Abstract
Phosphate limitation is an important environmental stress that affects the metabolism of various organisms and, in particular, can trigger the virulence of numerous bacterial pathogens. Clostridium perfringens, a human pathogen, is one of the most common causes of enteritis necroticans, gas gangrene and food poisoning. Here, we focused on the high affinity phosphate-binding protein (PBP-1) of an ABC-type transporter, responsible for cellular phosphate uptake. We report the crystal structure (1.65 Å resolution) of the protein in complex with phosphate. Interestingly, PBP-1 does not form the short, low-barrier hydrogen bond with phosphate that is typical of previously characterized phosphate-binding proteins, but rather a canonical hydrogen bond. In its unique binding configuration, PBP-1 forms an unusually high number of hydrogen bonds (14) with the phosphate anion. Discrimination experiments reveal that PBP-1 is the least selective PBP characterised so far and is able to discriminate phosphate from its close competing anion, arsenate, by ~150-fold.
Phosphorus is essential for living organisms because it is involved in the composition of critical biomolecules (e.g., nucleic acids) and in key biological processes1. Although phosphorus is relatively abundant on our planet, it is mostly trapped in rocks2. Therefore, the bioavailability of phosphorus is limited, and living organisms have had to develop efficient mechanisms to extract phosphate from the environment3. In prokaryotes, two main systems have been identified so far: (i) the Phosphate inorganic transport (Pit) and (ii) the Phosphate specific transport (Pst) systems. The Pit system is a single transmembrane component4,5, that is fueled by the proton-motive force and transports a divalent metal cation complexed with a phosphate anion6. Pst, the predominant phosphate transporter7, is a classical ABC-transporter, composed of 5 proteins: 2 membrane permeases (PstA and PstC), 2 ATPases (PstB) and the high-affinity phosphate binding protein PstS (or phosphate-binding protein, PBP)8. The Pst system is a high-affinity, high-specificity transporter that enables the extraction of phosphate from the environment9,10, including niches where there are plethora of competing anions, such as arsenate11. Moreover, the Pst system, as well as other components of the phosphate (Pho) regulon, is directly involved in bacterial phosphate homeostasis8. Indeed, phosphate starvation may be sensed by the Pst system, which subsequently signals for the regulation of expression of numerous genes, including genes encoding virulence factors. Therefore, the Pst system has been linked to bacterial virulence12,13,14,15.
The crystal structures of several PstS (or PBP) proteins from various sources have been determined16,17,18,19. These proteins possess a venus-flytrap topology, composed of two globular domains comprising a central β-sheet core flanked by an α-helix and, linked together with a flexible hinge20. At the interface of both domains lies a phosphate-binding cleft which comprises 8 residues, forming 12 hydrogen bonds with the phosphate molecule in several PBPs (e.g., PfluDING19, and in Mycobacterium tuberculosis, Escherichia coli, Yersinia pestis17,19,21), and 14 hydrogen bonds in the Borrelia burgdorferi B31 PBP18. Most of the PBPs (e.g., PfluDING, M. tuberculosis, E. coli and Y. pestis) characterised so far possess a key aspartate residue involved in a short, low barrier hydrogen bond (LBHB) with the bound phosphate anion17. This bond was shown to be highly selective, and is responsible for the extremely high selectivity of PBPs over various competing anions such as arsenate (discriminated by ~3 orders of magnitude11) or sulphate (discriminated by ~5 orders of magnitude17).
Here, we focus on PBP-1 from Clostridium perfringens, a Gram-positive bacteria belonging to Clostridia. Mainly anaerobic and able to form heat-resistant endospores, most Clostridia strains are pathogenic for both humans and animals, and produce potent extracellular toxins such as tetanus or botulinum neurotoxins22. Among them, C. perfringens is responsible for more than 42% of clostridial human infection23 and causes numerous cases of enteritis necroticans, gas gangrene (also called clostridial myonecrosis) and food poisoning22,24,25,26. In such pathogenic bacteria, the phosphate ABC transporter components, involved in bacterial pathogenicity and virulence, constitute interesting putative targets for vaccine development27.
We report the crystal structure of a PBP-1 from the C. perfringens strain ATCC 13124 in the F222 space group, and compare it to other known PBP structures. We highlight that, as for B. burgorferi B31 PBP, C. perfringens PBP-1 forms 14 hydrogen bonds, but no LBHB, with the bound phosphate anion. Using an in vitro discrimination assay, we show that PBP-1 is less discriminating towards arsenate (~200-fold) than previously reported PBPs (~700 to 5000-fold11), possibly relevant to function.
Methods
Production and purification of the C. perfringens PBP-1
The gene encoding the phosphate-binding protein of Clostridium perfringens ATCC 13124 (C. perfringens) was optimized for heterologous expression in Escherichia coli (E. coli) and synthetised by Genwiz (South Plainfield, New Jersey, USA). The gene was cloned into a pET22b (+) plasmid, using NcoI and NotI as cloning sites. Protein production and purification was performed in the E. coli strain BL21(DE3)-pGro7/GroEL (TaKaRa) in ZYP medium28 (complemented with 100 μg.ml−1 ampicillin and 34 μg.ml−1 chloramphenicol). The cultures were grown at 37°C to reach OD600nm = 0.6 and were then induced by the lactose consumption of ZYP medium coupled with a temperature transition to 17°C over 16 h. Cells were harvested by centrifugation (4 500 g, 4°C, 15 min) and pellets were suspended in lysis buffer (20 mM TRIS, pH 8, 100 mM NaCl, Lysozyme 0.25 mg.ml−1, DNAse I 10 μg.ml−1, PMSF 0.1 mM, MgSO4 20 mM and EDTA-free anti-protease (Roche)) and stored at −80°C for 2 h. Partially lysed cells were thawed at 37°C for 15 min and disrupted by 3 30 sec sonication steps (QSonica sonicator; amplitude 30). Debris was removed by centrifugation (12 500 g, 4°C, 30 min). The supernatant was then loaded onto a Nickel affinity column (HisTrap 5 ml, FFCrude from GE Healthcare) at a flow rate of 5 ml.min−1. Proteins attached to the column were eluted by imidazole competition using an elution buffer (20 mM TRIS, pH 8, 100 mM NaCl and 250 mM imidazole). Then, a size exclusion chromatography step (Superdex 75 16/60, GE Healthcare) was performed using a 20 mM TRIS, pH 8 and 100 mM NaCl buffer. Protein production and purity were checked by 15% SDS-PAGE analysis. Protein concentrations were quantified using Nanodrop 2000c (Thermo-scientific, Illkirch, France) by measuring the absorbance ε0.1% = 0.547 g/l at 280 nm (based on the calculation from ProtParam29).
Crystallisation of the C. perfringens PBP-1
C. perfringens PBP-1 was concentrated up to 60 mg.ml−1 using a centrifugation device (Vivaspin 500, MWCO 3 kDa, Sartorius, Germany). The best conditions for crystallisation were screened for using a commercial kit (Wizard I and II, Molecular Dimension, England) at 298 K with a Mosquito instrument (TTP Labtech, England). The sitting-drop vapour diffusion method setup in a 96-well plate was used with 2 protein:reservoir ratios (1:1 and 2:1). Drops were monitored with a Discovery V8 binocular microscope (Zeiss, Germany). Crystals of 150–200 μm were obtained in the condition containing 0.1 M Sodium Acetate, 0.2 M Zinc Acetate pH 4.5 and 10% PEG 3000 at both ratios. To obtain bigger crystals, an optimisation of this condition was performed using the hanging drop method and a ratio of 1:1 (protein:reservoir, 500 nL:500 nL). This led to the creation of reproducible crystals of 200–250 μm after 3 days at 298 K.
Data collection, molecular replacement and refinement
Crystals were transferred for a few seconds in a drop (1 μL) containing a cryo-protectant solution made out of the crystallisation solution and 10% (v/v) of glycerol. After being mounted on a CryoLoop (Hampton research), crystals were flash-frozen in liquid nitrogen. X-ray diffraction intensities were collected on the ID29-2 beamline at the ESRF (Grenoble, France) with 0.1 s exposure times. Diffraction data were collected from 2000 images; each frame consisted of 0.1° step oscillations, over a range of 200° (Table 1). The molecular replacement was performed using Phaser30 and PDB 4GD5 as a starting model. Refinement was carried out with REFMAC5 and Phenix31,32. The model was improved using Coot33. The model and structure factors were deposited in the Protein Data Bank code, ID 4Q8R.
Table 1. Data collection and refinement statistics.
| Data collection | |
|---|---|
| PDB ID | 4Q8R |
| Beamline | ID29 |
| Wavelength (Å) | 0.8000 |
| Detector | PILATUS 6M |
| Oscillation (°) | 0.1 |
| Number of frames | 2000 |
| Resolution (Å) (last bin) | 1.65 (1.75 − 1.65) |
| Space group | F222 |
| Unit-cell parameters (Å) | a = 70.78; b = 105.73; c = 146.45 |
| No. of observed reflections (last bin) | 240770 (38721) |
| No. of unique reflections (last bin) | 33133 (5276) |
| Completeness (%)(last bin) | 99.9 (100) |
| Rmeas (%) (last bin) | 4.3 (62.8) |
| CC (1/2) (last bin) | 100 (89.7) |
| I/σ(I) (last bin) | 25.76 (3.37) |
| Redundancy (last bin) | 7.26 (7.33) |
Discrimination assay
The discrimination assay was performed as previously described11. Briefly, the ability of PBP-1 to discriminate phosphate from arsenate was tested by dialysing PBP-1 against a competition buffer, made of 50 mM Tris-HCl buffer pH 8.0, containing 140 nM phosphate (with 1 μCi of radiolabelled phosphate 32P; Perkin Elmer NEX053011MC), and varying concentrations of arsenate (from 0.1 μM to 100 μM). The protein was diluted to 5 μM into a buffer made of 50 mM Tris-HCl buffer pH 8.0 and 0.1% BSA (Sigma), and placed in dialysis units (Thermo Scientific). These units were placed in 50 mL tubes containing the competition buffer and incubated 24 hours, at room temperature, with gentle shaking. The intake of radiolabelled phosphate by PBP-1 was measured by the radioactivity of 250 μL of the protein sample and of the outer buffer. These samples were added to 4 mL of scintillation liquid (Ultima Gold, Perkin Elmer) and measured in a liquid scintillation analyser (Packard TRE-CARB 2100TR). The experiment was performed in four independent replicates, and included a control without PBP-1. The background adsorption by 0.1% BSA was ≤1000-fold relative to the PBP-1 signal. The measured level of radioactivity of the protein solution (PBP bound to radiolabelled phosphate) in conditions containing no arsenate corresponded to 100% binding (or 0% replacement). Conversely, the background level of radioactivity in the dialysis tube corresponded to 0% binding (or 100% replacement). The replacement values were plotted using GraphPad Prism software (v.5) and fitted to the sigmoidal equation (1):
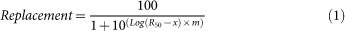 |
R50 is the ratio [arsenate]/[phosphate] yielding 50% replacement, and m is the sigmoidal slope factor.
Points that do not coincide with the fitted slope (e.g., for 100% or 0% replacement) are due to experimental errors of the system.
Results
C. perfringens PBP-1 crystal structure
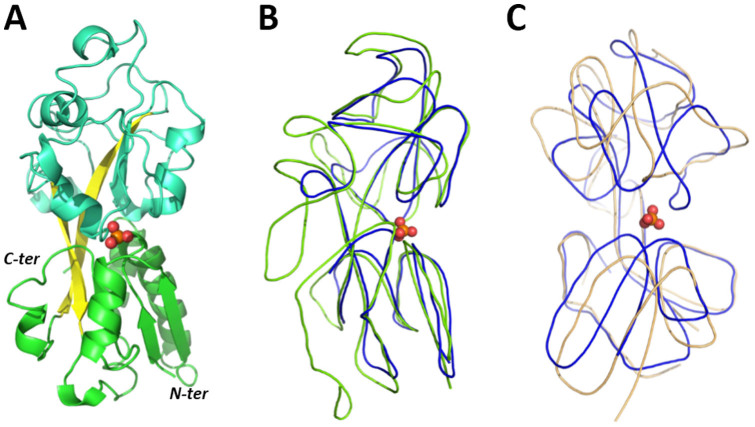
C. perfringens PBP-1 is a relatively distant protein from the known P. fluorescens PBP (PfluDING) and E. coli PstS (41 and 52% sequence similarity; 13 and 25% sequence identity, respectively). C. perfringens PBP-1 is closer to B. burdgorferi B31 PBP (70% sequence similarity; 28% sequence identity) (Fig. 1). C. perfringens PBP-1 was overexpressed in E. coli strain BL21(DE3)-pGro7/EL and purified (Fig. S1) with a significant yield, ~30 mg of protein per litre of culture. The crystal structure of C. perfringens PBP-1 was determined at 1.65 Å resolution in the F222 space group (Fig. 2A; Table 1). The PBP-1 structure exhibits two globular domains, each one composed of a central core β-sheets flanked by α-helices. Both domains are linked together with a flexible hinge. This topology, known as a Venus flytrap, is believed to adopt an open state while empty and to close upon ligand binding20,34.
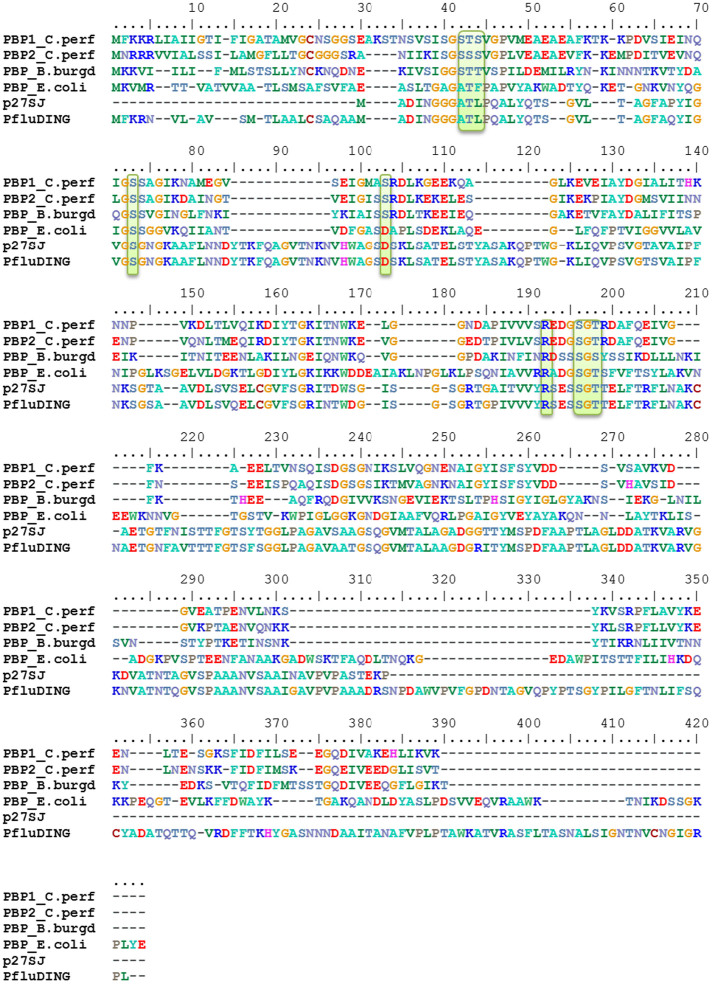
Figure 1. Sequence alignment of PBPs.
Sequences from Hypericum perforatum PBP (p27SJ, gb|AY866430.1), Pseudomonas fluorescens SBW 25 PBP (PfluDING, YP_002872028.1), Escherichia coli PBP (PBP_E. coli, WP_000867147.1), B. burgdorferi B31 PBP (PBP_B.burd, NP_212349.2), and both C. perfringens PBPs (PBP1_C.per, YP_695070.1 and PBP2_C.per, YP_695071.1) were aligned using T-COFFEE Expresso41 and were manually refined. Residues involved in phosphate binding are highlighted in green.
Figure 2. Structural analysis of C. perfringens PBP-1 and structural comparison with other PBPs.
(A). Structure of the C. perfringens PBP-1. Domains are colourised in cyan and green, whereas β-sheets from the hinge are colourised in yellow. (B). Structures of C. perfringens and E. coli PBPs are represented by lines and colourised in blue and green, respectively. The phosphate molecule is represented as a sphere. (C). Structures of PBP-1 from C. perfringens and B. burgdorferi are represented as lines and colourised in blue and orange, respectively. The phosphate molecule is represented as a sphere.
The structure of C. perfringens PBP-1 was also solved in another space group (P21) at a similar resolution (1.7 Å) by the Center for Structural Genomics of Infectious Diseases (PDB ID: 4GD5). As expected, both structures are nearly superimposed (RSMD on α-carbon = 0.275 Å); interestingly, however, some regions adopt significantly different conformations, e.g., two surface helices are shifted by >1 Å between both structures (Fig. S2). The structural cause for these different conformations is not obvious from the structures, and may be due to the different crystallisation conditions and/or to the different crystal packings.
Structural comparison with other PBPs
C. perfringens PBP-1 is shorter (246 residues) than most of PBPs characterised thus far: the PBPs from E. coli and PfluDING are indeed much larger (321 and 371 residues, respectively). This discrepancy is strikingly visible while superposing their respective structures. In particular, while the “frontside” of the protein (right, in Fig. 2B), i.e. the part that binds phosphate and putatively interacts with the transporter is similar, the “backside” (left, in Fig. 2B) of the C. perfringens PBP-1 seems to be missing. In fact, a significant loop fragment and C-terminus fragment are “missing” compared with the E. coli or P. fluorescens PBPs. Remarkably, C. perfringens PBP-1 is very similar in size to p27SJ, a biologically active albeit truncated form of a PBP called p38SJ (38 kDa) isolated from St John's Wort35. Interestingly, this truncated form modulates the cell cycle of human cells and inhibits HIV-1 replication35,36. The structure of C. perfringens PBP-1 is closer to that of B. burgdorferi B31 PBP, although it is important to note that both structures are not in the same state: the B. burgdorferi B31 PBP was crystallized in the absence of ligand, and is therefore in the “open” state, whereas C. perfringens PBP-1 is bound to phosphate and adopts a “closed” conformation (Fig. 2C). B. burgdorferi B31 PBP and C. perfringens PBP-1 shares only 28% sequence identity, but the structural comparison of both structures suggests that these PBPs undergoes dramatic conformational changes upon phosphate binding/release (Fig. S3A&B). Indeed, the binding site residue Arg125 in PBP-1 (bound state) is about 10 Å away from its corresponding residue in the B. burgdorferi B31 PBP structure (free state). Interestingly, the conformational changes in the binding/release motion may not only relate to a rigid body motion of the two globular domains along the flexible hinge (“venus flytrap” motion) but might also involve some secondary structure reorganisations, as seen for Helix 7 (Fig. S3B).
The phosphate-binding site
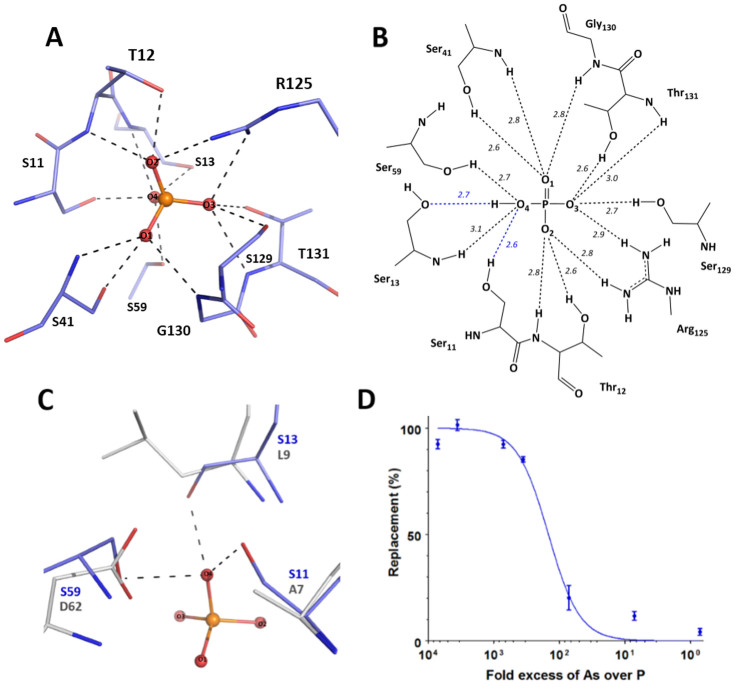
The structure of C. perfringens PBP-1 bound to phosphate reveals that the protein forms 14 hydrogen bonds with the anion, via 9 different residues (Fig. 3A). The phosphate anion is completely sequestered, buried between the two protein domains, with no solvent accessibility. The phosphate anion interacts with 5 backbone N-H group (Thr12, Ser13, Ser41, Gly130, Thr131), 6 side chain O-H groups (Ser11, Thr12, Ser41, Ser59, Thr131, Ser129) and 2 side chain N-H2 groups (Arg125) (Fig. 3B). Although the structure resolution (1.65 Å) does not allow for the direct location of hydrogen atoms from the electron density maps, a careful analysis of the position of the residues interacting with the phosphate anion, as well as of the neighbouring residues, enables us to establish a hypothetical hydrogen bond network. Assuming that, similarly to what was previously observed, including at acidic pH (4.5)19, C. perfringens PBP-1 binds dibasic phosphate, the only proton carried by the phosphate moiety may be accepted by Ser13 side chain O-H: indeed it appears to be the only group with a riding hydrogen atom that does not already unambiguously accept a hydrogen atom from another protein residue (Fig. S4). Moreover, the case of phosphate O4 is intriguing: in addition to the hydrogen bond it forms with Ser13, it is located at distances (between 2.6 to 3.1 Å) and angles (between 108 to 131°) that are compatible with 3 more hydrogen bonds, with Ser13 (main chain NH group), Ser59 (side chain –OH), and Ser11 (side chain –OH). Phosphate O2 only has two free electron doublets and, is likely to form two hydrogen bonds out of these three possible interactions. We surmise that this is due to the tremendous tightness and contact density within the phosphate-binding site, as previous described in PBPs11.
Figure 3. The phosphate-binding site of the C. perfringens PstS.
(A). Phosphate-binding cleft: The phosphate anion is sequestered via 14 hydrogen bonds (black dashes) formed with the protein (shown as blue sticks). (B). Hydrogen bonding network. The 14 hydrogen bonds between the phosphate anion and the protein (in dashes) and the interaction distances (in Å), are highlighted. Blue dashes indicate hydrogen bonds that are unique to C. perfringens PBP-1. (C). Superposition of the phosphate-binding sites from PBP-1 (blue sticks) and PfluDING (grey sticks; PDB ID 4F1V). Only the interactions with the phosphate anion (in ball and sticks) in PBP-1, which differ from those in PfluDING, are shown (black dashes). (D). The phosphate-arsenate selectivity of PBP-1. Increasing arsenate concentrations were used to compete with a constant, radiolabelled phosphate concentration for binding to PBP-1. The level of measured radioactivity (or protein-bound radiolabelled phosphate) corresponds to 0% replacement when no arsenate is bound to the protein. When all the protein is bound to arsenate, replacement is 100%.
Compared with other PBPs (from E. coli, Y. pestis, M. tuberculosis PBPs or PfluDING), the C. perfringens PBP-1 forms 2 additional hydrogen bonds with the phosphate anion (14 versus 12 in other PBPs). These two additional bonds are made possible by 2 amino acids substitutions in the phosphate binding pocket: a substitution of an alanine into serine (Ala11Ser according to the numbering of C. perfringens PBP-1) and a substitution of a leucine into serine (Leu13Ser) (Fig. 3C). Notably, these two substitutions occur in residues putatively involved in repulsive interactions with the bound anion11, replacing them by hydrogen bonds. A third substitution also replaces the short (2.50 Å)17, Low barrier Hydrogen Bond (LBHB) formed between the phosphate anion and an aspartate residue, with a serine residue (Asp59Ser). The distance between the Ser11 O-H and the phosphate' O2 of 2.7 Å, suggests a classical hydrogen bond.
PBP-1 is less discriminant than other PBPs
A previous work on PBPs highlighted the link between the bonds involved in phosphate binding, and the ability of PBPs to discriminate phosphate and close competing anions such as arsenate11. The case of C. perfringens PBP-1 is indeed interesting, as it forms 14 hydrogen bonds with the phosphate anion, whereas the highly discriminating PBPs form 12. Discrimination experiments reveal that C. perfringens PBP-1 discriminates between phosphate and the very close arsenate, albeit only by ~150-fold (Fig. 3D). This result is interesting, as it reveals that C. perfringens PBP-1 is the least discriminating PBP tested thus far.
Discussion
C. perfringens PBP-1 exhibits a reduced selectivity
The crystal structure of C. perfringens PBP-1 reveals an interesting phosphate binding cleft, that significantly differs from that of other known PBPs. C. perfringens PBP-1 binds phosphate via 9 residues and 14 hydrogen bonds, whereas most of previously characterised PBPs utilise 8 residues, 11 hydrogen bonds and 1 short, Low Barrier H-bond (LBHB). A recent work highlighted the role of the LBHB in the tremendous anion selectivity of PBPs11 and suggested that the LBHB is the main determinant for the discrimination of PBPs.
Interestingly, C. perfringens PBP-1 does not form a LBHB with phosphate, but rather 2 additional classical hydrogen bonds (14 total). Here, we show that PBP-1 is the least discriminating PBP isolated thus far, capable of discriminating phosphate from the closest competing anion, arsenate by ~150-fold, whereas other tested PBPs all discriminate by >500-fold11.
Notably, the selectivity of C. perfringens PBP-1 is only slightly higher than that of PBP mutants where the LBHB was suppressed (discrimination factors of 50 to 75-fold11). Therefore, we surmise that the observed reduced discrimination capacity of PBP-1 is the result of its unique phosphate-binding configuration. This hypothesis requires further investigations.
Relationship between selectivity and environment
The use of a different phosphate binding strategy than the other characterized PBPs is intriguing, especially because the differences include a key, unique bond formed between the phosphate anion and the PBP. The reduced ability of C. perfringens PBP-1 to discriminate phosphate from arsenate might be linked to the life cycle of this organism. Indeed, C. perfringens, which lives in sediments and soils37, as well as in the intestinal tracks of humans and other vertebrates26, might not have as critical a need as other soils or water-living organisms to prevent arsenate uptake.
However, the absence of a LBHB, and/or the relatively low selectivity of C. perfringens PBP-1 might also relate to a functional adaptation, providing or improving upon a property of the PBP that is beneficial for C. perfringens. Given the recently reported involvement of PBPs (or pstS) in bacterial virulence8,38,39, future studies should assess, among other features, the phosphate transport rates and the phosphate binding affinity of PBP-1. In particular, given the facts that the LBHB of PBPs have been previously reported not to be critical for phosphate binding11,40, and that substitutions Ala11Ser and Leu13Ser putatively replace the repulsive interactions by hydrogen bonds, the affinity of PBP-1 for phosphate might differ from that of other PBPs.
Supplementary Material
Supplementary Information
Acknowledgments
This research was supported by a grant to EC from “Agence Nationale pour la Recherche sur le Sida et les Hépatites Virales” (grant N°12264). M.R. and D.G. were supported by grants from AP-HM (Marseille, France).
Footnotes
The authors declare no competing financial interests.
Author Contributions E.C., D.G. and M.E. designed experiments. D.G., M.R., C.B. and M.E. performed the experiments. D.G. and M.E. analysed the results. D.G., E.C. and M.E. wrote the paper. All of the authors offered a critical review of the paper.
References
- Westheimer F. H. Why nature chose phosphates. Science 235, 1173–1178 (1987). [DOI] [PubMed] [Google Scholar]
- Cordell D., Drangert J.-O. & White S. The story of phosphorus: Global food security and food for thought. Global Environ. Chang. 19, 292–305 (2009). [Google Scholar]
- Cordell D. W. S. Peak phosphorus: clarifying the key issues of a vigorous debate about long-term phosphorus security. Sustainability 3, 2027–2049 (2011). [Google Scholar]
- Willsky G. R. & Malamy M. H. Characterization of two genetically separable inorganic phosphate transport systems in Escherichia coli. J. Bacteriol. 144, 356–365 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris R. M., Webb D. C., Howitt S. M. & Cox G. B. Characterization of PitA and PitB from Escherichia coli. J. Bacteriol. 183, 5008–5014 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Veen H. W., Abee T., Kortstee G. J., Konings W. N. & Zehnder A. J. Translocation of metal phosphate via the phosphate inorganic transport system of Escherichia coli. Biochemistry 33, 1766–1770 (1994). [DOI] [PubMed] [Google Scholar]
- Hsieh Y.-J. & Wanner B. L. Global regulation by the seven-component Pi signaling system. Curr. Opin. Microbiol. 13, 198–203 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamarche M. G., Wanner B. L., Crepin S. & Harel J. The phosphate regulon and bacterial virulence: a regulatory network connecting phosphate homeostasis and pathogenesis. FEMS Microbiol. Rev. 32, 461–473, 10.1111/j.1574-6976.2008.00101.x (2008). [DOI] [PubMed] [Google Scholar]
- Hsieh Y. J. & Wanner B. L. Global regulation by the seven-component Pi signaling system. Curr. Opin. Microbiol. 13, 198–203, 10.1016/j.mib.2010.01.014 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner B. L. Signal transduction in the control of phosphate-regulated genes of Escherichia coli. Kidney Int. 49, 964–967 (1996). [DOI] [PubMed] [Google Scholar]
- Elias M. et al. The molecular basis of phosphate discrimination in arsenate-rich environments. Nature 491, 134–137, 10.1038/nature11517 (2012). [DOI] [PubMed] [Google Scholar]
- Lamarche M. G. et al. Inactivation of the pst system reduces the virulence of an avian pathogenic Escherichia coli O78 strain. Infect. Immun. 73, 4138–4145 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bains M., Fernandez L. & Hancock R. E. Phosphate starvation promotes swarming motility and cytotoxicity of Pseudomonas aeruginosa. Appl. Environ. Microbiol. 78, 6762–6768, 10.1128/AEM.01015-12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J., Zaborina O., Holbrook C., Zaborin A. & Alverdy J. Depletion of intestinal phosphate after operative injury activates the virulence of P aeruginosa causing lethal gut-derived sepsis. Surgery 144, 189–197, 10.1016/j.surg.2008.03.045 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaborin A. et al. Red death in Caenorhabditis elegans caused by Pseudomonas aeruginosa PAO1. Proc. Natl. Acad. Sci. USA 106, 6327–6332, 10.1073/pnas.0813199106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas N. K., Vyas M. N. & Quiocho F. A. Crystal structure of M tuberculosis ABC phosphate transport receptor: specificity and charge compensation dominated by ion-dipole interactions. Structure 11, 765–774 (2003). [DOI] [PubMed] [Google Scholar]
- Luecke H. & Quiocho F. A. High specificity of a phosphate transport protein determined by hydrogen bonds. Nature 347, 402–406, 10.1038/347402a0 (1990). [DOI] [PubMed] [Google Scholar]
- Brautigam C. A., Ouyang Z., Deka R. K. & Norgard M. V. Sequence, biophysical, and structural analyses of the PstS lipoprotein (BB0215) from Borrelia burgdorferi reveal a likely binding component of an ABC-type phosphate transporter. Protein Sci. 23, 200–212, 10.1002/pro.2406 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebschner D. et al. Elucidation of the phosphate binding mode of DING proteins revealed by subangstrom X-ray crystallography. J. Am. Chem. Soc. 131, 7879–7886, 10.1021/ja901900y (2009). [DOI] [PubMed] [Google Scholar]
- Felder C. B., Graul R. C., Lee A. Y., Merkle H. P. & Sadee W. The Venus flytrap of periplasmic binding proteins: an ancient protein module present in multiple drug receptors. AAPS pharmSci 1, E2 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe M. et al. Structures of OppA and PstS from Yersinia pestis indicate variability of interactions with transmembrane domains. Acta Crystallogr. D 63, 1185–1193 (2007). [DOI] [PubMed] [Google Scholar]
- Stiles B. G., Barth G., Barth H. & Popoff M. R. Clostridium perfringens epsilon toxin: a malevolent molecule for animals and man? Toxins (Basel) 5, 2138–2160, 10.3390/toxins5112138 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leal J., Gregson D. B., Ross T., Church D. L. & Laupland K. B. Epidemiology of Clostridium species bacteremia in Calgary, Canada, 2000–2006. J. Infect. 57, 198–203, 10.1016/j.jinf.2008.06.018 (2008). [DOI] [PubMed] [Google Scholar]
- Scallan E. et al. Foodborne illness acquired in the United States--major pathogens. Emerg. Infect. Dis. 17, 7–15, 10.3201/eid1701.091101p1 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. et al. Toxin plasmids of Clostridium perfringens. Microbiol. Mol. Biol. Rev. 77, 208–233, 10.1128/MMBR.00062-12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers G. S. et al. Skewed genomic variability in strains of the toxigenic bacterial pathogen, Clostridium perfringens. Genome Res. 16, 1031–1040, 10.1101/gr.5238106 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falero-Diaz G. et al. Intranasal vaccination of mice against infection with Mycobacterium tuberculosis. Vaccine 18, 3223–3229 (2000). [DOI] [PubMed] [Google Scholar]
- Graslund S. et al. Protein production and purification. Nature methods 5, 135–146, 10.1038/nmeth.f.202 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins M. R. et al. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 112, 531–552 (1999). [DOI] [PubMed] [Google Scholar]
- McCoy A. J. et al. Phaser crystallographic software. J. Appl. Cryst. 40, 658–674, 10.1107/S0021889807021206 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin A. A. et al. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr. D 60, 2184–2195, 10.1107/S0907444904023510 (2004). [DOI] [PubMed] [Google Scholar]
- Adams P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221, 10.1107/S0907444909052925 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W. G. & Cowtan K. Features and development of Coot. Acta Crystallogr. D 66, 486–501, 10.1107/S0907444910007493 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn S. et al. Structure-function relationships in a bacterial DING protein. FEBS Lett. 581, 3455–3460 (2007). [DOI] [PubMed] [Google Scholar]
- Darbinian N. et al. Evidence for phosphatase activity of p27SJ and its impact on the cell cycle. J. Cell. Biochem. 107, 400–407, 10.1002/jcb.22135 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darbinian-Sarkissian N. et al. p27(SJ), a novel protein in St John's Wort, that suppresses expression of HIV-1 genome. Gene Ther. 13, 288–295 (2006). [DOI] [PubMed] [Google Scholar]
- Rood J. I. & Cole S. T. Molecular genetics and pathogenesis of Clostridium perfringens. Microbiol. Rev. 55, 621–648 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blus-Kadosh I., Zilka A., Yerushalmi G. & Banin E. The effect of pstS and phoB on quorum sensing and swarming motility in Pseudomonas aeruginosa. PloS one 8, e74444, 10.1371/journal.pone.0074444 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crepin S. et al. The Pho regulon and the pathogenesis of Escherichia coli. Vet. Microbiol. 153, 82–88, 10.1016/j.vetmic.2011.05.043 (2011). [DOI] [PubMed] [Google Scholar]
- Wang Z., Luecke H., Yao N. & Quiocho F. A. A low energy short hydrogen bond in very high resolution structures of protein receptor--phosphate complexes. Nat. Struct. Biol. 4, 519–522 (1997). [DOI] [PubMed] [Google Scholar]
- Notredame C., Higgins D. G. & Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302, 205–217 (2000). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information