Abstract
Flavivirus includes a large group of human pathogens with medical importance. Especially, neurotropic flaviviruses capable of invading central and peripheral nervous system, e.g. Japanese encephalitis virus (JEV) and Zika virus (ZIKV), are highly pathogenic to human and constitute major global health problems. However, the dynamic dissemination and pathogenesis of neurotropic flavivirus infections remain largely unknown. Here, using JEV as a model, we rationally designed and constructed a recombinant reporter virus that stably expressed Renilla luciferase (Rluc). The resulting JEV reporter virus (named Rluc-JEV) and parental JEV exhibited similar replication and infection characteristics, and the magnitude of Rluc activity correlated well with progeny viral production in vitro and in vivo. By using in vivo bioluminescence imaging (BLI) technology, we dissected the replication and dissemination dynamics of JEV infection in mice upon different inoculation routes. Interestingly, besides replicating in mouse brain, Rluc-JEV predominantly invaded the abdominal organs in mice with typical viscerotropism. Further tests in mice deficient in type I interferon (IFN) receptors demonstrated robust and prolonged viral replication in the intestine, spleen, liver, kidney and other abdominal organs. Combined with histopathological and immunohistochemical results, the host type I IFN signaling was evidenced as the major barrier to the viscerotropism and pathogenicity of this neurotropic flavivirus. Additionally, the Rluc-JEV platform was readily adapted for efficacy assay of known antiviral compounds and a live JE vaccine. Collectively, our study revealed abdominal organs as important targets of JEV infection in mice and profiled the unique viscerotropism trait controlled by the host type I IFN signaling. This in vivo visualization technology described here provides a powerful tool for testing antiviral agents and vaccine candidates for flaviviral infection.
Keywords: Flavivirus, Japanese encephalitis virus, Bioluminescence imaging, Mouse, Interferon signaling.
Introduction
Neurotropic flavivirus includes a large family of important human pathogens such as Japanese encephalitis virus (JEV), Zika virus (ZIKV), yellow fever virus (YFV), West Nile virus (WNV), St Louis encephalitis virus (SLEV), Murray Valley encephalitis virus (MVEV) and tick-borne encephalitis virus (TBEV). These pathogens are maintained in a transmission cycle in a zoonotic cycle between vectors and vertebrates, and humans get infected by bites of infected vectors. These neurotropic flaviviruses have been the most important causative agents of viral encephalitis in the world 1. For example, approximately 50,000-175,000 cases of JEV infection are reported annually with a high fatality rate of 30% 2, and up to 50% of survivors live with severe residual neurological sequelae 3-5. Over the past decades, JEV has extended its geographic range to Australia 6, Pakistan 7, and Saipan 8. JEV is now recognized as the most significant cause of mosquito-borne encephalitis worldwide, with over 3 billion people at risk of infection 9. WNV, first introduced into the Western hemisphere in 1999, has caused over 41,700 cases, with more than 1,700 death in the USA alone 10. Recently, WNV has disseminated broadly in the Western hemisphere and now poses a significant public health risk in the USA, Europe, the Middle East and Africa. The ongoing ZIKV outbreak in the Americas has aroused global concerns due to its unexpected association with birth defects and Guillain-Barre ´ syndrome 11.
The clinical manifestations of neurotropic flavivirus infections varied, ranging from non-specific mild symptoms to severe forms, including microcephaly, Guillain-Barre ´ syndrome, meningoencephalitis, aseptic meningitis, and a polio-like acute flaccid paralysis 12-14. Following a subcutaneous bite of a mosquito, the neurotropic flavivirus is believed to firstly replicate in resident dendritic cells 15, 16 or epidermal keratinocytes 17, and disseminate to local lymph nodes where further replication in monocyte lineage cells takes place. Then viremia occurs followed by transport of the virus to the brain and hosts develop central nervous system (CNS) disease 18, 19. The neuroinvasion mechanism of JEV remains largely unknown, and is likely preceded by viral replication in peripheral tissues. Innate cellular antiviral mechanisms mediated by type I interferon (IFN) are potentially the most important pathways of the host defense against initial viral replication. Small animal models infected with JEV or WNV have been established and the viruses could be isolated from the kidney, liver and spleen, indicating that these pathogens were capable of infecting peripheral tissues in vivo 20-22. Mice deficient in type I IFN receptor (IFNAR-/-) infected with JEV or WNV showed sustained high viremia and fulminant disease 23, 24. However, the kinetics of neurotropic flavivirus dissemination and pathogenesis in mouse parenchymal organs have not been characterized.
The bioluminescence imaging (BLI) technique has emerged as a powerful tool for studying of both viral and bacterial pathogenesis in animal models 25-30. Using engineered viruses carrying specific reporter genes, this technology permits the real-time monitoring of the spatial and temporal progression of viral infection within the same animal. The time course of viral infection can be easily visualized, and the relative magnitude of bioluminescence correlates well with viral replication. Besides, BLI has the potential to detect viral spread without a priori knowledge of tissue tropism. BLI models for many viral infections, including herpes simplex virus, vaccinia virus, adenovirus, human hepatitis C virus, influenza virus and respiratory syncytial virus have been described 29, 31-35. Recently, BLI results of a non-neurotropic flavivirus dengue virus (DENV) infection in mice have been reported by Schoggins et al and our group 36, 37.
Like other flaviviruses, JEV has an approximately 11 kb single-stranded positive-sense RNA genome containing a single open reading frame (ORF) flanked by untranslated regions (UTRs) at both terminals. The ORF encodes three structural proteins (capsid [C], the membrane [prM/M], and envelope [E]) and seven nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) that are required for the complete viral life cycle. Here, we rationally designed and constructed a recombinant JEV carrying Renilla (Renilla reniformis) luciferase (Rluc) gene. The stable reporter virus (named Rluc-JEV) replicated well in various cells with similar properties to the parental JEV, and Rluc activity had a good correlation with viral infectious titer in vitro and in vivo. BLI results in wild type (WT) and IFNAR-/- mice upon various injection routes were characterized in details. Furthermore, Rluc-JEV was used to test known antiviral compounds in vitro and protective efficiency of a live JEV vaccine in mice. To our knowledge, this is the first real time noninvasive detection of neurotropic flavivirus infection in vivo. The novel reporter virus Rluc-JEV described here provide a powerful tool for studying pathogenesis of neurotropic flavivirus and this technology can be easily expanded to other important viruses.
Materials and Methods
Cells and virus
BHK-21 (baby hamster kidney) and C6/36 (Aedes albopictus) cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 units/mL of penicillin and 100 µg/mL of streptomycin. BHK-21 and C6/36 cells were maintained in 5% CO2 at 37 °C and 28 °C, respectively. The parental JEV was produced by electroporation of BHK-21 cells with transcribed RNA from the full-length cDNA clone, pACYC-JEV-SA14, as previously described 38.
Plasmid construction
As shown in Fig. 1A and Fig. S1, the full-length cDNA clone pACYC-JEV-SA14 was used as a backbone to construct the cDNA clone of the reporter virus Rluc-JEV. First, three fragments covering “KpnI-T7pro-5'UTR-C34”, “Rluc-2A” and “C-prM-E-BsrGI” were amplified with the indicated primer pairs in Table S1 using pACYC-JEV-SA14 and WNV-Rluc-2A replicon 39 as a template, respectively. The two fragments, “KpnI-T7pro-5'UTR-C34” and “Rluc-2A” were fused together by overlap PCR, resulting in the cassette containing “KpnI-T7pro-5'UTR-C34-Rluc-2A”. The purified product was then fused with “C-prM-E-BsrGI” by a second-step overlap PCR, generating the fragment of “KpnI-T7pro-5'UTR-C4-Rluc-2A-C-prM-E-BsrGI”. Finally, the fragment from KpnI to BsrGI was engineered at the corresponding site into pACYC-JEV-SA14 to generate the full-length cDNA clone of Rluc-JEV. Bacterial strain HB101 (Promega) was used as the host to propagate the above-mentioned cDNA clones. The complete sequence of the cDNA clone of Rluc-JEV was validated by DNA sequencing analysis before the subsequent experiments. All restriction endonucleases were purchased from New England Biolabs.
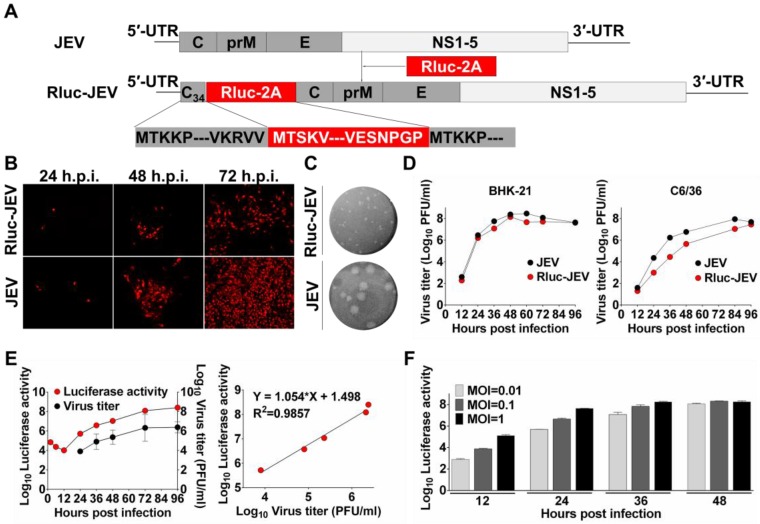
Figure 1.
Construction and characterization of Rluc-JEV. (A) Strategy for the construction of the infectious cDNA clone of Rluc-JEV. The Rluc gene preceded by N-terminal 34 amino acids of capsid protein (C34) of JEV is inserted at 5'UTR-C junction of JEV genome. Rluc gene fused with the FDMV-2A sequence (designated as “Rluc-2A”) is shown in red box. (B) IFA analysis of viral protein expression in BHK-21 cells transfected with in vitro transcribed genome-length RNAs of the parental JEV and Rluc-JEV. (C) Plaque morphology of JEV and Rluc-JEV in BHK-21 cells. (D) Growth curve of Rluc-JEV and the parental JEV in BHK-21 and C6/36 cells. Cells were infected with viruses at an MOI of 1, and viral titer in the culture supernatant was determined by plaque assay on BHK-21 cells. (E) Luciferase activity of Rluc-JEV in BHK-21 cells and correlation of viral titer to Rluc activity. BHK-21 cells were infected Rluc-JEV at an MOI of 0.01. Viral titers in the culture supernatant and luciferase activity in the cells at indicated time points were determined by plaque assay and luciferase assay, respectively. (F) Luciferase signals derived from virus-infected cells at MOI of 0.01, 0.1 or 1.
In vitro transcription and RNA transfection
The genome-length RNAs of parental JEV and Rluc-JEV were transcribed from the corresponding XhoI-linearized cDNA plasmids using T7 mMESSAGE mMACHINE Kit (Ambion). The transcription reactions were performed according to the manufacturer's protocols. For transfection, approximately 5 μg RNA was electroporated into 8 × 106 BHK-21 cells in 0.8 ml of ice cold PBS buffer (pH 7.5) in a 0.4 cm cuvette with the GenePulser apparatus (Bio-Rad) at 0.85 kV and 25 μF, pulsing three times at 3s intervals. After a 10-min recovery at room temperature, the transfected cells were mixed with 25 ml pre-warmed DMEM supplemented with 10% FBS, and were transferred into a T-75 flask and incubated at 37 °C with 5% CO2.
Virus titer and plaque morphology were determined by plaque assay. Briefly, a series of 1:10 dilutions were prepared, and 1 ml of viruses for each dilution were seeded onto each well of 6-well plates containing confluent BHK-21 cells (5× 105 cells/well, plated 1 day in advance). The plates were incubated at 37 °C with 5% CO2 for 1 h before the first layer of agar was added. After 3 days of incubation at 37 °C with 5% CO2, a second layer of agar containing neutral red was added. Plaques were photographed and counted after incubation of the plates for another 12 to 24 h.
Immunofluorescence Assay (IFA)
BHK-21 cells transfected with genome-length RNAs of the parental JEV or Rluc-JEV were seeded on a Chamber Slide (Nalge Nunc). At 24, 48, and 72 h post-transfection, the cells were fixed by 5% cold acetic acid in methanol for 10 min at room temperature. The fixed cells were washed three times with PBS and then incubated with mouse anti-JEV envelope protein monoclonal antibody (1:250 dilution with PBS, Chemicon, USA) for 1 h. After washing with PBS for three times, the cells were incubated with goat anti-mouse IgG conjugated with Texas-Red (ProteinTech Group Inc., USA) at room temperature for 1 h. After three times of washing with PBS, the slide was mounted with 95% glycerol and examined under a fluorescent microscope. Cell images were taken at 200× magnification.
Luciferase assay
BHK-21 cells were infected with the Rluc-JEV reporter virus or transfected with the Rluc-JEV viral RNA, and cell lysis were reclaimed at indicated time points and stored at -80°C for subsequent luciferase assay. Luciferase activity was measured in a Microplate Reader (Varioskan Flash, Thermo Fisher, Finland) by mixing 20 μl lysates with 50 μl substrate (Promega).
Antiviral assay
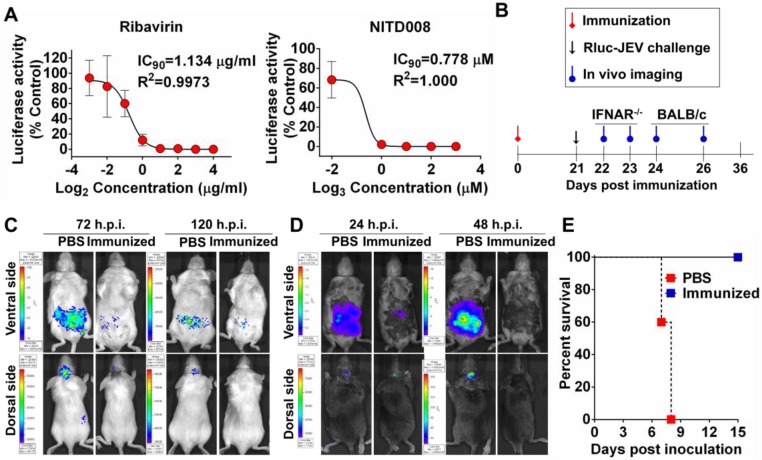
BHK-21 cells were seeded into 12-well plates at a density of 1×105 cells/well. After 24 h incubation, the cells were infected with Rluc-JEV at an MOI of 0.01 and incubated with various concentrations of Ribavirin (0 to 16 μg/ml, Sigma) or NITD008 (0 to 27 μM) 40. Each assay was done in triplicate. The Rluc activity was measured by Multimode Microplate Reader (Varioskan Flash, Thermo Fisher, Finland) at 48 h post infection (h.p.i.). The IC90 was calculated by using GraphPad Prism software 5.0.
BLI
BLI imaging in mice was performed using a charge-coupled device camera (Xenogen Corp., Alameda, CA, USA) as described previously 36. Briefly, ViviRen substrate (Promega) was administered intraperitoneally (i.p.) or intracranially (i.c.) to each animal 5 min before the imaging, and images were acquired for 90 s. Anesthesia was maintained during the imaging through nose-cone delivery of an anesthetic. To quantify the amount of light emitted from different organs, regions of interest (ROIs) were manually defined around specific organs, and photon flux (photons/second/square centimeter/steradian) was calculated using Living Image 3.0 (Caliper Life Sciences, Alameda, CA, USA).
Animal experiments
All animal experiments were performed in strict accordance with the guidelines of the Chinese Regulations of Laboratory Animals (Ministry of Science and Technology of People's Republic of China) and Laboratory Animal-Requirements of Environment and Housing Facilities (National Laboratory Animal Standardization Technical Committee). The experimental protocols were approved by the Animal Experiment Committee of Beijing Institute of Microbiology and Epidemiology, Beijing, China. Mouse strains used in this study included 3 to 4-week-old wild type 129/Sv/Ev mice (Beijing Vital River Co. Ltd, China) and 129/Sv/Ev mice deficient in type I IFN receptors (IFNAR-/-) which were kindly provided by Prof. Qi-Bin Leng (Shanghai Institute for Pasteur, CAS, China).
For correlation studies, the animals were i.c. inoculated with 102.5 PFU of Rluc-JEV. At 24 and 72 h.p.i., the infected animals were euthanized by CO2 asphyxiation immediately after imaging, and the brains were collected, homogenized, and subjected to virus titration by quantitative reverse transcriptase PCR (qRT-PCR) or plaque assay. For the survival study, groups of WT and IFNAR-/- mice were infected via the footpad with 107 PFU of the WT JEV or Rluc-JEV, and monitored daily for 15 days to assess morbidity and mortality. For the tissue distribution study, the animals were inoculated with 106 PFU of Rluc-JEV via footpad. At 72 h.p.i., the infected animals were euthanized by CO2 asphyxiation immediately after imaging. Then the brain, liver, spleen, kidney, intestine, heart and lung were collected for in vitro BLI. All the tissues were stored at -80 °C for viral RNA detection.
For evaluation of efficacy of a live attenuated JE vaccine, groups of 3 to 4-week-old BALB/c and IFNAR-/- mice were immunized s.c. with 105 PFU of a live JE vaccine strain SA14-14-2 or PBS as control. On day 21 post immunization, the immunized mice were challenged i.p. with 107 PFU of Rluc-JEV. Imaging was performed for 90 s at the indicated times shown in Fig. 8B.
Figure 8.
Application of Rluc-JEV for antiviral and vaccine efficacy evaluation. (A) Antiviral assay using Ruc-JEV with known antiviral agents. BHK-21 cells were infected with Rluc-JEV at an MOI of 0.01 and incubated with various concentrations of Ribavirin or NITD008. The Rluc activity was measured by Multimode Microplate Reader at 48 h.p.i.. One representative experiment of three is shown. (B) Graphic depiction of assessment of JE vaccine efficacy by using BLI. (C) and (D) BLI of the immunized BALB/c mice or IFNAR-/- mice challenged with Rluc-JEV. Group of BALB/c (C) or IFNAR-/- mice (D) were immunized s.c. with 105 PFU of live JE vaccine SA14-14-2. On 21 days p.i., the immunized mice were challenged with 107 PFU of Rluc-JEV by the i.p. route. BLI of the challenged mice was performed at indicated time points. (E) Survival curve of the immunized IFNAR-/- mice challenged with Rluc-JEV described in (D).
Titration of virus from tissue samples
All the tested tissue samples were snap frozen for JEV-RNA detection or PFU assay. In brief, each tissue was weighted and homogenized in 1ml of DMEM with 2% FBS and penicillin/streptomycin in an Eppendorf tube for 30 s. The homogenate was clarified by centrifugation for 5 min at 5000 rpm. The supernatants of tissues were collected for RNA extraction and virus titration. Viral RNA in tested tissues homogenates was determined by qRT-PCR as described previously 41. Briefly, viral genomic RNA was extracted from 200 μl of the supernatant of tissue by using the PureLinkTM RNA minikit (Invitrogen, USA) according to the manufacturer's instructions. RNA was eluted in 60 μl of RNase-free water, aliquoted, and stored at -80°C until use. qRT-PCR was performed by using a One Step PrimeScript RT-PCR kit (TaKaRa, Japan). The 20 μl reaction mixtures were set up with a 0.5 μM concentration of each primer (qJEVF: AGAGCACCAAGGGAATGAAATAGT; qJEVR: AATAGGTTGTAGTTGGGCACTCTG), 0.25 μM probe (FAM-CCACGCCACTCGACCCATAGACTG-TAMRA), and 2 μl of RNA. Thermocycling programs consisted of 42°C for 5 min, 95°C for 10 s, and 40 cycles of 95°C for 5 s and 60 °C for 20 s.
Histology and immunohistochemistry assay
Mice were inoculated with 107 PFU of Rluc-JEV via the footpad. At 120 h.p.i., the brain, liver, kidney, spleen and intestine were collected, fixed in 4% (v/v) phosphate-buffered paraformaldehyde overnight. After fixation, the tissues were embedded in paraffin, sectioned, and stained with hematoxylin and eosin (H&E) for the examination of histological changes by light microscopy.
For immunohistochemistry, sections were deparaffinized with xylene, rehydrated through successive bathes of ethanol/water (from 100% ethanol successively to 50% ethanol till pure water) and incubated in 3% H2O2 at room temperature. The sections were then put in 10 mM sodium citrate buffer for 1 h at 96 °C for antigen retrieval and blocked with BSA at saturation for 20 min. For detection of JEV antigen, JEV E-specific monoclonal antibody JE1 (ab41671, Abcam) was used. Horse radish peroxidase-conjugated anti-rabbit polyclonal antibody was used as a secondary antibody.
Statistical analysis
For survival analysis, Kaplan-Meier survival curves were analyzed by log-rank test using standard GraphPad Prism software 5.0. The results of bioluminescence level were expressed with error bars representing the standard deviation. Statistical analysis was completed using a two-way analysis of variance (ANOVA) *, P<0.05; ****, P<0.0001.
Results
Construction and characterization of the JEV reporter virus
To design and generate a recombinant JEV reporter virus, the bioluminescence gene Rluc was rationally engineered into the full-length infectious cDNA clone of JEV 38. As shown in Fig. 1A, the cDNA clone of Rluc-JEV encompasses the 5'UTR, the N terminal 34 amino acids of C protein (named C34), the Rluc gene followed by an FMDV-2A sequence, the structural protein genes, the nonstructural protein genes and the 3'UTR. The C34 that contains cis-acting elements required for viral genome cyclization and replication was duplicated and placed in frame preceding the Rluc gene. The FMDV-2A autoproteolytic cleavage sequence in the downstream of the reporter gene was engineered to ensure proper processing of the Rluc protein from the genome translated polyprotein.
Following standard virus recovery procedure, we examined viral envelope protein synthesis, plaque morphology and viral replication dynamics of the recovered Rluc-JEV. IFA showed the reporter virus was capable of expressing JEV-specific E protein, although the percentage of IFA-positive cells transfected with Rluc-JEV RNA was slightly less than that of the parental JEV at each time point (Fig. 1B). We also found Rluc-JEV produced a smaller plaque in BHK-21 cells than that of the parental virus (Fig. 1C), and exhibited patterns of replication in BHK-21 or C6/36 cells similar to that of parental JEV (Fig. 1D). These results demonstrate the successful recovery of replication-competent JEV reporter virus, which exhibits similar infectivity with a minor reduction in replication efficacy in comparison with the parental JEV.
We then determined whether Rluc-JEV expressed Rluc and further whether the Rluc activity correlated with viral replication in vitro. As shown in Fig. 1E, the luciferase signal increased in proportion to virus titer in culture. Linear regression analysis showed a good correlation between luciferase values and viral titers (Fig. 1E). Additionally, the Rluc signal correlated well with the MOI in the Rluc-JEV-infected BHK-21 cells (Fig. 1F). Collectively, these observations validate that the presence of Rluc gene is not likely to significantly affect the virus replication in culture, and the luciferase activity accurately reflects the proliferation of Rluc-JEV in vitro.
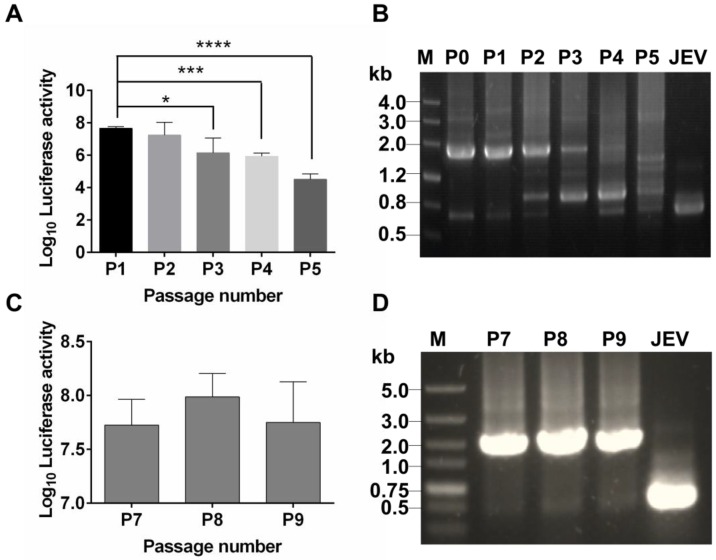
To test stability of the recovered Rluc-JEV, we passaged Rluc-JEV in BHK-21 cells for five rounds. Viruses from each passage (P0-P5) were used to infect BHK-21 cells, and the infected cells were assayed for luciferase activities. As shown in Fig. 2A, comparable Rluc activity was maintained for the first two rounds of passage, while luciferase signals decreased from P3 to P5. RT-PCR assay confirmed that different length of sequences deletion within Rluc gene occurred during the passaging history (Fig. 2B). To improve the genetic stability of Rluc-JEV, the recovered Rluc-JEV (P0) was plaque-purified for six times in BHK-21 cells. Then the resulted plaque purified virus (P6) was serially passaged in BHK-21 cells for three times (P7-P9), followed by evaluation of the Rluc activity of each passaged virus. Comparable luciferase signals were observed in the cells infected with P7 to P9 viruses (Fig. 2C). The full length Rluc gene from viral RNA at difference passages were detectable by RT-PCR (Fig. 2D), and full genome sequencing of the P6 virus identified a point nucleotide mutation (T142C) at the full C protein coding region, which leads to an amino acid substitution M16T of C protein. This kind of mutation has been demonstrated to enhance the stability of other reporter flavivivruses 37, 42. Since the plaque-purified Rluc-JEV virus (P6) showed a superior stability in cell culture, we used this viral stock for the following experiments.
Figure 2.
Genetic stability of Rluc-JEV in BHK-21 cells. (A) The Rluc-JEV virus (P0) was blindly passaged on BHK-21 cells for five rounds (P1-P5). Luciferase activity of each passaged viruses was detected as described in Materials and methods. Error bars represent the standard deviation of triplicate measurement. *, P<0.05; ***, P<0.001; ****, P<0.0001. (B) Detection of the Rluc gene during blind passage. Viral RNA was extracted from the supernatants of BHK-21 cells infected with each passaged virus, and RT-PCR assay was performed to amplify the region from 5'UTR to prM which includes the Rluc gene. (C) Individual Rluc-JEV viruses were isolated by six rounds of plaque purification, designated as P6 viruses. P6 viruses were then serially passaged on BHK-21 cells for another three rounds (P7-P9). Luciferase activity in the cells infected with P7-P9 viruses was detected. Error bars represent the standard deviation of triplicate measurement. (D) Detection of the Rluc gene of P7-P9 viruses by RT-PCR using the same primers in (B). The expected band, corresponding to 1.6 kb, indicates the presence of the Rluc gene in the passaged plaque-purified reporter viruses.
Noninvasive imaging of JEV infection in mouse
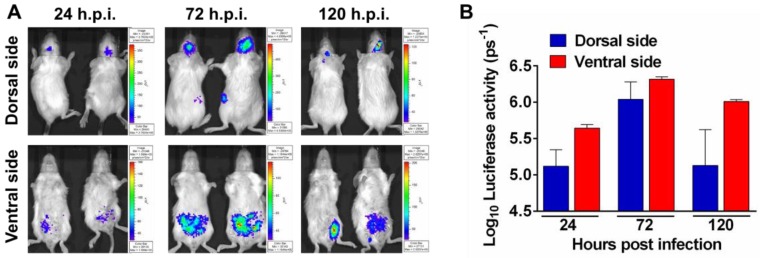
To determine the feasibility of Rluc-JEV for monitoring viral replication dynamics and spread in vivo with BLI, we inoculated BALB/c mice intraperitoneally (i.p.) with 107 PFU of Rluc-JEV, and monitored the luminescent signal over time via the Xenogen IVIS200 imaging system. Animals infected with the parental JEV were used as controls. As shown in Fig. 3A, 3B and Fig. S2A, photon flux was readily seen in the brain of infected mice as early as 24 h.p.i., peaked at 72 h.p.i., and decreased at 120 h.p.i.. Meanwhile, strong bioluminescence signal was also detected in the abdomen of infected mice, suggesting visceral organs may be potential targets of Rluc-JEV. No bioluminescence could be detected above background levels in mice infected with parental JEV (Fig. S2B).
Figure 3.
Noninvasive imaging to monitor JEV replication in mouse. (A) BLI of JEV infection in BALB/c mice. Groups of 3 to 4-week-old BALB/c mice were inoculated i.p. with 107 PFU of Rluc-JEV. Imaging was performed for 90 s at the indicated times. Representative images from two mice are shown. (B) Photon flux was quantified from ROI analysis of the dorsal and ventral side. The data are representative of at least three independent experiments, and error bars indicate the standard deviation.
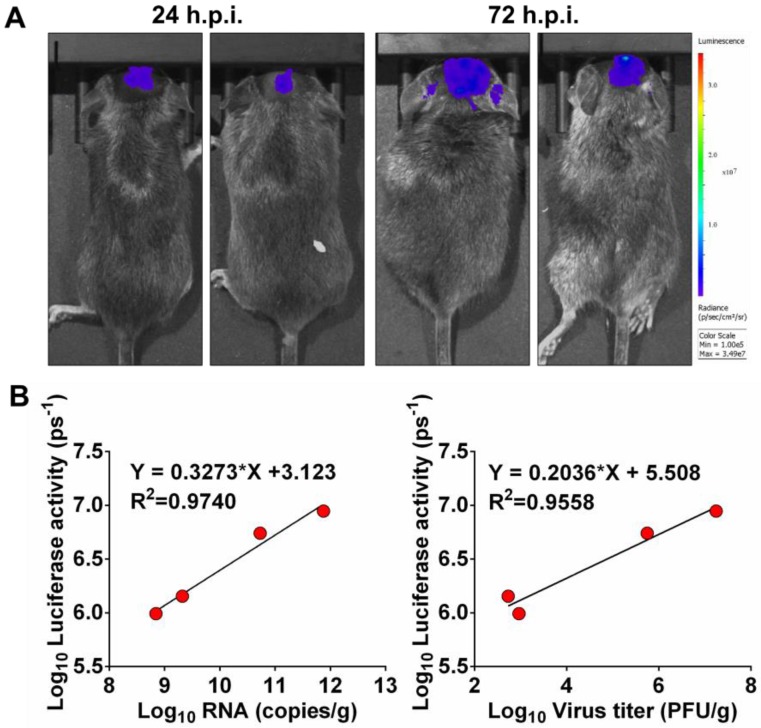
Next, to determine whether luminescence signal in vivo directly correlated to the amount of virus replicating in vivo, mice inoculated intracranially (i.c.) with 102.5 PFU of Rluc-JEV were subjected to BLI. Viral RNA and viral titers in mouse brains were measured by qRT-PCR and plaque assay, respectively. As shown in Fig. 4A, obvious luminescence signal could be detected in infected mice at 24 h.p.i., and level of the signal elevated at 72 h.p.i.. Linear regression analyses showed the viral RNA and infectious viral particles measured in vitro directly correlated with the amount of light emitted in vivo (Fig. 4B, R2=0.9740 and 0.9558, respectively). The good correlation between Rluc activity and viral load in tissues validates that the luminescent signal can reliably represent the replication of Rluc-JEV in vivo.
Figure 4.
Correlation between fluorescence signal and viral replication. (A) IFNAR-/- mice were infected i.c. with 102.5 PFU of Rluc-JEV and subjected to imaging at 24 and 72 h.p.i.. BLI was performed for 90 s, and fluorescence signals were quantified using Living Image software 3.0. (B) Viral RNA and viral titers in excised brains were quantified by real time RT-PCR and plaque assay, respectively. The photon flux determined as described in the legend to Fig. 3 was plotted against the RNA copies or viral titer (PFU/g).
Type I IFN is critical for viral dissemination to visceral organs
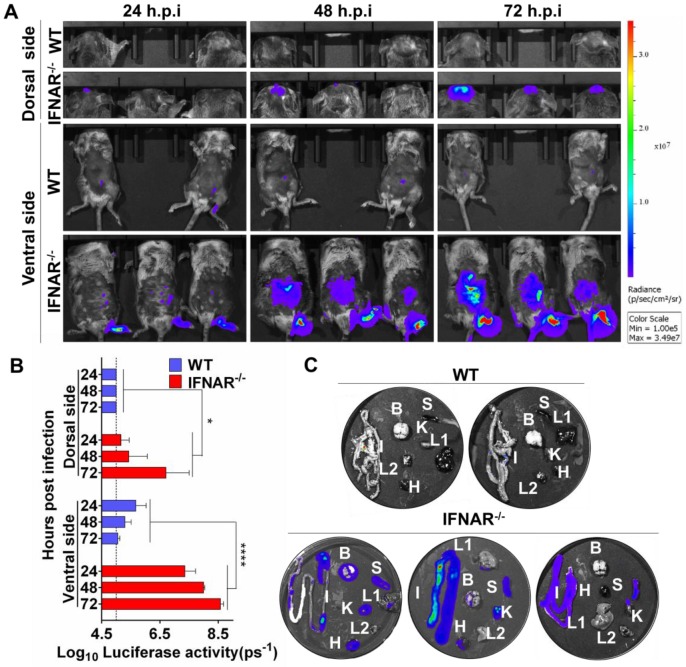
Our preliminary BLI results suggested abdomen organs were potential targets for JEV infection. This unexpected observation urged us to characterize the viscerotropism of JEV in vivo, and examine the role of type I IFN in controlling JEV infection. To this end, WT and IFNAR-/- mice were infected via footpad with 106 PFU of Rluc-JEV or the parental JEV as control, and BLI of infected mice was performed at 24, 48 and 72 h.p.i.. In infected IFNAR-/- mice, following footpad injection, the signals were primarily detected at the local site of inoculation and the brain at 24 h.p.i., and further disseminated to the peritoneal cavity at 48 h.p.i., and finally peaked in the brain and peritoneal cavity at 72 h.p.i. (Fig. 5A), suggesting the Rluc-JEV may target the brain and various abdominal organs. In contrast, only slight Rluc activity were observed in the injection site and peritoneal cavity of the Rluc-JEV-infected WT mice at indicated time points (Fig. 5A), and no signal was detected in mice infected with the parental JEV (Fig. S3A). Overall, the bioluminescence in the dorsal and ventral sides of IFNAR-/- mice was significantly greater than that of the WT mice by ROI analysis (Fig. 5B and Fig. S3B).
Figure 5.
Type I IFN plays a critical role in control of viral dissemination in mice. (A) BLI of Rluc-JEV-infected WT or IFNAR-/- mice. The mice were inoculated via the footpad with 106 PFU of Rluc-JEV. Imaging was performed for 90 s at the indicated times. Representative images from three or two mice are shown. (B) The photon flux was quantified from ROI analysis of the dorsal and ventral side. The data are representative of three independent experiments, and error bars indicate the standard deviation. *, P<0.05; ****, P<0.0001. (C) WT or IFNAR-/- mice were inoculated via the footpad with 106 PFU of Rluc-JEV. At 72 h.p.i., the infected mice was subjected to BLI. Immediately after being imaged, the animals were sacrificed and parenchymal organs including brain(B), liver (L1), kidney (K), spleen (S), intestine (I), heat (H) and lung(L2) were isolated for in vitro BLI.
To further clarify the potential target organs of Rluc-JEV infection, in vitro BLI of the harvested tissues from the IFNAR-/- and WT mice infected with 106 PFU of Rluc-JEV were performed at 72 h.p.i. The results showed that strong luminescence signals could be detected in the abdominal organs of the infected IFNAR-/- mice, including the heart, liver, spleen, kidney and intestine (Fig. 5C). By comparison, albeit slight Rluc activity could be detected in abdominal organs of Rluc-JEV infected WT mice (Fig. 5A), luciferase signal was undetectable in all tested organs, and only weak signal was seen in the intestines (Fig. 5C). These results indicate the critical role of type I IFN for the viscera dissemination of Rluc-JEV in mice.
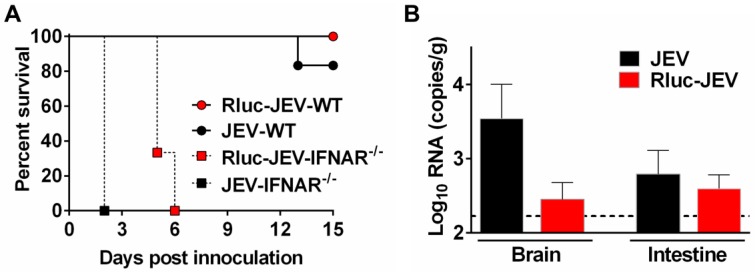
Then, to make sure Rluc-JEV retained the same virulence phenotype to the parental virus JEV, we compared the mortality and organ tropism of Rluc-JEV and JEV in both WT and IFNAR-/- mice. As shown in Fig. 6A, Rluc-JEV was slightly attenuated in both WT and IFNAR-/- mice compared with the parental JEV, yet there was no significant difference based on survival curves. Especially, viral RNA could be detected in brains and intestines in both JEV and Rluc-JEV infected WT mice (Fig. 6B). These results indicate Rluc-JEV exhibited similar viscerotropism to the wild type virus, whereas with a slightly attenuated phenotype.
Figure 6.
Susceptibility of WT or IFNAR-/- mice to parental JEV and Rluc-JEV. (A) Survival curve of mice infected with viruses. WT or IFNAR-/- mice (n=6 or 7) were injected via the footpad with of 107 PFU of Rluc-JEV or the parental JEV, and monitored daily for 15 days to assess morbidity and mortality. Rluc-JEV-WT, WT mice infected with Rluc-JEV; JEV-WT, WT mice infected with parental JEV; Rluc-JEV-IFNAR-/-, IFNAR-/- mice infected with Rluc-JEV; JEV-IFNAR-/-, IFNAR-/- mice infected with parental JEV. The data are representative of at least three independent experiments. (B) Viral load in tissues of WT mice infected with viruses. Groups of WT mice (n=2) were injected via the footpad with of 106 PFU of Rluc-JEV or the parental JEV. At 72 h.p.i., brains and intestines were collected, homogenized, and subjected to virus titration by qRT-PCR. The dotted line indicates the limit of detection. The data are representative of at least three independent experiments.
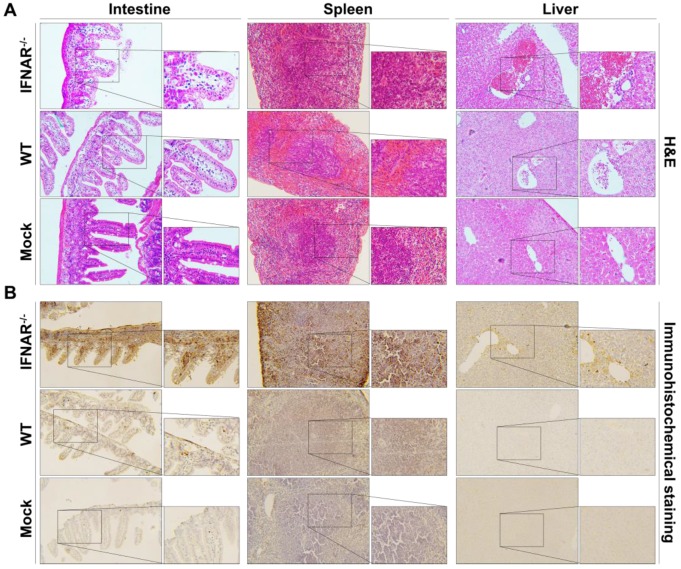
Furthermore, we performed histopathological and immunohistochemistrical analyses to validate our BLI results. In the spleen section from the Rluc-JEV infected IFNAR-/- mice, we observed the red pulp expansion with white pulp reduction and severe necrosis with markedly reduced numbers of small lymphocytes in the mantle and marginal zones (Fig. 7A). Liver tissues from the Rluc-JEV infected IFNAR-/- mice were injured as evidenced by lymphocytic infiltrate, sinusoidal congestion, hyperplasia of Kupffer cells and abundant extramedullary erythropoiesis foci. (Fig. 7A). While the spleen and liver section from Rluc-JEV infected WT mice and the uninfected mice showed normal. Notably, excessive amounts of epithelial cell debris and heavy cellular infiltration of lamina propria were observed in the lumen and mucosa of the ileum from the intestine section from both IFNAR-/- and WT mice infected with Rluc-JEV (Fig. 7A). As expected, immunohisto-chemistrical analyses using JEV specific antibody revealed that JEV E protein were present in spleens, livers and intestines of the infected IFNAR-/- mice, while viral antigens were only detected in intestines in WT mice infected with Rluc-JEV (Fig. 7B). No viral antigen was detected in any of the tested tissues from the uninfected mice (Fig. 7B). Together, these combined results highlight the protective role of type I IFN in controlling JEV viscerotropism and virulence in mice.
Figure 7.
Histology and immunohistochemistry of tissue sections from mice infected with Rluc-JEV. WT or IFNAR-/- mice were inoculated via the footpad with 107 PFU of Rluc-JEV. At 5 days p.i., parenchymal organs including brain, liver, kidney, spleen and intestine were collected for histology (A) and immunohistochemistry (B).
BLI application of Rluc-JEV
Next, we utilized Rluc-JEV to test known antiviral compounds. Two known flavivirus inhibitors Ribavirin and NITD008 were used here, former of which is a competitive inhibitor of inosine monophosphate (IMP) dehydrogenase 43, 44, latter of which is an adenosine analogue inhibitor of DENV 40, 45. BHK-21 cells were infected with Rluc-JEV and treated with different concentrations of the inhibitors. As shown in Fig. 8A, the IC90 of Ribavirin and NITD008 was 1.134 μg/ml and 0.778 μM, respectively.
To adapt the BLI platform for vaccine efficacy in living mice, BALB/c or IFNAR-/- mice were s.c. immunized with 105 PFU of the live JE vaccine SA14-14-2 (Fig. 8B). Mice immunized with PBS were used as control. Upon i.p. challenge with 107 PFU of Rluc-JEV, a strong signal was observed in both the peritoneal cavity and brains of the PBS-immunized mice at 72 h.p.i., and became subdued at 120 h.p.i., whereas only a slight signal was emitted from the peritoneal cavity in the vaccine-immunized BALB/c mice (Fig. 8C). In the PBS-immunized IFNAR-/- mice, strong luminescence signals were emitted from the peritoneal cavity at 24 h.p.i., followed by a significantly increased signal at 48 h.p.i. (Fig. 8D). By comparison, only weak luminescence signal was detected in the peritoneal cavity of the vaccine-immunized mice at 24 h.p.i., and became undetectable at 48 h.p.i. (Fig. 8D and Fig. S4). Additionally, vaccine-immunized IFNAR-/- mice were resistant to infection of Rluc-JEV, as indicated by the observation that all the mice survived during the observation period, while all PBS-immunized IFNAR-/- mice succumbed to Rluc-JEV challenge within 7-8 days post infection (Fig. 8E). Overall, these results show that Rluc-JEV can be used to evaluate antiviral compounds in vitro and monitor vaccine efficacy in living mice.
Discussion
Here, we report the generation of a novel replication-competent Rluc-reporter JEV. The luciferase activity produced by the reporter virus can well serve as a readout of viral replication in culture and viral load in mice, which makes possible detection of real-time JEV infection in living mice. Importantly, by using BLI of the Rluc-JEV, we reveal the critical role of type I IFN in control of viscerotropic infection of JEV in mice, and identify the intestine as an important target visceral organ of JEV infection. Finally, we confirm the feasibility of the reporter virus for efficacy assays for antivirals and vaccine candidates.
Although multiple versions of reporter viruses have been previously described, no neurotropic flavivirus reporter virus has ever been developed and used for in vivo BLI. We obtained the Rluc-JEV by insertion of the Rluc gene appended to the first 34 amino acids of C protein at the 5'UTR-C junction of JEV genome. This strategy for construction of Rluc-JEV is based on the findings that the 5' end of the flavivirus genome harbors RNA elements essential for viral replication such as stem-loop A 46, C-coding region hairpin 47 and downstream of 5' cyclization sequence (CS) pseudoknot 48. A similar strategy has been applied to successfully generate DENV reporter viruses, in which the coding region for the first 25 or 38 amino acids of C protein was duplicated upstream of the reporter gene 37, 45. As expected, Rluc-JEV is infectious in cell culture illustrated by the expression of viral protein, formation of plaque in cells and replication in mammalian and mosquito cells. The reporter virus has a slower growth rate and smaller plaque than that of the parental JEV, indicating some levels of attenuation of Rluc-JEV in cell culture. This observation is unsurprising because previous studies showed that insertion of a reporter gene into the viral genome always attenuates virus replication in cell culture 32, 45, 49, 50. Despite this, Rluc-JEV achieves robust expression of Rluc whose activity attains a peak value of 108 light unit, about over 30-fold greater than that of DENV reporter virus 45. Importantly, the magnitude of Rluc activity produced by Rluc-JEV correlates directly with increasing titer of virus in cell culture and in vivo. Notably, consistent with previous study on dengue reporter virus 37, the Rluc gene was found unstable in the Rluc-JEV genome during virus passage. We obtained a new Rluc-JEV virus strain stably expressing Rluc gene after six rounds of plaque purification. The plaque-purified Rluc-JEV virus displayed a high stability and full genome sequencing identified a single nucleotide mutation (T142C), leading to a M16T mutation in the full C protein. This mutation locates within the 5' CS, and is supposed to facilitate the genome cyclization between 3'CS and 5' CS in the C34 coding region, not with the full C protein coding region. Such mutation has been observed in WNV and DENV reporter viruses 37, 42, and well demonstrated to contribute to the stability of the flaviviral reporter viruses. Overall, these phenotypes allow us to further characterize the dynamics of JEV infection in living mice by using BLI of the stable Rluc-JEV reporter virus.
A major finding reported here by utilizing BLI system of the reporter virus is that JEV spreads to a set of parenchymal tissues in IFNAR-/- mice that are not normally infected in WT mice, demonstrating an essential function for type I IFN in preventing systemic infection with JEV. To our knowledge, this is the first study that characterizes the real-time JEV infection in the absence of type I IFN response in living mice. Type I IFN comprises an important innate immune system against viral infections. By studying genetically deficient animals, type I IFN has been shown to play a protective role for flaviviruses with different degree. For example, Lee et al found that IFN-αR-/- mice infected with JEV showed early and high viremia, rising virus titers in examined parenchymal tissues, and 100% mortality by day 6 p.i., which confirms the importance of IFN-α/β in the resistance of mice to JEV infection 24. Additionally, IFN-α/β receptor pathway plays a role in controlling initial viral replication and/or spread during primary DENV infection 51. In contrast, mice absolutely require functional type I IFN response for survival following WNV or MVEV infection 52, 53. It is believed that neurotropic viruses entry CNS following virus replication in extraneural tissues. Type I IFN inhibits JEV growth in primary infected tissues, which probably plays an important role in preventing virus entry into the CNS. It still remains unclear how type I IFN restricts tissue tropism in JEV infection. Our study found that significantly increased viral burden in the spleen of infected IFNAR-/- mice compared to infected WT mice. This finding is consistent to previous studies in which absence of the IFN-α/β receptor resulted in colocalization of viral antigen with macrophage and/or dendritic cells in the spleen 53, 54. We propose that IFN-α/β may modulate tropism by restricting infection in myeloid-derived cells. Further studies are needed to identify infected cell types in IFNAR-/- mice and distinguish between these non-mutually exclusive functions of IFNs.
An important finding of this study is that the intestine might be a novel target of JEV infection in mice. Intestines could be infected with DENV in the absence of type I and type II IFN. For example, DENV-infected mice deficient in type I and II IFN showed viral RNA could be detected in the small intestines of infected animals, and DENV prM protein was detected in a fraction of isolated laminar propria macrophages 55. Schoggins et al also found the presence of luciferase in the intestines of IFN-αβγR-/- mice infected with reporter virus DENV-Fluc, suggesting DENV can localize to gut-associated lymphoid tissues 37. The encephalitis is the most common presentation in syndrome caused by JEV infection. Status epilepticus, brain hypoxia and increased intracranial pressure are the most common complications associated with poor outcome and death 56. However, little evidence of the intestine as a target tissue of neurotropic JEV infection in immunocompetent mice exists. Our data in this study showed Rluc signal and viral RNA as well as viral antigen could be detected in the intestine of mice infected with either Rluc-JEV or JEV, suggesting the intestine as an important potential target organ of JEV infection, which further expanded our understanding of JEV pathogenesis. A previous study found that mice inoculated in the footpad with JEV showed viremia and presence of infectious virus the brain and liver 24. Recently, yellow fever vaccine-associated viscerotropic and neurotropic diseases have been reported 57, 58, resulting in deaths in Brazil 59 and Australia 60. Our finding of this novel target tissue of JEV infection may give insight into the contribution of tissue distribution to eventual neuronal dysfunction of the neurotropic flavivirus infection.
Finally, we used the BLI system based on the reporter virus to evaluate two antiviral compounds in cell culture as well as efficacy of the known live JE vaccine in mice. Similar to other reporter viruses, Rluc-JEV is sensitive to treatment with antiviral drugs, allowing us to screen for anti-JEV compounds. As expected, BLI of the immunized mice challenged with Rluc-JEV showed significant decrease in magnitude of Rluc signal compared with PBS-immunized mice, suggesting a good efficacy of the JE live vaccine in protecting against JEV infection. Conventional studies of viral pathogenesis need to have sufficient animals to generate statistically significant data. Noninvasive imaging allows the same group of animals to be studied over time, which reduces the number of animals used in experiments and minimizes the impact of individual differences. Additionally, BLI has been validated to be substitute for lethality as an endpoint in testing the efficacies of new vaccines and prophylactic interventions against poxvirus infections in the mouse model 35. Based on these findings, the BLI system of the reporter virus Rluc-JEV will be optimized to be a substitute for conventional measures for vaccine evaluation.
In summary, by using a novel JEV reporter virus suitable for in vivo imaging, we observe the in vivo replication kinetics and viral dissemination model in real-time. Importantly, the intestine, besides the brain, is identified as a potential target organ of JEV infection, and host type I interferon plays critical roles in controlling the viscerotropism of a neurotropic flavivirus. Recently, another neurotropic flavivirus ZIKV reporter virus has been developed which would also facilitate the tracking of viral replication in vivo 61. The well-established BLI system of reporter virus will enhance greatly the pathogenesis studies of neurotropic flavivirus and facilitate evaluation of candidate vaccines and antiviral compounds in vivo.
Supplementary Material
Figures S1-S4 and Table S1.
Acknowledgments
We are grateful to Prof. Jing An (Capital Medical University, China), Prof. Qi-Bin Leng (Shanghai Institute of Pasteur, CAS, China), and Prof. Pei-Yong Shi (University of Texas Medical Branch, USA) for providing key reagents and instructive comments. We thank the Core Facility and Technical Support, Wuhan Institute of Virology and Wuhan Key Laboratory on Emerging Infectious Diseases and Biosafety for effective support during the course of the work.
Funding
This work was supported by the National Key Research and Development Program of China (No.2016YFD0500400 and No.2016YFD0500304), and the National Natural Science Foundation of China (No.81572003, No.31300151, No.81621005 and No.31470265). Cheng-Feng Qin was supported by the Excellent Young Scientist Foundation from NSFC (No. 81522025) and the Newton Advanced Fellowship from the UK Academy of Medical Sciences and NSFC (No. 81661130162). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author Contributions
Conceived and designed the experiments: X.F.L., X.D.L., B.Z. and C.F.Q.. Performed the experiments: X.F.L., X.D.L., C.L.D, H.L.D., Q.Y.Z., Q.Y., H.Q.Y., and X.Y.H. Analyzed the data: X.F.L., X.D.L., Y.Q.D., B.Z. and C.F.Q.. Wrote the paper: X.F.L., X.D.L., B.Z. and C.F.Q..
References
- 1.Peltier DC, Lazear HM, Farmer JR, Diamond MS, Miller DJ. Neurotropic arboviruses induce interferon regulatory factor 3-mediated neuronal responses that are cytoprotective, interferon independent, and inhibited by Western equine encephalitis virus capsid. Journal of virology. 2013;87:1821–33. doi: 10.1128/JVI.02858-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Campbell GL, Hills SL, Fischer M, Jacobson JA, Hoke CH, Hombach JM. et al. Estimated global incidence of Japanese encephalitis: a systematic review. Bulletin of the World Health Organization. 2011;89:766–74. doi: 10.2471/BLT.10.085233. 74A-74E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ghosh D, Basu A. Japanese encephalitis-a pathological and clinical perspective. PLoS neglected tropical diseases. 2009;3:e437. doi: 10.1371/journal.pntd.0000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Le Flohic G, Porphyre V, Barbazan P, Gonzalez JP. Review of climate, landscape, and viral genetics as drivers of the Japanese encephalitis virus ecology. PLoS neglected tropical diseases. 2013;7:e2208. doi: 10.1371/journal.pntd.0002208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Solomon T. Control of Japanese encephalitis-within our grasp? The New England journal of medicine. 2006;355:869–71. doi: 10.1056/NEJMp058263. [DOI] [PubMed] [Google Scholar]
- 6.Mackenzie JS, Johansen CA, Ritchie SA, van den Hurk AF, Hall RA. Japanese encephalitis as an emerging virus: the emergence and spread of Japanese encephalitis virus in Australasia. Current topics in microbiology and immunology. 2002;267:49–73. doi: 10.1007/978-3-642-59403-8_3. [DOI] [PubMed] [Google Scholar]
- 7.Igarashi A, Tanaka M, Morita K, Takasu T, Ahmed A, Ahmed A. et al. Detection of west Nile and Japanese encephalitis viral genome sequences in cerebrospinal fluid from acute encephalitis cases in Karachi, Pakistan. Microbiology and immunology. 1994;38:827–30. doi: 10.1111/j.1348-0421.1994.tb01866.x. [DOI] [PubMed] [Google Scholar]
- 8.Paul WS, Moore PS, Karabatsos N, Flood SP, Yamada S, Jackson T. et al. Outbreak of Japanese encephalitis on the island of Saipan, 1990. The Journal of infectious diseases. 1993;167:1053–8. doi: 10.1093/infdis/167.5.1053. [DOI] [PubMed] [Google Scholar]
- 9.Mackenzie JS, Gubler DJ, Petersen LR. Emerging flaviviruses: the spread and resurgence of Japanese encephalitis, West Nile and dengue viruses. Nature medicine. 2004;10:S98–109. doi: 10.1038/nm1144. [DOI] [PubMed] [Google Scholar]
- 10.West Nile virus disease cases and deaths reported to CDC by year and clinical presentation, 1999-2014. CDC; http://www.cdc.gov/westnile/statsmaps/cummapsdata.html. [Google Scholar]
- 11.Petersen LR, Jamieson DJ, Powers AM, Honein MA. Zika Virus. N Engl J Med. 2016;374:1552–63. doi: 10.1056/NEJMra1602113. [DOI] [PubMed] [Google Scholar]
- 12.Rasmussen SA, Jamieson DJ, Honein MA, Petersen LR. Zika Virus and Birth Defects-Reviewing the Evidence for Causality. N Engl J Med. 2016;374:1981–7. doi: 10.1056/NEJMsr1604338. [DOI] [PubMed] [Google Scholar]
- 13.Solomon T, Dung NM, Kneen R, Gainsborough M, Vaughn DW, Khanh VT. Japanese encephalitis. Journal of neurology, neurosurgery, and psychiatry. 2000;68:405–15. doi: 10.1136/jnnp.68.4.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Solomon T, Kneen R, Dung NM, Khanh VC, Thuy TT, Ha DQ. et al. Poliomyelitis-like illness due to Japanese encephalitis virus. Lancet. 1998;351:1094–7. doi: 10.1016/S0140-6736(97)07509-0. [DOI] [PubMed] [Google Scholar]
- 15.Welte T, Reagan K, Fang H, Machain-Williams C, Zheng X, Mendell N. et al. Toll-like receptor 7-induced immune response to cutaneous West Nile virus infection. The Journal of general virology. 2009;90:2660–8. doi: 10.1099/vir.0.011783-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu SJ, Grouard-Vogel G, Sun W, Mascola JR, Brachtel E, Putvatana R. et al. Human skin Langerhans cells are targets of dengue virus infection. Nature medicine. 2000;6:816–20. doi: 10.1038/77553. [DOI] [PubMed] [Google Scholar]
- 17.Lim PY, Behr MJ, Chadwick CM, Shi PY, Bernard KA. Keratinocytes are cell targets of West Nile virus in vivo. Journal of virology. 2011;85:5197–201. doi: 10.1128/JVI.02692-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Samuel MA, Diamond MS. Pathogenesis of West Nile Virus infection: a balance between virulence, innate and adaptive immunity, and viral evasion. Journal of virology. 2006;80:9349–60. doi: 10.1128/JVI.01122-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dumpis U, Crook D, Oksi J. Tick-borne encephalitis. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 1999;28:882–90. doi: 10.1086/515195. [DOI] [PubMed] [Google Scholar]
- 20.Mathur A, Arora KL, Chaturvedi UC. Congenital infection of mice with Japanese encephalitis virus. Infection and immunity. 1981;34:26–9. doi: 10.1128/iai.34.1.26-29.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mathur A, Kulshreshtha R, Chaturvedi UC. Evidence for latency of Japanese encephalitis virus in T lymphocytes. The Journal of general virology. 1989;70( Pt 2):461–5. doi: 10.1099/0022-1317-70-2-461. [DOI] [PubMed] [Google Scholar]
- 22.Huang CH, Wong C. Relation of the peripheral multiplication of Japanese B encephalitis virus to the pathogenesis of the infection in mice. Acta Virol. 1963;7(16):322–30. [Google Scholar]
- 23.Lee E, Hall RA, Lobigs M. Common E protein determinants for attenuation of glycosaminoglycan-binding variants of Japanese encephalitis and West Nile viruses. Journal of virology. 2004;78:8271–80. doi: 10.1128/JVI.78.15.8271-8280.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee E, Lobigs M. Mechanism of virulence attenuation of glycosaminoglycan-binding variants of Japanese encephalitis virus and Murray Valley encephalitis virus. Journal of virology. 2002;76:4901–11. doi: 10.1128/JVI.76.10.4901-4911.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andreu N, Zelmer A, Wiles S. Noninvasive biophotonic imaging for studies of infectious disease. FEMS microbiology reviews. 2011;35:360–94. doi: 10.1111/j.1574-6976.2010.00252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang MH, Cirillo SL, Cirillo JD. Using luciferase to image bacterial infections in mice. Journal of visualized experiments: JoVE; 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harmache A, LeBerre M, Droineau S, Giovannini M, Bremont M. Bioluminescence imaging of live infected salmonids reveals that the fin bases are the major portal of entry for Novirhabdovirus. Journal of virology. 2006;80:3655–9. doi: 10.1128/JVI.80.7.3655-3659.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hutchens M, Luker GD. Applications of bioluminescence imaging to the study of infectious diseases. Cellular microbiology. 2007;9:2315–22. doi: 10.1111/j.1462-5822.2007.00995.x. [DOI] [PubMed] [Google Scholar]
- 29.Luker GD, Bardill JP, Prior JL, Pica CM, Piwnica-Worms D, Leib DA. Noninvasive bioluminescence imaging of herpes simplex virus type 1 infection and therapy in living mice. Journal of virology. 2002;76:12149–61. doi: 10.1128/JVI.76.23.12149-12161.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mandl S, Schimmelpfennig C, Edinger M, Negrin RS, Contag CH. Understanding immune cell trafficking patterns via in vivo bioluminescence imaging. Journal of cellular biochemistry Supplement. 2002;39:239–48. doi: 10.1002/jcb.10454. [DOI] [PubMed] [Google Scholar]
- 31.Luker KE, Hutchens M, Schultz T, Pekosz A, Luker GD. Bioluminescence imaging of vaccinia virus: effects of interferon on viral replication and spread. Virology. 2005;341:284–300. doi: 10.1016/j.virol.2005.06.049. [DOI] [PubMed] [Google Scholar]
- 32.Pan W, Dong Z, Li F, Meng W, Feng L, Niu X. et al. Visualizing influenza virus infection in living mice. Nature communications. 2013;4:2369. doi: 10.1038/ncomms3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rameix-Welti MA, Le Goffic R, Herve PL, Sourimant J, Remot A, Riffault S. et al. Visualizing the replication of respiratory syncytial virus in cells and in living mice. Nature communications. 2014;5:5104. doi: 10.1038/ncomms6104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang L, Fu Q, Dong Y, Zhou Y, Jia S, Du J. et al. Bioluminescence imaging of Hepatitis C virus NS3/4A serine protease activity in cells and living animals. Antiviral research. 2010;87:50–6. doi: 10.1016/j.antiviral.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 35.Zaitseva M, Kapnick SM, Scott J, King LR, Manischewitz J, Sirota L. et al. Application of bioluminescence imaging to the prediction of lethality in vaccinia virus-infected mice. Journal of virology. 2009;83:10437–47. doi: 10.1128/JVI.01296-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li XF, Deng YQ, Zhao H, Ye Q, Wang HJ, Li SH. et al. Noninvasive bioluminescence imaging of dengue virus infection in the brain of A129 mice. Applied microbiology and biotechnology. 2013;97:4589–96. doi: 10.1007/s00253-013-4799-8. [DOI] [PubMed] [Google Scholar]
- 37.Schoggins JW, Dorner M, Feulner M, Imanaka N, Murphy MY, Ploss A. et al. Dengue reporter viruses reveal viral dynamics in interferon receptor-deficient mice and sensitivity to interferon effectors in vitro. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:14610–5. doi: 10.1073/pnas.1212379109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li XD, Li XF, Ye HQ, Deng CL, Ye Q, Shan C. et al. Recovery of a chemically synthesized Japanese encephalitis virus reveals two critical adaptive mutations in NS2B and NS4A. The Journal of general virology. 2014;95:806–15. doi: 10.1099/vir.0.061838-0. [DOI] [PubMed] [Google Scholar]
- 39.Tilgner M, Shi PY. Structure and function of the 3' terminal six nucleotides of the west nile virus genome in viral replication. J Virol. 2004;78:8159–71. doi: 10.1128/JVI.78.15.8159-8171.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yin Z, Chen YL, Schul W, Wang QY, Gu F, Duraiswamy J. et al. An adenosine nucleoside inhibitor of dengue virus. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:20435–9. doi: 10.1073/pnas.0907010106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li XF, Dong HL, Huang XY, Qiu YF, Wang HJ, Deng YQ. et al. Characterization of a 2016 Clinical Isolate of Zika Virus in Non-human Primates. EBioMedicine. 2016;12:170–7. doi: 10.1016/j.ebiom.2016.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang PT, Shan C, Li XD, Liu SQ, Deng CL, Ye HQ. et al. Generation of a recombinant West Nile virus stably expressing the Gaussia luciferase for neutralization assay. Virus Res. 2016;211:17–24. doi: 10.1016/j.virusres.2015.09.015. [DOI] [PubMed] [Google Scholar]
- 43.Leyssen P, Balzarini J, De Clercq E, Neyts J. The predominant mechanism by which ribavirin exerts its antiviral activity in vitro against flaviviruses and paramyxoviruses is mediated by inhibition of IMP dehydrogenase. Journal of virology. 2005;79:1943–7. doi: 10.1128/JVI.79.3.1943-1947.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Benarroch D, Egloff MP, Mulard L, Guerreiro C, Romette JL, Canard B. A structural basis for the inhibition of the NS5 dengue virus mRNA 2'-O-methyltransferase domain by ribavirin 5'-triphosphate. The Journal of biological chemistry. 2004;279:35638–43. doi: 10.1074/jbc.M400460200. [DOI] [PubMed] [Google Scholar]
- 45.Zou G, Xu HY, Qing M, Wang QY, Shi PY. Development and characterization of a stable luciferase dengue virus for high-throughput screening. Antiviral research. 2011;91:11–9. doi: 10.1016/j.antiviral.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 46.Filomatori CV, Lodeiro MF, Alvarez DE, Samsa MM, Pietrasanta L, Gamarnik AV. A 5' RNA element promotes dengue virus RNA synthesis on a circular genome. Genes & development. 2006;20:2238–49. doi: 10.1101/gad.1444206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clyde K, Harris E. RNA secondary structure in the coding region of dengue virus type 2 directs translation start codon selection and is required for viral replication. J Virol. 2006;80:2170–82. doi: 10.1128/JVI.80.5.2170-2182.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu ZY, Li XF, Jiang T, Deng YQ, Zhao H, Wang HJ. et al. Novel cis-acting element within the capsid-coding region enhances flavivirus viral-RNA replication by regulating genome cyclization. Journal of virology. 2013;87:6804–18. doi: 10.1128/JVI.00243-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Manicassamy B, Manicassamy S, Belicha-Villanueva A, Pisanelli G, Pulendran B, Garcia-Sastre A. Analysis of in vivo dynamics of influenza virus infection in mice using a GFP reporter virus. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:11531–6. doi: 10.1073/pnas.0914994107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tokusumi T, Iida A, Hirata T, Kato A, Nagai Y, Hasegawa M. Recombinant Sendai viruses expressing different levels of a foreign reporter gene. Virus research. 2002;86:33–8. doi: 10.1016/s0168-1702(02)00047-3. [DOI] [PubMed] [Google Scholar]
- 51.Shresta S, Kyle JL, Snider HM, Basavapatna M, Beatty PR, Harris E. Interferon-dependent immunity is essential for resistance to primary dengue virus infection in mice, whereas T- and B-cell-dependent immunity are less critical. Journal of virology. 2004;78:2701–10. doi: 10.1128/JVI.78.6.2701-2710.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lobigs M, Mullbacher A, Wang Y, Pavy M, Lee E. Role of type I and type II interferon responses in recovery from infection with an encephalitic flavivirus. The Journal of general virology. 2003;84:567–72. doi: 10.1099/vir.0.18654-0. [DOI] [PubMed] [Google Scholar]
- 53.Samuel MA, Diamond MS. Alpha/beta interferon protects against lethal West Nile virus infection by restricting cellular tropism and enhancing neuronal survival. Journal of virology. 2005;79:13350–61. doi: 10.1128/JVI.79.21.13350-13361.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ryman KD, Klimstra WB, Nguyen KB, Biron CA, Johnston RE. Alpha/beta interferon protects adult mice from fatal Sindbis virus infection and is an important determinant of cell and tissue tropism. Journal of virology. 2000;74:3366–78. doi: 10.1128/jvi.74.7.3366-3378.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zellweger RM, Prestwood TR, Shresta S. Enhanced infection of liver sinusoidal endothelial cells in a mouse model of antibody-induced severe dengue disease. Cell host & microbe. 2010;7:128–39. doi: 10.1016/j.chom.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Solomon T, Vaughn DW. Pathogenesis and clinical features of Japanese encephalitis and West Nile virus infections. Curr Top Microbiol Immunol. 2002;267:171–94. doi: 10.1007/978-3-642-59403-8_9. [DOI] [PubMed] [Google Scholar]
- 57.Martin M, Tsai TF, Cropp B, Chang GJ, Holmes DA, Tseng J. et al. Fever and multisystem organ failure associated with 17D-204 yellow fever vaccination: a report of four cases. Lancet. 2001;358:98–104. doi: 10.1016/s0140-6736(01)05327-2. [DOI] [PubMed] [Google Scholar]
- 58.Centers for Disease C Prevention. Adverse events associated with 17D-derived yellow fever vaccination-United States, 2001-2002. MMWR Morb Mortal Wkly Rep. 2002;51:989–93. [PubMed] [Google Scholar]
- 59.Vasconcelos PF, Luna EJ, Galler R, Silva LJ, Coimbra TL, Barros VL. et al. Serious adverse events associated with yellow fever 17DD vaccine in Brazil: a report of two cases. Lancet. 2001;358:91–7. doi: 10.1016/s0140-6736(01)05326-0. [DOI] [PubMed] [Google Scholar]
- 60.Chan RC, Penney DJ, Little D, Carter IW, Roberts JA, Rawlinson WD. Hepatitis and death following vaccination with 17D-204 yellow fever vaccine. Lancet. 2001;358:121–2. doi: 10.1016/S0140-6736(01)05341-7. [DOI] [PubMed] [Google Scholar]
- 61.Shan C, Xie X, Muruato AE, Rossi SL, Roundy CM, Azar SR. et al. An Infectious cDNA Clone of Zika Virus to Study Viral Virulence, Mosquito Transmission, and Antiviral Inhibitors. Cell Host Microbe. 2016;19:891–900. doi: 10.1016/j.chom.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1-S4 and Table S1.