal and explain this mechanism, with the goal of contributing to the development of new diagnostic biomarkers and individualized therapeutic tools.
Methods: The expression of miR-93 was determined in NSCLC cell lines A549, H1975, and H1299. The cells were transfected with control plasmids (Mock group), miR-93 overexpression plasmids (miR-93 Up group), or miR-93 inhibitor plasmids (miR-93 Down group) to generate stable miR-93-overexpressing or -depleted cells. The effects of miR-93 on proliferation, migration, and invasion of these cells were determined. The in vivo effects of miR-93 on tumor metastasis were determined in an NSCLC xenograft mouse model. The molecular mechanisms underlying these effects were investigated via dual luciferase reporter assay and western blotting.
Results: MiR-93 expression levels were significantly greater in the NSCLC cell lines than in normal lung epithelial cells. Cell proliferation, migration, and invasion were significantly stimulated by miR-93 upregulation (all P<0.05) and inhibited by miR-93 downregulation. Dual luciferase reporter assay demonstrated that miR-93 directly bound with the 3′-untranslated region of the tumor suppressor gene LKB1. Western blotting analysis indicated that miR-93 activated the PI3K/Akt pathway by inhibiting LKB1, PTEN, and p21. Increased expression of miR-93 induced significant hepatic metastasis of lung cancer in the xenograft mouse model.
Conclusion: Overexpression of miR-93 facilitates tumorigenesis and metastasis of NSCLC. These findings provide novel insight into the mechanism of miR-93 involvement in NSCLC, suggesting that miR-93 may serve as a potential therapeutic target.
Keywords: non-small cell lung cancer, miR-93, LKB1, PI3K/Akt, metastasis.
Introduction
Lung cancer is the most common cancer and the leading cause of cancer death in the USA and China 1, 2. Lung carcinoma is generally classified as either small-cell lung carcinoma (about 20% of all lung carcinomas) or non-small cell lung carcinoma (NSCLC; about 80% of all lung carcinomas and about 85% of clinical lung cancer cases) 3. Although new molecular targeted therapies have shown promising results in some types of cancer, no known targeted therapy can be applied to a large proportion of lung cancer patients. Despite improvements in early diagnosis of lung cancer, most lung cancers are still diagnosed at an advanced stage. Therefore, the identification of novel diagnostic biomarkers or treatment strategies is critical and essential for the control of lung cancer.
Because of their characteristic nature, microRNAs (miRNAs) have potential for development as diagnostic, prognostic, and targeted therapeutic tools 4. Specific miRNAs, such as miR-21, miR-93, and the miR-17-92 cluster (miR-17, miR-18a, miR-19a, miR-19b-1, miR-20a, and miR-92a-1), have been proven to have important regulatory effects on cell proliferation in a variety of human cancers, including NSCLC 5-10. MiR-93, which belongs to the miR-106b-25 cluster, is an oncomiR in many types of human cancers 11, 12, including NSCLC 13-18. The aberrant expression and dysfunction of miR-93 have been associated with tumor progression, metastasis, and poor prognosis in hepatocellular carcinoma 7, lung cancer 15, breast cancer 19, gastric carcinoma 14, and nasopharyngeal carcinoma 18; these effects are mediated through downregulation of various tumor suppressor genes, such as PTEN, CDKN1A, NEDD4L, SMAD7, PDCD4, and TGFβR2 7, 14, 15, 20. MiR-93 promotes cancer progression by targeting PTEN in osteosarcoma, ovarian cancer, and glioma 21-23. MiR-93 expression has been shown to be increased in tissues and plasma of NSCLC patients and to be a biomarker for early-stage NSCLC 9, 24. The mechanism of miR-93 involvement in NSCLC is unknown, however. Our purpose was to define the mechanism of miR-93 involvement in NSCLC and thus provide a novel potential strategy for molecular targeted therapy for NSCLC.
Materials and Methods
Cell lines and cell culture
NSCLC cell lines A549, NCI-H1975, and NCI-H1299 were purchased from the cell bank of the Chinese Academy of Sciences (Shanghai, China). Each cell line's genetic background is shown in Table 1. The normal lung epithelial cell line BEAS-2B was originally purchased from ATCC (Manassas, VA, USA) and was a gift from Mr. Yilin, Pang, a researcher in our laboratory. All cell lines were authenticated by the cell bank of the Chinese Academy of Sciences or ATCC by using the DNA-fingerprinting method. Cells were cultured in Dulbecco minimal essential medium (DMEM) supplemented with 10% (v/v) heat-inactivated fetal bovine serum (FBS) (Natocor, Cordoba, Argentina) and antibiotics (100 U/mL penicillin and 100 μg/mL streptomycin) in a humidified atmosphere of 5% CO2 at 37°C.
Table 1.
Genetic background of NSCLC cell lines
| NSCLC cell line | Histology | PTEN status | KRAS/LKB1 status | p53 status |
|---|---|---|---|---|
| NCI-H1299 | AD | wt | wt | loss |
| A549 | AD | wt | c,35G>A;p,G12S109C>T; Q37Ter | wt |
| NCI-H1975 | AD | L858R and T790M | wt | R273H |
Note: NSCLC, non-small cell lung carcinoma; AD, adenocarcinoma; wt, wild type.
MiR-93 expression in NSCLC and BEAS-2B cell lines
MiRNAs were extracted from cells by using the method described in our previous study 5. Taqman probes (miR-93, U6) were acquired from the Taqman miRNA assay (Life Technologies, Carlsbad, CA, USA). Subsequent detection by qPCR was performed using the TaqMan PCR Master Mix (TAKARA, Dalian, China) with the StepOne Plus Real Time PCR Detection System (Life Technologies). MiR-93 expression was quantified by the relative quantification method with U6 as the reference gene and was verified by comparing with the control cells.
MiR-93 transfection in NSCLC cell lines
NSCLC cells (2×105) were transfected with 1 μg of plasmids expressing miR-93 inhibitor sponge (miR-93 Down group), miR-93 (miR-93 Up group), or control miRNA (Mock group) (Genepharma, Shanghai, China). Transfection into subconfluent cells was carried out according to the manufacturer's instructions using Lipofectamine3000 for 48 h (Invitrogen-Life Technologies). Proliferation, migration, and invasion were determined in these plasmid-transfected cells.
NSCLC cells were transfected with lentivirus containing the miR-93 inhibitor (miR-93 Down group) or control sequence (Mock group) (Genechem, Shanghai, China). Stable miR-93-deficient or control cell lines were obtained after 10 days of incubation in complete DMEM with puromycin (5 µM). We pooled the successful clones with green fluorescence for the xenograft tumor mouse model experiments.
NSCLC cell proliferation, colony formation, migration, and invasion
Cell proliferation was quantified by measurement of absorbance of water-soluble tetrazolium salt at 450 nm, using the Cell Counter Kit-8 (Dojindo, Kumamoto, Japan) as described previously. Target cells (500 cells) were seeded into 6-well plates and incubated with DMEM with 10% FBS for 15 days. Cells were then fixed with 4% formaldehyde for 20 min, stained with crystal violet for 20 min at ambient temperature, and photographed. The colony-formation capacity of each group of cells was assessed by counting the colonies formed.
Cell migration was assessed by a transwell migration assay. In brief, transfected cells were resuspended in serum-free DMEM and seeded (2.5×104 cells per well) into a 24-well transwell plate with an 8-µm pore membrane insert (Corning, Corning, NY, USA). DMEM with 10% FBS was placed in the lower chambers as a chemoattractant, and cells were incubated for 24 h. Cells that penetrated the membrane were fixed with 4% paraformaldehyde for 20 min, stained with crystal violet for 20 min at ambient temperature, photographed under a microscope (Nikon, Tokyo, Japan), and counted by ImagePro Plus 6.0 software.
For the cell invasion assay, transfected cells were resuspended in serum-free DMEM and seeded (2.5×104 cells per well) into a 24-well transwell plate with an 8-µm pore membrane insert, each well coated with matrigel (1:10 diluted with DMEM; BD Biosciences, Franklin Lakes, NJ, USA). DMEM with 10% FBS was placed in the lower chambers as a chemoattractant, and cells were incubated for 24 h. Cells that penetrated the membrane were fixed with 4% paraformaldehyde for 20 min, stained with crystal violet for 20 min at ambient temperature, photographed under a microscope (Nikon, Tokyo, Japan), and counted.
Western blotting
Cell lysates were prepared for western blotting in Protein Lysis Buffer (Beyotime, Shanghai, China) according to the manufacturer's directions. Protein concentrations were determined by the BCA Assay (Beyotime). An aliquot of 20 µg of denatured protein from each sample was subjected to 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane, followed by incubation at ambient temperature with 3% skimmed milk for 1 h, and then with primary antibody for 1 h (1:1000~10000 dilution, Abcam, Cambridge, MA, USA), then with horseradish peroxidase-conjugated secondary antibody (1:3000 dilution; Beyotime) for 1 h. The blots were then incubated with enhanced chemiluminescence solution for 1 min. The signals were detected by the ChemiDoc XRS+ Chemiluminescence imaging system (Bio-Rad, Hercules, CA, USA) and quantified by densitometry using ImageJ software. β-actin was used as an endogenous control to normalize expression data.
Luminescent reporter gene transfection and luciferase assays
MiRNA targets were predicted by using the TargetScan database (http://www.targetscan.org/). For the luciferase activity assay, ~200-nucleotide sequences (the complete predicted miR-93 target site or the mutant miR-93 target site) of the LKB1 3′-untranslated region (UTR) were inserted downstream of the Renilla luciferase gene in a Renilla/firefly luciferase reporter plasmid, psiCHECK-2 (GenePharma). The psiCHECK-2 plasmids and miR-93 mimic were synthesized by GenePharma. 293T cells were transfected with 0.5 µg of reporter plasmids containing the miR-93 target site or the mutant miR-93 target site, or empty plasmid, followed by dealing with miR-93 mimic. Twenty-four hours after transfection, Renilla/firefly luciferase activity was measured by a dual luciferase reporter assay (Promega, Fitchburg, WI, USA) in an automatic microplate reader (Thermo Fisher Scientific, Waltham, MA, USA).
Xenograft tumor model in mice
Twenty male BALB/c nu/nu nude mice (aged 3 weeks, 17-22 g) were purchased from the Shanghai Medical Experimental Animal Care Commission (Shanghai, China). The mice were housed and manipulated in specific pathogen-free conditions using a laminar airflow rack with continuous access to sterilized food and autoclaved water and controlled light/dark cycle, temperature, and humidity. All experiments were performed in accordance with the Guide for The Care and Use of Laboratory Animals of the U.S. National Institutes of Health. The protocol was approved by the Laboratory Animal Ethics Committee of Wenzhou Medical University, Wenzhou, China (Permit Number: wydw2015-0120).
Experiments commenced after 1 week of mouse acclimatization. The mice were divided randomly into two groups (Mock group and miR-93 Down group). Each mouse was injected with A549 cells transfected with miR-93 inhibitor (miR-93 Down) or control (Mock) miRNA (4×106 cells per mouse in 50 µL of serum-free medium mixed with 50 µL matrigel) into the right middle lobe of the lung. On day 35, five mice of each group were anesthetized with chloral hydrate and killed by decapitation, and their lungs and liver with tumors were removed and photographed. The rest five mice of each group were raised until they dead naturally.
Statistical analysis
The one-way ANOVA and Student t-tests were used, with the Welch correction for variables, to assess the significance of differences between groups. All statistical analyses used GraphPad 6.0 software. A P-value <0.05 was considered statistically significant. For the in vitro studies, data (mean±SD; n=3) and images shown are representative of one experiment. Similar results were obtained in three independent experiments.
Results
MiR-93 is overexpressed in NSCLC cell lines and promotes their proliferation
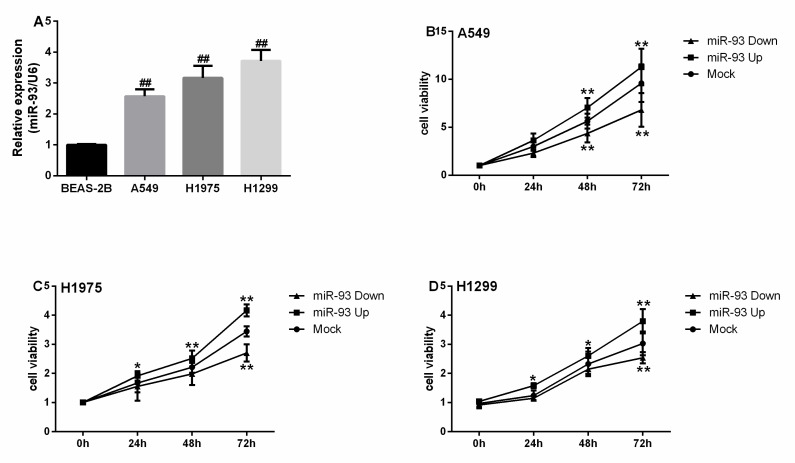
MiR-93 expression in A549, H1975, and H1299 cells was 2.57-, 3.17-, and 3.72-fold higher, respectively, than in BEAS-2B cells (all P<0.001, Figure 1A). The proliferation of A549 cells in the miR-93 Up group was significantly greater than that of the Mock group after culture for 48 h (P<0.01, Figure 1B). The proliferation of the H1975 and H1299 cells in the miR-93 Up groups were significantly greater than that in the Mock groups after culture for 24 h (all P<0.05, Figures 1C-D). The proliferation of A549 cells in the miR-93 Down group was significantly attenuated compared with the Mock group after culture for 48 h (P<0.01, Figure 1B). The proliferation of H1975 and H1299 cells in miR-93 Down groups was attenuated compared with the Mock groups after culture for 72 h (all P<0.01, Figures 1C-D).
Figure 1.
miR-93 is overexpressed in NSCLC cell lines and promotes NSCLC cell proliferation in vitro. (A) miR-93 levels were determined in NSCLC cell lines (A549, H1975, H1299) and normal lung epithelial cells (BEAS-2B). (B-D) Cells were transfected with miR-93 mimic (miR-93 Up), miR-93 inhibitor sponge (miR-93 Down), or control miRNA (Mock) for 72 h and subjected to a proliferation assay. (B) A549 cells. (C) H1975 cells. (D) H1299 cells. ##, P<0.01, compared with BEAS-2B. *P<0.05, ** P<0.01, compared with Mock.
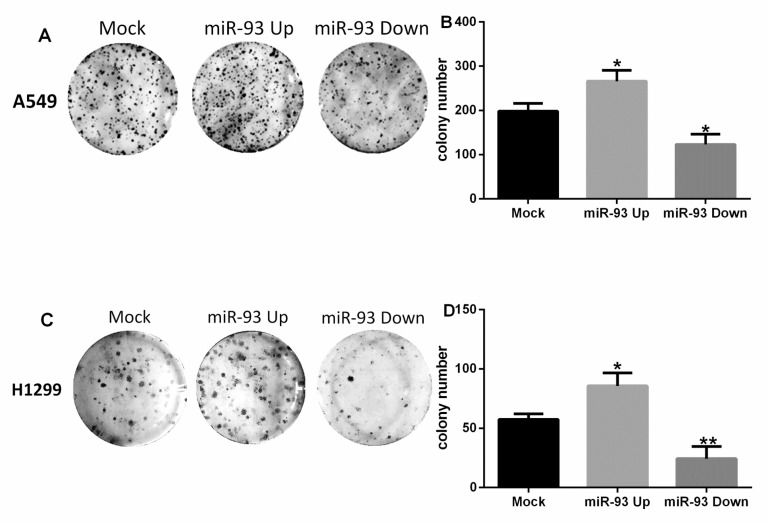
Similar results were observed in the colony-formation assay. The number of colonies formed by A549 cells in the Mock group, the miR-93 Up group, and the miR-93 Down group were 198±18, 266±25, and 123±23, respectively (Figures 2A-B). Compared with the Mock group, the colony-forming capacity of the miR-93 Up group was significantly enhanced (P<0.05) and that of the miR-93 Down group was significantly attenuated (P<0.05). The colonies formed by H1299 cells in the Mock group, the miR-93 Up group, and the miR-93 Down group numbered 57±5, 86±11, and 24±10, respectively (Figures 2C-D). Compared with the Mock group, the colony-forming capacity of the miR-93 Up group was enhanced (P<0.05), and that of the miR-93 Down group was attenuated (P<0.01).
Figure 2.
miR-93 promotes colony formation by NSCLC cells in vitro. Cells were transfected with miR-93 mimic (miR-93 Up), miR-93 inhibitor sponge (miR-93 Down), or control miRNA (Mock) and incubated in 6-well culture plates. Colonies were counted after 15 days. (A-B) A549 cells. (C-D) H1299 cells. *P<0.05, **P<0.01, compared with Mock.
MiR-93 overexpression promotes migration and invasion of NSCLC cells
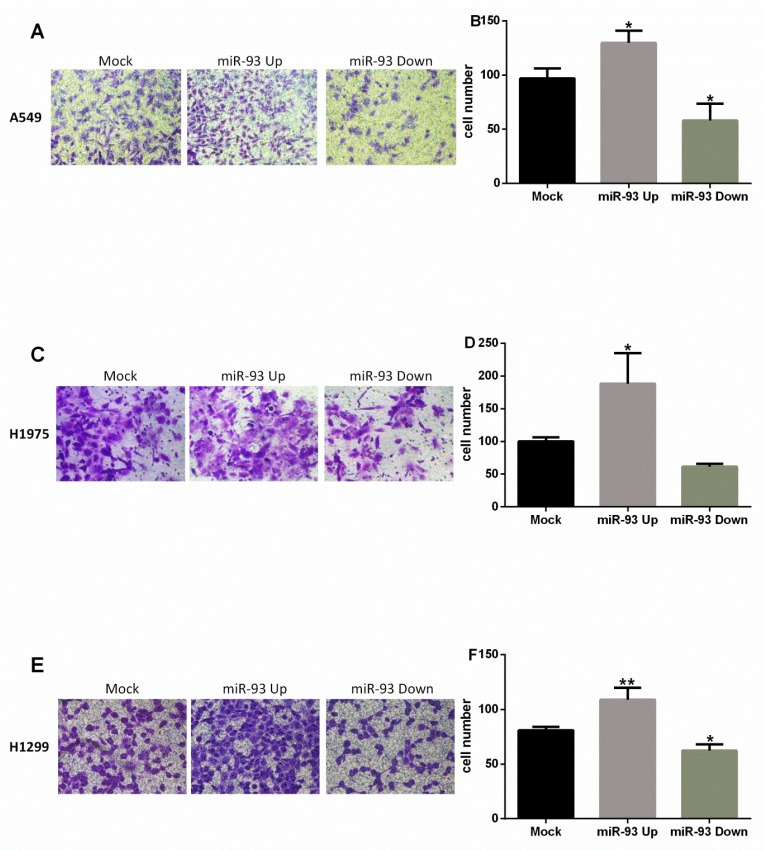
The migration of NSCLC cells was promoted by overexpression of miR-93 and attenuated by inhibition of miR-93. Compared with Mock group, A549 cells in the miR-93 Up group had greater penetration (130±11 vs 97±9, P<0.05), whereas those in the miR-93 Down group had reduced penetration (58±16 vs 97±9, P<0.05; Figures 3A-B). In H1975 cells, the miR-93 Up group also had significantly greater penetration than the Mock group (188±47 vs 100±7, P<0.05); the miR-93 Down group had reduced penetration, but the difference was not significant (61±5 vs 100±7, P>0.05; Figures 3C-D). In H1299 cells, the miR-93 Up group had significantly greater penetration than the Mock group (109±11 vs 81±3, P<0.01), and the miR-93 Down group had significantly less penetration (62±6 vs 81±3, P<0.05; Figures 3E-F).
Figure 3.
miR-93 promotes NSCLC cell migration in vitro. Cell migration was assessed by determining the number of cells that migrated through the transwell chamber. Cells were transfected with miR-93 mimic (miR-93 Up), miR-93 inhibitor sponge (miR-93 Down), or control miRNA (Mock) and placed in the upper chambers. Cells that penetrated to the lower chambers were counted. (A-B) A549 cells. (C-D) H1975 cells. (E-F) H1299 cells. *P<0.05, **P<0.01, compared with Mock.
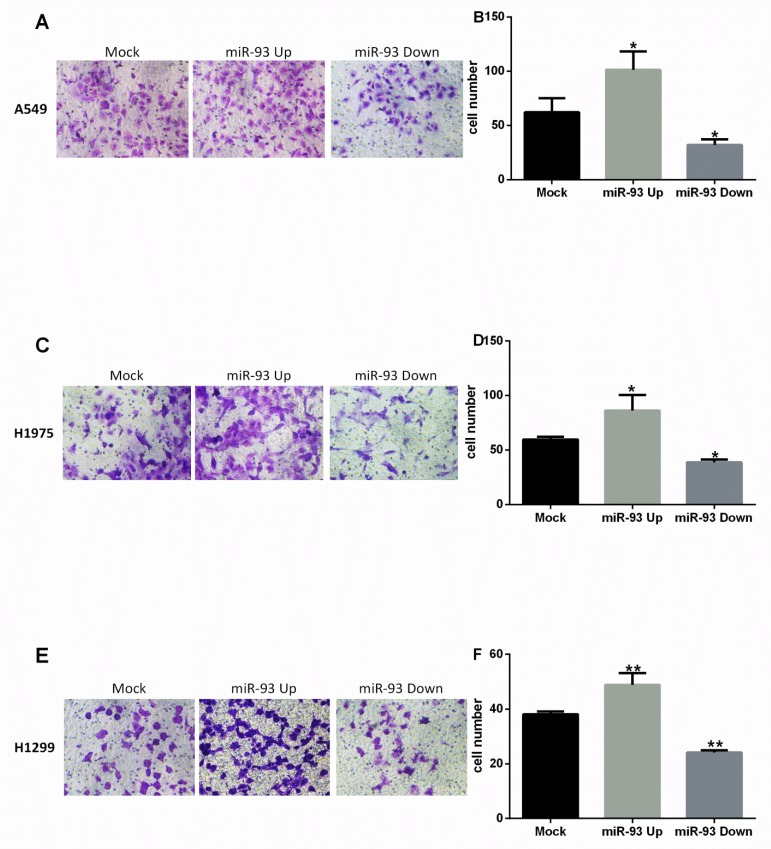
Similar results were observed in the transwell invasion assay. Invasion was enhanced in NSCLC cells overexpressing miR-93 and inhibited in NSCLC cells in which miR-93 was downregulated. In A549 cells, invasion was significantly greater in the miR-93 Up group than in the Mock group (101±17 vs 62±13, P<0.05), while invasion was decreased in the miR-93 Down group (32±5 vs 62±13, P<0.05; Figures 4A-B). In H1975 cells, invasion also was greater in the miR-93 Up group than in the Mock group (86±14 vs 60±3, P<0.05) and decreased in the miR-93 Down group (39±3 vs 60±3, P<0.05; Figures 4C-D). In H1299 cells, similarly, invasion was significantly greater in the miR-93 Up group than in the Mock group (86±14 vs 60±3, P<0.01) and decreased in the miR-93 Down group (39±3 vs 60±3, P<0.05; Figures 4E-F).
Figure 4.
miR-93 promotes NSCLC cell invasion in vitro. Cell invasiveness was assessed by determining the number of cells that migrated through a matrigel coating in a transwell chamber. Cells transfected with miR-93 mimic (miR-93 Up), miR-93 inhibitor sponge (miR-93 Down), or control miRNA (Mock) were placed in the upper transwell chambers and allowed to incubate for 24 h. Cells that penetrated to the lower chambers were counted. (A-B) A549 cells. (C-D) H1975 cells. (E-F) H1299 cells. *P<0.05, **P<0.01, compared with Mock.
MiR-93 activates the PI3K-Akt pathway by targeting LKB1/PTEN/CDKN1A
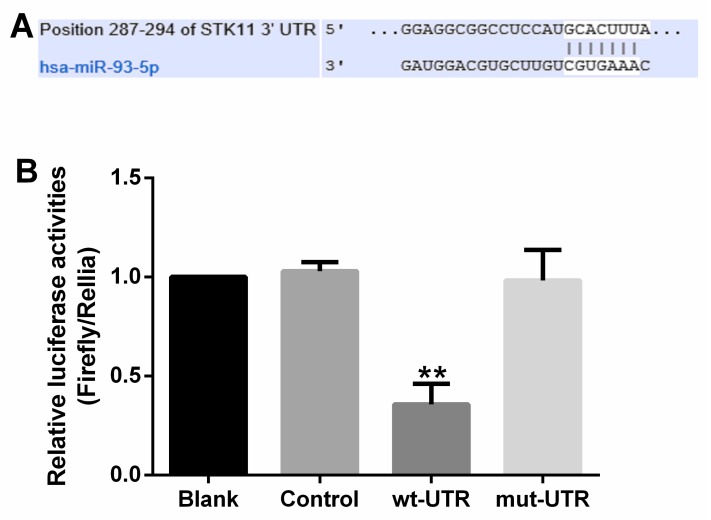
Analysis in silico indicated that LKB1, PTEN, and CDKN1A were candidate targets of miR-93. Katsuya et al. showed, using a dual luciferase assay, that PTEN and CDKN1A are direct targets of miR-93 7. Results of our dual luciferase assay showed that LKB1 is a novel candidate target of miR-93 with a 7mer binding site (Figure 5A). To demonstrate direct binding of miR-93 to this gene, we co-transfected plasmids containing miR-93 mimic, control miRNA, or control together with a vector expressing LKB1 wild-type 3′-UTR, LKB1 mutant 3′-UTR, or empty vector into 293T cells and analyzed their luciferase activity. 293T cells transfected with miR-93 mimic had 64% lower luciferase activity than parental 293T cells, 293T cells transfected with the control plasmid, or 293T cells transfected with the plasmid expressing LKB1 mutant 3′-UTR (all P<0.01; Figure 5B), with no obvious difference among these three groups. This suggested that LKB1 is a target of miR-93.
Figure 5.
miR-93 directly targets LKB1. (A) Sequence alignment of miR-93 and its conserved target site in the 3′-UTR of LKB1. (B) Luciferase activity was measured in 293T cells with a dual luciferase reporter assay. The cells were transfected with plasmids containing the wild-type LKB1 3′-UTR (wt-3′UTR), mutant LKB1 3′-UTR (mut-3′UTR), empty vector (Mock), or nothing (Blank) and co-transfected with miR-93 mimic. Firefly luciferase activity was normalized to Renilla luciferase activity, and the value for the empty vector was used as the control. **P<0.01, compared with all other groups.
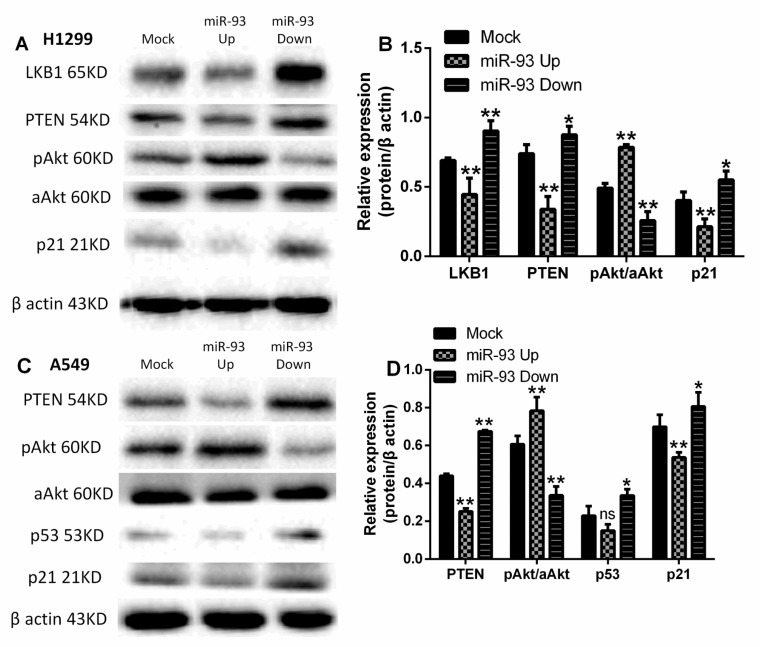
In H1299 cells, the levels of LKB1, PTEN, and p21 proteins were lower in the miR-93 Up group than in the Mock group (all P<0.01) and increased in the miR-93 Down group (P<0.01, P<0.05, P<0.05; Figures 6A-B). Conversely, the phosphorylation of Akt was increased in the miR-93 Up group (P<0.01) and decreased in the miR-93 Down group (P<0.01). Because of the mutation of LKB1 in A549 cells, we determined the levels of PTEN/p53/p21, phosphorylated Akt (p-Akt), and all-Akt in A549 cells. In A549 cells, similarly, the expression of PTEN, p53, and p21 was lower in the miR-93 Up group than in the Mock group (P<0.01, P>0.05, P<0.01) and increased in the miR-93 Down group (P<0.01, P<0.05, P<0.05; Figures 6C-D). Again, the phosphorylation of Akt was increased in the miR-93 Up group (P<0.01) and decreased in the miR-93 Down group (P<0.01).
Figure 6.
miR-93 regulates expression of LKB1/PTEN/p21 and PI3K/Akt pathway proteins. Levels of the indicated PI3K/Akt pathway proteins were analyzed in (A-B) H1299 and (C-D) A549 cells by western blotting; differences in expression are represented in the bar graphs. Cells were first transfected with miR-93 mimic (miR-93 Up), miR-93 inhibitor sponge (miR-93 Down), or control miRNA (Mock). *P<0.05, **P<0.01, compared with Mock.
Downregulation of miR-93 inhibits metastasis and prolongs the survival time in a NSCLC xenograft mouse model
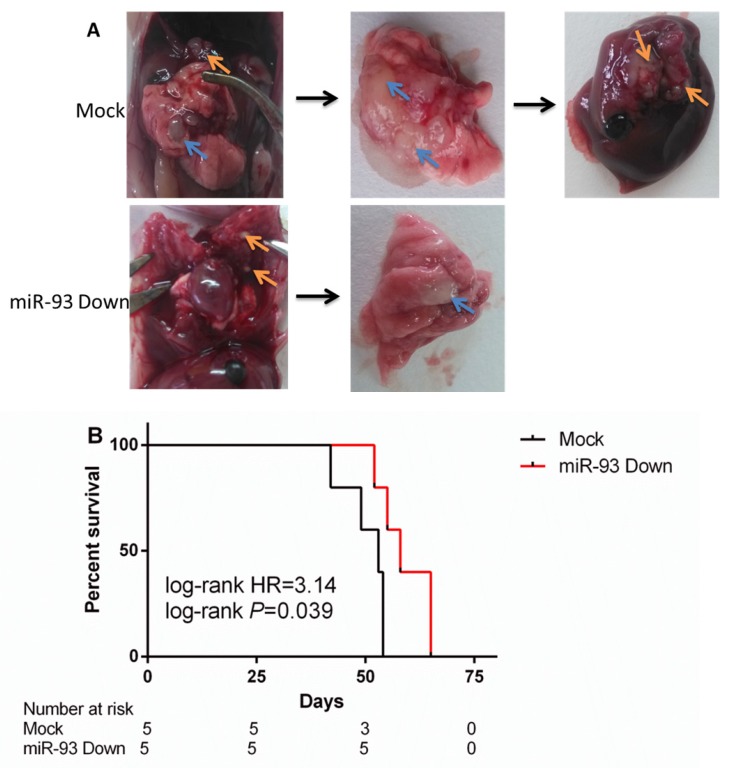
The tumors that developed from the miR-93 Down A549 cells were associated with significantly lower rates of pleural dissemination and hepatic metastasis than those that developed from controls (Figure 7A). The rate of tumorigenesis was not significantly different between the Mock group and the miR-93 Down group (100% vs 100%). The miR-93 Down group had a lower rate of pleural dissemination than the Mock group (80% vs 100%, P<0.05) and no hepatic metastasis (0 vs 80%, P<0.01). The survival time of miR-93 Down group was significantly longer than that of Mock group (P=0.039, Figure 7B), with the median survival time was 58 days vs 53 days.
Figure 7.
Downregulation of miR-93 inhibits lung cancer hepatic metastasis and prolongs the survival time in vivo. The lung cancer mouse model was constructed by injecting mice with A549 cells transfected with empty lentivirus (Mock) or miR-93 inhibitor lentivirus (miR-93 Down). (A) Five mice of each group were killed on day 35 and their tumors were removed. The blue arrows indicate the primary tumor location. The orange arrows indicate the metastatic tumor location. (B) Survival curve: the rest five mice of each group were raised until dead naturally.
Discussion
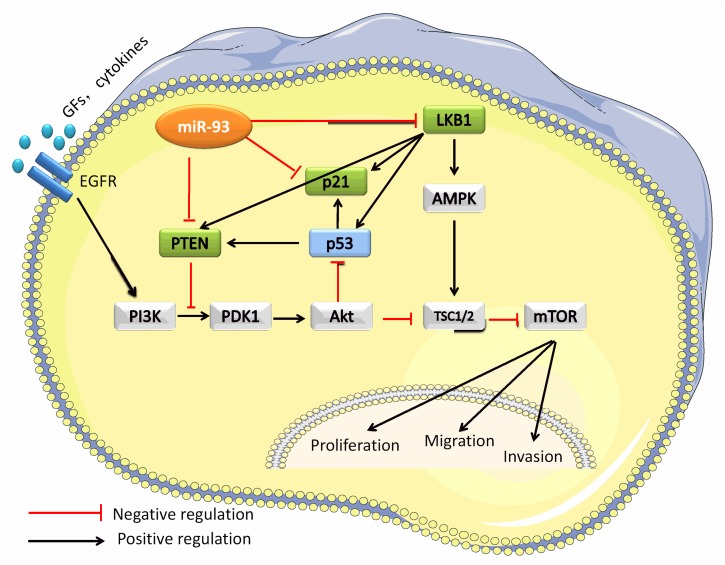
With this study, we demonstrated that miR-93 promotes lung cancer proliferation and metastasis both in vitro and in vivo and identified LKB1 as a novel target of miR-93 in NSCLC. We further demonstrated that miR-93 inhibits LKB1, PTEN, and CDKN1A, thereby activating proliferation and metastases through the PI3K/Akt pathway in NSCLC. Previous studies have shown that miR-93 promotes proliferation in liver cancer by directly targeting PTEN and CDKN1A 7. Activation of miR-93 not only blocked LKB1, PTEN, and p21 directly but also blocked PTEN and p21 indirectly by blocking LKB1, because LKB1 is known to induce the expression of PTEN, p53, and p21 25. When LKB1, PTEN, and p21 were blocked by miR-93, the PI3K/Akt pathway was activated to promote proliferation, migration, and invasion (Figure 8).
Figure 8.
MiR-93 activates the PI3K/Akt pathway by targeting LKB1/PTEN/p21. This schematic figure represents the mechanism whereby miR-93 activates the PI3K/Akt pathway. MiR-93 binds directly binds to the 3′-UTRs of LBK1, PTEN, and CDKN1A.
Our results indicate that the proliferation, migration, and invasion of A549, H1975, and H1299 NSCLC cells were markedly promoted by overexpression of miR-93 and abolished by downregulation of miR-93 in vitro. The hepatic metastasis of A549 tumors in a nude mouse model was significantly inhibited by miR-93 downregulation and the survival time was prolonged. Even though the three cell lines have different genetic backgrounds, the functional changes mediated by miR-93 expression were the same in all three. We speculate that the potential mechanism of miR-93 involvement in NSCLC is based on a multiple target effect.
LKB1, also known as STK11, is a tumor suppressor gene located in chromosome 19p13 and on translation takes on a type kind of serine-threonine kinase function as an inhibitor of mTOR 26. LKB1 is involved in many biological processes, such as cell cycle control, chromatin remodeling, cell polarity, and energy metabolism. LKB1 mutation is common in Peutz-Jeghers syndrome 27. In lung cancer, LKB1 is truncated and deactivated via somatic mutation or deletion of LKB1 28. The incidence of this is 11%-30% in lung adenocarcinoma 29. In NSCLC, LKB1 is somatically inactivated in 25% to 30% of cases, often concurrently with activating KRAS mutations 30. Therefore, LKB1 mutation is the third most common mutation in lung adenocarcinoma, after TP53 and KRAS.
Because of the induction of PTEN, p53, and p21 proteins by LKB1 25, 31, 32, loss of LKB1 function activates the PI3K/Akt/mTOR pathway to promote proliferation and metastasis 33. PI3K is the center of a variety of signaling pathways, and it has important off-target effects. Because of the many molecular changes in NSCLC, such as amplification and mutation of PIK3CA, PTEN, and AKT, loss of LKB1, and KRAS mutation, targeting the PI3K/Akt pathway is a rational and promising therapeutic approach in this cancer. Mk-2206, a type of oral protease inhibitor, was combined with Akt inactivation to inhibit NSCLC progression in clinical trials 34.
Our results indicate that miR-93 activates the PI3K/Akt pathway by inhibiting LKB1, PTEN, and p21. We conclude that miR-93 is not only a potential diagnostic biomarker for NSCLC but also a candidate target for treatment. A number of clinical trials using miRNA profiling for patient prognosis and clinical response are currently underway 35-37. To complete transition of miRNA research from basic to bedside, however, for application of miRNAs as novel biomarkers and treatment targets, several obstacles remain.
Conclusion
Consistent with the previous studies, we have confirmed that miR-93 is an oncomiR in NSCLC. Moreover, we demonstrate that miR-93 is not only inducing proliferation, migration, and invasion in vitro, but also promoting tumor metastases in vivo, in NSCLC. We demonstrate for the first time that LKB1 is a target of miR-93 in NSCLC. The tumorigenic activity of miR-93 is mediated by activation of the oncogenic PI3K/Akt pathway through inhibition of LKB1, PTEN, and p21 expression. We speculate that miR-93 could be a promising therapeutic target in NSCLC.
Acknowledgments
We thank Huaibin Zhou, Jindan Wang, Binjiao Zheng, Danli Xie, and Chaowei Wen for technical assistance.
Funding
This work was supported by grants from the National Natural Science Foundation of China (81170257). This study was also partially supported by the MD Anderson Cancer Center Startup Fund. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Authors' contribution
QHM, JL, and CL conceived and designed the study. CL carried out most of the experiments. CL, JL, and QHM prepared the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All animal procedures and experimental protocols were approved by Laboratory Animal Ethics Committee of Wenzhou Medical University.
Abbreviations
- Akt
protein kinase B
- CDKN1A
cyclin-dependent kinase inhibitor 1A
- FBS
fetal bovine serum
- LKB1
liver kinase B1
- miR-93
microRNA-93
- mTOR
mammalian target of rapamycin
- NSCLC
non-small cell lung cancer
- PI3K
phosphatidyl inositol 3-kinase
- PTEN
phosphatase and tensin homolog
- RT-PCR
real-time quantitative polymerase chain reaction.
References
- 1.Chen W, Zheng R, Baade PD. et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;2:115–32. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;1:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 3.Inamura K, Ishikawa Y. MicroRNA In Lung Cancer: Novel Biomarkers and Potential Tools for Treatment. J Clin Med. 2016;3:36. doi: 10.3390/jcm5030036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;2:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Li C, Zhao L, Chen Y. et al. MicroRNA-21 promotes proliferation, migration, and invasion of colorectal cancer, and tumor growth associated with down-regulation of sec23a expression. BMC Cancer. 2016;16:605. doi: 10.1186/s12885-016-2628-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guinot A, Oeztuerk-Winder F, Ventura JJ. MiR-17-92/p38α Dysregulation Enhances Wnt Signaling and Selects Lgr6+ Cancer Stem-like Cells during Lung Adenocarcinoma Progression. Cancer Res. 2016;76:4012–22. doi: 10.1158/0008-5472.CAN-15-3302. [DOI] [PubMed] [Google Scholar]
- 7.Ohta K, Hoshino H, Wang J. et al. MicroRNA-93 activates c-Met/PI3K/Akt pathway activity in hepatocellular carcinoma by directly inhibiting PTEN and CDKN1A. Oncotarget. 2015;5:3211–24. doi: 10.18632/oncotarget.3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang Y, Meng H, Peng Q. et al. Downregulation of microRNA-21 expression restrains non-small cell lung cancer cell proliferation and migration through upregulation of programmed cell death 4. Cancer Gene Ther. 2015;1:23–9. doi: 10.1038/cgt.2014.66. [DOI] [PubMed] [Google Scholar]
- 9.Du L, Zhao Z, Ma X. et al. MiR-93-directed downregulation of DAB2 defines a novel oncogenic pathway in lung cancer. Oncogene. 2014;34:4307–15. doi: 10.1038/onc.2013.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mendell JT. MiRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;2:217–22. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murphy BL, Obad S, Bihannic L. et al. Silencing of the miR-17~92 cluster family inhibits medulloblastoma progression. Cancer Res. 2013;23:7068–78. doi: 10.1158/0008-5472.CAN-13-0927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith AL, Iwanaga R, Drasin DJ. et al. The miR-106b-25 cluster targets Smad7, activates TGF-β signaling, and induces EMT and tumor initiating cell characteristics downstream of Six1 in human breast cancer. Oncogene. 2012;31:5162–71. doi: 10.1038/onc.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu J, Wang H, Wang Y. et al. Repression of the miR-93-enhanced sensitivity of bladder carcinoma to chemotherapy involves the regulation of LASS2. Onco Targets Ther. 2016;9:1813–22. doi: 10.2147/OTT.S97399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liang H, Wang F, Chu D. et al. MiR-93 functions as an oncomiR for the downregulation of PDCD4 in gastric carcinoma. Sci Rep. 2016;6:23772. doi: 10.1038/srep23772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qu MH, Han C, Srivastava AK. et al. MiR-93 promotes TGF-β-induced epithelial-to-mesenchymal transition through downregulation of NEDD4L in lung cancer cells. Tumour Biol. 2016;37:5645–51. doi: 10.1007/s13277-015-4328-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi J, Jiang X, Yu Z. et al. ZNRF3 contributes to the growth of lung carcinoma via inhibiting Wnt/β-catenin pathway and is regulated by miR-93. Tumour Biol. 2016;37:3051–7. doi: 10.1007/s13277-015-3949-2. [DOI] [PubMed] [Google Scholar]
- 17.Li C, Ding C, Chen T. et al. Micro ribonucleic acid-93 promotes proliferation and migration of esophageal squamous cell carcinoma by targeting disabled 2. Thorac Cancer. 2015;4:524–33. doi: 10.1111/1759-7714.12242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lyu X, Fang W, Cai L. et al. TGFβR2 is a major target of miR-93 in nasopharyngeal carcinoma aggressiveness. Mol Cancer. 2014;13:51. doi: 10.1186/1476-4598-13-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fang L, Du WW, Yang W. et al. MiR-93 enhances angiogenesis and metastasis by targeting LATS2. Cell Cycle. 2012;23:4352–65. doi: 10.4161/cc.22670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yeung ML, Yasunaga J, Bennasser Y. et al. Roles for microRNAs, miR-93 and miR-130b, and tumor protein 53-induced nuclear protein 1 tumor suppressor in cell growth dysregulation by human T-cell lymphotrophic virus 1. Cancer Res. 2008;21:8976–85. doi: 10.1158/0008-5472.CAN-08-0769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawano M, Tanaka K, Itonaga I. et al. MicroRNA-93 promotes cell proliferation via targeting of PTEN in Osteosarcoma cells. J Exp Clin Cancer Res. 2015;34:76. doi: 10.1186/s13046-015-0192-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen Q, Qin R, Fang Y. et al. Berberine Sensitizes Human Ovarian Cancer Cells to Cisplatin Through miR-93/PTEN/Akt Signaling Pathway. Cell Physiol Biochem. 2015;3:956–65. doi: 10.1159/000430270. [DOI] [PubMed] [Google Scholar]
- 23.Jiang L, Wang C, Lei F. et al. MiR-93 promotes cell proliferation in gliomas through activation of PI3K/Akt signaling pathway. Oncotarget. 2015;10:8286–99. doi: 10.18632/oncotarget.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu W, He J, Chen D. et al. Expression of miR-29c, miR-93, and miR-429 as potential biomarkers for detection of early stage non-small lung cancer. PLoS One. 2014;2:e87780. doi: 10.1371/journal.pone.0087780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jimenez AI, Fernandez P, Dominguez O. et al. Growth and molecular profile of lung cancer cells expressing ectopic LKB1: down-regulation of the phosphatidylinositol 3'-phosphate kinase/PTEN pathway. Cancer Res. 2003;6:1382–8. [PubMed] [Google Scholar]
- 26.Shaw RJ, Bardeesy N, Manning BD. et al. The LKB1 tumor suppressor negatively regulates mTOR signaling. Cancer Cell. 2004;1:91–9. doi: 10.1016/j.ccr.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 27.Shorning BY, Clarke AR. Energy sensing and cancer: LKB1 function and lessons learnt from Peutz-Jeghers syndrome. Semin Cell Dev Biol; 2016. pp. 21–9. [DOI] [PubMed] [Google Scholar]
- 28.Ding L, Getz G, Wheeler DA. et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;7216:1069–75. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koivunen JP, Kim J, Lee J. et al. Mutations in the LKB1 tumour suppressor are frequently detected in tumours from Caucasian but not Asian lung cancer patients. Br J Cancer. 2008;2:245–52. doi: 10.1038/sj.bjc.6604469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y, Marks K, Cowley GS. et al. Metabolic and functional genomic studies identify deoxythymidylate kinase as a target in LKB1-mutant lung cancer. Cancer Discov. 2013;8:870–9. doi: 10.1158/2159-8290.CD-13-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hou L, Liu T, Wang J. Isoflurane suppresses the self-renewal of normal mouse neural stem cells in a p53-dependent manner by activating the Lkb1-p53-p21 signalling pathway. Mol Med Rep. 2015;5:7412–8. doi: 10.3892/mmr.2015.4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu C, Fillmore CM, Koyama S. et al. Loss of Lkb1 and Pten leads to lung squamous cell carcinoma with elevated PD-L1 expression. Cancer Cell. 2014;5:590–604. doi: 10.1016/j.ccr.2014.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Faubert B, Vincent EE, Poffenberger MC. et al. The AMP-activated protein kinase (AMPK) and cancer: many faces of a metabolic regulator. Cancer Lett. 2015;2(Pt A):165–70. doi: 10.1016/j.canlet.2014.01.018. [DOI] [PubMed] [Google Scholar]
- 34.Hirai H, Sootome H, Nakatsuru Y. et al. MK-2206, an allosteric Akt inhibitor, enhances antitumor efficacy by standard chemotherapeutic agents or molecular targeted drugs in vitro and in vivo. Mol Cancer Ther. 2010;7:1956–67. doi: 10.1158/1535-7163.MCT-09-1012. [DOI] [PubMed] [Google Scholar]
- 35.Feng B, Zhang K, Wang R. et al. Non-small-cell lung cancer and miRNAs: novel biomarkers and promising tools for treatment. Clin Sci (Lond) 2015;10:619–34. doi: 10.1042/CS20140530. [DOI] [PubMed] [Google Scholar]
- 36.Lanford RE, Hildebrandt-Eriksen ES, Petri A. et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;5962:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Agostini M, Knight RA. MiR-34: from bench to bedside. Oncotarget. 2014;4:872–81. doi: 10.18632/oncotarget.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]