Abstract
Fancd2 is a component of the Fanconi anemia (FA) DNA repair pathway, which is frequently found defective in human cancers. The full repertoire of Fancd2 functions in normal development and tumorigenesis remains to be determined. Here we developed a Flag- and hemagglutinin-tagged Fancd2 knock-in mouse strain that allowed a high throughput mass spectrometry approach to search for Fancd2-binding proteins in different mouse organs. In addition to DNA repair partners, we observed that many Fancd2-interacting proteins are mitochondrion-specific. Fancd2 localizes in the mitochondrion and associates with the nucleoid complex components Atad3 and Tufm. The Atad3-Tufm complex is disrupted in Fancd2−/− mice and those deficient for the FA core component Fanca. Fancd2 mitochondrial localization requires Atad3. Collectively, these findings provide evidence for Fancd2 as a crucial regulator of mitochondrion biosynthesis, and of a molecular link between FA and mitochondrial homeostasis.
Fanconi anemia (FA) is a genetic disorder associated with congenital developmental defects, bone marrow failure and predisposition to cancers, particularly acute myelogenous leukemia1,2,3. In recent decades, the function of FA proteins has been extensively studied and very well documented. As a genetically heterogeneous disease, more than a dozen FA and FA-associated proteins (FANCA, −B, −C, −E, −F, −G, −L, −M, FAAP100, FAAP24, FAAP20, HES1, MHF1, and MHF2) form a core complex4,5. In response to DNA damage or DNA replication stress, this multimeric FA core complex monoubiquitinates two downstream FA proteins, FANCD2 and FANCI, which recruit other components of the FA/BRCA pathway to damaged DNA loci and consequently affect DNA replication, cell cycle control and DNA damage repair processes4,5,6,7,8.
While many studies point to an essential role for the FA pathway in DNA damage repair and genome maintenance, emerging evidence suggests that FA deficiency causes mitochondrion dysfunction, which may play roles in the pathogenesis of bone marrow failure and leukemia progression in FA. For example, several studies show that FA cells exhibit abnormalities in mitochondrial metabolism, Ca2+ homeostasis, and gene expression9,10,11. Additional studies revealed mitochondrion defects in FA patients and mice12,13,14,15. At the molecular level, it has been shown that FA deficiency causes altered mitochondrial morphology and mitochondrial complex defect, and decreased mitochondrial membrane potential and ATP production11,16,17. More recently, it has been reported that some of the FA proteins are required for removal of damaged mitochondria and reduction of mitochondrial reactive oxygen species (ROS)18. Furthermore, the utility of antioxidants shows in vivo protective effects against the onset of malignancies and bone marrow failure in FA knockout mice19,20. Although these studies have indicated a correlation between mitochondrion defects and FA deficiency, direct evidence for a mechanistic link is missing.
In this study, we developed a Fancd2 knock-in mouse model that enabled the identification of a non-canonical function of the FA pathway in mitochondrion biosynthesis. We further demonstrated that Fancd2 localizes in the mitochondrion and associates with the nucleoid complex components Atad3 and Tufm, thus providing a molecular link between FA and mitochondrial homeostasis.
Results
Generation and analysis of 3XFLAG/HA-tagged Fancd2 knock-in mice
To study the in vivo function of the FA pathway, we generated a Fancd2 knock-in mouse model, in which a duel tandem (3XFLAG and HA) tag was inserted at the C-terminus of the endogenous Fancd2 locus (Fig. 1A; Supplemental Fig. 1A,B). We confirmed that the tagged Fancd2 protein was expressed and could be pulled down by FLAG and HA antibodies using 2-step immunoprecipitation in Fancd2KI ES cells (Supplemental Fig. 1C). In addition, immunoblotting of mouse embryonic fibroblast (MEF) cells from wild-type (WT) and Fancd2KI/KI mice with anti-FLAG and anti-Fancd2 antibodies demonstrated that the tagged Fancd2 and the WT endogenous Fancd2 proteins were expressed at roughly the same levels (Supplemental Fig. 1D), indicating that the FLAG-HA tag does not affect the expression of the Fancd2 protein. Furthermore, treatment of Fancd2KI ES cells showed that both mitomycin C (MMC) and hydrourea (HU) induced monoubiquitination of the tagged Fancd2 protein (Supplemental Fig. 1E). Immunofluorescence microscopy for Fancd2KI/KI MEF cells with an anti-HA antibody showed that tagged Fancd2 retained the ability to form DNA damage foci in nuclei after MMC treatment (Supplemental Fig. 1F). These results indicate that the tagged Fancd2 protein retains its functionality in the context of the DNA damage response at the cellular level.
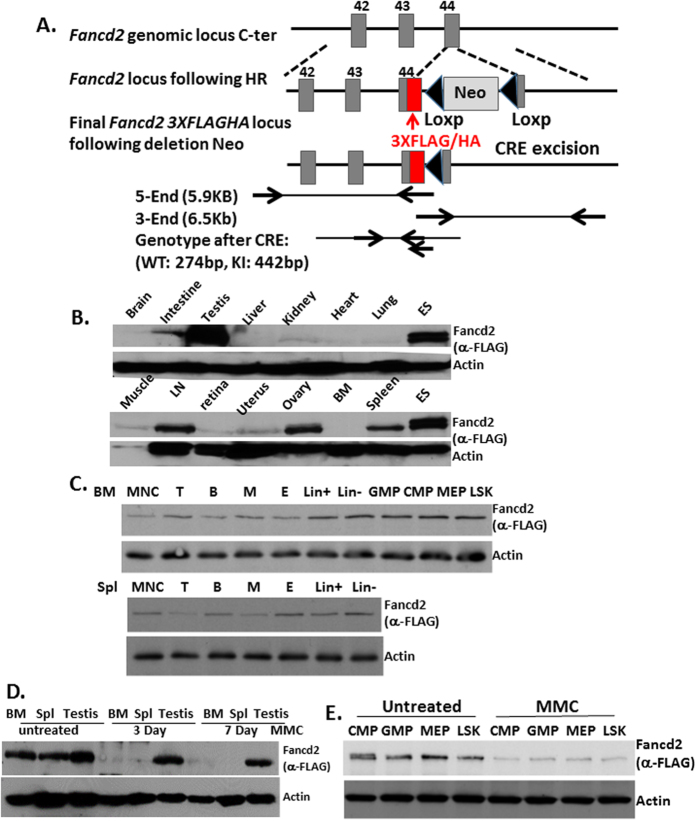
Figure 1. Generation of 3XFLAG/HA-tagged Fancd2 knock-in mice.
(A) Schematic diagram of Fancd23XFLAG/HA (Fancd2-KI) mouse knock-in strategy. Fancd2 genomic locus, Fancd2 locus following homologous recombination (targeted DNA) and the final Fancd23XFLAG/HA locus following excision of the loxP-flanked Neomycin resistance gene (Neo) is illustrated. Fancd2 exons are shown as gray boxes and are numbered. FLAG/HA (red box) tags are also indicated. (B) Tissues isolated from Fancd2-KI mice were analyzed by immunoblotting with the M2 anti-FLAG antibody. Fancd2 is highly expressed in ES cells, testes, spleen, lymph nodes and ovaries. (C) Bone marrow and spleen mononuclear cells isolated from Fancd2-KI mice were sorted and subjected to western blot. Fancd2 is highly expressed in B, erythroid, Lin−, MPP populations and relatively low in T, myeloid, Lin+ cells. (D) Fancd2 expression in bone marrow, spleen and testis is dramatically reduced after a single dose (3 mg/kg) of MMC IP injection in mice. (E) Fancd2 expression was analyzed in sorted population cells in bone marrow after MMC treatment. Full-length blot is presented in Supplementary Fig. 4. The data represent a summary of more than three mice of each genotype from two independent experiments.
To examine the function of the tagged Fancd2 alleles in vivo, we subjected Fancd2KI/KI animals to MMC injection. Unlike Fancd2-KO (Fancd2−/−) mice, which showed hypersensitivity to MMC and died within 10 days after injection, homozygous Fancd2KI/KI mice were resistant to MMC (Supplemental Fig. 2A). Histopathological analysis showed that the Fancd2KI/KI mice had normal testes and ovaries compared to Fancd2−/− mice, characteristic of severe gonadal defects (Supplemental Fig. 2B)2. In the context of hematopoiesis, the colony-forming activity of Fancd2KI/KI bone marrow progenitor cells was not affected by MMC at a dose that significantly inhibited the proliferation of Fancd2−/− progenitor cells (Supplemental Fig. 2C). We also examined primary MEF cells derived from the homozygous Fancd2KI/KI mice for other two FA cellular hallmarks: MMC-induced cell death and G2/M arrest21,22. Not surprisingly, unlike the Fanca−/− or Fancd2−/− cells, the Fancd2KI/KI cells were completely resistant to MMC-induced cell death (Supplemental Fig. 2D and G)2/M arrest (Supplemental Fig. 2E). Taken together, the Fancd2KI/KI mice display normal development and the tagged Fancd2 protein retains full functionality in the DNA damage response.
Tissue-specific expression of Fancd2 in mice
We next determined the expression of the Fancd2 protein in different tissues of Fancd2KI/KI mice. We found that Fancd2 was highly expressed in the testes, which is consistent with previously reported data23. We also observed high levels of Fancd2 expression in ES cells, lymph node, spleen and ovary tissue (Fig. 1B). Since bone marrow failure and later myeloid malignancies are the major symptoms in FA patients1,2,3, we analyzed Fancd2 expression in different hematopoietic lineages. Fancd2 was found highly expressed in B (B220+), erythroid (CD71+Ter119+), lineage-negative (Lin−), committed progenitors and Lin−Sca1+c-Kit+ (LSK) cell populations, and was relatively low in T (CD3e+), myeloid (Gr-1+CD11b+), Lin+ cell populations (Fig. 1C). Interestingly, little ubiquitinated (Ub) form of Fancd2 was detected in the fresh tissues or cells, unlike the obvious Ub-Fancd2 in ES cell lines (Fig. 1B).
Since hypersensitivity to MMC-induced DNA damage is the cellular hallmark of FA cells, we examined whether MMC treatment would alter the level of Fancd2 protein in vivo. We found that the levels of the Fancd2 protein were dramatically reduced in bone marrow, spleen, testis, committed progenitors and LSK cells after a single dose (3 mg/kg body weight) of MMC injection (Fig. 1D,E). Unexpectedly, MMC treatment did not induce robust Fancd2 monoubiquitination in the testis (Fig. 1D).
Proteomic analysis of Fancd2-associated proteins
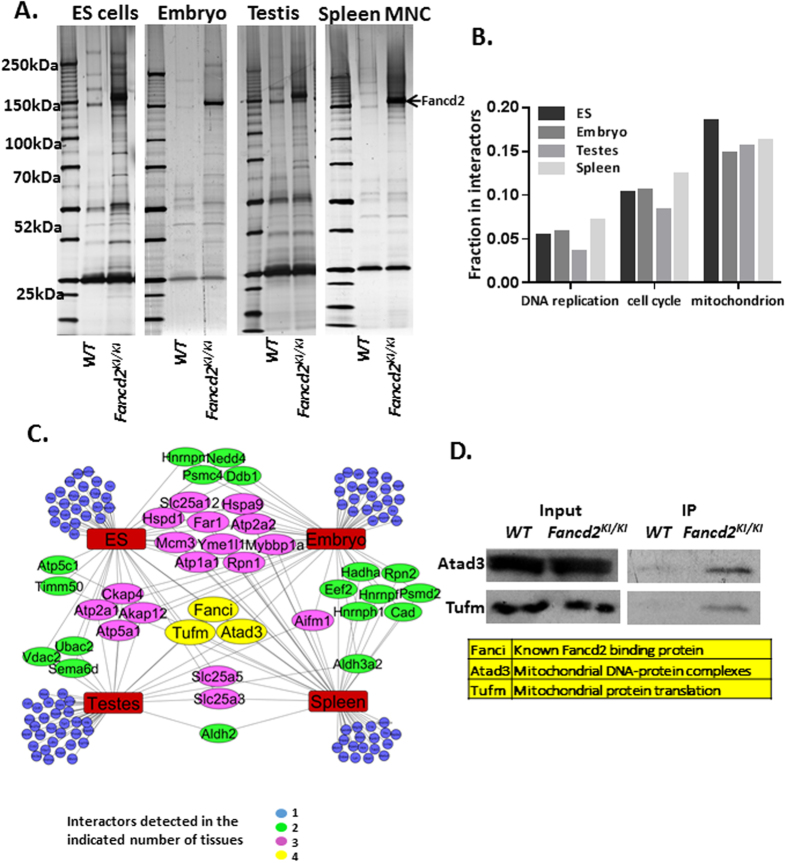
To identify Fancd2 interactors in vivo, we isolated Fancd2-containing complexes from ES cells, E11.5 mouse embryos, testes and spleen mononuclear cells (Fig. 2A), as these compartments express high levels of Fancd2 (Fig. 1B). The identities of Fancd2-associated proteins were determined by liquid chromatography and tandem mass spectrometry (LC–MS/MS; Supplementary Table 1). We validated the association of Fancd2 with some of the interactors using anti-Flag pull-down. Among them, we detected known Fancd2-binding partners, including Fanci, Msh2 and DDB1 (Supplemental Fig. 3). Screening the interactors for ontology categories showed enrichment for cell cycle (P < 0.001) and DNA replication (P < 0.001) proteins. Unexpectedly, proteins involved in mitochondrion homeostasis were enriched the most (P < 0.001) (Fig. 2B). The Fancd2 interaction network reveals different patterns of Fancd2-associated polypeptides in these four compartments (Fig. 2C), suggesting the presence of developmental and tissue-specific differences in the repertoire of Fancd2 interactors. Significantly, we observed three proteins, Fanci, Atad3, and Tufm that interacted with Fancd2 in all four tissues (Fig. 2C). We confirmed the interaction of Fancd2 with Atad3 and Tufm in the spleen (Fig. 2D). Fanci is a well-known Fancd2-binding partner7; whereas Atad3 (component of the mitochondrial nucleoid complex) and Tufm (component of mitochondrial translation machinery) are essential for mitochondrion biosynthesis24,25,26,27.
Figure 2. Proteomic analysis of Fancd2-associated proteins.
(A) Silver stained gels with Fancd2 containing complexes purified from indicated tissues (Fancd2-IP). WT: Mock-IP, mock purification from organs of wild-type mice. Bands corresponding to Fancd2 are marked by an arrow. (B) Fractions of proteins, among Fancd2 interactors, classified to indicate Gene Ontology categories show biological process/molecular function enrichment. (C) Diagram depicting Fancd2 interactors, grouped by tissue in which they were detected. Each color corresponds to number of occurrences across all four tissues. Blue nodes, only in one tissue, Green nodes, common to two tissues, Purple nodes, common to 3 tissues, Yellow nodes, common to all 4 tissues. (D) Interaction of endogenous with Fancd2 to Atad3 and Tufm, detected by anti-FLAG immunoprecipitation and anti-Atad3 and Tufm immunoblotting in spleen MNCs.
Loss of Fancd2 or Fanca causes dysregulation of mitochondrion genes
Since FA proteins have been shown to regulate oxidative stress and mitochondrion respiration complex enzymes9,12,13,16,17,18, and since mitochondrial Fancd2-interactors show the highest enrichment score in our proteomic analysis (Fig. 2 above), we decided to study the link between FA and the mitochondrion further. We first examined whether FA deficiency affected mitochondrion morphology. Transmission electron microscopy analysis revealed swollen mitochondria with disorganized cristae in spleen MNCs and MEFs of Fanca−/− and Fancd2−/− mice (Fig. 3A).
Figure 3. Dysregulation of mitochondrial genes in the absence of Fancd2 or Fanca.
(A) Transmission electron microscopy of spleen MNCs and MEFs from WT, Fanca−/− and Fancd2−/− mice. Swollen mitochondria with disorganized cristae are indicated with a white arrow. (B) The mtDNA copy number in bone marrow MNCs and spleen MNCs was analyzed by QPCR using primers against mtDNA and nuclear DNA (3 mice for each genotype). No significant difference was observed between the one-year old WT, Fanca−/− and Fancd2−/− mice with Student’s t-test analysis. (C) To analyze mtDNA integrity in FA mice, 16 kb mtDNA of 2 month old mice spleen MNCs was amplified. The PCR product was subjected to small genome sequencing. The Fancd2−/− mtDNA shows slightly higher mutation rates compared to WT, Fancd2KI/KI and Fanca−/− mice, especially in the 16 s/12 s rRNA region. (D) Quantitative PCR analysis shows up-regulation of genes associated with mitochondrial biogenesis (Atad3, Tufm, Tfam) and down-regulation of genes involved in mitochondrial oxidative phosphorylation (Atp1a1, Atp2a2, Scl25a5) in Fanca−/− and Fancd2−/− spleen MNCs compared to WT spleen MNC cells. The data represent a summary of more than three mice of each genotype from two independent experiments. The P values indicated were analyzed using Student’s t-test analysis. (E) For mtDNA-encoded genes, quantitative PCR analysis shows up-regulation of the 16 s rRNA and 12 s rRNA and down-regulation of Cox-1 and Nd2 expression in the Fanca−/− and Fancd2−/− spleen MNCs comparing to WT cells. (F) Western blot shows increased expression of Atad3, Tufm, Aifm in the mitochondrion fraction from Fanca−/− and Fancd2−/− spleen MNCs. (G) Western blot shows decreased Cox-1 expression in the mitochondrion fractions of Fanca−/− and Fancd2−/− spleen MNCs. Full-length blot is presented in Supplementary Fig. 5.
Since the FA pathway is essential for the response to replicative stress and repair of DSBs generated during DNA replication, we suspected that the FA proteins were directly involved in mtDNA replication and repair. To our surprise, the mtDNA copy number in the bone marrow and spleen MNCs showed no significant difference between the one year old WT, Fanca−/− and Fancd2−/− mice (Fig. 3B). To determine if mtDNA integrity was impaired in FA-deficient mice, we performed mtDNA sequencing and found no significant difference in mutation rates between WT and Fanca−/− or Fancd2−/− mtDNA of spleen MNCs (Fig. 3C). These results indicate that loss of Fanca or Fancd2 in adult mice has no effect on mtDNA copy number or the mutation frequency, suggesting that mtDNA variance was likely not the cause of the mitochondrion defect in FA.
To search for clues about Fancd2 function in mitochondria, we first analyzed nuclear- and mtDNA-encoded gene expression at the mRNA and protein levels. Quantitative PCR analysis of nuclear genes functioning in the mitochondrion showed specific up-regulation of genes associated with mitochondrial biogenesis (Atad3, Tufm, Tfam) and down-regulation of genes involved in oxidative phosphorylation (Atp1a1, Atp2a2, Scl25a5) in Fanca−/− and Fancd2−/− spleen MNCs comparing to WT cells (Fig. 3D). For mtDNA-encoded genes, up regulation of 16 s rRNA and 12 s rRNA and down regulation of ND2 and CoxI mRNA were detected in Fanca−/− and Fancd2−/− cells (Fig. 3E). Immunoblotting of mitochondrial proteins showed increased expression of nuclear-coded proteins Atad3, Tufm, Aifm and decreased mtDNA-coded CoxI in the spleen MNCs of Fanca−/− and Fancd2−/− mice (Fig. 3F,G). Collectively, these results indicate that loss of Fanca or Fancd2 leads to an alteration in mtDNA gene expression.
Fancd2 is required for the stability of the mitochondrion nucleoid complex
We then turned our attention to the two mitochondrial Fancd2-interactors, Atad3 and Tufm, which interacted with Fancd2 in all four tissues (Fig. 2C,D). Both Atad3 and Tufm are among the most frequently identified components of the mitochondrion nucleoid complex24,25,26,27, which is essential for mitochondrion biosynthesis. In addition, Atad3 and Tufm have been reported to be required for the mitochondrial transcription and translation process24,25,26,27,28. Since Atad3 and Tufm are localized exclusively in mitochondria and are present in the mitochondrial nucleoprotein complex25, we postulated that Fancd2 might play a role in the mtDNA transcription and translation process, probably by stabilizing the Atad3/Tufm/Tfam nucleoid complex. We first determined whether Fancd2 was localized to the mitochondria. Immunofluorescence staining of tagged-Fancd2 in Fancd2KI/KI MEFs with an anti-HA antibody showed that Fancd2 was predominantly localized in the nucleus with a small fraction found in the cytosol (Fig. 4A). Cytoplasmic Fancd2 exhibited co-localization with the mitochondrial maker Mito tracker RED (Fig. 4A), suggesting that Fancd2 is also localized to the mitochondria. To confirm this finding, we performed immunoblotting on fractionated lysates of Fancd2KI/KI MEFs cells. The mitochondrial, cytosol, and nuclear fractions were marked by Atp5a1, GAPDH and Histone2B, respectively. Consistent with the immunostaining result, the Fancd2 protein was detected in both the nuclear and cytosol fractions (Fig. 4B). Importantly, a significant amount of Fancd2 was co-fractionated with Atp5a1 in the mitochondria fraction, adding further support to mitochondrial localization of Fancd2.
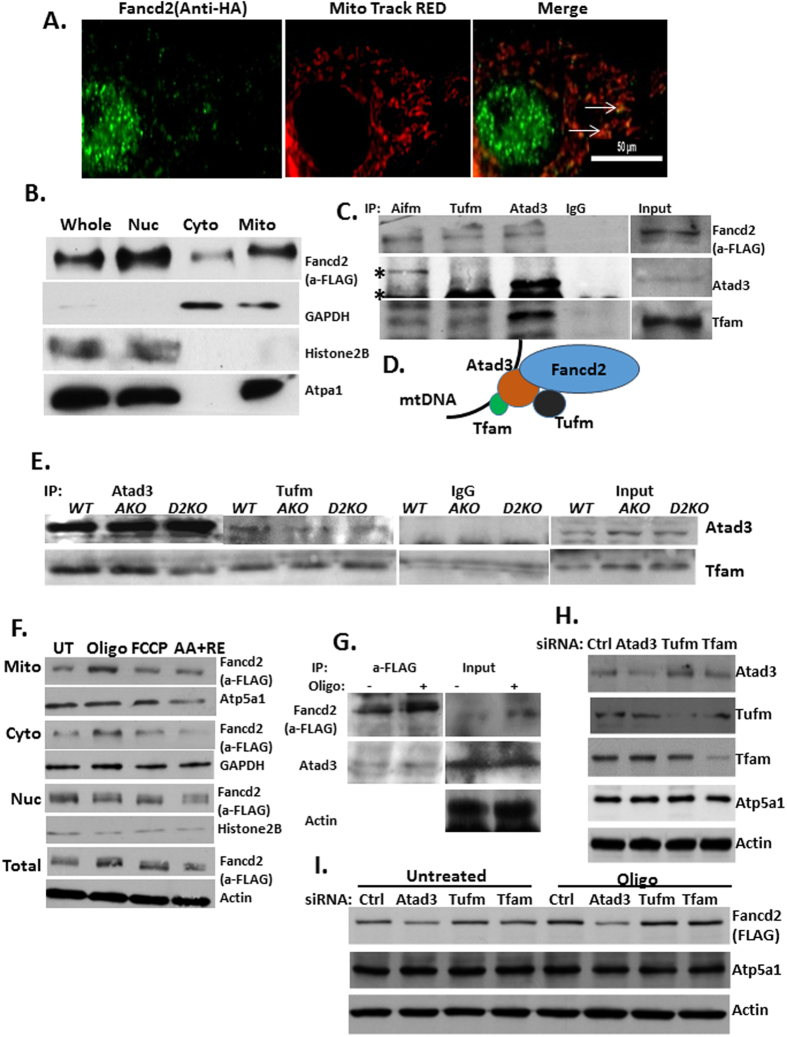
Figure 4. Fancd2 is a component of the mitochondrial nucleoid complex.
(A) Fancd2 is localized in mitochondria. Anti-HA antibody and FITC-labeled secondary antibody were used to detect tagged-Fancd2 protein in Fancd2-KI MEFs. Mito tracker RED was used to label mitochondria. (B) Western analysis of the FLAG-tagged Fancd2 protein in MEF cells of Fancd2-KI mice in whole cell lysates (Whole), nuclear (Nuc), cytoplasmic (Cyto) and mitochondrial (Mito) fractions. Anti-FLAG was used to probe Fancd2. GAPDH, ATP5A1, and Histone2B were probed as the markers for cytoplasmic, mitochondrial and nuclear controls, respectively. A significant fraction of the Fancd2 protein is localized in the mitochondrion. (C) Fancd2 co-immunoprecipitates with Atad3, Tufm and Aifm. Aifm, Atad3 and Tufm immunoprecipitation were performed on Fancd2KI/KI spleen MNCs. Nonspecific band is indicated by an asterisk. (D) A model of the Fanc2/Atad3/Tufm complex. (E) Effect of FA deficiency on the Atad3/Tufm/Tfam complex. Atad3 and Tufm immunoprecipitation in Fancd2KI/KI, Fanca−/− Fancd2KI/KI and Fancd2−/− spleen MNCs. (F) Fancd2 expression responds to mitochondrial stress. Fancd2KI/KI MEFs were treated with different mitochondrion respiration complex inhibitors: 4 uM oligomycin, 4 uM FCCP, or 2 uM antimycin A and 2 uM rotenone for 16 hr. Immunoblot of cell fraction lysates shows an increase in the levels of Fancd2 in the whole cell, mitochondrion and cytosol fraction, but not in the nuclear fractions. (G) Immunoblot showing that more Atad3 was co-immunoprecipitated with Fancd2 in oligomycin treated Fancd2KI/KI MEFs. (H) Immunoblot of Fancd2KI/KI MEFs lysates shows siRNA knockdown of control, Atad3, Tufm, and Tfam. (I) Immunoblot shows reduced Fancd2 in the mitochondrial fraction after Atad3 knockdown with or without oligomycin treatment. Full-length blot is presented in Supplementary Fig. 6.
We next determined whether Fancd2 was an integral component of the mitochondrion nucleoid complex in vivo. To this end, we first performed immunoprecipitation experiments using antibodies specific for Atad3, Tufm, and Aifm (Apoptosis-Inducing Factor, Mitochondrion-Associated), which is a Fancd2 interactor found in testes, spleen and embryos (Fig. 2C,D). Fancd2 co-immunoprecipitated with Atad3, Tufm and Aifm in the lysates of Fancd2KI/KI spleen MNCs (Fig. 4C). In the meantime, Tufm associated with Atad3, which co-immunoprecipitated with Tfam (Transcription Factor A, Mitochondrial) (Fig. 4C), another component of the mitochondrion nucleoid complex25. Collectively, these results suggest that Fancd2 is part of the mitochondrion nucleoid complex, probably through interacting with Atad3 and Tufm (Fig. 4D).
We then asked whether loss of FA proteins would affect the Fancd2/Atad3/Tufm complex by carrying out co-immunoprecipitation experiments using Fancd2KI/KI, Fanca−/−Fancd2KI/KI and Fancd2−/− spleen MNCs cells. We observed that the Atad3/Tufm interaction was reduced in the absence of Fanca and Fancd2 (Fig. 4E). Altogether, loss of Fanca or Fancd2 disrupted Fancd2/Atad3/Tufm mitochondrion nucleoid complex, which likely contributes to the mtDNA transcription and translation defect in FA cells.
Atad3 mediates mitochondrial import of Fancd2
Since the expression of numerous genes encoding mitochondrial proteins, including Atad3, is induced by mitochondrial dysfunction leading to a protective response29,30, we examined next whether the expression of Fancd2, as an Atad3-interacting partner, also responded to mitochondrial stress. We treated Fancd2KI/KI MEFs with different mitochondrion respiration complex inhibitors oligomycin (Complex V inhibitor), FCCP (Uncoupling agents), or antimycin A (Complex III inhibitor) combined with rotenone (complex I inhibitor). We observed that the level of Fancd2 was significantly increased 16hr after oligomycin treatment, in the mitochondrion and cytosol fractions but not in the nuclear fraction (Fig. 4F). In addition, more Atad3 was co-immunoprecipitated with Fancd2 in oligomycin treated MEFs (Fig. 4G). These results indicate that Fancd2 is involved in the response of cells to mitochondrial stress.
Atad3 has been found anchored in the inner mitochondrial membrane and to deliver substrate into mitochondria31. This prompted us to ask whether Atad3 mediated Fancd2 import to mitochondria. Fancd2KI/KI MEFs were transfected with siRNA targeting Atad3, Tufm or Tfam, and then analyzed for mitochondrial Fancd2. We obtained significant knockdown (>80%) for these proteins in the mitochondrion (Fig. 4H). Knocking down Atad3 but not Tufm or Tfam ablated Fancd2 in both resting (untreated) and stressed (oligomycin-treated) mitochondria (Fig. 4I). Thus, Atad3 is indispensable for the localization and importation of Fancd2 to the mitochondrion with or without stress.
Discussion
Most studies on FA cellular and molecular mechanisms have been conducted in cultured cell lines in vitro. For in vivo studies, several FA knockout mice have been reported, to date, with only a few phenotypic features recapitulating FA patients, including hypersensitivity to DNA-crosslinking agents, abnormal gonads and stressed hematopoiesis32,33. Studies on FA mice or human patient-derived primary cells identified additional phenotypes including apoptotic response deregulation, oxidative stress hypersensitivity, and mitochondrion deficiency, which all appear to fall outside of DNA damage repair9,10,11,12,13,14,15,16,17,18,19,20. Using an innovative Fancd2 knock-in mouse model, the recent study shows that most Fancd2-binding proteins do not have a recognized function in DNA damage response and repair. Particularly, many Fancd2-interactors are mitochondrion-specific. We also show that Fancd2 is localized in the mitochondrion and associates with the nucleoid complex components Atad3, Tufm and Tfam, all of which are required for mitochondrial biosynthesis. Moreover, the Atad3/Tufm/Tfam complex is disrupted in Fancd2−/− mice and those deficient for the FA core component Fanca. These findings add another facet to the non-repair function of FA proteins.
A major limitation for studying the molecular mechanisms in FA murine models is the lack of antibodies with high-affinity for mouse FA proteins. We generated a 3 × FLAG-HA tagged Fancd2 knock-in mouse model that overcomes this limitation and this has enabled us to identify Fancd2-binding proteins in different organs, thus gaining new insights into FA protein function in vivo. We have shown that the 3 × FLAG/HA tagged Fancd2 is essentially wild-type and that the homozygous Fancd2KI/KI mice are indistinguishable from their wild-type counterparts. The 3 tandem FLAG is ideal for detecting Fancd2 in a small amount or for immunoprecipitation. Indeed, this duel tag has enabled us to perform 2 step purification of the Fancd2-containing complex with limited amount of materials from different tissues.
A thorough examination of Fancd2KI/KI mice reveals tissue- and cell type-specific expression patterns that likely correspond to differential Fancd2 function, especially in the reproductive and hematopoietic organs. The general absence of Fancd2 monoubiquitination in the organs of the Fancd2KI/KI mice are particularly striking and suggest that a low level of DNA repair activity of Fancd2 may be sufficient for the maintenance of tissue homeostasis in vivo. Conversely, we detected a substantial amount of monoubiquitinated Fancd2 in ES cells and MEFs derived from Fancd2KI/KI embryos, suggesting that environmental exposure during in vitro cell culture may have induced Fancd2 monoubiquitination, likely by triggering cellular stress including DNA damage. These findings demonstrate a very different in vivo operation of the FA pathway with a majority of the Fancd2 protein exhibiting as non-ubiquitination form, which leads us to propose the existence of ubiquitination-independent functions outside of DNA repair. However, it is also possible that the status of Fancd2 monoubiquitination may reflect cell proliferation or tissue turnover.
Our proteomic study reveals that the most predominant Fancd2 interactors in all four tested tissues belong to those involved in mitochondrial homeostasis, suggesting FA deficiency as a potential contributing factor to mitochondrion deficiency. This intriguingly universal interaction between Fancd2 and mitochondrial proteins also lends support to previous observations that FA proteins regulate oxidative stress response and mitochondrion functions9,12,13,16,17,18. Remarkably, we identified two important mitochondrial proteins, Atad3 and Tufm, with which Fancd2 interacts in all four tissues. Atad3 and Tufm are among the most frequently identified components in the mitochondrion nucleoid complex27. Since the mitochondrion nucleoid complex participates in mitochondrial protein synthesis characteristic of uniquely coupled transcription and translation processes34,35, both Atad3 and Tufm proteins have been demonstrated to be required for mitochondrial transcription and translation processes24,27. In contrast to the assumption that Fancd2 impacts mtDNA replication and integrity, we found that Fancd2 interferes with mtDNA-encoded gene transcription and translation, by interacting with, and stabilizing, the Atad3/Tufm/Tfam mitochondrial nucleoid complex. Indeed, our results indicate that Atad3 mediates Fancd2 localization and importation to mitochondria. Additionally, the deregulation of nuclear-encoded mitochondrial proteins in the Fanca- and Fancd2-deficient cells appear to be part of the feedback loop ultimately leading to mitochondrial dysfunction. However, the exact mechanism underling the functional interaction between Fancd2, the Atad3, Tufm, Tfam mitochondrial nucleoid complex and nuclear-coded mitochondrial proteins remains to be further investigated. Also, how Fanci or Fanca regulates Fancd2 in mitochondrial function remains unknown. Nevertheless, these findings shed new light into the potential link between FA and mitochondrial defects.
Further investigation is needed to determine whether the Fancd2-Atad3-Tufm complex formation requires Fancd2 monoubiquitination or an intact FA core complex. Since we do not observe monoubiquitinated Fancd2 in the cytoplasm or mitochondria, we envision another intriguing scenario, where both a non-monoubiquintinated Fancd2 or the Fancd2 protein in cells lacking the key FA core component Fanca could still form complexes with Atad3-Tufm. However, given that mice deficient for several components of the FA core complex (e.g. Fanca, Fancb, Fancc, Fancg) show exactly the same phenotypes as those deleted for Fancd219,20,21,22,23,32,33, we also speculate that both Fancd2/Fanci monoubiquitination and FA core complex have a crucial role in Fancd2-Atad3-Tufm complex formation.
In summary, the present study employs an innovative Fancd2 knock-in mouse model to demonstrate that, in addition to its well-established role in DNA repair, Fancd2 influences gene expression and post-translational function of mitochondrial proteins Atadf3, Tufm, and Tfam.
Methods
Generation of 3XFLAG/HA-tagged Fancd2 knock-in mice
The C-terminally tagged allele (Fancd23XFlag/HA) was generated by inserting sequences encoding 3XFLAG/HA tags immediately upstream of the STOP codon (TGA) within the forty-forth exon of the Fancd2 gene (Fig. 1A). Fancd2 targeting vectors were constructed by recombineering as previously described36. The linearized targeting plasmid was electroporated into 129/B6 F1 hybrid V6.5 embryonic stem cells (Novus biologicals) and DNA from G418-resistant ES cell clones were analyzed. The floxed Neomycin selection marker was then excised by transient expression of Cre recombinase. Positive ES cell lines were then injected into mouse blastocysts. Chimeric animals were obtained and crossed to C57BL/6J mice, and homozygous Fancd2KI/KI mice on a mixed 129S4, C57BL/6J background were generated. Founder animals were also backcrossed to pure C57BL/6J mice for six generations to generate a population of homozygous Fancd2KI/KI mice on the C57BL/6J background.
Purification of Fancd2-containing complexes, mass spectrometry and proteomic analysis
For one purification round of Fancd2-containing complexes, we pooled around 40 10 cm dishes of cultured ES cells, or 20 11.5D embryos, or 8 adult testes, or 24 adult spleens. The same number of cells or organs from WT mice was used for mock purifications. Organs were lysed using a Homogenizer, on ice, in 3 volumes of ice-chilled Lysis buffer with Roche Complete proteinase inhibitor. Lysates were cleared by centrifugation (40,000 g, 20 min, at 4 °C), and the supernatant was incubated with 100 ul of agarose beads conjugated to anti-FLAG antibody (M2, Sigma) at 4 °C. After a few washing steps with ice-cold lysis buffer, Fancd2, together with the associated proteins was eluted in 400 ul of the lysis buffer containing 200ug/ml FLAG peptide (Sigma). The eluate was incubated with 20 ul of anti-HA beads (Roche) for 4 hrs at 4 °C. After several washes with the lysis buffer, Fancd2 together with the associated proteins was boiled and run on a NuPAGE Bis-Tris gel (Invitrogen). In parallel, we performed identical “mock” purifications from the same number of organs or tumors dissected from wild-type animals (which do not express tagged Fancd2). The final samples were submitted for Liquid chromatography and tandem mass spectrometry (LC-MS/MS) at the Taplin Biological Mass Spectrometry Facility, Harvard Medical School.
All peptides and corresponding spectral count results are shown in Supplementary Table 1. Each protein was evaluated for mock or Fancd2 immunoprecipitated samples. All proteins that were present in mock samples were excluded from further analysis for each tissue. We observed about 200–300 proteins for each tissue. Proteins with more than 1 or 2 peptides were subjected to further study. Toppgene was used for Gene Ontology (GO) categories enrichment analysis. STRING and Cytoscape were used for protein interaction study.
Mice
Fanca+/− and Fancd2 +/− mice were provided by Dr. Madeleine Carreau (Laval University) and Dr. Markus Grompe (Oregon Health & Sciences University)33,37, respectively. All animals were sacrificed 2–3 weeks after the last injection for experiments. Mice were maintained on a C57BL/6J background in the animal barrier facility at Cincinnati Children’s Hospital Medical Center. Animals were kept in accordance with the protocol approved by the CCHMC Institutional Animal Care and Use Committees. All animal experiments were performed in accordance with the institutional guidelines and were approved by the Institutional Animal Care and Use Committee of Cincinnati Children’s Hospital Medical Center (IACUC2013-0159).
Flow cytometry
All antibodies were obtained from eBioscience or BD Pharmingen, unless otherwise indicated. Samples were examined on Fortessa or LSRII flow cytometers, and the cytometry data was analyzed with BD FASC Diva or FlowJo software. For lineage cell analysis: Anti-CD71 and anti-Ter119 for erythroid, anti-Gr1 and anti-CD11b for granulocytic/monocytes, anti-CD45R (B220) for B-lymphoid, and anti-CD3e for T-lymphoid. For quantification of CMP, MEP, GMP, CLP, LK and LSK progenitors, mono-nucleated cell were separated by HISTOPAQUE 1083 (sigma) spin, then labeled with a cocktail of anti-mouse lineage markers (CD3e, B220, Ter119, Mac1, and Gr1), anti-mouse CD117 (cKit), and anti-Ly-6A/E (Sca1), anti-CD34, anti-CD16/32, anti-CD150, anti-CD48 and anti-IL-7Ra. Common myeloid progenitors (CMPs) were defined as Lin-Sca1-cKit + CD34 + CD16/32low, and granulocyte-macrophage progenitors (GMPs) were defined as Lin-Sca1-cKit + CD34 + CD16/32+. Megakaryocyte-erythroid progenitors (MEPs) were defined as Lin-Sca1-cKit + CD34-CD16/32-. Cell sorting was performed with a BD FACSAria II at the Flow Cytometry Core facility in CCHMC.
Western blotting and immunoprecipitation
The Thermo Scientific Mitochondria Isolation Kit and Nucleus Isolation Kit were used for cell fraction isolation. The extracts were obtained and proteins were solubilized for immunoblotting. The following antibodies were used: HA.11 Clone 16B12 (Covance, MMS-101P), FLAG M2-Peroxidase (HRP) (sigma, M8592), beta-actin (sigma), Anti-ATP5A1 antibody (abcam, ab14748), MYBBP1A Antibody (santa cruz, sc-133800), Anti-PP2A Antibody (Millipore,05-421), Hsp60 (Pierce, MA3-012), Cdk1 Antibody (santa cruz, sc-53219), MSH2 Antibody (Novus, NB100-56428), Anti-Histone H2B antibody (Abcam, ab18977), GAPDH (14C10) (cell signaling, 3683), FANCD2 (novusbio, NB100-182), AIF Antibody (santa cruz, sc-5586), ATAD3A/B/C (santa cruz, sc-292156), EF-Tu (santa cruz, sc-367739), Anti-ALDH2 (sigma, SAB2501484), mtTFA (santa cruz, sc-23588), Anti-MTCO1 antibody (abcam, ab14705). EZview™ Red Anti-HA Affinity Gel (sigma, E6779-1ML), anti-HA beads (Roche), ANTI-FLAG® M2 Affinity Gel (Sigma, A2220-1ML), Protein G Agarose (Roche, 11719416001) were used for IP.
MtDNA copy number
Total DNA was extracted from cell samples using the DNeasy Blood and Tissue kit (Qiagen, USA). To quantify the mtDNA copy number, real-time PCR was performed using iTaq Universal SYBR Green Supermix (Bio-rad, USA) with a Bio-Rad CFX system. For determination of the amount of nuclear DNA, the apoB gene and B2M gene were used as a reference. The quantification using the COXII or ND4 region in mtDNA showed similar results. These real-time qPCRs were carried out in quadruplicate for all measurements. All the primers are listed in Supplementary Table S2.
MtDNA sequencing
The integrity of mtDNA was analyzed by long range PCR, using TaKaRa PrimeSTAR GXL DNA polymerase (Qiaqen) to generate an approximately 16 k bp molecule from primers spanning the entire 16 kb mouse mitochondrial genome (forward 5′-CCCAGCTACTACCATCATTCAAGT-3′ and reverse 5′-GAGAGATTTTATGGGTGTAATGCGG-3′). The PCR product was gel purified and submitted to small genome sequencing at the DNA sequencing facility of CCHMC. The NGS result was analyzed by Nextgene software.
siRNA transfection
siRNA for Tufm (Qiagen, S101458919), Atad3 (Qiagen, S100905723), Tfam (Qiagen, S100183974), and Ubr5 (Sigma, EMU08461) was transfected to the MEFs using Lipofectamine™ RNAiMAX reagent. The effect of targeted siRNA sequences was compared with negative control siRNA (Qiagen).
Statistical analysis
Student’s t-test and ANOVA test, were performed using Microsoft EXCEL software or Prism 6.0 software (GraphPad Software Inc). Error bars indicate SD. Differences were judged as significant if the P value was <0.05 (*), <0.01 (**) or <0.001 (***).
Additional Information
How to cite this article: Zhang, T. et al. Fancd2 in vivo interaction network reveals a non-canonical role in mitochondrial function. Sci. Rep. 7, 45626; doi: 10.1038/srep45626 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We thank Dr. Madeleine Carreau (Laval University) for Fanca +/− mice, Dr. Markus Grompe (Oregon Health & Sciences University) for Fancd2 +/− mice, Dr. Taosheng Huang (Cincinnati Children’s Hospital Medical Center) for mitochondrion study, the Transgenic Animal and Genome Editing Core (Cincinnati Children’s Hospital Medical Center) for ES cells microinjection service, the DNA sequencing Core (Cincinnati Children’s Hospital Medical Center) for mtDNA sequencing service, the Pathology Research Core (Cincinnati Children’s Hospital Medical Center) for H&E staining, EM tissue processing and sectioning, and Taplin Biological Mass Spectrometry Facility of Harvard Medical School for Mass Spec service. We also thank Ms. Samantha Losekamp for editing the manuscript. This investigation was supported by NIH grants R01 HL076712, R01 CA157537 and T32 HL091805. Q.P. was supported by a Leukemia and Lymphoma Scholar award.
Footnotes
The authors declare no competing financial interests.
Author Contributions T.Z., designed research, performed research, analyzed data and wrote the paper; W.D., performed research, analyzed data and wrote the paper; A.F.W., performed research; S.H.N., contributed vital new reagents and designed research; P.R.A., contributed vital new reagents and designed research; A.R.M., contributed vital new reagents and designed research; Q.P., designed research and wrote the paper.
References
- Bagby G. C. Jr. Genetic basis of Fanconi anemia. Curr Opin Hematol 10, 68–76 (2003). [DOI] [PubMed] [Google Scholar]
- Auerbach A. D. & Allen R. G. Leukemia and preleukemia in Fanconi anemia patients. A review of the literature and report of the International Fanconi Anemia Registry. Cancer Genet Cytogenet 51, 1–12 (1991). [DOI] [PubMed] [Google Scholar]
- Tischkowitz M. & Dokal I. Fanconi anaemia and leukaemia - clinical and molecular aspects. Br J Haematol 126, 176–191 (2004). [DOI] [PubMed] [Google Scholar]
- Kim H. & D’Andrea A. D. Regulation of DNA cross-link repair by the Fanconi anemia/BRCA pathway. Genes Dev 26, 1393–1408 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossan G. P. & Patel K. J. The Fanconi anaemia pathway orchestrates incisions at sites of crosslinked DNA. J Pathol 226, 326–337 (2012). [DOI] [PubMed] [Google Scholar]
- Kee Y. & D’Andrea A. D. Expanded roles of the Fanconi anemia pathway in preserving genomic stability. Genes Dev 24, 1680–1694 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Andrea A. D. & Grompe M. The Fanconi anaemia/BRCA pathway. Nat Rev Cancer 3, 23–34 (2003). [DOI] [PubMed] [Google Scholar]
- D’Andrea A. D. Susceptibility pathways in Fanconi’s anemia and breast cancer. N Engl J Med 362, 1909–1919 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano G. et al. Bone marrow cell transcripts from Fanconi anaemia patients reveal in vivo alterations in mitochondrial, redox and DNA repair pathways. Eur J Haematol 91, 141–151 (2013). [DOI] [PubMed] [Google Scholar]
- Usai C. et al. Dysregulated Ca2+ homeostasis in Fanconi anemia cells. Sci Rep 5, 8088 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari U., Ya Jun W., Huat Bay B. & Lyakhovich A. Evidence of mitochondrial dysfunction and impaired ROS detoxifying machinery in Fanconi anemia cells. Oncogene 33, 165–172 (2014). [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay S. S. et al. Defective mitochondrial peroxiredoxin-3 results in sensitivity to oxidative stress in Fanconi anemia. J Cell Biol 175, 225–235 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanier R., Briot D., Dugas du Villard J. A., Sarasin A. & Rosselli F. Fanconi anemia C gene product regulates expression of genes involved in differentiation and inflammation. Oncogene 23, 5004–5013 (2004). [DOI] [PubMed] [Google Scholar]
- Rousset S., Nocentini S., Rouillard D., Baroche C. & Moustacchi E. Mitochondrial alterations in fanconi anemia fibroblasts following ultraviolet A or psoralen photoactivation. Photochem Photobiol 75, 159–166 (2002). [DOI] [PubMed] [Google Scholar]
- Pagano G., Shyamsunder P., Verma R. S. & Lyakhovich A. Damaged mitochondria in Fanconi anemia - an isolated event or a general phenomenon? Oncoscience 1, 287–295 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravera S. et al. Mitochondrial respiratory chain Complex I defects in Fanconi anemia complementation group A. Biochimie 95, 1828–1837 (2013). [DOI] [PubMed] [Google Scholar]
- Cappelli E. et al. Mitochondrial respiratory complex I defects in Fanconi anemia. Trends Mol Med 19, 513–514 (2013). [DOI] [PubMed] [Google Scholar]
- Sumpter R. Jr. et al. Fanconi Anemia Proteins Function in Mitophagy and Immunity. Cell 165, 867–881 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q. S. et al. Fancd2−/− mice have hematopoietic defects that can be partially corrected by resveratrol. Blood 116, 5140–5148 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. et al. Fanconi anemia links reactive oxygen species to insulin resistance and obesity. Antioxid Redox Signal 17, 1083–1098 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M. et al. Inactivation of Fac in mice produces inducible chromosomal instability and reduced fertility reminiscent of Fanconi anaemia. Nat Genet 12, 448–451 (1996). [DOI] [PubMed] [Google Scholar]
- Wong J. C. et al. Targeted disruption of exons 1 to 6 of the Fanconi Anemia group A gene leads to growth retardation, strain-specific microphthalmia, meiotic defects and primordial germ cell hypoplasia. Hum Mol Genet 12, 2063–2076 (2003). [DOI] [PubMed] [Google Scholar]
- Parmar K. et al. Hematopoietic stem cell defects in mice with deficiency of Fancd2 or Usp1. Stem Cells 28, 1186–1195 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S. et al. ATAD3 is a limiting factor in mitochondrial biogenesis and adipogenesis of white adipocyte-like 3T3-L1 cells. Mol Cell Biol(2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J. et al. Mitochondrial nucleoid interacting proteins support mitochondrial protein synthesis. Nucleic Acids Res 40, 6109–6121 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente L. et al. Infantile encephalopathy and defective mitochondrial DNA translation in patients with mutations of mitochondrial elongation factors EFG1 and EFTu. Am J Hum Genet 80, 44–58 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeitink J. A. et al. Distinct clinical phenotypes associated with a mutation in the mitochondrial translation elongation factor EFTs. Am J Hum Genet 79, 869–877 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonicka H., Sasarman F., Kennaway N. G. & Shoubridge E. A. The molecular basis for tissue specificity of the oxidative phosphorylation deficiencies in patients with mutations in the mitochondrial translation factor EFG1. Hum Mol Genet 15, 1835–1846 (2006). [DOI] [PubMed] [Google Scholar]
- Ryan M. T. & Hoogenraad N. J. Mitochondrial-nuclear communications. Annu Rev Biochem 76, 701–722 (2007). [DOI] [PubMed] [Google Scholar]
- Haynes C. M., Fiorese C. J. & Lin Y. F. Evaluating and responding to mitochondrial dysfunction: the mitochondrial unfolded-protein response and beyond. Trends Cell Biol 23, 311–318 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issop L. et al. Mitochondria-associated membrane formation in hormone-stimulated Leydig cell steroidogenesis: role of ATAD3. Endocrinology 156, 334–345 (2015). [DOI] [PubMed] [Google Scholar]
- Parmar K., D’Andrea A. & Niedernhofer L. J. Mouse models of Fanconi anemia. Mutat Res 668, 133–140 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houghtaling S. et al. Epithelial cancer in Fanconi anemia complementation group D2 (Fancd2) knockout mice. Genes Dev 17, 2021–2035 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestwick M. L. & Shadel G. S. Accessorizing the human mitochondrial transcription machinery. Trends Biochem Sci 38, 283–291 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonawitz N. D., Clayton D. A. & Shadel G. S. Initiation and beyond: multiple functions of the human mitochondrial transcription machinery. Mol Cell 24, 813–825 (2006). [DOI] [PubMed] [Google Scholar]
- Zhou D., Ren J. X., Ryan T. M., Higgins N. P. & Townes T. M. Rapid tagging of endogenous mouse genes by recombineering and ES cell complementation of tetraploid blastocysts. Nucleic Acids Res 32, e128 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J. C. et al. Targeted disruption of exons 1 to 6 of the Fanconi Anemia group A gene leads to growth retardation, strain-specific microphthalmia, meiotic defects and primordial germ cell hypoplasia. Hum Mol Genet 12, 2063–2076 (2003). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.