Abstract
A facultatively anaerobic, spore-forming Bacillus strain, FSL W8-0169T, collected from raw milk stored in a silo at a dairy powder processing plant in the north-eastern USA was initially identified as a Bacillus cereus group species based on a partial sequence of the rpoB gene and 16S rRNA gene sequence. Analysis of core genome single nucleotide polymorphisms clustered this strain separately from known B. cereus group species. Pairwise average nucleotide identity blast values obtained for FSL W8-0169T compared to the type strains of existing B. cereus group species were <95 % and predicted DNA–DNA hybridization values were <70 %, suggesting that this strain represents a novel B. cereus group species. We characterized 10 additional strains with the same or closely related rpoB allelic type, by whole genome sequencing and phenotypic analyses. Phenotypic characterization identified a higher content of iso-C16 : 0 fatty acid and the combined inability to ferment sucrose or to hydrolyse arginine as the key characteristics differentiating FSL W8-0169T from other B. cereus group species. FSL W8-0169T is psychrotolerant, produces haemolysin BL and non-haemolytic enterotoxin, and is cytotoxic in a HeLa cell model. The name Bacillus wiedmannii sp. nov. is proposed for the novel species represented by the type strain FSL W8-0169T (=DSM 102050T=LMG 29269T).
Keywords: Bacillus cereus group, Bacillus, whole genome sequence, cytotoxic, psychrotolerant
The Bacillus cereus group, also called Bacillus cereus sensu lato (s.l.), is a group of closely related micro-organisms of diverse ecotypes, which include pathogenic and non-pathogenic species (Ceuppens et al., 2013). At the time of writing, the B. cereus group comprises eight species with validly published names: B. anthracis (Logan et al., 1985) B. cereus (Smith et al., 1952), B. cytotoxicus (Guinebretière et al., 2013), B. mycoides (Lechner et al., 1998) B. pseudomycoides (Nakamura, 1998), B. thuringiensis (Nakamura, 1994), B. toyonensis (Jiménez et al., 2013) and B. weihenstephanensis (Lechner et al., 1998). In addition to these, three B. cereus group strains have been proposed as representing novel species [i.e. ‘B. gaemokensis’ (Jung et al., 2010), ‘B. manliponensis’ (Jung et al., 2011) and ‘B. bingmayongensis’ (Liu et al., 2014)], but have not yet been validly published. B. cereus group species are facultatively anaerobic, spore-forming bacteria that are ubiquitously distributed throughout a number of environments (Huck et al., 2007; Ivy et al., 2012; Ceuppens et al., 2013). Depending on virulence gene presence and expression, select strains of these species can cause anthrax or gastrointestinal illness in humans or insects (Kim et al., 2015; Moayeri et al., 2015). Members of the B. cereus group are also known food spoilage organisms (Lücking et al., 2013). Therefore, their presence in the food production chain is often monitored (e.g. Miller et al., 2015a).
Strain FSL W8-0169T was obtained from silo raw milk collected from a dairy powder processing plant in the north-eastern USA in 2012 (Watterson et al., 2014; Miller et al., 2015b). The strain was initially identified as a member of B. cereus s.l. based on analysis of a partial sequence of the rpoB gene, which encodes the β-subunit of RNA polymerase. Strain FSL W8-0169T has rpoB allelic type (AT) 61, which was also identified for 12 other B. cereus group dairy-associated strains deposited in the Food Microbe Tracker database (http://www.foodmicrobetracker.com). Additionally, two closely related ATs (AT 410 and AT 417) were found in the database, representing 12 and two strains, respectively. Nine strains representing rpoB AT 61, one strain representing rpoB AT 417 and one strain representing rpoB AT 194 were characterized in detail in this study (Table 1). Phenotypic, phylogenetic and whole genome sequence (WGS) data failed to classify these 11 strains into existing B. cereus group species. These 11 strains are presented as representing a novel species within the B. cereus group, for which the name Bacillus wiedmannii sp. nov. is proposed. Strain FSL W8-0169T is the type strain of B. wiedmannii sp. nov.
Table 1. Characteristics of the 11 Bacillus wiedmannii sp. nov. strains characterized in this study.
| FSL W8-0169T | FSL H7-0353 | FSL H8-0032 | FSL J3-0113 | FSL M7-0044 | FSL M7-0938 | FSL M7-1251 | FSL P2-0415 | FSL P2-0558 | FSL P4-0569 | FSL K6-0069 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Source | Raw milk | Ready to eat dairy food product | Dairy food in process | Ready to eat dairy food product | Raw milk | Raw milk | Raw milk | Dairy food in process | Dairy food in process | Dairy food in process | Raw milk |
| Year of isolation | 2012 | 2005 | 2005 | 2012 | 2011 | 2011 | 2011 | 2009 | 2009 | 2010 | 2012 |
| rpoB AT | 61 | 61 | 61 | 417 | 61 | 61 | 61 | 61 | 61 | 61 | 194 |
| MLST | 1081* | 1272* | 1272* | 1094 | 1271* | 1269* | 1270* | 1268* | 644 | 1266* | 1080* |
| Draft genome length (Mbp) | 5.3 | 5.6 | 5.6 | 5.6 | 5.4 | 5.4 | 5.4 | 5.6 | 5.6 | 5.4 | 5.6 |
| Draft genome G+C content (mol%) | 35.3 | 35.1 | 35.1 | 35.1 | 35.2 | 35.2 | 35.2 | 35.2 | 35.1 | 35.2 | 35.3 |
| No. of contigs | 104 | 75 | 74 | 60 | 73 | 63 | 97 | 93 | 110 | 56 | 158 |
| Draft genome coverage (×) | 49 | 123 | 121 | 101 | 134 | 142 | 118 | 112 | 114 | 130 | 58 |
| Contig N50 | 189870 | 248295 | 247812 | 542359 | 233954 | 301752 | 206020 | 203426 | 196369 | 329376 | 77402 |
| BioSample accession no. | SAMN03800026 | SAMN04909723 | SAMN04909724 | SAMN04909725 | SAMN04909726 | SAMN04909727 | SAMN04909728 | SAMN04909729 | SAMN04909730 | SAMN04909731 | SAMN03800020 |
| WGS SRA accession no. | SRR2541651 | SRR3458441 | SRR3458442 | SRR3458443 | SRR3458444 | SRR3458445 | SRR3458446 | SRR3458447 | SRR3458448 | SRR3458449 | SRR2541606 |
| WGS GenBank accession no. | LOBC00000000 | LXFL00000000 | LXFM00000000 | LXFN00000000 | LXFO00000000 | LXFP00000000 | LXFQ00000000 | LXFR00000000 | LXFS00000000 | LXFT00000000 | LOBB00000000 |
*Novel MLST sequence type.
AT, allelic type; MLST, multi-locus sequence typing; WGS, whole genome sequencing; SRA, Sequence Read Archive.
Phylogenetic analyses
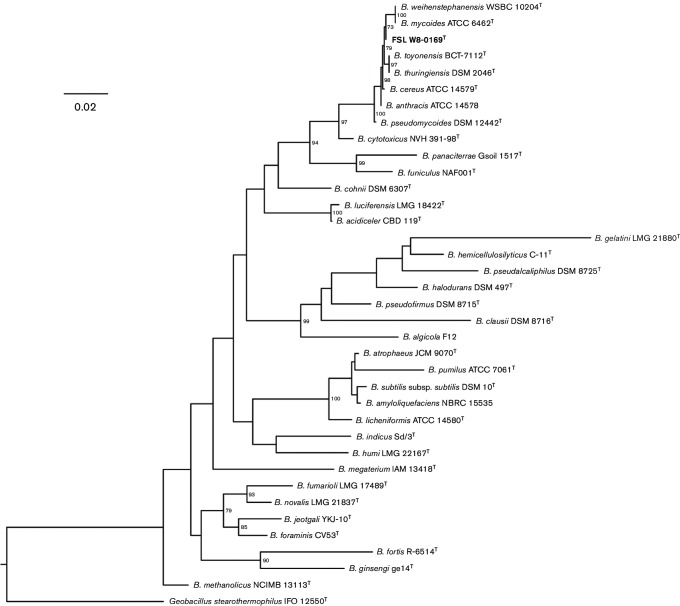
Sequences of the 1471 bp 16S rRNA gene (Fig. 1) and a 632 bp internal fragment of the rpoB gene (Fig. S1, available in the online Supplementary Material; Miller et al., 2015a) were aligned using the muscle algorithm; pairwise distance matrices were calculated in mega version 6.06 (Tamura et al., 2013). Maximum-likelihood phylogenetic trees were reconstructed using RaxML version 8.2.3 (Stamatakis, 2014) under the Generalized Time Reversible (GTR) model with gamma and invariant site parameters (GTRGAMMAI) and 1000 bootstrap repetitions. The 16S rRNA gene sequence of B. wiedmannii sp. nov. strain FSL W8-0169T was checked for the presence of chimeras using decipher (Wright et al., 2012) and submitted to GenBank (accession number listed in Table S1). The 16S rRNA gene sequence phylogeny (Fig. 1) supports the close relatedness of B. wiedmannii sp. nov. with existing members of the B. cereus group, as indicated by the ≥98.2 % sequence similarity and high bootstrap values. It is known that B. cereus group species cannot be delineated based on 16S rRNA gene sequences (Liu et al., 2015), and therefore an rpoB gene phylogeny was reconstructed to allow for a more discriminatory analysis.
Fig. 1.
Maximum-likelihood tree reconstructed in RaxML based on 1471 bp 16S rRNA gene sequences using the generalized time reversible evolutionary model (GTR) with gamma distributed and invariant sites, rooted using Geobacillus stearothermophilus IFO 12550T as an outgroup. Bootstrap values above ≥70 are displayed on branches. Bar, 0.02 substitutions per site. The B. wiedmannii sp. nov. type strain FSL W8-0169T is indicated in bold type.
Based on the rpoB gene sequence, the 11 B. wiedmannii sp. nov. strains characterized formed a monophyletic, well-supported (bootstrap value of 97) cluster within the B. cereus group (Fig. S1). The rpoB gene sequences of all 11 B. wiedmannii sp. nov. strains were deposited in the Food Microbe Tracker database where they can be found under strain name records.
Genomes of FSL W8-0169T and 10 other characterized B. wiedmannii sp. nov. isolates were sequenced on an Illumina MiSeq or HiSeq platform, respectively. The Nextera XT adapters were trimmed from 250 or 100 bp paired-end reads with Trimmomatic version 0.32, respectively (Bolger et al., 2014). Quality of reads was assessed using FastQC version 0.11.2. Sequences were assembled de novo using SPAdes version 3.0.0 or 3.6.2, respectively (Bankevich et al., 2012). Draft genome quality was verified by quast version 3.2 (Gurevich et al., 2013) and sufficient coverage was confirmed using SAMtools version 1.2 (Li et al., 2009). Short reads were submitted to the Sequence Read Archive (SRA) and draft genome assemblies to NCBI GenBank and Prokaryotic Genome Annotation Pipeline (Angiuoli et al., 2008). Assembled draft genomes ranged from 5.3 to 5.6 Mbp (Table 1). The NCBI accession numbers for WGS and draft genome assembly statistics are listed in Table 1.
Whole genome sequences of FSL W8-0169T and the other 10 strains representative of B. wiedmannii sp. nov. were queried against the B. cereus PubMLST database (http://pubmlst.org/bcereus/). Two (gmk 136, pta 234) new ATs and a novel sequence type ST-1081 were identified for FSL W8-0169T. New ATs were identified also for strains FSL M7-0044 (ilv 267, pur 236), FSL M7-0044 and FSL H8-0032 (pyc 193) and FSL K6-0069 (glp 224, gmk 135, ilv 237, pta 233).
Four novel STs were identified as a result of finding a novel AT and four novel STs were identified as a result of new combinations of existing ATs (Table 1). Strains FSL J3-0113 and FSL P2-0558 carried STs that were previously observed in the PubMLST database.
Pairwise average nucleotide blast identities (ANIb) between the genomes for the 11 B. wiedmannii sp. nov. strains, representative strains of the other eight validly published and three effectively published B. cereus group species were calculated (Fig. 2; Richter & Rosselló-Móra, 2009). The representative strains of other B. cereus group species were type strains, except in the case of B. anthracis where the WGS was not available for the type strain; therefore, the WGS data for the widely used B. anthracis strain Ames were used. ANIb was computed using the calculate_ani.py program, which is available on Github (https://github.com/widdowquinn/scripts/blob/master/bioinformatics/calculate_ani.py). A cladogram (Fig. 2) was built based on the pairwise ANIb similarity matrix using the ‘hclust’ package in R (R Core Team, 2014). Using a 95 % cut-off for species delineation, these analyses confirmed B. wiedmannii sp. nov. as a novel species (Richter & Rosselló-Móra, 2009). The B. cereus group species closest to B. wiedmannii sp. nov. FSL W8-0169T is B. anthracis (strain Ames with an ANIb of 93.62); the species most distant is B. cytotoxicus (strain NVH 391-98T with an ANIb of 82.73). The minimum pairwise ANIb value between B. wiedmannii sp. nov. type strain FSL W8-0169T and 10 other strains representative of this species was 95.92 %, confidently classifying them in the same species. The ANIb values of the effectively, but not validly, published B. cereus group species (i.e. ‘B. manliponensis’, ‘B. bingmayongensis’, ‘B. gaemokensis’) support their recognition as species. Resolving the taxonomic status of these proposed new species would help in minimizing the occurrence of species misclassification of organisms in the B. cereus group.
Fig. 2.
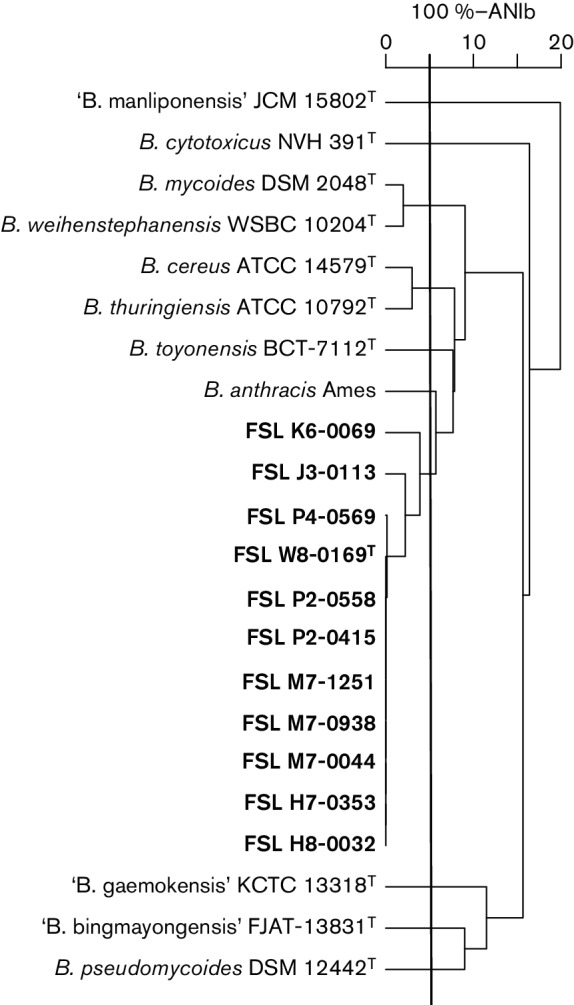
UPGMA cladogram built based on ANIb values in R. The horizontal rule denotes 100 %−ANIb. The vertical line denotes 100–95 %-ANIb, which correlates to 70 % traditional DNA–DNA hybridization, set as a species cut-off (Chan et al., 2012). The 11 representative strains of B. wiedmannii sp. nov. are shown in bold type.
ANIb results were confirmed by in silico DNA–DNA hybridization (DDH) conducted using the Genome-to-Genome Distance Calculator (GGDC; http://ggdc.dsmz.de/distcalc2.php; Meier-Kolthoff et al., 2013). Predicted pairwise DDH values between B. wiedmannii sp. nov. FSL W8-0169T and representative strains of other B. cereus group species were substantially below the species cut-off of 70 % (Table S2; Richter & Rosselló-Móra, 2009), with the exception of FSL K6-0069, which was classified as a borderline member of B. wiedmannii sp. nov., as it had a predicted DDH of 57.9 %, and with a 45.24 % probability of DDH being 70 % or above (Table S2). This strain was deposited with the DSMZ (DSM 102051) and BCCM (LMG 29270).
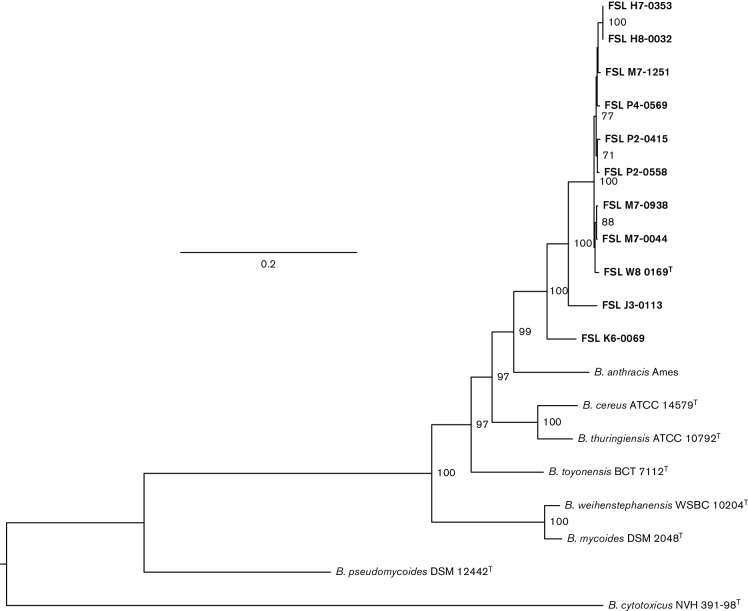
To further confirm B. wiedmannii sp. nov. as a novel taxon, the core genome single nucleotide polymorphisms (SNPs) were identified with kSNP v2 (Gardner & Hall, 2013) using a kmer size of 21 as identified by Kchooser and used to reconstruct a maximum-likelihood phylogenetic tree (Fig. 3). The B. wiedmannii sp. nov. type strain FSL W8-0169T and 10 other representatives of this species formed a monophyletic, robust clade with a bootstrap value of 99. Overall, comparative ANIb and DDH analyses, as well as WGS phylogenetic analysis provide strong evidence in favour of recognizing B. wiedmannii sp. nov. as a novel species in the B. cereus group.
Fig. 3.
Maximum-likelihood tree reconstructed in RaxML based on core SNPs using the GTR model with gamma distributed sites and 1000 bootstrap repetitions, rooted by mid-point. Bootstrap values above ≥70 are displayed on branches. Bar, 0.2 substitutions per site. The representative strains of B. wiedmannii sp. nov. are shown in bold type.
To examine whether strains belonging to B. wiedmannii sp. nov. were found in previous studies, we have classified the 11 B. wiedmannii sp. nov. strains into previously defined phylogenetic groups (Guinebretière et al., 2010). We extracted panC gene sequences from the WGS and used them for phylogenetic classification with an online tool (Guinebretière et al., 2010; https://www.tools.symprevius.org/bcereus/english.php). All 11 strains were affiliated to phylogenetic group II, which is known for psychrotolerance (Guinebretière et al., 2008) and cytotoxicity (Guinebretière et al., 2010). The combination of these two characteristics sets phylogenetic group II apart from other phylogenetic groups within the B. cereus group. This, together with genomic evidence, supports the novel species delineation.
Phenotypic characteristics
Phenotypic characterizations were performed for B. wiedmannii sp. nov. FSL W8-0169T and the 10 additional B. wiedmannii sp. nov. strains (Table S3). Microscopic evaluation of FSL W8-0169T revealed cells were rod-shaped with an average length of 2.8 µm and average width of 1.2 µm. Transmission electron microscopy (negative staining with 2 % uranyl acetate) images revealed spores in the centre of the vegetative cell (Fig. 4). Colonies grown on brain heart infusion medium (BHI, Becton Dickinson) were large, round and off-white. Colonies grown on Bacara (bioMérieux) plating medium were orange, with an off-white precipitate surrounding colonies, indicative of lecithinase activity. All tested B. wiedmannii sp. nov. strains were positive for phosphoinositide phospholipase C based on blue pigmentation on BC/BT agar (R & F Products) plates after 24 h of incubation at 35 °C.
Fig. 4.
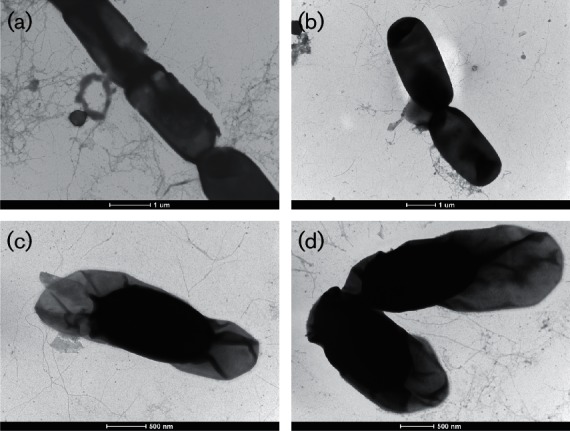
Transmission electron microscopy (negative staining with 2 % uranyl acetate) images of B. wiedmannii sp. nov. FSL W8-0169T. Panels (a) and (b) show vegetative cells, while (c) and (d) show spores with exosporium. Bars, 1 µm (a, b); 500 nm (c, d).
B. wiedmannii sp. nov. strain FSL W8-0169T and the 10 additional strains were haemolytic on sheep’s blood trypticase soy agar, were Gram-stain-positive and were positive for catalase activity, as determined according to the FDA BAM protocols (U.S. Food and Drug Administration, 2015). All strains were oxidase-negative (BBL Dry Slide Oxidase; Becton Dickinson). All strains hydrolysed starch and casein, as determined by plating on starch agar and skimmed milk agar using standard methods (Logan & De Vos, 2009). All strains were facultative anaerobes, as determined by the BAM method (U.S. Food and Drug Administration, 2015). All tested strains were motile at 30 °C, with the exception of one strain (FSL H7-0353), as determined by observing growth outside of the inoculation stab on motility agar.
The range of growth temperatures was assessed by plating overnight cultures onto BHI agar (Becton Dickinson) plates and subsequently incubating plates at 5, 10, 15, 20, 25, 30, 40, 45 and 55 °C, for incubation periods as defined by Logan & De Vos (2009). The minimum growth temperature for five out of 11 B. wiedmannii sp. nov. strains was 5 °C and all strains grew between 10 and 40 °C. Six out of 11 strains were able to grow at 43 °C. None of the strains grew at 45 °C or above. Sodium tolerance was assessed by inoculating strains into tryptic soy broth (TSB) containing 2, 5, 7, 8, 9 or 10 % (w/v) NaCl, followed by incubation at 30 °C for up to 14 days. Growth of seven out of 11 tested strains was observed at NaCl concentrations up to 5 % (w/v) (for four strains growth was inhibited at concentrations >2 % NaCl), but all strains were inhibited by NaCl concentrations of 7–10 % (w/v). Growth in pH-adjusted TSB incubated at 30 °C for 14 days demonstrated that all strains were capable of growing at pH 5–10.
API CH 50 kits were used to characterize acid production from catabolism of carbohydrates according to the manufacturer’s instructions (bioMérieux); incubation was performed at 30 °C for 48 h (Table 2). Of the 11 strains tested, all were able to utilize (acid production from catabolism) d-ribose, d-glucose, d-fructose, arbutin, aesculin ferric citrate, salicin, cellobiose, maltose, trehalose, amidon and glycogen. Acid production from d-mannose (positive for one out of 11 strains), amygdalin (positive for three out of 11 strains), potassium gluconate (positive for two out of 11 strains) and N-acetylglucosamine (positive for 10 out of 11 strains) was variable. All 11 B. wiedmannii sp. nov. strains were also tested with API 20E kits, which were used as per the manufacturer’s instructions; incubation was performed at 30 °C for 48 h (see Table 2 for results). Strain FSL W8-0169T and the 10 additional strains were positive for acetoin production [Voges-Proskauer (VP) reaction] and gelatinase activity. Utilization of citrate as a carbon source was variable (six out of 11 strains were able to utilize citrate). All other reactions included in the API 20E kit were negative (Table 2). Nitrate reduction was determined according to the FDA BAM method (U.S. Food and Drug Administration, 2015) and was variable. Strain FSL W8-0169T and seven out of the other 10 strains were able to reduce nitrate to nitrite, while three strains (FSL H8-0032, FSL K6-0069 and FSL H7-0353) were not able to reduce nitrate.
Table 2. Characteristics of the B. cereus group strains from this study and previously published studies.
Species: 1, B. wiedmannii sp. nov. FSL W8-0169T (n=11 strains); 2, B. cytotoxicus NVH 391-98T (n=5 strains, data from Guinbretière et al., 2013); 3, B. pseudomycoides DSM 12442T (n=7 strains, data from Guinbretière et al., 2013); 4, B. cereus ATCC 14579T (n=13 strains, data from Guinbretière et al., 2013 and this study); 5, B. thuringiensis CIP 53137T (n=8 strains; data from Guinbretière et al., 2013); 6, B. weihenstephanensis WSBC 10204T (n=5 strains, data from Guinbretière et al., 2013); 7, B. mycoides CIP 102472T (n=3 strains data from Guinbretière et al., 2013); 8, B. anthracisNCTC 10340 (n=37 strains; data from Logan et al., 1985 and Guinbretière et al., 2013); 9, B. toyonensis BCT-7112T (data from Jiménez et al., 2013). All species are catalase-positive, Gram-positive rods. All species/strains were positive for acid production from utilization of d-glucose, d-fructose, aesculin and maltose and were negative for acid production from utilization of erythritol, d-arabinose, l-arabinose, d-xylose, l-xylose, d-adonitol, methyl β-d-xylopyranoside, l-sorbose, l-rhamnose, dulcitol, inositol, d-mannitol, d-sorbitol, methyl α-d-mannopyranoside, melibiose, inulin, melezitose, raffinose, xylitol, gentiobiose, d-lyxose, d-tagatose, d-fucose, l-fucose, d-arabitol, l-arabitol, 2-ketogluconate and 5-ketogluconate. All strains were negative for lysine decarboxylase, ornithine decarboxylase, hydrogen sulfide production, urease, tryptophan deaminase and indole production, as confirmed by API 20E strips. +, Positive result; –, negative result; v, variable trait (i.e. some strains gave a positive reaction while others gave a negative reaction); + w, weak positive result. Data are given for the type strains, with the proportion of positive reactions for all tested strains of the same species in parentheses.
| Characteristic | Species | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
| Cell diameter >1.0 µm | + | + | + | + | + | + | + | + | + |
| Anaerobic growth | + | + w | + | + | + | + | + | + | + |
| Motile | + (91) | + | + | + | + | + | + | − | + |
| Rhizoid colony | − | − | + | − | − | − | + | − | − |
| Starch hydrolysis | + | − | − | + (67) | + | + | + | + (97) | + |
| Egg yolk lecithinase | + | + w | + w | + | + | + | + | + | + |
| Nitrate reduction | + (73) | + | + | + (81) | + | + | + | + | + |
| Arginine dihydrolase | − | + | + (71) | + (70) | + (67) | + (40) | − (36) | − | + |
| Citrate | + (55) | − (33) | − (11) | + (34) | + (33) | − | − (60) | − | + |
| Gelatin | + | + | + (86) | + | + | + (80) | + (67) | + (70) | + |
| Voges–Proskauer reaction | + | + w | + | + | + | + (60) | + (67) | + | + |
| Glycerol | − | − | + w (17) | − (90) | − (92) | + w (33) | − (96) | − | − |
| Ribose | + | + (67) | + | + | − (81) | + w | + w (67) | + | + |
| Galactose | − | − | − | − | − (38) | − | − (32) | − | − |
| d-Mannose | − | + | − | − (18) | + (25) | − (67) | − | − | − |
| Methyl α-d-glucoside | − | − | − | − | − | − (33) | − (12) | − (3) | + |
| N-Acetylglucosamine | + | + | + | + | + (69) | + | + | + | + |
| Amygdalin | − (36) | + w (67) | − | + w (23) | − | + w (83) | − (50) | − | + w |
| Arbutin | + | + | − (33) | + (82) | + (63) | + | + (84) | v (32) | + |
| Salicin | + | + | + (67) | + (64) | + (75) | + | + (80) | - | + |
| Cellobiose | + | + | − (33) | + (55) | + (63) | + w (83) | + w (75) | − | − |
| Trehalose | + | − | + | + | + | + | + | + | + |
| Glycogen | + | − | + | + (73) | + (88) | + (67) | + | + (92) | + |
| Maltose | + | + | + | + | + | + | + | + | + |
| Sucrose | − | − | − (33) | + (64) | + (38) | − (33) | + (50) | + | + |
| Gentiobiose | − | − | − | − (9) | − | − (33) | − | − | − |
| Potassium gluconate | − (18) | − | − | − (36) | − | − | − (12) | − (3) | − |
| Minimum growth temperature (°C) | 10 (5–10) |
20 | 10 (8–10) |
10 (7–15) |
10 (7–15) |
5 (5–7) |
5 (5–7) |
>10 | 10–45 |
| Maximum growth temperature (°C) | 40 (40–43) |
50 | 40 (40–43) |
45 (40–45) |
45 (40–45) |
37 (35–37) |
37 (35–37) |
<50 | 45 |
Fatty acid methyl ester extraction and analysis were performed according to the MIDI protocol by MIDI Labs. The content of iso-C13 : 0 in B. wiedmannii sp. nov. FSL W8-0169T was comparable to that in B. cytotoxicus and B. toyonensis, but was substantially lower than in other B. cereus group species (Table 3). The content of iso-C16 : 0, which proved to be the most discriminatory, was higher in B. wiedmannii sp. nov. compared to all other validly published B. cereus group species. The peptidoglycan type was determined by DSMZ as described previously (Schumman, 2011). meso-Diaminopimelic acid was identified as the diagnostic diamino acid for the peptidoglycan type A1γ in B. wiedmannii sp. nov. FSL W8-0169T.
Table 3. Cellular fatty acid composition of B. wiedmannii sp. nov. FSL W8-0169T and the type strains of other B. cereus group species.
Strains: 1, B. wiedmannii sp. nov. FSL W8-0169T; 2, B. mycoides KCTC 3453T; 3, B. thuringiensis KCTC 3452T; 4, B. weihenstephanensis KCTC 3975T; 5, B. cereus KCTC 3624T; 6, B. pseudomycoides KCTC 3862T; 7, B. cytotoxicus NVH 391-98T; 8, B. toyonensis BCT-7112T. Data represent the per cent of the total fatty acids as determined by the Microbial Identification System software. t, Trace amount detected; nd, not detected; nr, not reported. Data for strain 1 are from this study, for strains 2–6 from Jung et al. (2011), for strain 7 from Guinebrètiere et al. (2013) and for strain 8 from Jiménez et al. (2013).
| Fatty acid | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
|---|---|---|---|---|---|---|---|---|
| iso-C12 : 0 | 0.8 | t | t | 1.6 | t | 6.4 | 0.4 | nd |
| C12 : 0 | 0.2 | t | t | t | t | 3.5 | 0.6 | nd |
| iso-C13 : 0 | 6.9 | 11.8 | 11.2 | 16.6 | 13.9 | 10.6 | 7.0 | 7.1 |
| anteiso-C13 : 0 | 1.0 | t | 1.1 | 1.7 | 1.7 | 4.9 | 1.8 | nd |
| iso-C14 : 0 | 5.1 | 2.8 | 5.1 | 4.7 | 4.3 | 2.4 | 5.0 | 2.3 |
| C14 : 0 | 3.3 | 3.2 | 3.6 | 4.5 | 3.1 | 3.4 | 2.4 | 3.2 |
| iso-C15 : 0 | 27.6 | 33.5 | 31.3 | 29.2 | 33.2 | 33.2 | 36.5 | 38.6 |
| anteiso-C15 : 0 | 4.0 | 2.1 | 3.4 | 2.7 | 4 | 1.9 | 10.8 | 3.1 |
| C15 : 0 | nd | nd | t | nd | nd | 1.1 | 0.1 | nd |
| C16 : 1ω7c alcohol | 1.9 | 2.1 | t | 1.3 | t | nd | nd | nd |
| iso-C16 : 0 | 9.1 | 5.5 | 4.5 | 5.8 | 4.2 | 6.8 | 6.7 | 5.1 |
| C16 : 1ω11c | 1.1 | 2.9 | nd | 1.2 | nd | nd | nd | nd |
| C16 : 0 | 7.3 | 5.3 | 3.1 | 7.7 | 3.1 | 8.2 | 10.8 | 5.6 |
| C15 : 0 2-OH | 0.4 | t | t | nd | 1.1 | nd | nr | nd |
| iso-C17 : 1ω10c | 4.7 | 10.6 | 2 | 5.7 | 2.6 | nd | nr | 5.8 |
| iso-C17 : 1ω5c | 2.6 | 2 | 4.2 | 2.1 | 5.3 | nd | nr | 4.9 |
| anteiso C17 : 1 A | 0.5 | nd | nd | nd | t | nd | 0.1 | nd |
| iso-C17 : 0 | 10.1 | 6.8 | 13.7 | 7.6 | 5.9 | 8.9 | 8.2 | 11.4 |
| anteiso C17 : 0 | 1.5 | t | nd | t | 1.5 | t | 3.4 | nr |
| C18 : 0 | 0.5 | nd | nd | nd | nd | 2.6 | 0.5 | nr |
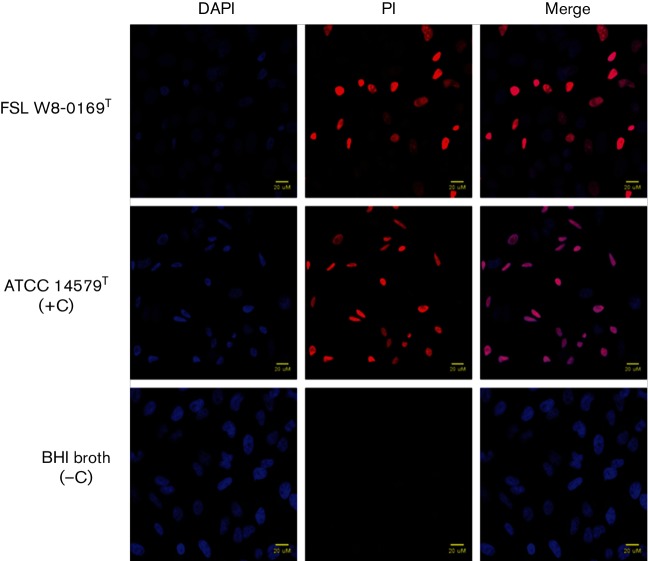
The presence of detectable levels of haemolysin BL (HBL) and non-haemolytic enterotoxin (NHE) in cultures grown in BHI for 20 h at 32 °C was evaluated using Duopath Cereus Enterotoxins kits (Merck Millipore). Both HBL and NHE were detected for FSL W8-0169T and seven additional strains. For three strains (FSL P4-0569, FSL K6-0069 and FSL J3-0113), HBL was not detected, but NHE was. Cytotoxicity was determined by measuring the influx of propidium iodide (PI) into HeLa cells exposed to supernatants (5 %, v/v) of B. wiedmannii sp. nov. cultures grown in BHI at 32 °C for 20 h. B. wiedmannii sp. nov. supernatants were added to HeLa cells maintained in Eagle’s minimal essential medium (EMEM) containing 5 µg PI ml−1 and incubated for 30 min at 37 °C with 5 % CO2. HeLa cells were fixed with 4 % paraformaldehyde (PFA) at room temperature for 10 min. Cells were permeabilized with 1 % Triton X-100 and incubated with 4′,6-diamidino-2-phenylindole (DAPI; 1 µg ml−1) at room temperature for 1–5 min, prior to fixing coverslips and gluing onto microscope slides. Slides were imaged using a Zeiss 710 confocal microscope (Bio-imaging Facility, Cornell University). Images were processed using FIJI software (Schindelin et al., 2012). Bacterial strains were considered cytotoxic if the proportion of PI-positive cells was greater than the average proportion of PI-positive cells for the BHI-negative control. FSL W8-0169T and all additional strains were cytotoxic (Fig. 5).
Fig. 5.
Cytotoxicity of B. wiedmannii sp. nov. FSL W8-0169T supernatants. HeLa cells grown on coverslips were co-incubated with 5 % (v/v) culture supernatants of FSL W8-0169T and B. cereus ATCC 14579T (included as a positive control), for 30min with 5 µg PI ml−1 (red). After fixation with 4 % PFA, HeLa cells were stained with 1 µg DAPI ml−1 (blue). All cells stained blue with DAPI, while only cells with compromised membrane integrity stained red. Incubation with BHI media was included as a negative control. The merged images demonstrate the proportion of cells with damaged membrane due to cytotoxic activity of bacterial supernatant.
Description of Bacillus wiedmannii sp. nov.
Bacillus wiedmannii (wied.mann′i.i; N.L. gen. masc. n. wiedmannii named in honour of Martin Wiedmann, for his contribution to the understanding of the biology of Bacillus).
Cells are Gram-stain-positive rods with an average length of 2.8 µm and average width of 1.2 µm. Endospores are ellipsoidal and are present in the centre of vegetative cells; cells have a non-swollen sporangia. Colonies are positive for egg-yolk lecithinase, with the ability to hydrolyse casein and starch. Facultative anaerobe. Colonies grown on BHI agar at 37 °C for 24 h appear cream-coloured, round and flat, with a rough surface. Growth temperature ranges from 5 and 43 °C, with optimum growth between 20 and 40 °C. Most strains are motile at 30 °C. Citrate utilization and acid production from potassium gluconate and mannose are variable. Can be differentiated from other strains in the B. cereus group by the combination of an inability to produce acid from fermentation of sucrose and inability to hydrolyse arginine (arginine dihydrolase-negative). Produces toxins HBL and the non-haemolytic toxin NHE. Cells are cytotoxic in a HeLa cell culture model. Strains can be differentiated from other B. cereus group species by panC gene sequence comparisons, ANIb and WGS.
The type strain, FSL W8-0169T (=DSM 102050T=LMG 29269T), was isolated from a silo raw milk sample collected from a dairy powder processing plant in the north-eastern USA. The genomic DNA G+C content of the type strain is 35.3 mol%, as determined by genome sequencing.
Acknowledgements
Bacara chromogenic plating media and API CH 50 and API 20E kits were donated by bioMérieux. BCBT chromogenic plating medium was donated by R&F laboratories. We would like to thank the staff at the Bio-imaging facility (Cornell University) and John L. Grazul from Cornell Center for Materials Research (CCMR, Cornell University) for their assistance with microscopic analyses and Avery Becker, Miquela Hanselman and Jiahui Jian for their assistance with media preparation and experimental analyses.
Supplementary Data
Abbreviations:
- ANIb
average nucleotide identity blast
- AT
allelic type
- DDH
DNA–DNA hybridization
- GTR
generalized time-reversible model
- HBL
haemolysin BL
- MLST
multilocus sequence typing
- NHE
non-haemolytic enterotoxin
- PI
propidium iodide
- SNP
single nucleotide polymorphism
- SRA
sequence read archive
- WGS
whole genome sequence
References
- Angiuoli S. V., Gussman A., Klimke W., Cochrane G., Field D., Garrity G., Kodira C. D., Kyrpides N., Madupu R., et al. (2008). Toward an online repository of standard operating procedures (SOPs) for (meta)genomic annotation. OMICS 12137–141. 10.1089/omi.2008.0017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M., Kulikov A. S., Lesin V. M., Nikolenko S. I., Pham S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19455–477. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A. M., Lohse M., Usadel B.(2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 302114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceuppens S., Boon N., Uyttendaele M.(2013). Diversity of Bacillus cereus group strains is reflected in their broad range of pathogenicity and diverse ecological lifestyles. FEMS Microbiol Ecol 84433–450. 10.1111/1574-6941.12110 [DOI] [PubMed] [Google Scholar]
- Chan J. Z., Halachev M. R., Loman N. J., Constantinidou C., Pallen M. J.(2012). Defining bacterial species in the genomic era: insights from the genus Acinetobacter. BMC Microbiol 12302. 10.1186/1471-2180-12-302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner S. N., Hall B. G.(2013). When whole-genome alignments just won't work: kSNP v2 software for alignment-free SNP discovery and phylogenetics of hundreds of microbial genomes. PLoS One 8e81760. 10.1371/journal.pone.0081760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinebretière M. H., Thompson F. L., Sorokin A., Normand P., Dawyndt P., Ehling-Schulz M., Svensson B., Sanchis V., Nguyen-The C., et al. (2008). Ecological diversification in the Bacillus cereus group. Environ Microbiol 10851–865. 10.1111/j.1462-2920.2007.01495.x [DOI] [PubMed] [Google Scholar]
- Guinebretière M. H., Velge P., Couvert O., Carlin F., Debuyser M. L., Nguyen-The C.(2010). Ability of Bacillus cereus group strains to cause food poisoning varies according to phylogenetic affiliation (groups I to VII) rather than species affiliation. J Clin Microbiol 483388–3391. 10.1128/JCM.00921-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinebretière M. H., Auger S., Galleron N., Contzen M., De Sarrau B., De Buyser M. L., Lamberet G., Fagerlund A., Granum P. E., et al. (2013). Bacillus cytotoxicus sp. nov. is a novel thermotolerant species of the Bacillus cereus Group occasionally associated with food poisoning. Int J Syst Evol Microbiol 6331–40. 10.1099/ijs.0.030627-0 [DOI] [PubMed] [Google Scholar]
- Gurevich A., Saveliev V., Vyahhi N., Tesler G.(2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 291072–1075. 10.1093/bioinformatics/btt086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huck J. R., Hammond B. H., Murphy S. C., Woodcock N. H., Boor K. J.(2007). Tracking spore-forming bacterial contaminants in fluid milk-processing systems. J Dairy Sci 904872–4883. 10.3168/jds.2007-0196 [DOI] [PubMed] [Google Scholar]
- Ivy R. A., Ranieri M. L., Martin N. H., den Bakker H. C., Xavier B. M., Wiedmann M., Boor K. J.(2012). Identification and characterization of psychrotolerant sporeformers associated with fluid milk production and processing. Appl Environ Microbiol 781853–1864. 10.1128/AEM.06536-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez G., Urdiain M., Cifuentes A., López-López A., Blanch A. R., Tamames J., Kämpfer P., Kolstø A. B., Ramón D., et al. (2013). Description of Bacillus toyonensis sp. nov., a novel species of the Bacillus cereus group, and pairwise genome comparisons of the species of the group by means of ANI calculations. Syst Appl Microbiol 36383–391. 10.1016/j.syapm.2013.04.008 [DOI] [PubMed] [Google Scholar]
- Jung M. Y., Paek W. K., Park I. S., Han J. R., Sin Y., Paek J., Rhee M. S., Kim H., Song H. S., et al. (2010). Bacillus gaemokensis sp. nov., isolated from foreshore tidal flat sediment from the Yellow Sea. J Microbiol 48867–871. 10.1007/s12275-010-0148-0 [DOI] [PubMed] [Google Scholar]
- Jung M. Y., Kim J. S., Paek W. K., Lim J., Lee H., Kim P. I., Ma J. Y., Kim W., Chang Y. H.(2011). Bacillus manliponensis sp. nov., a new member of the Bacillus cereus group isolated from foreshore tidal flat sediment. J Microbiol 491027–1032. 10.1007/s12275-011-1049-6 [DOI] [PubMed] [Google Scholar]
- Kim M. J., Han J. K., Park J. S., Lee J. S., Lee S. H., Cho J. I., Kim K. S.(2015). Various enterotoxin and other virulence factor genes widespread among Bacillus cereus and Bacillus thuringiensis strains. J Microbiol Biotechnol 25872–879. 10.4014/jmb.1502.02003 [DOI] [PubMed] [Google Scholar]
- Lechner S., Mayr R., Francis K. P., Prüss B. M., Kaplan T., Wiessner-Gunkel E., Stewart G. S., Scherer S.(1998). Bacillus weihenstephanensis sp. nov. is a new psychrotolerant species of the Bacillus cereus group. Int J Syst Bacteriol 481373–1382. 10.1099/00207713-48-4-1373 [DOI] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup (2009). The sequence alignment/Map format and SAMtools. Bioinformatics 252078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Liu G. H., Hu G. P., Sengonca C., Cetin S., Lin N. Q., Tang J. Y., Tang W. Q., Lin Y. Z.(2014). Bacillus bingmayongensis sp. nov., isolated from the pit soil of Emperor Qin's Terra-cotta warriors in China. Antonie Van Leeuwenhoek 105501–510. 10.1007/s10482-013-0102-3 [DOI] [PubMed] [Google Scholar]
- Liu Y., Lai Q., Göker M., Meier-Kolthoff J. P., Wang M., Sun Y., Wang L., Shao Z.(2015). Genomic insights into the taxonomic status of the Bacillus cereus group. Sci Rep 514082. 10.1038/srep14082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan N. A., Carman J. A., Melling J., Berkeley R. C.(1985). Identification of Bacillus anthracis by API tests. J Med Microbiol 2075–85. 10.1099/00222615-20-1-75 [DOI] [PubMed] [Google Scholar]
- Logan N. A., De Vos P.(2009). Genus I. Bacillus Cohn 1872, 174AL. Bergey’s Manual of Systematic Bacteriology, 2nd edn,vol. 3(The Firmicutes), 21–128. Edited by Vos P., Garrity G., Jones D., Krieg N. R., Ludwig W., Rainey F. A., Schleifer K.-H., Whitman W.New York: Springer. [Google Scholar]
- Lücking G., Stoeckel M., Atamer Z., Hinrichs J., Ehling-Schulz M.(2013). Characterization of aerobic spore-forming bacteria associated with industrial dairy processing environments and product spoilage. Int J Food Microbiol 166270–279. 10.1016/j.ijfoodmicro.2013.07.004 [DOI] [PubMed] [Google Scholar]
- Meier-Kolthoff J. P., Auch A. F., Klenk H. P., Göker M.(2013). Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 1460. 10.1186/1471-2105-14-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R. A., Kent D. J., Watterson M. J., Boor K. J., Martin N. H., Wiedmann M.(2015a). Spore populations among bulk tank raw milk and dairy powders are significantly different. J Dairy Sci 988492–8504. 10.3168/jds.2015-9943 [DOI] [PubMed] [Google Scholar]
- Miller R. A., Kent D. J., Boor K. J., Martin N. H., Wiedmann M.(2015b). Different management practices are associated with mesophilic and thermophilic spore levels in bulk tank raw milk. J Dairy Sci 984338–4351. 10.3168/jds.2015-9406 [DOI] [PubMed] [Google Scholar]
- Moayeri M., Leppla S. H., Vrentas C., Pomerantsev A. P., Liu S.(2015). Anthrax pathogenesis. Annu Rev Microbiol 69185–208. 10.1146/annurev-micro-091014-104523 [DOI] [PubMed] [Google Scholar]
- Nakamura L. K.(1994). DNA relatedness among Bacillus thuringiensis serovars. Int J Syst Bacteriol 44125–129. 10.1099/00207713-44-1-125 [DOI] [PubMed] [Google Scholar]
- Nakamura L. K.(1998). Bacillus pseudomycoides sp. nov. Int J Syst Bacteriol 481031–1035. 10.1099/00207713-48-3-1031 [DOI] [PubMed] [Google Scholar]
- R Core Team (2014). R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing. [Google Scholar]
- Richter M., Rosselló-Móra R.(2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA 10619126–19131. 10.1073/pnas.0906412106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Method 9676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann P.(2011). Peptidoglycan structure. Method Microbiol 38101–129. [Google Scholar]
- Smith N. R., Gordon R. E., Clarck F. E.(1952). Aerobic Spore-Forming Bacteria Monograph No 16. Washington, DC: US Department of Agriculture. [Google Scholar]
- Stamatakis A.(2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 301312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S.(2013). mega6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 302725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. Food and Drug Administration (2015). Bacteriological analytical manual (BAM). http://www.fda.gov/Food/FoodScienceResearch/LaboratoryMethods/ucm2006949.htm.
- Watterson M. J., Kent D. J., Boor K. J., Wiedmann M., Martin N. H.(2014). Evaluation of dairy powder products implicates thermophilic sporeformers as the primary organisms of interest. J Dairy Sci 972487–2497. 10.3168/jds.2013-7363 [DOI] [PubMed] [Google Scholar]
- Wright E. S., Yilmaz L. S., Noguera D. R.(2012). DECIPHER, a search-based approach to chimera identification for 16S rRNA sequences. Appl Environ Microbiol 78717–725. 10.1128/AEM.06516-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.