Abstract
Receptor tyrosine kinase EGFR usually is localized on plasma membrane to induce progression of many cancers including cancers in children (Bodey et al. In Vivo. 2005, 19:931-41), but it contains a nuclear localization signal (NLS) that mediates EGFR nuclear translocation (Lin et al. Nat Cell Biol. 2001, 3:802-8). Here we report that NLS of EGFR has its old evolutionary origin. Protein-protein interaction maps suggests that nEGFR pathways are different from membrane EGFR and EGF is not found in nEGFR network while androgen receptor (AR) is found, which suggests the evolution of prostate cancer, a well-known AR driven cancer, through changes in androgen- or EGF-dependence. Database analysis suggests that nEGFR correlates with the tumor grades especially in prostate cancer patients. Structural predication analysis suggests that NLS can compromise the differential protein binding to EGFR through stretch linkers with evolutionary mutation from N to V. In experiment, elevation of nEGFR but not membrane EGFR was found in castration resistant prostate cancer cells. Finally, systems analysis of NLS and transmembrane domain (TM) suggests that NLS has old origin while NLS neighboring domain of TM has been undergone accelerated evolution. Thus nEGFR has an old origin resembling the cancer evolution but TM may interfere with NLS driven signaling for natural selection of survival to evade NLS induced aggressive cancers. Our data suggest NLS is a dynamic inducer of EGFR oncogenesis during evolution for advanced cancers. Our model provides novel insights into the evolutionary role of NLS of oncogenic kinases in cancers.
Keywords: NLS, EGFR, prostate cancer.
Introduction
One of the prominent features of eukaryotic cells is the spatial separation of genetic materials from the translational machinery by the nuclear envelope 1,2. Molecules smaller than 40 kDa can passively diffuse through the nucleus envelope, whereas compounds greater than 40 kDa require an active transport 3 which is mediated by the signal peptide named as nuclear localization signal or sequence (NLS). NLS guided transport is mediated by assisting proteins such as importins 1. NLS can be classified into two major categories: monopartite and bipartite 4. The monopartite NLS contains only one cluster of basic amino acids, while the bipartite NLS has two clusters of basic amino acids separated by a linker region of 10-12 residues 2, 5, 6.
Dysregulation of NLS-mediated transport has been found in many cancers 7. For example, dysregulated nuclear transport of NF- κB is associated with cancers 2, 7, 8. Recent studies showed that nuclear epidermal growth factor receptor (nEGFR) is essential in cancer 9. EGFR is a receptor tyrosine kinase which is normally localized on the plasma membrane 9. Upon binding to EGF, membrane EGFR dimerizes leading to kinase activation 10 to stimulate several downstream signaling pathways such as PLC-γ-PKC, PI-3K-Akt-mTOR, and JAK2-STAT3 11. These oncogenic pathways are essential in tumorigenesis, cancer progression, survival, resistance to chemotherapy, and metastasis 11, 12.
Nuclear localized EGFR has been found in regenerated hepatocytes, benign and cancerous tumors 9, 11, 13, 14, 15. nEGFR is induced by chemo- and radiotherapy 10, indicating its association with the drug resistance. Sequencing analysis uncovered that nEGFR translocation is mediated by its tripartite NLS that contains three clusters of basic amino acids separated by linker regions at 2-3 residues in length 9,16. The EGFR NLS has the sequence of amino acids as 645 RRRHIVRKRTLRR 657 9.
nEGFR plays essential roles in transcriptional regulation, protein kinase, and physical interactions with other molecules 9, 14. As a transcription regulator, nEGFR can activate the expression of various genes related to chromosome instability, cell proliferation, inflammation, and resistance to cancer drugs that include cyclin D1, iNOS, b-Myb, Aurora-A, cyclooxygenase-2, c-Myc, thymidylate synthase, BCRP, and STAT1 9,11,13,14,16,17. Interestingly, nEGFR does not contain a DNA-binding domain, but can form a complex with other proteins to activate transcription 11, 16. The information on nEGFR and membrane EGFR interaction with other pathways during evolution is not well analyzed. Studying the protein-protein interaction maps during evolution would reveal the whole networks of nEGFR and membrane EGFR in differential signaling pathways for natural selection.
In clinical studies, enhanced expression of nEGFR has been found in many cancers such as breast carcinoma 18, 19, ovarian cancer 20, non-small cell lung cancer 21, gallbladder carcinoma 22, and oropharyngeal 23. Recently, nEGFR elevation has been found to promote bone metastasis of prostate cancer through repression of miR-1 thereby elevating of miR-1 target of TWIST1, an EMT inducer 25. In clinical samples of prostate cancer, nEGFR levels were inversely correlated with the miR-1 tumor suppressor 25. Xia et al. found nEGFR levels were inversely related to the overall survival of ovarian cancer patients 26. It is worth mentioning that prognostic value of the high levels of nEGFR still remains unclear in many cancers 27. This study aims to investigate the original or evolved form (nuclear or membrane) of EGFR during cancer development and EGFR contributions to the evolution of cancer using systems biology approach.
Materials and Methods
Nuclear localization signal reference and sequence search
The NLS sequence in human EGFR, 645 RRRHIVRKRTLRR 657, was used as a reference for the comparison of the NLS 9 in randomly selected species. The sequences of selected species were obtained from GenBank database.
Correlation analysis of nuclear EGFR and cancer progression using database sets
The PubMed, google scholar were used for searching literatures on nuclear EGFR in cancer progression. Several studies were reviewed to find correlations between nuclear EGFR and cancer development 19, 25, 26, 29. Membrane EGFR expression data were used as control. Pearson correlation was used to measure a linear relationship between expressions of EGFR with cancer stages.
Construction of protein-protein interaction maps and predications of protein structures
STRING software was used to construct interaction maps of membrane EGFR and nuclear EGFR respectively (http://version10a.string-db.org/). Protein structures were predicated by inputting sequences to software (http://raptorx.uchicago.edu/StructurePrediction/) and NLS region were marked.
Cell culture, treatment and western blot
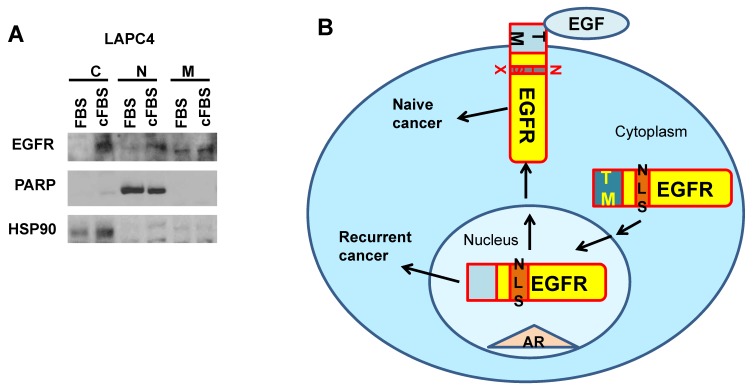
LAPC4 cells were cultured in DMEM medium (Life Technologies) with or without androgen depletion treatments by charcoal striped FBS (Life Technologies) for 7 days and were passaged twice. Cells were lysed in buffer provided by the Qproteome Cell Compartment Kit (Qiagen), and cell lysates from nucleus, cytosol, and membrane were subjected to western blot analysis (protocol provided by BioRad) with antibodies of EGFR (Cell signaling), PARP (Cell signaling), and HSP90 (Cell signaling).
Analysis of nuclear localization signal conservation and construction of phylogenetic trees
Protein BLAST was used to obtain identity percentage of the full length EGFR protein and NLS sequence in the randomly selected species. A cladogram was used to estimate overall organisms' relatedness and conservation of NLS in different species 25. All animals which have discovered EGFR sequences were used for further construction of phylogenetic trees with tools and protein sequences of EGFR taken from database as described in supplementary figure legend.
Results
EGFR is highly conserved among species closely related to human
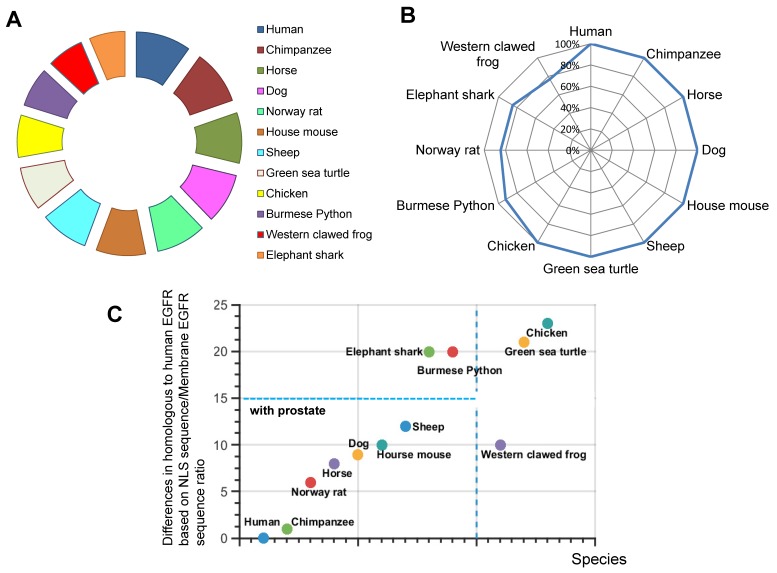
The presence and evolutionary relationships of EGFR among various species was studied. The hypothesis is that the possession of high identity in EGFR protein will indicate that species are closely related to each other during EGFR evolution. To test this, we analyzed 11 randomly selected species using BLAST database. We found the percentage identity in the full length EGFR in relative to human EGFR sequence (Fig.1A). Our analyses showed that EGFR is conserved throughout various species. For example, Pygmy chimpanzee, Horse, Dog, Norway rat and House mouse of EGFR sequence are 100% identity with human EGFR (Fig. 1A). Thus, EGFR is conserved in mammalian species closely related to human.
Figure 1.
Homologues comparisons of EGFR and NLS of EGFR in different species. A. Similarity of EGFR. B. Similarity of NLS of EGFR. C. Percentage difference in NLS of EGFR and full length EGFR.
Nuclear localization signal of EGFR is highly conserved among species with the prostate
We were interested in understanding how EGFR is translocated into the nucleus. By taking the advantage of recent finding about NLS sequence (RRRHIVRKRTLRR) 9, we decided to determine the evolutionary origin of this nuclear event. Therefore, we used the sequence of human EGFR NLS as a reference to compare with previously chosen species. To our surprise, compared to the full-length EGFR, more organisms have a 100 % identity with human NLS, namely, Pygmy chimpanzee, Horse, Dog, House mouse, Sheep, and Green sea turtle, Chicken (Fig. 1B). By contrast, NLS sequences in Burmese python, Norway rat, Elephant shark and Western clawed frog were less identical to human NLS. We found incidences of different percentages in identity (Fig. 1B). Hence, we focused on the establishment of phylogenetic characterization of selected species.
We next investigated whether there was any correlation between the full length EGFR conservation and the presence of NLS. To estimate the extent of NLS conservation in comparison with full length EGFR sequence, the relative percentage difference for similarity was obtained by a ratio of similarities of NLS vs full length EGFR. Our results demonstrate that Human, Chimpanzee, Norway rat, Horse, Dog, Western clawed frog, House mouse and Sheep have less than 10% difference in full length EGFR sequence and NLS sequence (Fig. 1C). Most of these animals have prostate organs (Fig. 1C). Meanwhile, Elephant shark, Burmese python, Green sea turtle, and Chicken account for more than 20% difference between NLS sequence and full length EGFR sequence (Fig. 1C). Thus, our results suggest that the conservation and evolution of the full length EGFR and NLS are independent events.
Nuclear localization signal of EGFR has old evolutionary origin
To determine whether nEGFR is an old or modern evolutionary event, phylogenetic analysis was performed. First we used cladogram to estimate overall organism's relatedness and conservation of NLS in different species (data not shown) 25, 30. Human has a common ancestor with elephant Shark, Jaws, as well as Western Clawed frog and Four-Legged 31. NLS similarities of human with these species above are 84.61% and 76.92%, respectively. Furthermore, Green sea turtle and Burmese Python showing NLS similarities of 100% and 92.31%, respectively, were found to come from Amniotic Egg ancestor. From the previously mentioned examples, we can see that some species from older periods, such as Elephant Shark, show higher identity percentages of NLS of EGFR. Thus, our analysis suggests that EGFR nuclear localization sequence has old evolutionary origin.
The mutations of nuclear localization signal of EGFR remain the similar character of amino acids
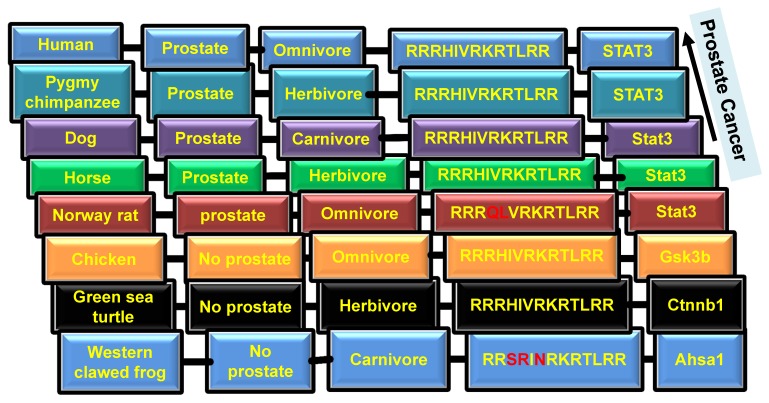
To further examine NLS of EGFR sequence in evolution, amino acid analysis of NLS sequences of 11 randomly selected species was performed. Our analysis reveals that human NLS of EGFR (RRRHIVRKRTLRR) has three upstream stretches of basic amino acids (RRR, RKR and RR, here R is arginine, K is lysine, which are known to be positive and basic polar amino acids). In addition, threonine and leucine amino acid in the RRRHIVRKRTLRR sequence is conserved among all species 32. The combination of isoleucine and valine amino acids is present in the majority of species. Moreover, some specific changes of amino acids are present in Western clawed frog, Burmese python, and Norway rat. Specifically, Western clawed frog has SR amino acids instead of RH in human; Burmese python has R amino acid instead of H; Norway rat has QL instead of HI. In the most cases, amino acids are evolved in or changed to the same character for NLS. For example, in the Western clawed frog and Burmese python histidine was substituted to arginine, which is also basic amino acid. The analysis suggests that NLS is conserved in the component of amino acids to allow the conservation of nuclear EGFR. We next asked whether the differences in NLS among species correspond with their tendency to acquire cancers. To clarify, whale and rat have significantly low rates of cancer, so maybe they have evolved evolutionary mechanisms that help them to escape from cancers. It is known that Dog has a similar tendency to develop cancer as Human, and Green sea turtle has cancer too 33. As shown in Figure 2, Elephant shark and Norway rat have a low identity with Human NLS, which correlates to the low chance of developing cancer.
Figure 2.
Presence of prostate and types of diet in different species. Columns from left to right: 1. Name of species; 2. Presence of prostate; 3. Type of diet; 4. NLS sequence; 5. Interaction of EGFR with differential proteins.
Nuclear localization signal of EGFR reflects the macro-environmental factors for prostate cancer in evolution
To further examine the role of NLS of EGFR in the development of cancer, we investigated animals' tendency to develop prostate cancer. It is known that only mammals have the prostate organ. As shown in Figure 1 and 3, NLS of EGFR reflects the prostate organ formation during evolution. Although there are variations in NLS, 100% conserved NLS are found in majority of species having prostates.
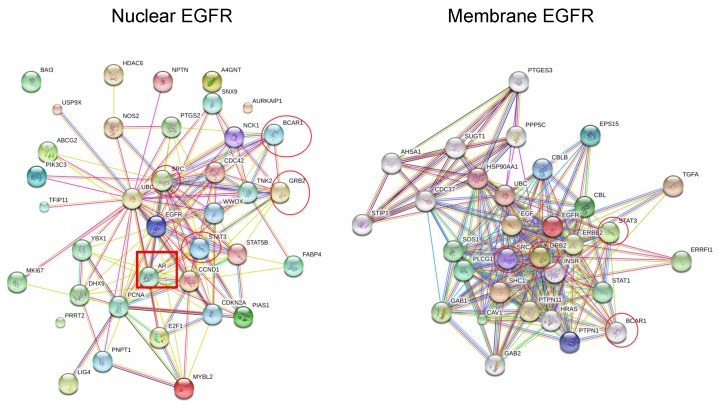
Figure 3.
The protein-protein interaction maps of nuclear EGFR and membrane EGFR in Human. The maps were generated by the SRTING 10.0a (http://version10a.string-db.org).
Coffey proposed that among mammals, human and dog develop prostate cancer due to the consumption of meat 34 (Fig.2). It is well known that blood serum contains EGF. Thus raw meat eater or carnivore animals may have more chances to evolve cancer because if EGF/EGFR signaling activates cancer through bloody raw meat eating, herbivore animals may have low chances to get cancer (Fig.2). Moreover, consumption of green tea, isoflavones, lycopenes, and cruciferous vegetables can reduce the chances of prostate cancer 35. Plant phenolics, including curcumin, genistein and catechins, are known as potential inhibitors of cancer 36. For example, curcumin inhibits EGFR activity and EGF expression, thus decreases the invasiveness of cancer 37. To conclude, the diet diversity of plant foods and meat most likely has the effect on prostate cancer evolution through reflection of NLS evolution and cooperation with environmental factors.
Nuclear EGFR correlates to advanced prostate cancer
To further test our predication on role of nEGFR in prostate cancer through evolutionary analysis, we searched database to analyze the correlation between nEGFR and prostate cancer development. Clinical studies revealed that nEGFR levels had a correlation with the poor survival of cancer patients. Literatures in PubMed support the correlations between expression of nEGFR with cancer grade or metastasis stage 19, 25, 26, 29. Furthermore, there is a direct correlation between nEGFR expression and Gleason score 38 in prostate cancer patients with metastasis 25 (data not shown). Analysis of other three types of cancer also shows the variations in the effect of nEGFR expression on the survival of patients (data not shown). For example, nEGFR expression in breast cancer patients is associated with poor prognosis, but the expression of nEGFR leads to an inverse correlation with the survival of patients in some cohorts studied in ovarian cancer 19, 25, 26, 29. In addition, the effect of membrane EGFR on the progression of prostate cancer was investigated using the study of Peraldo-Neia 39. However, the expression of membrane EGFR cannot be used to predict prostate cancer, because there is poor correlation between the expression of membrane EGFR and cancer progression 39. Moreover, natural selection may play an essential role in nEGFR-mediated evolution of cancer metastasis. Many types of cancers show the negative correlation between nEGFR and cancer stages even nEGFR expressed in all stages of cancers (data not shown). Negative correlation between nEGFR and cancer stages in some types of cancer may suggest that metastatic cancer cells express low levels nEGFR which may produce the heritable genetic networks of nEGFR to resistant the nEGFR loss due to natural selection. In summary, the expression of nEGFR as a prognosis marker of a few types of cancer such as prostate cancer suggests the organ specific evolution of cancer driven by nEGFR.
Protein-protein interaction maps suggest the evolutionary networks of nuclear EGFR
To fully understand the signaling pathways associated with nEGFR in cancer evolution, the protein-protein interaction maps of membrane EGFR and nEGFR were compared. STRING software and previous studies on EGFR pathways were used to construct interaction maps of membrane EGFR and nEGFR 40-44. The results show that common frequent interaction nodes for both nEGFR and membrane EGFR are STAT 3, GRB 2, BCAR1, and STAT 1 (Fig. 3). In addition, the protein interaction maps of membrane EGFR were constructed in different species. We used maps of Zebra Fish and Bos Taurus instead of Elephant shark and Sheep respectively, since they could still give approximate analyzes of EGFR interaction in fish and ruminant domesticated animal. Moreover, the maps for Burmese python and Killer whale were not found in the database of the software. Overall, we observed that EGFR in some species possesses the same interaction proteins as that of human EGFR. For instance, STAT3 protein is presented in all of species which have close ancestor to human and Pygmy chimpanzee, while Chicken, Green sea turtle, and Western clawed frog does not show interactions between STAT3 and EGFR (string-db.org; supplementary Figure 1). Our data suggest that EGFR interacts with STAT3 in the species closely related to human, which leads to evolutionary transition in the signaling pathway. Figure 2 shows that Chicken, Green sea turtle and Western clawed frog's EGFR does not interact with STAT3 pathways but interact with other proteins such as Gsk3b, Ctnnb1 and Ahsa1. Thus, the data suggest that evolutionary origin of EGFR and its interaction with other signaling pathways can be evolved through cooperation with NLS to induce cancer evolution. Interestingly, as shown in Fig. 3, EGF is not found in nEGFR network nodes while androgen receptor (AR) is found as a node, which suggests the evolution of prostate cancer, a well-known AR driven cancer, through original androgen dependence to androgen independence but EGF dependence and to EGF independence with a low androgen reactivation. In summary, protein-protein interaction maps suggest the distinct evolutionary pathways between nEGFR and membrane EGFR even they have common interaction networks. STAT3 and AR may be transition pathways that correlate with the prostate organ formation and accelerate the evolution.
The protein structure predicates the driving force of nuclear localization signal in protein-protein interactions during evolution
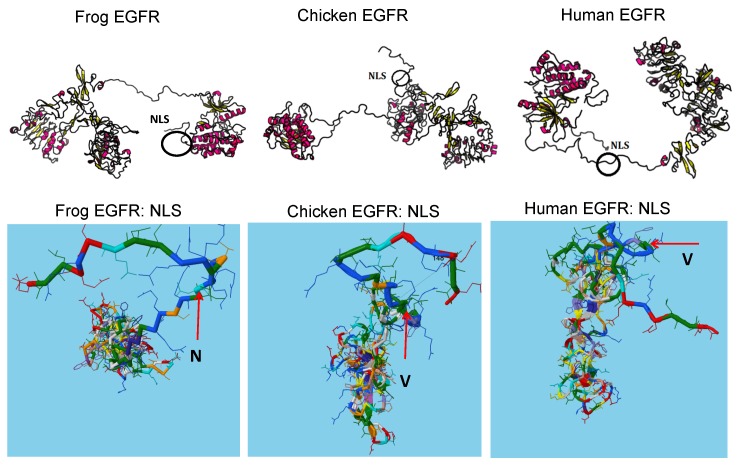
To fully understand how NLS evolution can affect protein-protein interactions for signaling pathway transition, we investigated the structural changes upon NLS evolution. As shown in Figure 4, naturally occurring variations of NLS between frog, chicken, mouse and human can affect the linker region adjacent to the catalytic kinase domain. The stretch structures adjacent to NLS allow NLS to provide the structural diversity for evolution such as rotation inside peptides. For example, amino acid N in NLS is evolved into V which loses the open space and allows more twisted structure of linker region to interfere more interactions between NLS with other proteins or peptide. Thus, consistent with the protein-protein interaction maps analysis, structural predication analysis suggests the NLS may act to compromise the differential protein binding to EGFR during evolution. In summary, our data suggest NLS is a dynamic driver for EGFR evolution in advanced cancer especially recurrent or therapy resistant cancer. Our model provides novel insights into the evolutionary role of oncogenic NLS in cancer.
Figure 4.
The protein structures predicate the driving force of NLS in protein-protein interactions during evolution. Protein structures were predicated by inputting sequences to software (http://raptorx.uchicago.edu/StructurePrediction/) and NLS region were marked.
Nuclear EGFR elevation during prostate cancer evolution from androgen sensitive to androgen insensitive
To further test whether nEGFR indeed plays essential roles in prostate cancer evolution involving in AR pathways, we applied an androgen responsive cell line LAPC4 to monitor the changes of prostate cancer from androgen sensitive to insensitive status. We used charcoal striped FBS (cFBS) to remove androgen and selected cells after 2 passages. Cells survived from androgen depletion were further tested for expression levels of EGFR in different cellular compartment. As shown in Fig.5, nuclear EGFR but not membrane EGFR was elevated in cells treated with cFBS medium. Cytosolic EGFR was also accumulated, which might be inefficient nuclear translocation in short terms as 7 days during cancer evolution. However, our data supports the induction of nuclear EGFR from transition of androgen-dependence to androgen-independence during prostate cancer evolution.
Figure 5.
Nuclear EGFR elevation during prostate cancer evolution from androgen sensitive to androgen insensitive. A. LAPC4 prostate cancer cells were androgen deprived for 7 days and expression levels of EGFR in cellular compartment were analyzed by western blot. C: cytosol; N: nucleus; M: membrane. B. Model of EGFR in broad cancer evolution by NLS reactivation through EGF independent pathways.
The systems analysis of nuclear localization signal and transmembrane domain suggests the old origin and accelerated evolution
To further explore the mechanism of NLS in evolution, we compared the sequences NLS and its nearby domain of TM in 93 organisms. To our surprise, the systems analysis showed that NLS is much more conserved than TM domain (Supplementary Fig. 2). As EGFR predominately localizes at plasma membrane TM domain should be conserved. However, we found TM has been undergone accelerated evolution in ancestors. Polygenetic trees showed that TM has more branches than NLS. Thus systems analysis further confirms that NLS has an older origin than TM. In summary, our data suggest that nEGFR has an old origin resembling the cancer evolution. Modern TM may prohibit NLS driven pathways to evade NLS induced aggressive cancers. Natural selection for survival may play an important role to against the cancer induced death. Thus reactivation of nEGFR may allow historic aggressive cancer to appear.
Discussion
Recent literature reviewed that tyrosine kinases such as EGFR can localize into the nucleus due to its NLS 9. Thus, it is important to analyze NLS in different organisms to understand the mechanism of nEGFR and the evolution of cancer. The significant similarity among Green sea turtle, Horse, Sheep, Dog, Human, Chimpanzee, Chicken, and House mouse is found in their putative NLS sequences. Less similar, but still significant similarity is found among Elephant shark, Western clawed frog, Norway rat and Burmese python (Fig. 1). Although the reduced similarity in amino acid sequence of NLS among species, they possess a similar structure. For example, all organisms possess three stretches of basic amino acids and two sets of nonpolar amino acids (IV and L). The results suggest that EGFR protein retained ability to translocate into the nucleus despite some changes of NLS in amino acid. The difference in amino acid sequence might affect the efficiency and rate of nuclear import and mobility of EGFR. It is likely that the degree of nuclear localization is affected by the number of arginine residues in the basic stretch. Figure 2 demonstrates that NLS of EGFR is rich in arginine (R), suggesting that the length of arginine stretch in NLS sequence might affect the internalization of EGFR into nucleus. It can be seen that various organisms have different numbers of arginine. For example, Burmese python's NLS sequence is the richest in arginine residues, whereas Western clawed frog has the least number of arginine in its NLS sequence. The effect of arginine on nuclear localization can be explained by ionic interactions of positively charged arginine with the negatively charged membrane phosphate. In addition, arginine might bind with the extranuclear components such as importin, which results in the nuclear import of EGFR.
To understand how the changes in a single amino acid can affect the nuclear events of EGFR, it will be worth to look at the phylogenetic origin of the analyzed species in relation to Human. Some species showed higher percentages of similarity possessing ancestry with Amniotic Egg from older period being Carboniferous era 359 Mya (million years ago). Whereas, species having ancestor from Silurian period 443 Mya and Devonian period about 416 Mya showed lower similarity percentages of NLS of EGFR with human. Therefore, it was predicted that NLS of EGFR has old evolutionary origin based on the results. The finding of percentage difference of NLS and full length EGFR sequence is consistent with the evolutionary species (Fig. 1C). In particular, Human, Pygmy chimpanzee, Dog, Horse, Sheep, Norway rat and House mouse are closely related and have common ancestor, which is also seen from the percentage difference.
To investigate the development of cancer, it was important to examine animals which have tendency to develop prostate cancer. In the last 10,000-15,000 years human and dog have significantly changed their diets. This time was a turnover in the diet of Human from plant-feeding to meat-eating, which possibly affects their tendency to develop prostate cancer. Several studies were conducted to further investigate what type of food causes prostate cancer 45. The studies of Kristal et al. figured out that high-grade prostate cancer was correlated with the consumption of polyunsaturated fats, n-6 fatty acids, vitamins D and E, and selenium, whereas nutrition with calcium decreases the chances to develop prostate cancer 46. Another research highlighted that intake of plant food can prevent prostate cancer 35. Moreover, consumption of high total fat, meat, and multivitamin may increase the chances of prostate cancer 47. It can be concluded that prostate cancer is modern event in evolution, which possibly is resulted from dietary changes.
Since EGFR could promote tumor progression from various interactions involving signaling pathways, we generated the protein-protein interaction maps on EGFR 48. The analysis of protein interaction maps revealed some proteins interact with both forms of EGFRs, such as STAT1, STAT3, GRB2, and BCAR1 17, 48-50. It was found that the interaction of nEGFR with STAT3 results in the transcriptional activation of iNOS 18. Moreover, IGFBP2 has a role in activation of EGFR/STAT3 signaling and facilitating EGFR accumulation in the nucleus 51. Also, nEGFR interaction maps showed the interaction with androgen receptor (AR). Chen and his colleagues suggested that AR in Sertoli cells regulates the double strand breaks repair and chromosomal synapsis of spermatocytes via EGF-EGFR signaling 52. AR is the key factor in the progression and metastasis of prostate cancer. Taplin et al. found that 5 of 10 patients with distant metastases failed to undergo endocrine therapy, and the authors also found that AR mutations influence the metastatic development of prostate cancer 53. The amplified AR in some cases of prostate cancer are mostly not mutated and therefore retained specificity for androgen 50, 54. In addition, AR modifications such as phosphorylation may promote metastatic prostate cancer 55, 56.
Protein-protein interaction analysis of membrane EGFR in different species showed that STAT3, which can act as regulator of genes responsible for cell migration and apoptosis, was found in frequent interaction nodes of species closed to Human such as Pygmy chimpanzee, Norway rat, Horse, and Dog. Other species showed other interacting proteins with EGFR. For instance, Chicken and Green sea turtle both had interaction of EGFR with CBLB, which is known as E3 ubiquitin-protein ligase regulating T cell activation, and SHC (Src Homology 2 Domain Containing) Transforming Protein 1 involved in different signaling pathways 57, 58. STAT3 protein interacts with EGFR in only closely related species to Human, which suggests that Chicken is the evolutionary transition for different signaling pathways. Chicken and Green sea turtle's EGFR interact with other proteins, CBLB and SHC-1, which can be explained by same amino acid sequence in NLS of EGFR.
To conclude, we found that NLS of EGFR is conserved among different species. Phylogenetic tree analysis shows that NLS of EGFR has an old evolutionary origin. The connection between nEGFR and cancer progression was observed from several clinical studies by database search. The effect of nEGFR expression on cancer development varies with cancer types. Protein-protein interaction maps of nEGFR and membrane EGFR in different species revealed several proteins being involved in same pattern in closely related species. In addition, it will be essential to further investigate how structure of EGFR in the different species may cooperate with NLS to induce the dynamic signaling and cancer evolution.
Supplementary Material
Supplementary figures.
Acknowledgments
This work was supported in part by Kazakhstan-China collaboration grant, China-Kazakhstan collaboration grant (No. CK-07-09) to Yingqiu Xie and Lixia Miao. We would like to thank Charles Sawyers and Brett S. Carver from Memorial Sloan Kettering Cancer Center for providing the LAPC4 cells and technical support. The authors would like to thank the Department of Biology, SST, and Nazarbayev University for the support of acquired learning, flipped classroom and research integrated teaching innovations of Biochemistry I course for undergraduate students. We would like to thank all the Biochemistry I course students of Nazarbayev University due to their support for the publication of assignment of the Biochemistry I course including this paper and other publications.
References
- 1.Lange A, McLane L, Mills R, Devine S, Corbett A. Expanding the Definition of the Classical Bipartite Nuclear Localization Signal. Traffic. 2010;11:311–23. doi: 10.1111/j.1600-0854.2009.01028.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McLane L, Corbett A. Nuclear localization signals and human disease. IUBMB Life. 2009;61:697–706. doi: 10.1002/iub.194. [DOI] [PubMed] [Google Scholar]
- 3.Marfori M, Mynott A, Ellis J, Mehdi A, Saunders N, Curmi P. et al. Molecular basis for specificity of nuclear import and prediction of nuclear localization. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2011;1813:1562–77. doi: 10.1016/j.bbamcr.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 4.Chang C, Couñago R, Williams S, Boden M, Kobe B. The distribution of different classes of nuclear localization signals (NLSs) in diverse organisms and the utilization of the minor NLS-binding site inplantnuclear import factor importin-α. Plant Signaling & Behavior. 2013;8:e25976. doi: 10.4161/psb.25976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christie M, Chang CW, Róna G, Smith KM, Stewart AG, Takeda AA, Structural biology and regulation of protein import into the nucleus. Journal of Molecular Biology; 2015. pp. 1–31. [DOI] [PubMed] [Google Scholar]
- 6.Cautain B, Hill R, Pedro N, Link W. Components and regulation of nuclear transport processes. FEBS Journal. 2015;282:445–62. doi: 10.1111/febs.13163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hung MC, Link W. Protein localization in disease and therapy. Journal of Cell Science. 2011;124:3381–92. doi: 10.1242/jcs.089110. [DOI] [PubMed] [Google Scholar]
- 8.Kau TR, Way JC, Silver PA. Nuclear transport and cancer: from mechanism to intervention. Nature Reviews Cancer. 2004;4:106–17. doi: 10.1038/nrc1274. [DOI] [PubMed] [Google Scholar]
- 9.Brand TM, Iida M, Li C, Wheeler DL. The nuclear epidermal growth factor receptor signaling network and its role in cancer. Discovery Medicine. 2011;12:419–32. [PMC free article] [PubMed] [Google Scholar]
- 10.Huang WC, Chen YJ, Hung MC. Implication of nuclear EGFR in the development of resistance to anticancer therapies. BioMedicine. 2011;1:2–10. [Google Scholar]
- 11.Han W, Lo HW. Landscape of EGFR signaling network in human cancers: biology and therapeutic response in relation to receptor subcellular locations. Cancer Letters. 2012;318:124–34. doi: 10.1016/j.canlet.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang YN, Yamaguchi H, Hsu JM, Hung MC. Nuclear trafficking of the epidermal growth factor receptor family membrane proteins. Oncogene. 2010;29:3997–4006. doi: 10.1038/onc.2010.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lo HW. Nuclear mode of the EGFR signaling network: biology, prognostic value, and therapeutic implications. Discovery Medicine. 2010;10:44–51. [PMC free article] [PubMed] [Google Scholar]
- 14.Dittmann K, Mayer C, Paasch A, Huber S, Fehrenbacher B, Schaller M. et al. Nuclear EGFR renders cells radio-resistant by binding mRNA species and triggering a metabolic switch to increase lactate production. Radiotherapy and Oncology. 2015;116:431–7. doi: 10.1016/j.radonc.2015.08.016. [DOI] [PubMed] [Google Scholar]
- 15.Pereira NB, do Carmo AC, Diniz MG, Gomez RS, Gomes DA, Gomes CC. Nuclear localization of epidermal growth factor receptor (EGFR) in ameloblastomas. Oncotarget. 2015;6:9679–85. doi: 10.18632/oncotarget.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee HH, Wang YN, Hung MC. Non-canonical signaling mode of the epidermal growth factor receptor family. American Journal of Cancer Research. 2015;5:2944–58. [PMC free article] [PubMed] [Google Scholar]
- 17.Brand TM, Iida M, Luthar N, Starr MM, Huppert EJ, Wheeler DL. Nuclear EGFR as a molecular target in cancer. Radiotherapy and Oncology. 2013;108:370–7. doi: 10.1016/j.radonc.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 18.Lo H, Hsu S, Ali-Seyed M, Gunduz M, Xia W, Wei Y. et al. Nuclear interaction of EGFR and STAT3 in the activation of the iNOS/NO pathway. Cancer Cell. 2005;7:575–589. doi: 10.1016/j.ccr.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 19.Lo H, Xia W, Wei Y, Ali-Seyed M, Huang S, Hung M. Novel prognostic value of nuclear epidermal growth factor receptor in breast cancer. Cancer Research. 2005;65:338–348. [PubMed] [Google Scholar]
- 20.Xia W, Wei Y, Du Y, Liu J, Chang B, Yu Y. et al. Nuclear expression of epidermal growth factor receptor is a novel prognostic value in patients with ovarian cancer. Molecular Carcinogenesis. 2009;48:610–617. doi: 10.1002/mc.20504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Traynor A, Weigel T, Oettel K, Yang D, Zhang C, Kim K. et al. Nuclear EGFR protein expression predicts poor survival in early stage non-small cell lung cancer. Lung Cancer. 2013;81:138–141. doi: 10.1016/j.lungcan.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li C, Fang F, Wang J, Tzeng C, Tai H, Wei Y. et al. EGFR nuclear import in gallbladder carcinoma: nuclear phosphorylated EGFR upregulates iNOS expression and confers independent prognostic impact. Annals of Surgical Oncology. 2012;19:443–454. doi: 10.1245/s10434-011-1942-6. [DOI] [PubMed] [Google Scholar]
- 23.Psyrri A, Yu Z, Weinberger P, Sasaki C, Haffty B, Camp R. et al. Quantitative Determination of Nuclear and Cytoplasmic Epidermal Growth Factor Receptor Expression in Oropharyngeal Squamous Cell Cancer by Using Automated Quantitative Analysis. Clinical Cancer Research. 2005;11:5856–5862. doi: 10.1158/1078-0432.CCR-05-0420. [DOI] [PubMed] [Google Scholar]
- 24.Hoshino M, Fukui H, Ono Y, Sekikawa A, Ichikawa K, Tomita S. et al. Nuclear Expression of Phosphorylated EGFR Is Associated with Poor Prognosis of Patients with Esophageal Squamous Cell Carcinoma. Pathobiology. 2007;74:15–21. doi: 10.1159/000101047. [DOI] [PubMed] [Google Scholar]
- 25.Chang YS, Chen WY, Yin JJ, Sheppard-Tillman H, Huang J, Liu YN. EGF receptor promotes prostate cancer bone metastasis by downregulating miR-1 and activating TWIST1. Cancer Research. 2015;75:3077–86. doi: 10.1158/0008-5472.CAN-14-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xia W, Wei Y, Du Y, Liu J, Chang B, Yu YL. et al. Nuclear expression of epidermal growth factor receptor is a novel prognostic value in patients with ovarian cancer. Molecular Carcinogenesis. 2009;48:610–7. doi: 10.1002/mc.20504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ahel J, Dordevic G, Markic D, Mozetic V, Spanjol J, Grahovac B. et al. Nuclear EGFR characterize still controlled proliferation retained in better differentiated clear cell RCC. Medical Hypotheses. 2015;85:183–5. doi: 10.1016/j.mehy.2015.04.033. [DOI] [PubMed] [Google Scholar]
- 28.Fletcher WA. Computational statistics in molecular phylogenetics. Doctoral dissertation, UCL (University College London)
- 29.Dekanic A, Dintinjan RD, Budisavljevic I, Pecanic S, Butorac MZ. et al. Strong nuclear EGFR expression in colorectal carcinomas is associated with cyclin-D1 but not with gene EGFR amplification. Diagnostic Pathology. 2011;6:108–15. doi: 10.1186/1746-1596-6-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Phylogenetic systematics, a.k.a. evolutionary trees. Evolution.berkeley.edu. 2016. http://evolution.berkeley.edu/evolibrary/article/phylogenetics_01.
- 31.2016. Tree of Life; http://abacus.gene.ucl.ac.uk/will/files/TreeOfLife.pdf. [Google Scholar]
- 32.Nelson DL, Cox MM. Lehninger Principles of Biochemistry. New York: W.H. Freeman and Company; 2013. [Google Scholar]
- 33.Merlo DF, Rossi L, Pellegrino C, Ceppi M, Cardellino U, Capurro C. et al. Cancer incidence in pet dogs: findings of the Animal Tumor Registry of Genoa, Italy. Journal of Veterinary Internal Medicine. 2008;22:976–84. doi: 10.1111/j.1939-1676.2008.0133.x. [DOI] [PubMed] [Google Scholar]
- 34.Coffey DS. Similarities of prostate and breast cancer: evolution, diet, and estrogens. Urology. 2001;57:31–8. doi: 10.1016/s0090-4295(00)00938-9. [DOI] [PubMed] [Google Scholar]
- 35.Hori S, Butler E, McLoughlin J. Prostate cancer and diet: food for thought? BJU International. 2011;107:1348–59. doi: 10.1111/j.1464-410X.2010.09897.x. [DOI] [PubMed] [Google Scholar]
- 36.Wahle KW, Brown I, Rotondo D, Heys SD. Plant phenolics in the prevention and treatment of cancer. InBio-Farms for Nutraceuticals; 2010. pp. 36–51. Springer US. [DOI] [PubMed] [Google Scholar]
- 37.Kaneuchi M, Sasaki M, Tanaka Y, Yamamoto R, Sakuragi N, Dahiya R. Resveratrol suppresses growth of Ishikawa cells through down-regulation of EGF. International Journal of Oncology. 2003;23:1167–72. [PubMed] [Google Scholar]
- 38.Kweldam CF, Wildhagen MF, Steyerberg EW, Bangma CH, van der Kwast TH, van Leenders GJ. Cribriform growth is highly predictive for postoperative metastasis and disease-specific death in Gleason score 7 prostate cancer. Modern Pathology. 2015;28:457–64. doi: 10.1038/modpathol.2014.116. [DOI] [PubMed] [Google Scholar]
- 39.Peraldo-Neia C, Migliardi G, Mello-Grand M, Montemurro F, Segir R, Pignochino Y, Cavalloni G, Torchio B, Mosso L, Chiorino G, Aglietta M. Epidermal Growth Factor Receptor (EGFR) mutation analysis, gene expression profiling and EGFR protein expression in primary prostate cancer. BMC cancer. 2011;11:1–12. doi: 10.1186/1471-2407-11-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lo HW, Hung MC. Nuclear EGFR signalling network in cancers: linking EGFR pathway to cell cycle progression, nitric oxide pathway and patient survival. British Journal of Cancer. 2006;94:184–8. doi: 10.1038/sj.bjc.6602941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roßner F, Gieseler C, Morkel M, Royer HD, Rivera M, Bläker H. et al. Uncoupling of EGFR-RAS signaling and nuclear localization of YBX1 in colorectal cancer. Oncogenesis. 2016;5:e187. doi: 10.1038/oncsis.2015.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Z, Hu P, Tang F, Xie C. HDAC6-mediated EGFR stabilization and activation restrict cell response to sorafenib in non-small cell lung cancer cells. Medical Oncology. 2016;33:1–9. doi: 10.1007/s12032-016-0765-5. [DOI] [PubMed] [Google Scholar]
- 43.Savio MG, Wollscheid N, Cavallaro E, Algisi V, Di Fiore PP, Sigismund S. et al. USP9X Controls EGFR Fate by Deubiquitinating the Endocytic Adaptor Eps15. Current Biology. 2016;26:173–83. doi: 10.1016/j.cub.2015.11.050. [DOI] [PubMed] [Google Scholar]
- 44.Dayde D, Guerard M, Perron P, Hatat AS, Barrial C, Eymin B, Nuclear trafficking of EGFR by Vps34 represses Arf expression to promote lung tumor cell survival. Oncogene; 2015. pp. 1–9. [DOI] [PubMed] [Google Scholar]
- 45.Masko EM, Allott EH, Freedland SJ. The relationship between nutrition and prostate cancer: is more always better? European Urology. 2013;63:810–20. doi: 10.1016/j.eururo.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kristal AR, Arnold KB, Neuhouser ML, Goodman P, Platz EA, Albanes D, Thompson IM. Diet, supplement use, and prostate cancer risk: results from the prostate cancer prevention trial. American Journal of Epidemiology. 2010;172:566–77. doi: 10.1093/aje/kwq148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davies NJ, Batehup L, Thomas R. The role of diet and physical activity in breast, colorectal, and prostate cancer survivorship: a review of the literature. British Journal of Cancer. 2011;105:52–73. doi: 10.1038/bjc.2011.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tomas A, Futter CE, Eden ER. EGF receptor trafficking: consequences for signaling and cancer. Trends in Cell Biology. 2014;24:26–34. doi: 10.1016/j.tcb.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin SY, Makino K, Xia W, Matin A, Wen Y, Kwong KY. et al. Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nature Cell Biology. 2001;3:802–8. doi: 10.1038/ncb0901-802. [DOI] [PubMed] [Google Scholar]
- 50.Tan X, Lambert PF, Rapraeger AC, Anderson RA. Stress-Induced EGFR Trafficking: Mechanisms, Functions, and Therapeutic Implications. Trends in Cell Biology. 2016;26:352–66. doi: 10.1016/j.tcb.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chua CY, Liu Y, Granberg KJ, Hu L, Haapasalo H, Annala MJ. et al. IGFBP2 potentiates nuclear EGFR-STAT3 signaling. Oncogene. 2015;35:738–47. doi: 10.1038/onc.2015.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen SR, Hao XX, Zhang Y, Deng SL, Wang ZP, Wang YQ. et al. Androgen receptor in Sertoli cells regulates DNA double-strand break repair and chromosomal synapsis of spermatocytes partially through intercellular EGF-EGFR signaling. Oncotarget. 2016;7:18722–35. doi: 10.18632/oncotarget.7916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Taplin ME, Bubley GJ, Shuster TD, Frantz ME, Spooner AE, Ogata GK. et al. Mutation of the androgen-receptor gene in metastatic androgen-independent prostate cancer. New England Journal of Medicine. 1995;332:1393–8. doi: 10.1056/NEJM199505253322101. [DOI] [PubMed] [Google Scholar]
- 54.Koivisto P, Kononen J, Palmberg C, Tammela T, Hyytinen E, Isola J. et al. Androgen receptor gene amplification: a possible molecular mechanism for androgen deprivation therapy failure in prostate cancer. Cancer Research. 1997;57:314–9. [PubMed] [Google Scholar]
- 55.Guo Z, Dai B, Jiang T, Xu K, Xie Y, Kim O. et al. Regulation of androgen receptor activity by tyrosine phosphorylation. Cancer Cell. 2006;10:309–19. doi: 10.1016/j.ccr.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 56.Xie Y, Lu W, Liu S, Yang Q, Carver BS, Li E. et al. Crosstalk between nuclear MET and SOX9/β-catenin correlates with castration-resistant prostate cancer. Molecular Endocrinology. 2014;28:1629–39. doi: 10.1210/me.2014-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.CBLB - E3 ubiquitin-protein ligase CBL-B - Homo sapiens (Human) - CBLB gene & protein. Uniprot.org. http://www.uniprot.org/uniprot/Q13191.
- 58.SHC1 - SHC-transforming protein 1 - Homo sapiens (Human) - SHC1 gene & protein. Uniprot.org. http://www.uniprot.org/uniprot/P29353.
- 59.UniProt Consortium. UniProt: a hub for protein information. Nucleic acids research. 2014 Oct;27:gku989. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamada KD, Tomii K, Katoh K. Application of the MAFFT sequence alignment program to large data-reexamination of the usefulness of chained guide trees. Bioinformatics; 2016. Jul 4. pii: btw412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2-a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures.