Abstract
Background and aims
The Hippo pathway controls organ size through a negative regulation of the transcription co-activator Yap1. The overexpression of hyperactive mutant Yap1 or deletion of key components in the Hippo pathway leads to increased organ size in different species. Analysis of interactions of this pathway with other cellular signals corroborating organ size control is limited in part due to the difficulties associated with development of rodent models.
Methods
Here, we develop a new model of reversible induction of the liver size in mice using siRNA-nanoparticles targeting two kinases of Hippo pathway, namely, mammalian Ste20 family kinases 1 and 2 (Mst1 and Mst2), and an upstream regulator, neurofibromatosis type II (NF2).
Results
The triple siRNAs nanoparticle-induced hepatomegaly in mice phenocopies one observed with Mst1-/- Mst2-/- liver-specific depletion, as shown by extensive proliferation of hepatocytes and activation of Yap1. The simultaneous co-treatment with a fourth siRNA nanoparticle against Yap1 fully blocked the liver growth.
Hippo pathway-induced liver enlargement is associated with p53 activation, evidenced by its accumulation in the nuclei and upregulation of its target genes. Moreover, injections of the triple siRNAs nanoparticle in p53LSL/LSL mice shows that livers lacking p53 expression grow faster and exceed the size of livers in p53 wild type animals, indicating a role of p53 in controlling Yap1-induced liver growth.
Conclusion
Our data show that siRNA-nanoparticulate manipulation of gene expression can provide the reversible control of organ size in adult animals, which presents a new avenue for the investigation of complex regulatory networks in liver.
Keywords: Hippo pathway, p53, bile acids, siRNA, nanoparticles
Introduction
Maintenance of proper organ size in an organism is a fundamentally important process. The proper function of organs depends on coordinated control of their size during developmental and adult stages. In contrast, loss of organ size control contributes to a number of diseases, including hypertrophy and degenerative diseases(1). Organ size control integrates various factors, including local mechanical, autocrine/paracrine stimuli as well as soluble circulating cues and environmental factors(1-3).
One of the cues critical in controlling organ size and cellular proliferation is the Hippo signaling pathway (Reviewed in(4, 5)). This pathway is composed of an evolutionarily conserved core kinase cassette and upstream modulators (reviewed in(6)). In mammals, the core kinases consist of mammalian Ste20 family kinases 1 and 2 (Mst1 and Mst2), Salvador homolog 1 (Sav1), Large tumor suppressor 1 and 2 (Lats1 and Lats2), and Mps one binder 1 (Mob1)(4, 5). The core cascade is tightly regulated by multiple upstream modulators(6). Neurofibromatosis type II (NF2, also known as Merlin) can activate the core kinase cassette(7), likely through direct binding and recruiting Lats1/2 to the plasma membrane(8). The Hippo pathway negatively regulates its major downstream effector Yes-associated protein 1 (Yap1) through phosphorylation, provoking its degradation and cytoplasmic retention(9, 10). As a potent transcription co-activator, Yap1 can induce genes involved in cellular growth and apoptosis inhibition by association with the TEAD family and other transcription factors(9, 11, 12). Overexpression of S127 mutant Yap1 in the adult liver causes massive hepatomegaly(10). It has been reported that while one allele of either Mst1 or Mst2 is sufficient to maintain embryonic development, Mst1-/-Mst2-/- mice are embryonically lethal (13-15). One copy of either Mst1 or Mst2 can maintain quiescence of hepatocytes, but liver-specific genetic removal of both Mst1 and Mst2 leads to significant liver enlargement(14-16).
Hippo signaling has been shown to interact with several other pathways, including PI(3)K–mTOR(17), Wnt/beta-catenin(18), Insulin/IGF(19, 20), and Sonic hedgehog (Shh) signaling pathway(21) in normal tissue and tumorigenesis. However, deciphering its complex interactions in vivo requires sophisticated approaches allowing to gradually and simultaneous manipulate multiple genes.
Recent advances in synthetic siRNA delivery nanoparticles make it possible to specifically suppress one or more genes simultaneously in a range of species, from rodents to primates(22-25), including humans(26). Through manipulating the compositions of nanoparticles, siRNA delivery systems have shown potent and reversible silencing effects in vivo with high specificity in multiple tissues and cell types, including hepatocytes(22, 23), macrophages(27) and endothelial cells(28). Here we develop a siRNA nanoparticle-based approach to manipulate organ size through inhibition of the Hippo pathway. The deep reduction of the expression levels of Mst1/Mst2/Nf2 is critical to elucidate the role of p53 pathway in the control of liver growth.
Materials and Methods
siRNA synthesis, screening and lipid nanoparticles (LNP) formulation
siRNAs targeting mouse Mst1, Mst2, NF2, Yap1 were designed and screened as previously described(24). The sequences and IC50 values of each siRNA are provided in Supplementary Table 1. The RNA strands were synthesized, characterized and duplexed by Alnylam Pharmaceuticals as previously described(24). Individual siRNAs were formulated into lipid nanoparticles and mixed prior to injections(23).
Cell culture
Hepa1-6, NIH3T3, AML-12 cells were obtained from ATCC and were propagated in DMEM supplemented with 10% FBS. Cells were transfected with siRNA using either Lipofectamine RNAiMAX (Invitrogen) or LNP with siRNA as described elsewhere(24).
AML-12 cells obtained from ATCC were grown on BD BioCoat collagen I coated plasticware. Cells were treated with taurocholic acid (Sigma-Aldrich, time and dose as indicated). Cells were collected in RIPA buffers supplemented with proteases and phosphatases inhibitors (Pierce Bio) for western blot, or fixed with 2% buffered paraformaldehyde solution, followed by permeabilization with 1% Triton X-100 in PBS for immunocytochemistry.
Animals
C57BL/6 mice were purchased from Charles River laboratories. p53LSL/LSL mice were published elsewhere(29). All animals received humane care, and animal protocols were approved by the Committee on Animal Care at MIT and the Institutional Animal Care and Use Committee of Alnylam Pharmaceuticals, certified by the American Association for Accreditation of Laboratory Animal Care. After 3 days of acclimatization in the animal facility, 7-9 week-old mice were injected via tail vein (i.v.) with either PBS or siRNA in LNP formulations at various concentrations. To restore p53 expression in p53LSL/LSL mice, animals were treated with tamoxifen (two intraperitoneal doses) before the siRNA treatment. Animals were sacrificed by CO2 overdose; tissues were harvested at different time points as indicated. Hepatocytes, stellate cells and Kupffer cells were isolated from C57BL/6 mice via collagenase perfusion, density centrifugation and antibody selections as described previously(30-32).
Histological, immunohistological and immunocytochemical analysis
Mouse tissues were fixed in 4% paraformaldehyde. Tissue sections were stained according to standard immunohistochemistry protocols as previously described(33) or with secondary antibodies labelled with Alexa 488, Alexa 555 and Alexa 647 (Invitrogen) to visualize antigen localization. We have used the following primary antibodies: anti-Cytokeratin 18 (Progen), anti-Cytokeratin 19 (Abcam), anti-E-cadherin (BD Biosciences), anti-F4/80 (Biolegends), anti-glutamine synthase (BD Biosciences), anti-Ki67 (Neomarkers), anti-p21 (Santa Cruz), anti-p53 (Leica Biosystems), anti-Yap1 (Cell Signaling), phospho-pH2A.X (Cell Signaling). Phalloidin coupled with Alexa555 was used to visualize F-actin. Analysis of H&E, immunohistochemistry and immunocytochemical images was performed using ImageJ package (NIH).
Western blots and quantification
The liver tissues were homogenized in RIPA buffer (Thermo Fisher) to harvest proteins. Total protein was resolved on TGX gradient gels (BioRad). The following primary antibodies were used: anti-β-actin (Sigma), anti-a-catenin (Cell Signaling), anti-cyclin D1 (Millipore), anti-Mst1 (Cell Signaling), anti-Mst2 (Cell Signaling), anti-NF2 (Sigma), anti-p21 (Santa Cruz), anti-pYap1 (Cell Signaling), anti-Yap1, phospho-pH2A.X. (Abcam) Licor Odyssey Imaging system was used to visualize protein bands. Gray scale images were quantified with ImageJ as described previously(24).
Gene expression analysis
The levels of mRNAs were measured by branched DNA assay (Affymetrix) or quantitative PCR using TaqMan probes and Roche LightCycler 480. Levels of mRNA of the genes-of-interest were normalized to the levels of GAPDH mRNA and then to the average arbitrary value of the control group.
Serum chemistry
Albumin, total protein, glucose, HDL, LDL, direct and total bilirubin, AST, ALT, total bile acids were measured in serum using Beckman Coulter reagents and Olympus Au400 autoanalyser.
Statistical analysis
P values were calculated using the Student's t-tests and the One-Way ANOVA in Prism 5 (GraphPad).
Results
siRNA mediated knockdown of Mst1, Mst2 and NF2 leads to hepatomegaly
We designed and screened sets of chemically modified siRNAs(34) targeting Mst1 and Mst2 (Supplementary Fig. 1). Specifically, 27 siRNAs targeting Mst1 and 28 siRNAs targeting Mst2 were synthesized. The gene silencing efficiency for each siRNA was examined in NIH3T3 cells at 5nM (Supplementary Fig. 1a, c). We found that nine siRNAs targeting Mst1 and nine siRNAs targeting Mst2 can achieve deep knockdown at this dose (Supplementary Fig. 1a, c). A dose response study was performed using the 3 most potent siRNAs for each gene. The IC50 for the most potent siRNA targeting Mst1 and Mst2 were ~0.05nM and 0.2nM, respectively (Supplementary Fig. 1b, d). The most efficient siRNAs were chosen and formulated into lipid nanoparticles (LNP), which have shown hepatocyte specific targeting with high potency(23). Approximately 90% knockdowns of both Mst1 and Mst2 mRNAs were shown in hepatocytes isolated from mice with treatment of LNP-formulated siRNAs against Mst1 (termed as si-Mst1) or Mst2 (termed as si-Mst2), respectively (Supplementary Fig. 2). Animals treated with si-Mst1 alone, si-Mst2 alone, or their combination showed no liver growth (Supplementary Fig. 3). PBS and LNP-formulated luciferase siRNA (termed as si-Control) treated mice served as controls. This result suggested that the remaining expression of Mst1 and Mst2 (~10%) was sufficient to maintain Yap1 inactivation. To further inhibit the Hippo pathway, specific siRNAs against NF2, which is a key protein for maintaining activity of this pathway(7, 8), were developed and screened (Supplementary Fig. 4). We synthesized 27 siRNAs targeting NF2. The gene silencing efficiency for individual siRNA was examined, and five efficient siRNAs were identified (Supplementary Fig. 4a). The IC50 for the most potent siRNA targeting NF2 was ~0.02nM (Supplementary Fig. 4b). LNP-formulated siRNA against NF2 (termed as si-NF2) showed efficient knockdown in vivo (Supplementary Fig. 5). Animals treated with a combination of si-Mst1, si-Mst2 and si-NF2 twice per week for two weeks at the dose of 0.67 mg/kg for each siRNA (hereafter, designated as triple siRNAs nanoparticle) showed 2.5 folds induction of liver size at Day 15 after the first injections of triple siRNAs nanoparticle (Fig 1). Similar results were obtained with alternative siRNAs targeting Mst1/Mst2/NF2 (Supplementary table 1, and data not shown).
Figure 1. Treatment of lipid-based nanoparticles (LNP)-formulated siRNAs targeting Mst1, Mst2 and NF2 (Triple siRNAs) induced hepatomegaly in mice.
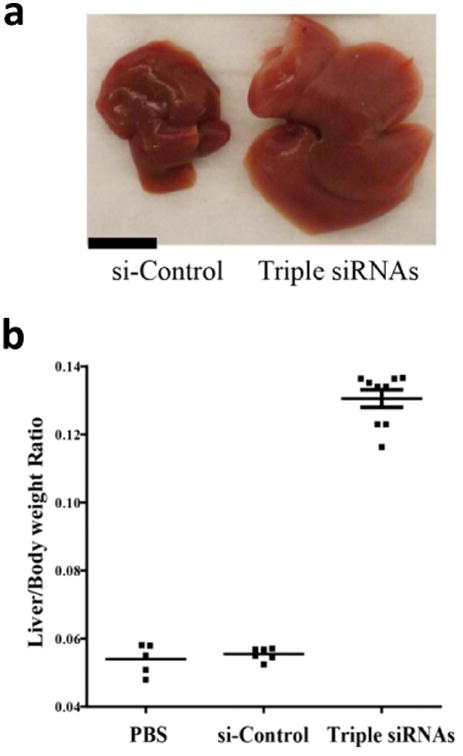
C57BL/6 female mice were treated with triple siRNAs, dose matched LNP-siRNA against the luciferase gene (si-Control), or PBS as controls. Livers were taken 15 days after the first injection. a, Macroscopic views of livers. Scale bar is equal to 1 cm. b, Maximal induction of liver size in triple siRNA treated mice. Error bars represent standard error (SEM), p<0.01.
To characterize this model, different combinations and doses of these siRNAs were examined. Combinations of si-Mst1 and si-NF2, or si-NF2 alone did not induce growth, while the combination of si-Mst2 and si-NF2 induced only minor liver growth (Supplementary Fig. 6a). Once we confirmed that all three siRNAs were necessary for induction of the hepatomegaly, we performed dose titration for each siRNA in vivo (Supplementary Fig. 6b-d). Based on these results, we optimized the dose for triple siRNA treatment as 0.3 mg/kg for si-Mst1, 0.6 mg/kg for si-Mst2 and 0.2 mg/kg for si-NF2 (Supplementary Fig. 6e). Our experiments described below used this combined dose.
We further characterized the time course of liver growth induced by siRNA. We found that the growth started at Day 9 and peaked at Day 15 after the first injection of triple siRNAs nanoparticle (4 injections in total) (Fig. 2a). After withdrawal of triple siRNAs nanoparticle treatment, the liver shrank back to its original size at Day 40 (Fig. 2a). In order to examine whether apoptosis is a key mechanism of liver shrinkage, we performed TUNEL assay and western blots of caspases. We found that in contrast with normal livers and enlarged livers, the shrinking livers had a significantly higher number of TUNEL-positive cells (Supplementary Fig. 7a). Furthermore, cleaved caspase 3 and 8 were significantly upregulated in the shrinking livers (Supplementary Fig. 7b). Our data is consistent with a previous study showing that conditional inactivation of Yap1 in enlarged mouse liver led to apoptosis of hepatocytes(10).
Figure 2. In vivo treatment with triple siRNAs induced proliferation of hepatocytes and activation of Yap1.
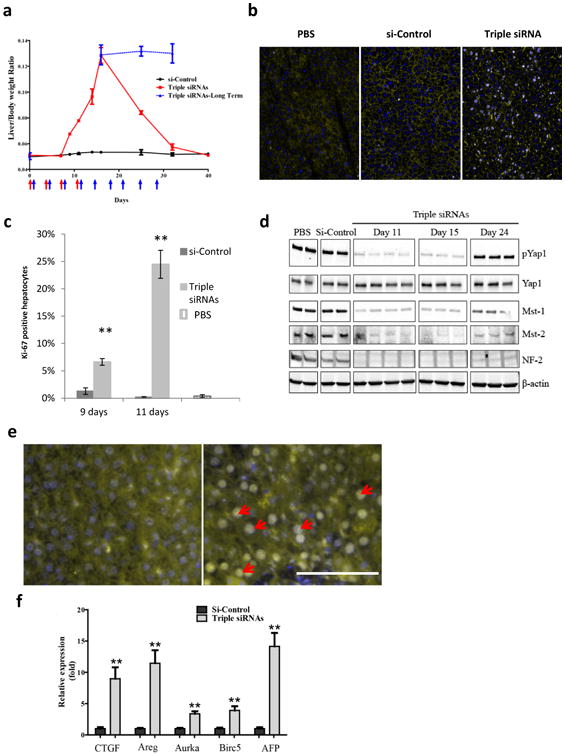
C57BL/6 female mice were treated with triple siRNAs or dose matched si-Control. Livers were taken at Day 7, 9, 11, 13, 15, 24, 32 or 40 days after the first injection. a, Liver/body weight ratio at different time points. Error bars represent SEM, (N≥3). b, Immunohistochemical analysis of hepatocyte proliferation (Day 11). Ki-67 (grey), Phalloidin (yellow), Hoechst (blue). c, Quantification of (b). Error bars represent SEM, **, p<0.01 (N=3). d, Western blot analysis of Hippo pathway signalling. e, Triple siRNA treatment changed Yap1 localization in liver (Day 11). Yap1 (yellow), nuclei (blue). f, Expression of Yap1-dependent genes were induced by triple siRNAs (Day 11). Error bars represent SEM, **, p<0.01 (N=3).
Remarkably, liver growth induced by the triple siRNAs nanoparticle treatment has an upper limit: prolonged treatment with siRNAs for an additional one or two weeks (up to 9 injections in total, 2 injections per week, 1.1 mg/kg per injection) did not induce further growth of the liver (Fig. 2a), indicating the possibility of mechanism(s) to restrict Yap1-induced hepatomegaly.
To clarify the targeted cell types in liver, mice were treated with siRNA nanoparticles against triple siRNAs. Hepatocytes, Kupffer cells, stellate cells and hepatic mononuclear cells were isolated from the livers. We found that with the formulation (C12-200) and siRNA dose (1.1mg/kg in total) we used, the knockdown effect is efficient (~90%) in hepatocytes (Supplementary Fig. 8a). In contrast, no significant knockdown was observed in Kupffer cells, stellate cells and hepatic mononuclear cells. We previously showed that our siRNA lipid formulation causes deep silencing of integrin β1 in hepatocytes, while no significant knockdown was found in CD31+, aSMA+, F4/80+ cells(35). Because it is difficult to get a specific commercially available antibody for the separation of biliary epithelial cells, we performed an alternative approach to explore whether our lipid nanoparticle could target biliary epithelial cells. We designed and screened a highly potent siRNA against Cytokeratin 19 (CK19), a specific marker for biliary epithelial cells. We found that this potent siRNA knockdown of CK19 has an IC50 of 0.07nM in vitro (Supplementary Fig. 8b). We formulated it into lipid nanoparticles and treated mice. We found it could not induce any silencing of CK19 in mouse liver (Supplementary Fig. 8c). These data indicate that our lipid nanoparticles cannot induce gene knockdown in biliary epithelial cells.
Knockdown of Mst1, Mst2 and NF2 reproduces major features of Yap1 hyperactivation
To explore whether triple siRNAs nanoparticle-induced liver growth was due to cell division, we analyzed the proliferation of hepatocytes using ki-67 staining. We found that proliferation started at Day 7 and peaked at Day 11, at which point about 25% of total hepatocytes were ki-67-positive (Fig. 2b and c). We consistently found that expression of related genes to cell cycle was significantly induced (Supplementary Fig. 9a). Furthermore, we determined whether the proliferation of hepatocytes is due to loss of expression of components in the Hippo pathway and activation of Yap1. The protein levels of Mst1, Mst2 and NF2 in livers of triple siRNAs nanoparticle treated animals were examined by immunoblotting. We observed deep reduction of each protein at Day 11 and 15 (Fig. 2d). In consequence, triple siRNAs nanoparticle treatment diminished phospho-Yap1 (p-Yap1) by about 95% in total liver lysates (Fig. 2d and Supplementary Figure 10), while the combination of si-Mst1 and si-Mst2 treatment only decreased p-Yap1 level by about 50% (Supplementary Fig. 10). Consistently, we observed Yap1 accumulation in the nuclei of hepatocytes in liver sections after treatment with triple siRNAs nanoparticle (Fig. 2e). Interestingly, 12 days after withdrawal of triple siRNAs nanoparticle treatment (as Day 24), Mst1, Mst2, and NF2, as well as p-Yap1 recovered, indicating the feasibility of reversible manipulation of Hippo pathway (Fig. 2d). Furthermore, we found the up-regulation of Yap signature genes including CTGF, Areg, Aurka, Birc5 and AFP (Fig. 2f and Supplementary Fig. 9b,c). In addition, we observed that the growth of the liver was zone dependent. The periportal area, which contains the highest levels of nutrients and oxygen(36), exhibited the most significant expansion, whereas the pericentral area had minimal expansion (Supplementary Fig. 11a). Finally, to rule out the possibility of the involvement of macrophages in liver growth(31), we depleted hepatic macrophages using Clodronate Liposomes(30). We found that triple siRNAs nanoparticle-induced similar growth of liver in control and macrophage-depleted mice (Supplementary Fig. 12).
To analyze functions of enlarged livers, we performed analysis of serum biochemistry (Table 1). We found no significant difference in levels of ALT and AST between PBS, control siRNAs and triple siRNA nanoparticle treatment, indicating integrity of hepatocytes. Meanwhile, other indicators showed pronounced hallmarks of cholestasis, including increased levels of alkaline phosphatase, cholesterol, low density lipoproteins, bilirubin, and bile acids. To further characterize the changes in the liver related to bile flow, we performed immunohistochemical staining of CK19 to visualize bile ducts and gene expression analysis of several genes related to bile acid metabolism and transport. The number of CK19 positive cells and the density of bile ducts were significantly increased after triple siRNAs treatment (Supplementary Fig. 14). This expansion, together with altered gene expression levels, e.g. NTCP, Abcg5, MDR1, MRP4, and Cyp7 (Supplementary Fig. 9d), indicates that triple siRNA treatment induced remarkable cholestasis.
Table 1. Parameters of serum chemistry in mice treated with different combinations of siRNA targeting Hippo pathway.
| PBS | si-Control | Triple siRNA | si-Mst2+si-NF2 | si-Mst1+si-NF2 | |
|---|---|---|---|---|---|
| Albumin, g/dl | 2.90±0.12 | 2.89±0.22 | 3.31±0.15* | 3.14±0.17# | 2.96±0.11 |
| Total protein, g/dl | 4.7±0.1 | 4.9±0.2 | 5.5±0.2** | 5.2±0.2## | 5.0±0.2 |
| Globulin, g/dl | 1.82±0.08 | 2.06±0.05 | 2.24±0.11* | 2.1±0.07# | 2.06±0.11 |
| Blood urine nitroqen, mg/dl | 19.2±2.3 | 20.5±2.5 | 18.5±1.9 | 21.4±2.8 | 19.4±1.1 |
| Alkaline phosphatase (ALP), IU/I | 101.5±14.4 | 116.3±17.9 | 209.2±87.4* | 125.8±31.5# | 108.7±10.9# |
| ALT (SGPT), IU/I | 19±2.4 | 24.1 ±8.9 | 50.9±15.7 | 52.9±28.7# | 19.8±1.6 |
| AST (SGOT), IU/I | 47.8±16.8 | 67.4±29.1 | 65.9±3.2 | 83.5±12.1 | 59.7±11.4 |
| Direct bilirubin, mg/dl | 0.04±0.024 | 0.023±0.015 | 0.228±0.189* | 0.042±0.017# | 0.023±0.011# |
| Total bilirubin, mg/dl | 0.178±0.046 | 0.140±0.036 | 0.448±0.244* | 0.186±0.032## | 0.132±0.013# |
| Total bile acids, mg/dl | 6.1±1 | 8.9±2.6 | 91.4±33*** | 25.5±5.6### | 15.1±1.8### |
| Cholesterol, mg/dl | 68.5±10.4 | 76.3±10.2 | 125.6±20.4*** | 87.2±6.6### | 88.7±5### |
| Triglycerides, mg/dl | 90.9±24.3 | 80.5±18.2 | 90.2±10.4 | 45.4±4.7*,# | 70.3±7.5 |
| HDL, mg/dl | 39.4±6.9 | 39.5±6.7 | 39.1 ±2.4 | 32.3±2 | 42.6±1.8 |
| LDL, mg/dl | 6.1±0.6 | 9.9±0.8 | 27.6±4.5*** | 23.3±1.9*** | 14.8±2.6### |
| Glucose, mg/dl | 231.9±46.8 | 233.4±22.4 | 152.5±15.7* | 208.3±34.8 | 214.3±48.1 |
- p<0.05, p<0.01, p<0.001 comparison vs si-Control
- p<0.05, p<0.01, p<0.001 comparison vs Triple siRNA
The triple siRNA induced hepatomegaly is Yap1 dependent
To validate Yap1-dependent mechanism of the observed phenotype, we depleted Yap1 in triple siRNAs nanoparticle treated animals. siRNAs against Yap1 (si-Yap1) were designed and screened (Supplementary Fig. 11). We synthesized 24 siRNAs targeting Yap1. The gene silencing efficiency for individual siRNA was tested and five efficient siRNAs were identified (Supplementary Fig. 13a). The IC50 for the most potent siRNA targeting Yap1 was ~0.03nM (Supplementary Fig. 13b). We co-treated animals with LNP-formulated si-Yap1 with triple siRNAs nanoparticle. We found that si-Yap1 completely blocked the liver growth induced by triple siRNAs nanoparticle (Fig. 3a), demonstrating that the hepatomegaly induced by triple siRNAs nanoparticles are Yap1-mediated. Co-treatment of si-Yap1 with triple siRNAs nanoparticles diminished both pYap1 and Yap1 expression (Fig. 3b), thereby blocking the proliferation of hepatocytes, as indicated by diminished expression of cell cycle related genes (Fig. 3b and c). Consistently, co-treatment with si-Yap1 also abolished the manifestation of cholestasis associated markers (Supplementary Table 2).
Figure 3. Additional siRNA treatment proved genetic interactions.
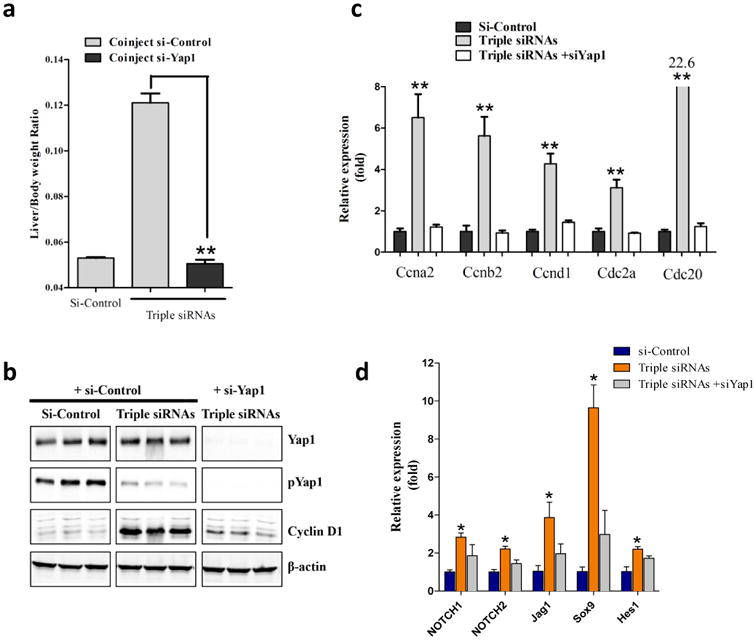
C57BL/6 female mice were treated with dose matched si-control, triple siRNAs plus si-Control, or triple siRNAs plus si-Yap1. Livers were taken 11 or 15 days after the first injection. a, weight at Day 15 was recorded. Error bars represent SEM, **, p<0.01 (N≥5). b, Liver lysate was made and western blot performed. c, Analysis of cell cycle related genes in liver tissues (Day 11) by qPCR. Error bars represent SEM, **, p<0.01 (N=3). d, YAP activation induces Notch pathway signaling mRNA expression of Notch pathway genes was determined by RT-qPCR in siLUC, Triple siRNA, or Triple siRNA+siYAP treatment groups,*p<0.05. N=4
A previous study showed that activation of Yap induces Notch pathway activation with expansion of hepatic progenitor cells(37). To explore whether activation of Yap induces Notch pathway activation in the triple siRNA-nanoparticles treated mouse liver, we performed a panel of qPCR analyses for Notch pathway. We found that Notch pathway signature genes including NOTCH1, NOTCH2, Jag1, SOX9, and HES were all significantly upregulated in triple siRNA treated livers (Fig 3d). Further, such induction was abolished by in vivo Yap1 siRNA treatment (Fig 3d). Notch pathway has been suggested as an inducer of biliary specification(38). This is consistent with the expansion of CK19 positive cells in triple siRNA-nanoparticles treated mouse livers (Supplementary Fig. 14).
p53 controls Yap1-induced liver growth
To further characterize the effects of hepatomegaly, we analyzed gene expression patterns of livers following long-term triple siRNAs nanoparticle treatment (Fig. 2a). Intriguingly, we observed the up-regulation of cell cycle checkpoint genes (Supplementary Fig. 7c) and apoptosis related genes (Supplementary Fig. 7e) with treatment of triple siRNAs nanoparticles. Importantly, we noticed significant induction of the cyclin-dependent kinase inhibitor 1a (p21) (Fig 4a and Supplementary Figure 15a). Withdrawal of treatment of triple siRNAs nanoparticle led to diminished p21 expression (Supplementary Fig. 15a). Moreover, co-treatment of si-Yap1 with triple siRNAs nanoparticle abolished p21 induction (Supplementary Fig. 15b), suggesting that such induction is mediated by Yap1, directly or indirectly. Based on these results, we hypothesized that the tumor suppressor protein p53, an essential regulator of p21 transcription(39), could be activated as a result of triple siRNAs nanoparticles treatment. Indeed, the pronounced nuclear staining of p53 indicated its activation with treatment of triple siRNAs nanoparticles (Fig. 4b). To investigate potential interaction between the Hippo pathway and p53, we compared the phenotype of triple siRNAs nanoparticles treatment between wildtype mice and p53LSL/LSL mice, which harbor a Lox-STOP-Lox cassette in the first intron of p53 and are phenotypically equivalent to p53 null strains(29). In contrast to that liver of wildtype mice, the livers of p53LSL/LSL mice grew much faster than wildtype mice and exceeded the upper limit of growth (Fig. 5a, b). Remarkably, more than 60% of hepatocytes were ki-67 positive in p53LSL/LSL mice at Day 14, which was near 3 folds higher than in wildtype mice (Fig. 5c, d). The H&E staining indicates more severe histological abnormalities in p53LSL/LSL mice than in wildtype animals with triple siRNAs treatment, such as a higher degree of pleomorphism, extreme polyploidy, dysplasia of hepatocytes (likely due to swelling with water/fluids), aneuploidy in a portion of hepatocytes, and intranuclear cytoplasmic inclusions (Fig 5e). To determine wehther a major part of ki-67 positive cells were indeed from hepatocytes, we performed double-staining for keratin 18 (CK18) and ki-67. We found that a major part of the ki-67 cells in both wildtype p53 and p53 KO were also CK18 positive (Fig 5f), indicating that hepatocytes were under active proliferation. To rule out the possibility of different levels of knockdown, we measured the levels of Mst1, Mst2, NF2, and pYap1 and found no differences between wildtype mice and p53LSL/LSL mice Supplementary Fig. 15c). To further prove that p53 plays a role in controlling liver growth, we restored p53 by Tamoxifen treatment in p53LSL/LSL; Cre-ER mice, whose stop cassette in p53 gene can be removed by Cre recombinase. We found that restoration of p53 significantly suppressed liver growth induced by triple siRNAs nanoparticle treatment (Fig. 5e and Supplementary Fig. 15d). Collectively, these results indicate that p53 is activated upon triple siRNAs nanoparticle induced hepatomegaly, and loss of p53 permits further expansion of liver size.
Figure 4. p53 was activated in livers of triple siRNAs treated animal.
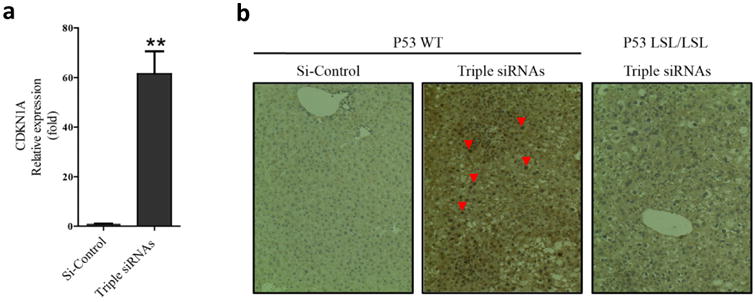
C57BL/6 mice and p53LSL/LSL were treated with triple siRNAs or dose matched si-Control. a, The levels of CDKN1A (P21) mRNA in livers of C57BL/6 mice treated with si-Control or triple siRNAs(Day 14). Error bars represent SEM, **, p<0.01 (N=5). b, p53 staining in liver sections. Red arrowheads indicate nuclear accumulation of p53.
Figure 5. p53 plays an important role in restraining liver growth.
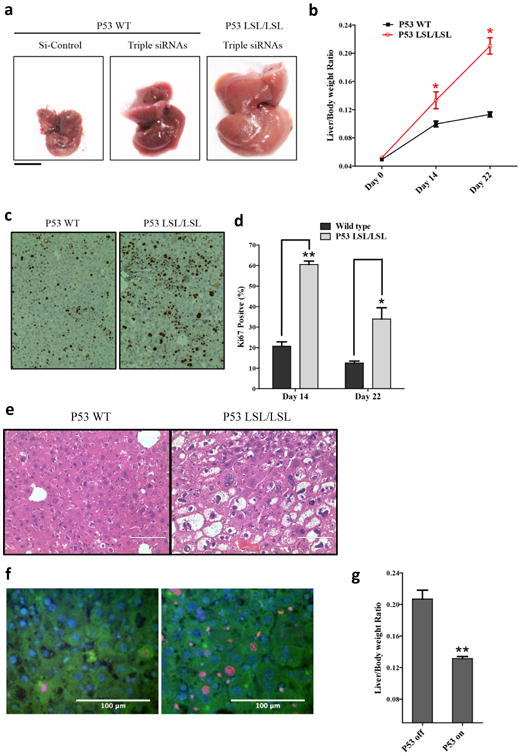
a, Macroscopic views of livers (Day 22). b, Liver/body weight ratio at different time points. Error bars represent SEM, *, p<0.05 (N=4). c, Hepatocyte proliferation evidenced by ki-67 staining (Day 14). d, Quantification of ki-67+ cells. Error bars represent SEM, **, p<0.01, *, p<0.05 (N=3). e, H&E staining of the liver sections (Day 22). f, immunofluorescent staining of ki-67 (red) and keratin 18 (green) of the liver sections (Day 22)., scale bars = 100µm. g, p53LSL/LSL; CreER mice treated with tamoxifen (p53 on) or vehicle controls (p53 off). Liver weight was measured 22 days after the first dose of triple siRNAs. Error bars represent SEM, **, p<0.01 (N≥3).
We have hypothesized that p53 may be activated due to DNA damage caused by the elevated levels of bile acids, a result of cholestasis associated with Yap1-induced hepatomegaly. Using the phosphorylated histone protein (H2A.X) as an indicator of DNA damage(40) we observed that triple siRNA-induced hepatomegaly led to DNA damage (Fig. 6). The intensity of the histone phosphorylation correlated to both the degree of hepatomegaly and p21 levels, i.e. maximal levels of phosphor-H2A.X were observed in the groups treated with triple siRNA; in contrast, co-treatment with triple siRNAs and si-Yap1 did not upregulate phospho-H2A.X. It suggests that p53 activation observed in this model is by activation of checkpoint cascades to DNA damage. To explore this mechanism further, we tested the direct effect of bile acids on p21 activation in murine hepatocyte-derived cell line AML-12. We treated cells with taurocholic acid, a conjugated primary bile salt which is abundant in mice with cholestasis(41). The taurocholic acid treatment led to, strong upregulation of p21 mRNA levels and nuclear accumulation of p21 as well as p53 (Fig 7a and b). To determine whether TCA treatment induced DNA damage in AML12 cells, we treated AML12 cells with TCA, and performed TUNEL assays. We found TUNEL-positive cells appeared in TCA treated cells but not control cells (Fig 7c), indicating severe DNA damage in TCA treated cells.
Figure 6. DNA damage in enlarged liver.
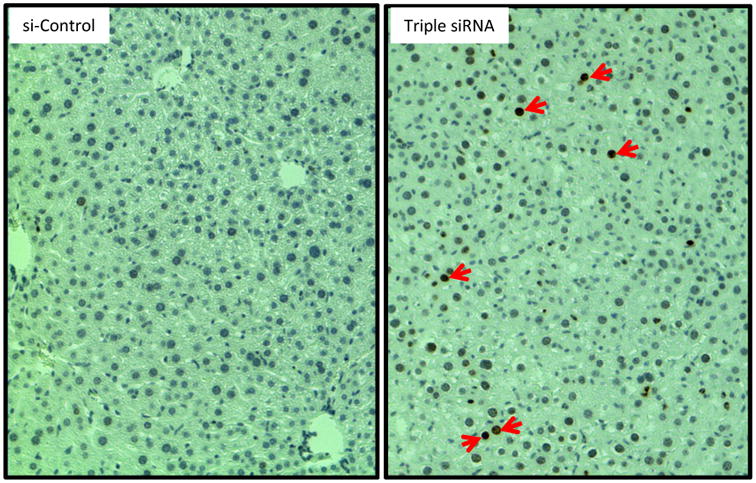
Livers were collected at Day 14 after the first injection of triple siRNAs. DNA damage was evidenced by phospho-H2A.X positive cells (indicated by red arrows).
Figure 7. Bile acids induce activity of p53-pathway in vitro.
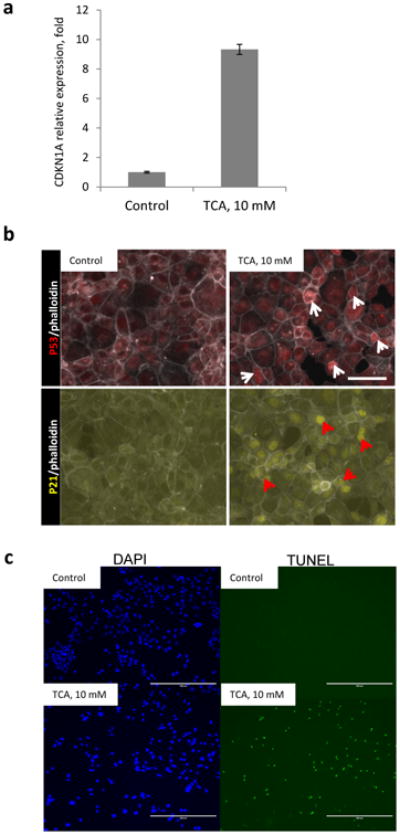
AML12 cells were treated with 10mM taurocholic acid (TCA) for 4 hours. Untreated cells serve as a control. a, mRNA levels of CDKN1A(p21) in AML12 cells. Error bars represent SEM. b, immunofluorescent staining, arrowheads indicate nuclear accumulation of p53 (white) and p21 (red). Bar indicates 100µm. C Cells were fixed and stained for TUNEL and counterstained with DAPI, scalebars = 400µm.
Discussion
Our data demonstrates that it is feasible to manipulate organ size reversibly with siRNA nanoparticles in vivo. Application of chemically modified synthetic siRNA does not lead to saturation of the RISC complex and induces minimal immunostimulatory response compared to viral delivery of shRNA(24, 42, 43). Inhibition at the mRNA level of multiple components of the Hippo pathway is sufficient to activate Yap1 in vivo, thereby inducing proliferation of hepatocytes and increasing liver size. Contrary to hepatomegaly induced by complete loss of Mst1/Mst2, one induced by siRNA treatment has an upper limit of growth (about 2.5 fold). Our data indicates that activation of p53 is a key regulator for limiting liver growth. While p53 signaling is essential in tumor suppression by coordinating multiple cellular processes, including activation of DNA repair proteins, arrest of cell cycle, and initiation of cellular apoptosis and senescence (reviewed in(44)), to our knowledge, our results provide the first demonstration that endogenous p53 plays a role in size regulation of the liver. Although, we cannot exclude completely direct activation of p53 by loss of Hippo pathway, e.g. feedback loop through Lats2(45, 46), our results suggest that siRNA mediated activation of Yap1 can lead to DNA damage response in the liver. Bile acids have been shown to induce DNA damage in a certain types of cells and cell death in hepatocytes(47-49). Concordant with previous observations, our data show that Yap1 activation leads to disruption of bile duct homeostasis(7) and provokes a cholestasis-like phenotype.
Crosstalk between Hippo and p53 pathways has been suggested in fly models as well as in mammalian cell line models(46, 50). Our approach allows investigation of pathway interactions in normal livers. It reveals indirect interactions between signaling pathways mediated by tissue remodeling and changes in composition of metabolites. Remarkably, not only is the proliferation rate of hepatocytes in p53LSL/LSL mice much higher than in the wildtype control, but also the size of hepatocytes in p53LSL/LSL mice is significantly enlarged (Fig. 5d; Supplementary Fig. 12c, d). It is noteworthy to investigate coordination of p53 and Hippo pathway during the process of regeneration and under the context of pathological conditions for future studies. It is known that abnormal bile acids homeostasis can activate p53 in the mouse liver (51). There is evidence indicating that Yap overactivation can lead to biliary epithelium over-proliferation and irregular shape(7). To our best knowledge, there is no report showing Yap overactivation provokes a cholestasis-like phenotype. Dysfunction of bile ducts may be mediated by Yap-dependent dysregulation of gene expression of bile acid transporters in hepatocytes(52) (Supplementary Fig. 9d) or indirect effects on the bile ducts function through modulation of paracrine/contact signaling between hepatocytes and cholangioytes (Fig. 3d). Extensive expansion of hepatocytes in the periportal zone may contribute to mechanical distortion of bile duct function. Our data suggests that Yap activation leads to abnormal bile acid homeostasis, which in turn triggers p53 activation (Fig 4, 6, 7). Furthermore, our data indicates that loss of p53 led to increased hepatocyte proliferation when Hippo pathway was suppressed (Fig 5). Previous studies reported that no obvious abnormal liver histology was found with short-term deletion of p53; in contrast, deletion of p53 induces liver cancer after 10-12 months in mouse model(53). Several studies showed that loss of p53 did not significantly accelerate phenobarbitone or diethylnitrosamine-induced hepatocellular carcinoma(54-56). Thus, our finding suggests an important connection between p53 and Hippo pathway, two key pathways for maintaining quiescence of hepatocytes. Although p21 is an important downstream effector of p53 and its induction is a recognized marker of p53 activation, the tumour suppressor p53 has a number of targets to exert its functions(57). The p21-deficient mice have a less obvious phenotype than p53-deficient animals(58).
We have shown that siRNA nanoparticle-mediated control of organ size is reversible, as indicated by the shrinking of liver size and restoration of pYap1 after withdrawal of triple siRNAs nanoparticle-treatment (Fig. 2a and d). Livers shrank back to their original size, indicating an intrinsic mechanism for regulating normal organ size. The technique presented in our manuscript provided a flexible way to induce transient gene inactivation and subsequent ability to study the mechanism of liver size control in a new manner. A number of questions remain unclear regarding the process of how the liver returns to the exact normal size, including which signals trigger and stop the apoptosis of hepatocytes, and why only part of cells are undergoing apoptosis. It is possible that multiple factors are involved in the process in the liver, including mechanical forces, oxygen levels and surface and nuclear receptors. The method described here allows dissection of these factors in vivo in a flexible and convenient way. A recent study indicated that a compensatory network can be activated to buffer against deleterious mutations rather than transcriptional knockdown(59). Thus, an in vivo genetic knockdown approach may reveal phenotypes which can not be observed by a knockout approach. As loss of the Hippo pathway is temporary, and the dose of siRNAs can be titrated to induce temporary proliferation in a portion of hepatocytes without substantial increase of organ size (data not shown), we believe that modulating liver size in vivo with siRNA-nanoparticles may provide an efficient tool to explore autonomous and non-autonomous mechanisms for organ size regulation.
Supplementary Material
Acknowledgments
We thank T. Jacks, A. Sachinidis, K. Meganathan and K. Natarajan for providing experimental materials and critical review of the manuscript. We thank technical support from Swanson biotechnology Center at Koch Institute for Integrative Cancer Research. This work was supported in part by the Koch Institute Support (core) Grant P30-CA14051 from the National Cancer Institute. This work was supported by Alnylam and NIH RO1-DE016516. H.Y. and R.L.B are supported by 5-U54-CA151884-04 NIH Centers for Cancer Nanotechnology Excellence and the Harvard-MIT Center of Cancer Nanotechnology Excellence. W.X. is supported by NIH 5R00CA169512 (to W.X.). V.K. acknowledges support from the Russian scientific fund, grant number 14-34–00017. The authors acknowledge the service to the MIT community of the late Sean Collier.
Footnotes
Author Contributions: H.Y., R.L.B., V.K. and D.G.A. directed the project. H.Y. and R.L.B. designed and performed experiments, and analyzed the data. H.Y. and R.L.B. wrote the manuscript. C.B., S.W, I.Z., V.R., H.N., S.K., L.N., A.A., W.X. and M.Z. performed experiments and analyzed data. All authors participated in data discussion and the manuscript editing.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tumaneng K, Russell RC, Guan KL. Organ size control by Hippo and TOR pathways. Curr Biol. 2012;22:R368–379. doi: 10.1016/j.cub.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stanger BZ. The biology of organ size determination. Diabetes Obes Metab. 2008;10(Suppl 4):16–22. doi: 10.1111/j.1463-1326.2008.00938.x. [DOI] [PubMed] [Google Scholar]
- 3.Shingleton AW. The regulation of organ size in Drosophila: physiology, plasticity, patterning and physical force. Organogenesis. 2010;6:76–87. doi: 10.4161/org.6.2.10375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pan D. The hippo signaling pathway in development and cancer. Dev Cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao B, Tumaneng K, Guan KL. The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nat Cell Biol. 2011;13:877–883. doi: 10.1038/ncb2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu FX, Guan KL. The Hippo pathway: regulators and regulations. Genes Dev. 2013;27:355–371. doi: 10.1101/gad.210773.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang N, Bai H, David KK, Dong J, Zheng Y, Cai J, Giovannini M, et al. The Merlin/NF2 tumor suppressor functions through the YAP oncoprotein to regulate tissue homeostasis in mammals. Dev Cell. 2010;19:27–38. doi: 10.1016/j.devcel.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yin F, Yu J, Zheng Y, Chen Q, Zhang N, Pan D. Spatial organization of Hippo signaling at the plasma membrane mediated by the tumor suppressor Merlin/NF2. Cell. 2013;154:1342–1355. doi: 10.1016/j.cell.2013.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 10.Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, et al. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yagi R, Chen LF, Shigesada K, Murakami Y, Ito Y. A WW domain-containing yes-associated protein (YAP) is a novel transcriptional co-activator. Embo j. 1999;18:2551–2562. doi: 10.1093/emboj/18.9.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vassilev A, Kaneko KJ, Shu H, Zhao Y, DePamphilis ML. TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes Dev. 2001;15:1229–1241. doi: 10.1101/gad.888601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oh S, Lee D, Kim T, Kim TS, Oh HJ, Hwang CY, Kong YY, et al. Crucial role for Mst1 and Mst2 kinases in early embryonic development of the mouse. Mol Cell Biol. 2009;29:6309–6320. doi: 10.1128/MCB.00551-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song H, Mak KK, Topol L, Yun K, Hu J, Garrett L, Chen Y, et al. Mammalian Mst1 and Mst2 kinases play essential roles in organ size control and tumor suppression. Proc Natl Acad Sci U S A. 2010;107:1431–1436. doi: 10.1073/pnas.0911409107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou D, Conrad C, Xia F, Park JS, Payer B, Yin Y, Lauwers GY, et al. Mst1 and Mst2 maintain hepatocyte quiescence and suppress hepatocellular carcinoma development through inactivation of the Yap1 oncogene. Cancer Cell. 2009;16:425–438. doi: 10.1016/j.ccr.2009.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu L, Li Y, Kim SM, Bossuyt W, Liu P, Qiu Q, Wang Y, et al. Hippo signaling is a potent in vivo growth and tumor suppressor pathway in the mammalian liver. Proc Natl Acad Sci U S A. 2010;107:1437–1442. doi: 10.1073/pnas.0911427107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tumaneng K, Schlegelmilch K, Russell RC, Yimlamai D, Basnet H, Mahadevan N, Fitamant J, et al. YAP mediates crosstalk between the Hippo and PI(3)K-TOR pathways by suppressing PTEN via miR-29. Nat Cell Biol. 2012;14:1322–1329. doi: 10.1038/ncb2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Varelas X, Miller BW, Sopko R, Song S, Gregorieff A, Fellouse FA, Sakuma R, et al. The Hippo pathway regulates Wnt/beta-catenin signaling. Dev Cell. 2010;18:579–591. doi: 10.1016/j.devcel.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 19.Xin M, Kim Y, Sutherland LB, Qi X, McAnally J, Schwartz RJ, Richardson JA, et al. Regulation of insulin-like growth factor signaling by Yap governs cardiomyocyte proliferation and embryonic heart size. Sci Signal. 2011;4:ra70. doi: 10.1126/scisignal.2002278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strassburger K, Tiebe M, Pinna F, Breuhahn K, Teleman AA. Insulin/IGF signaling drives cell proliferation in part via Yorkie/YAP. Dev Biol. 2012;367:187–196. doi: 10.1016/j.ydbio.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 21.Fernandez LA, Northcott PA, Dalton J, Fraga C, Ellison D, Angers S, Taylor MD, et al. YAP1 is amplified and up-regulated in hedgehog-associated medulloblastomas and mediates Sonic hedgehog-driven neural precursor proliferation. Genes Dev. 2009;23:2729–2741. doi: 10.1101/gad.1824509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dong Y, Love KT, Dorkin JR, Sirirungruang S, Zhang Y, Chen D, Bogorad RL, et al. Lipopeptide nanoparticles for potent and selective siRNA delivery in rodents and nonhuman primates. Proc Natl Acad Sci U S A. 2014 doi: 10.1073/pnas.1322937111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Love KT, Mahon KP, Levins CG, Whitehead KA, Querbes W, Dorkin JR, Qin J, et al. Lipid-like materials for low-dose, in vivo gene silencing. Proc Natl Acad Sci U S A. 2010;107:1864–1869. doi: 10.1073/pnas.0910603106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zeigerer A, Gilleron J, Bogorad RL, Marsico G, Nonaka H, Seifert S, Epstein-Barash H, et al. Rab5 is necessary for the biogenesis of the endolysosomal system in vivo. Nature. 2012;485:465–470. doi: 10.1038/nature11133. [DOI] [PubMed] [Google Scholar]
- 25.Baughman JM, Perocchi F, Girgis HS, Plovanich M, Belcher-Timme CA, Sancak Y, Bao XR, et al. Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter. Nature. 2011;476:341–345. doi: 10.1038/nature10234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coelho T, Adams D, Silva A, Lozeron P, Hawkins PN, Mant T, Perez J, et al. Safety and efficacy of RNAi therapy for transthyretin amyloidosis. N Engl J Med. 2013;369:819–829. doi: 10.1056/NEJMoa1208760. [DOI] [PubMed] [Google Scholar]
- 27.Leuschner F, Dutta P, Gorbatov R, Novobrantseva TI, Donahoe JS, Courties G, Lee KM, et al. Therapeutic siRNA silencing in inflammatory monocytes in mice. Nat Biotechnol. 2011;29:1005–1010. doi: 10.1038/nbt.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dahlman JE, Barnes C, Khan OF. In vivo endothelial siRNA delivery using polymeric nanoparticles with low molecular weight. 2014;9:648–655. doi: 10.1038/nnano.2014.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, Newman J, et al. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- 30.Yin H, Cheng L, Holt M, Hail N, Jr, Maclaren R, Ju C. Lactoferrin protects against acetaminophen-induced liver injury in mice. Hepatology. 2010;51:1007–1016. doi: 10.1002/hep.23476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.You Q, Holt M, Yin H, Li G, Hu CJ, Ju C. Role of hepatic resident and infiltrating macrophages in liver repair after acute injury. Biochem Pharmacol. 2013;86:836–843. doi: 10.1016/j.bcp.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jeong WI, Park O, Radaeva S, Gao B. STAT1 inhibits liver fibrosis in mice by inhibiting stellate cell proliferation and stimulating NK cell cytotoxicity. Hepatology. 2006;44:1441–1451. doi: 10.1002/hep.21419. [DOI] [PubMed] [Google Scholar]
- 33.Xue W, Meylan E, Oliver TG, Feldser DM, Winslow MM, Bronson R, Jacks T. Response and resistance to NF-kappaB inhibitors in mouse models of lung adenocarcinoma. Cancer Discov. 2011;1:236–247. doi: 10.1158/2159-8290.CD-11-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M, Elbashir S, et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 35.Bogorad RL, Yin H, Zeigerer A, Nonaka H, Ruda VM, Zerial M, Anderson DG, et al. Nanoparticle-formulated siRNA targeting integrins inhibits hepatocellular carcinoma progression in mice. Nat Commun. 2014;5:3869. doi: 10.1038/ncomms4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cunningham CC, Van Horn CG. Energy availability and alcohol-related liver pathology. Alcohol Res Health. 2003;27:291–299. [PMC free article] [PubMed] [Google Scholar]
- 37.Yimlamai D, Christodoulou C, Galli GG, Yanger K, Pepe-Mooney B, Gurung B, Shrestha K, et al. Hippo pathway activity influences liver cell fate. Cell. 2014;157:1324–1338. doi: 10.1016/j.cell.2014.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sparks EE, Huppert KA, Brown MA, Washington MK, Huppert SS. Notch signaling regulates formation of the three-dimensional architecture of intrahepatic bile ducts in mice. Hepatology. 2010;51:1391–1400. doi: 10.1002/hep.23431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, et al. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 40.Lu C, Zhu F, Cho YY, Tang F, Zykova T, Ma WY, Bode AM, et al. Cell apoptosis: requirement of H2AX in DNA ladder formation, but not for the activation of caspase-3. Mol Cell. 2006;23:121–132. doi: 10.1016/j.molcel.2006.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y, Hong JY, Rockwell CE, Copple BL, Jaeschke H, Klaassen CD. Effect of bile duct ligation on bile acid composition in mouse serum and liver. Liver Int. 2012;32:58–69. doi: 10.1111/j.1478-3231.2011.02662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.John M, Constien R, Akinc A, Goldberg M, Moon YA, Spranger M, Hadwiger P, et al. Effective RNAi-mediated gene silencing without interruption of the endogenous microRNA pathway. Nature. 2007;449:745–747. doi: 10.1038/nature06179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grimm D, Wang L, Lee JS, Schurmann N, Gu S, Borner K, Storm TA, et al. Argonaute proteins are key determinants of RNAi efficacy, toxicity, and persistence in the adult mouse liver. J Clin Invest. 2010;120:3106–3119. doi: 10.1172/JCI43565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 45.Ganem NJ, Cornils H, Chiu SY, O'Rourke KP, Arnaud J, Yimlamai D, Thery M, et al. Cytokinesis failure triggers hippo tumor suppressor pathway activation. Cell. 2014;158:833–848. doi: 10.1016/j.cell.2014.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aylon Y, Michael D, Shmueli A, Yabuta N, Nojima H, Oren M. A positive feedback loop between the p53 and Lats2 tumor suppressors prevents tetraploidization. Genes Dev. 2006;20:2687–2700. doi: 10.1101/gad.1447006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Woolbright BL, Dorko K, Antoine DJ, Clarke JI, Gholami P, Li F, Kumer SC, et al. Bile acid-induced necrosis in primary human hepatocytes and in patients with obstructive cholestasis. Toxicol Appl Pharmacol. 2015;283:168–177. doi: 10.1016/j.taap.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ohtaki Y, Hida T, Hiramatsu K, Kanitani M, Ohshima T, Nomura M, Wakita H, et al. Deoxycholic acid as an endogenous risk factor for hepatocarcinogenesis and effects of gomisin A, a lignan component of Schizandra fruits. Anticancer Res. 1996;16:751–755. [PubMed] [Google Scholar]
- 49.Ajouz H, Mukherji D, Shamseddine A. Secondary bile acids: an underrecognized cause of colon cancer. World J Surg Oncol. 2014;12:164. doi: 10.1186/1477-7819-12-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang W, Cohen SM. The Hippo pathway acts via p53 and microRNAs to control proliferation and proapoptotic gene expression during tissue growth. Biol Open. 2013;2:822–828. doi: 10.1242/bio.20134317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang H, Li TW, Ko KS, Xia M, Lu SC. Switch from Mnt-Max to Myc-Max induces p53 and cyclin D1 expression and apoptosis during cholestasis in mouse and human hepatocytes. Hepatology. 2009;49:860–870. doi: 10.1002/hep.22720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Halilbasic E, Claudel T, Trauner M. Bile acid transporters and regulatory nuclear receptors in the liver and beyond. J Hepatol. 2013;58:155–168. doi: 10.1016/j.jhep.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Katz SF, Lechel A, Obenauf AC, Begus-Nahrmann Y, Kraus JM, Hoffmann EM, Duda J, et al. Disruption of Trp53 in livers of mice induces formation of carcinomas with bilineal differentiation. Gastroenterology. 2012;142:1229–1239. doi: 10.1053/j.gastro.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 54.Gould S, Sidaway J, Sansom N, Betton G, Orton T. Phenobarbitone-induced liver response in wild type and in p53 deficient mice. Toxicol Lett. 2001;122:131–140. doi: 10.1016/s0378-4274(01)00349-6. [DOI] [PubMed] [Google Scholar]
- 55.Martin NC, McGregor AH, Sansom N, Gould S, Harrison DJ. Phenobarbitone-induced ploidy changes in liver occur independently of p53. Toxicol Lett. 2001;119:109–115. doi: 10.1016/s0378-4274(00)00299-x. [DOI] [PubMed] [Google Scholar]
- 56.Kemp CJ. Hepatocarcinogenesis in p53-deficient mice. Mol Carcinog. 1995;12:132–136. doi: 10.1002/mc.2940120304. [DOI] [PubMed] [Google Scholar]
- 57.Harris SL, Levine AJ. The p53 pathway: positive and negative feedback loops. Oncogene. 2005;24:2899–2908. doi: 10.1038/sj.onc.1208615. [DOI] [PubMed] [Google Scholar]
- 58.Martin-Caballero J, Flores JM, Garcia-Palencia P, Serrano M. Tumor susceptibility of p21(Waf1/Cip1)-deficient mice. Cancer Res. 2001;61:6234–6238. [PubMed] [Google Scholar]
- 59.Rossi A, Kontarakis Z, Gerri C, Nolte H, Holper S, Kruger M, Stainier DY. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature. 2015;524:230–233. doi: 10.1038/nature14580. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


