Abstract
Next-generation sequencing studies have revealed that a variety of transcripts are present in the prokaryotic transcriptome and a significant fraction of them are functional, being involved in various regulatory activities apart from coding for proteins. Identification of promoters associated with different transcripts is necessary for characterization of the transcriptome. Promoter regions have been shown to have unique structural features as compared with their flanking region, in organisms covering all domains of life. Here we report an in silico analysis of DNA sequence dependent structural properties like stability, bendability and curvature in the promoter region of six different prokaryotic transcriptomes. Using these structural features, we predicted promoters associated with different categories of transcripts (mRNA, internal, antisense and non-coding), which constitute the transcriptome. Promoter annotation using structural features is fairly accurate and reliable with about 50% of the primary promoters being characterized by all three structural properties while at least one property identifies 95%. We also studied the relative differences of these structural features in terms of gene expression and found that the features, viz. lower stability, lesser bendability and higher curvature are more prominent in the promoter regions which are associated with high gene expression as compared with low expression genes. Hence, promoters, which are associated with higher gene expression, get annotated well using DNA structural features as compared with those, which are linked to lower gene expression.
Keywords: promoter prediction, DNA structural properties, transcriptome, gene expression
1. Introduction
Advances in genome sequencing technology have resulted in a large amount of raw data in the form of whole genome sequences of organisms. This sequence data needs to be annotated, viz. identification of coding regions, non-coding regions and regulatory elements. Computational tools are the only viable option for fast and fairly reliable annotation of many genome sequences. Promoter prediction is an important step in the genome annotation process; not only for the validation of the predicted genes but also for the identification of novel genes, especially those associated with non-coding RNA, which are often missed by gene prediction programs. Identification of transcription factor-binding sites (TFBSs) and promoters is essential for understanding transcription and regulation of the genes. Regulatory regions that affect the transcription initiation process are beyond the target DNA sequence motifs.1–4 There are studies in which DNA structural features are integrated with motif search algorithms to identify TFBSs5,6. DNA shape determined by four distinct features—Minor groove width, Propeller twist, Roll and Helix twist has been found to be an important determinant in the identification of TFBSs and transcription start sites (TSSs).7–9 Physico-chemical properties of DNA double helix such as hydrogen bonding, stacking energy etc. show DNA sequence functional density signatures.10–12 Promoter regions have been found to be associated with some unique structural features (like low stability, lesser bendability and more curvature etc.) across organisms.13–16 Promoter annotation based on the relative stability of DNA duplex has been found to be more reliable and accurate for a wide range of prokaryotes, compared with sequence based approaches.17,18 PromBase is a comprehensive database which provides promoter predictions, based on relative stability, for whole genome sequences of bacteria and archaea with respect to their Translation Start Sites.19 Profiles of some other structural properties, such as bendability and curvature are also displayed in addition to stability. A previous study has shown that different categories of promoters (primary, secondary and internal promoters) display a gradation in their sequence dependent structural properties and which is mostly conserved for orthologous genes in ten strains of the bacterium Helicobacter pylori.20 Non-B DNA structural motifs are preferentially located in the regulatory regions of operons in E. coli.21The recent study in Mycoplasma pneumonia shows that free energy of promoter region is an important determinant in the identification of true promoter from abortive and non promoters, apart from sequence elements such as −10, extended −10 and −35 box.22
Whole genome transcriptome studies reveal the pervasive nature of transcription and suggest that transcription is not restricted to regions upstream of the annotated coding sequences (CDSs) but can initiate from almost any genomic location.23 The wide variety of transcripts like mRNA, internal, antisense and non-coding RNA have been found to exist in the prokaryotic genome. Large fractions of these non-coding RNAs are found to be functional and are involved in various regulatory activities in the cell.24 Antisense transcripts play an important role in gene regulatory networks and constitute a secondary layer of regulatory switch, which has great potential in genetic engineering.25,26 Hence, annotation of non-coding transcripts is crucial for a complete understanding of functional genomics and gene regulatory circuits.26,27 To address these issues, we have studied structural properties of the promoter regions in different categories of transcripts characterized in the transcriptome of six different model organisms, viz. H. pylori, Anabaena, Synechocystis, E. coli, Salmonella and Klebsiella. The genomic GC content for these organisms varies from 39% for H. pylori to 57% for Klebsiella (Table 1). We used the differences in these properties between promoter regions and the flanking sequences to predict promoters associated with the different categories of transcripts such as primary, internal, antisense and non-coding RNA. Ability to predict the strength of a promoter is the next challenge in promoter annotation process. Sequence motif based approaches have been reported for E. coli, which predict the strength of the promoter moderately well in an organism specific manner.28–30 DNA thermodynamics and supercoiling dynamics have been found to be associated with gene expression during different bacterial growth cycle.31,32 Following this up, we have analysed the relationship between DNA structural features and the strength of the promoter, in terms of gene expression in six different model organisms.
Table 1.
Genomic features and number of TSSs of various categories in six different model organisms used in this study
| Organisms | Genome size (in Mb) | Genome GC (in %) | Protein genes | RNA genes | gTSS | iTSS | aTSS | nTSS |
|---|---|---|---|---|---|---|---|---|
| H. pylori33 | 1.7 | 38.9 | 1,469 | 43 | 714 | 426 | 1,018 | NA |
| Anabaena34 | 6.4 | 41.4 | 5,365 | 66 | 2,517 | 1,878 | 2,196 | 1,266 |
| Synechocystis35 | 3.6 | 47.7 | 3,179 | 50 | 1,639 | 125 | 1,356 | 170 |
| E. coli36 | 4.6 | 50.8 | 4,145 | 175 | 1,333 | 1,184 | NA | NA |
| Salmonella37 | 4.9 | 52.2 | 4,446 | 109 | 981 | 238 | 171 | NA |
| Klebsiella36 | 5.3 | 57.5 | 4,776 | 111 | 1,329 | 837 | NA | NA |
Abbreviations used: gTSS, primary TSS; iTSS, internal TSS; aTSS, antisense TSS; nTSS, non-coding TSS.
2. Material and methods
2.1. Promoter sequence dataset preparation
TSS information of H. pylori strain 2669533 (H. pylori), Anabaena sp. PCC712034 (Anabaena), Synechocystis sp. PCC 680335 (Synechocystis), E. coli K-12 MG165536 (E. coli), Salmonella enterica serovar Typhimurium37 (Salmonella) and Klebsiella pneumoniae36 (Klebsiella) was obtained from the relevant published data. For each gene (protein/RNA), TSS with highest number of reads was selected for creating a unique dataset of primary/gTSS (transcribing mRNA), iTSS (TSSs present inside the gene), aTSS (corresponding to antisense transcripts, encompassed within the gene), whereas all available nTSS (non-coding transcripts) were taken (Table 1). Only those categories of TSSs, that had more than 100 TSSs, were selected for the study. Whole genome sequence and translation start site (TLS) information were downloaded from ftp site (ftp://ftp.ncbi.nih.gov/genomes/Bacteria/) of NCBI. Genome sequences of 1001 nucleotides length (spanning 500 nucleotides upstream and downstream of the TSS positioned at 0) were extracted from the whole genome sequence using TSS information as provided in published data.
Non-promoter sequence or negative dataset was created, by taking the CDS of the genes associated with gTSS (primary TSS).
2.2. Estimation of structural properties of promoter sequences
We have studied three sequence dependent structural properties stability, bendability and curvature. Stability of a piece of double stranded DNA sequence molecule can be calculated in terms of the free energy of its constituent dinucleotides. The free energy values of the 10 unique dinucleotides were taken from data based on melting studies of oligo and polynucleotides.38 Stability profile of 1001 nucleotides long promoter sequence was calculated by dividing each sequence in sliding windows of 15 base pairs each (or 14 dinucleotide steps) using the free energy values for the constituent dinucleotides. Average free energy (AFE) (in kcal/mol) was calculated at each nucleotide position after aligning all TSS at 0 position.39
Bendability of promoter sequences was calculated by using two different trinucleotide models, DNase I sensitivity40 (DNase I) and nucleosomal positioning preference (NPP).41 Bendability profile was calculated using a 30 nucleotides window size.
Curvature of promoter sequence was estimated by the calculated value of d/lmax (the ratio of minimum end to end distance ‘d’ to the length of a DNA fragment ‘lmax’)42 for the models generated using a set of dinucleotide values obtained from gel retardation mobility assay of multiple polymeric DNA sequences (BMHT).43 The DNA models were generated using 75 nucleotides window size and d/lmax vale was calculated by using in-house program NUCGEN.44 Values of d/lmax ranges from 0 (completely closed circle indicating high curvature) to 1 (perfectly linear DNA).
2.3. Promoter prediction methodology
Promoter sequences of 1001 nucleotide length from six different bacteria were categorized on the basis of their GC composition (at 5% GC intervals). Sequence information was translated to numerical information in terms of structural properties mentioned above. The training set of promoter sequences were selected from the primary TSS promoters, associated with gTSS. A 5-fold analysis was performed, by selecting 80% as training dataset and it was applied on full dataset. Two independent cycles of cutoff derivation and promoter predictions were performed using 75 and 100 nucleotide long windows, respectively, for all given structural features.
2.3.1. Cutoff derivations
The more prominent low stability peak was observed in the vicinity of TSS across all bacterium species. Hence, the region −55 to + 20 (with respect to TSS) was selected to derive one of the cutoff values (STB1) for the first cycle while −80 to + 20 region was taken for the second cycle. Promoters were predicted by comparing structural properties of neighboring regions; hence a 75 nucleotides window from +200 to + 275 with respect to TSSs and 100 nucleotides window from +200 to + 300 with respect to TSSs were chosen to derive second cutoff value (STB2) for stability for cycle one and cycle two respectively. Less bendability and high curvature span were observed in the upstream regions of TSSs across all the organisms. Hence −100 to −25 and −125 to −25 nucleotides with respect to TSSs were considered for derivation of first cutoff values for bendability (both models BDC1 and BNC1) and curvature (CBC1), for the first and second cycle of predictions respectively. Similarly, the second set of cutoff values for bendability (BDC2 and BNC2) and curvature (CBC2) were derived from +200 to + 275 and +200 to + 300 nucleotides with respect to TSSs for the first and second cycle of predictions, respectively.
2.3.2. Promoter prediction
The cutoff values derived at 5%GC interval for all DNA sequence dependent structural properties were applied within −200 to + 100 nucleotide region (R1) and +200 to + 500 nucleotide region (R2) with respect to TSS in the full dataset. A given sequence was identified as a promoter sequence if a particular sequence dependent structural property was found to be present and satisfies the following set of conditions.
R1 ≥ STB1 & R2 ≤ STB2 for stability
R1≤ BDC1 & R2 ≥ BDC2 for bendability (DNase I)
R1 ≥ BNC1 & R2 ≤ BNC2 for bendability (NPP)
R1 ≤ CBC1 & R2 ≥ CBC2 for curvature
Sequences satisfying the above conditions were assigned as true positive (TP) for the representative feature, else they were classified as false negative (FN). In the negative dataset, +200 to + 500 nucleotide region was scanned for conditions mentioned earlier. If a sequence from negative dataset was found to satisfy above criteria then it was designated as false positive (FP) for the respective property, else it was tagged as true negative (TN).
TP and TN rates were defined by calculating sensitivity (also known as recall) and specificity respectively, to measure the performance of binary classification test. Fidelity of the search was determined by calculating precision. Evaluation of classifier was done, by calculating balanced–accuracy and F-score, which are arithmetic and harmonic means of sensitivity and precision respectively.
2.4. Gene expression estimation
Microarray expression profiling data (RNA sample from wild type) H. pylori (GSM931753), Synechocystis (GSM1316725), E. coli (GSM1374996), Salmonella (GSM1102757) and Klebsiella (GSM877524) was downloaded from GEO (Gene Expression Omnibus), NCBI.45. Differential expression was calculated by using DESeq (R/Bioconductor package)46 to measure the change in the expression fold in the absence of Nitrogen from transcriptome data.34 Primary TSSs (gTSS) were sorted in descending order (in the terms of their expression level) and top 10 percentile were considered as high expression genes expected to be associated with strong promoters. The bottom 10 percentile was taken as a dataset for low expression genes with weak promoters. Structural features were calculated for the sequences extracted using TSS location and strand information from the whole genome sequence.
2.5. Statistical methods
Statistical significance of structural properties enrichment in the promoter regions of highly expressed genes as compared with lowly expressed genes were analyzed using the Kolmogorov-Smirnov test (KS test). One-sided two-sample KS test was performed to test the significance of the difference in feature value between the two classes (here null hypothesis was that the both datasets have similar CDF distribution). Null hypothesis was rejected at the significance level of 1%. MATLAB was used for statistical analysis.
3. Results and discussion
3.1 Promoter regions corresponding to prokaryotic transcriptomes show distinct structural properties
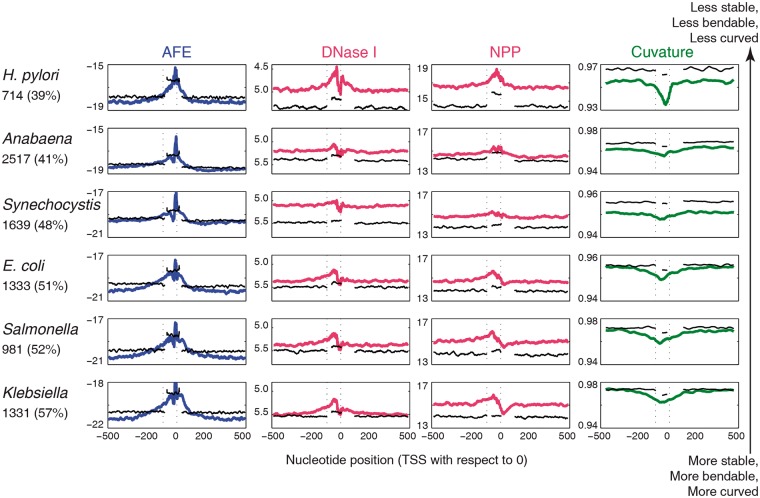
We have characterized DNA duplex stability (AFE), bendability using two different models DNase I sensitivity (DNase I) and NPP and intrinsic curvature in the promoter region of different categories of TSSs in six different model organisms. DNase I sensitivity model gives the bending propensity of each trinucleotide in terms of bending towards major groove while NPP model gives the rotational preference of each trinucleotide towards histone core. The DNA structural profiles show that promoter (primary) regions are in general less stable, less bendable and more curved as compared to neighboring regions (Fig. 1). DNA duplex stability, expressed in the terms of AFE (kcal/mol) profile, shows a low stability peak in the vicinity of the TSS across organisms with varying genomic GC content. With increase in genomic GC% from H. pylori with 39% to Klebsiella with 57% the baseline shifts to higher stability (lower AFE) values, while the low stability peak splits. The secondary low stability peak is likely to be associated with −35 element. Bendability profiles as calculated using two different trinucleotide models (DNase I and NPP), show similar low bendability peaks in the region upstream of the TSS. Curvature profile shows that the promoter region is more intrinsically curved as compared with flanking regions.
Figure 1.
DNA sequence dependent structural properties: AFE, DNase I sensitivity, NPP and curvature profiles for 1001 nucleotides long primary (gTSS) promoter sequences (extending from −500 to + 500 nucleotides with respect to TSS at 0). The black colour plots correspond to profiles of structural feature for shuffled sequences (upstream −500 to − 100, promoter region −80 to + 20 and downstream +100 to + 500 nucleotide position with respect to TSS at 0). The number of promoter sequences and whole genome GC% is mentioned along with the organism name, on the left of each row.
The shuffled promoter sequences obtained by the randomization of native promoter sequences do not show these properties, even when they were estimated separately for three different regions: upstream (−500 to −100 nucleotide), promoter region (−80 to +20 nucleotide) and downstream region (+100 to +500) nucleotide with respect to TSS at 0 position in order to retain the GC composition of these regions. Interestingly genomic sequences in the promoter, as well as the non-promoter region, are overall less bendable and more curved than the shuffled sequences. On the other hand, while genomic promoter sequences are less stable than their shuffled counterparts, the flanking genomic sequences are more stable. The loss of special structural features in shuffled sequences suggests that these features arise due to specific base sequence patterns.
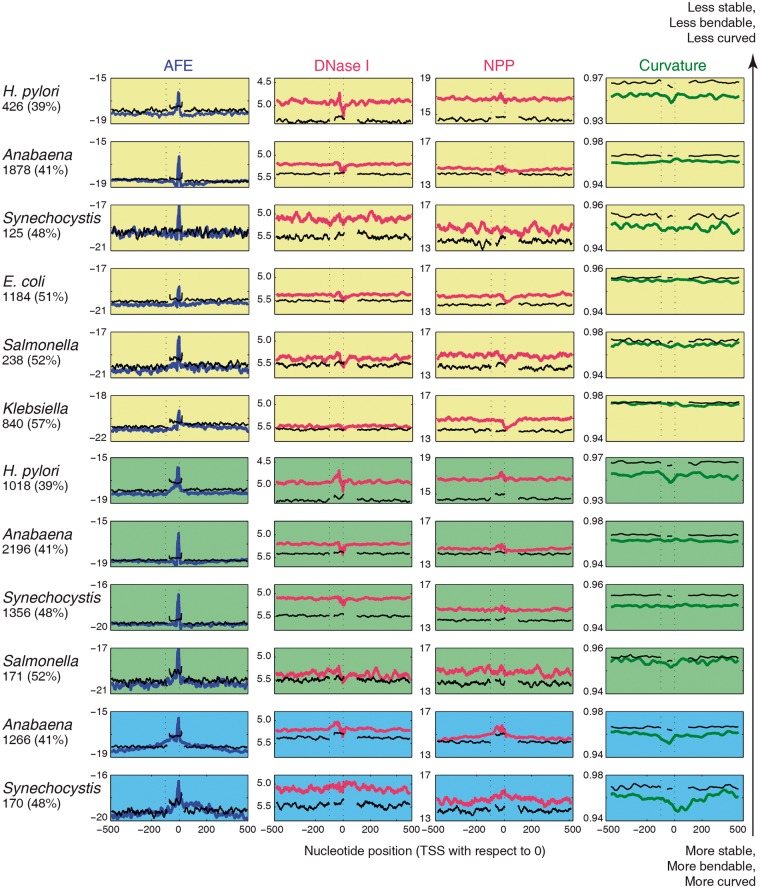
Similar structural profiles were observed for internal promoters, antisense promoters and non-coding RNA gene promoters (Fig. 2). Stability profiles show sharp low stability peaks in the case of internal, antisense and non-coding RNA promoters as compared with primary promoters across all organisms. Bendability and curvature profiles also show distinct features in their promoter regions, but they are less prominent for these categories of promoters as compared with primary promoters. Base composition analysis shows that primary category of promoters (−100 to 0 nucleotide with respect to TSS at 0) show preference for several AT rich tetramers as compared with background region (−500 to +500 nucleotide with respect to TSS at 0) (Supplementary Fig. S1). Internal and antisense categories of promoters show less preference for AT rich tetramers and TATA sequence motifs as compared with primary and non-coding RNA promoters. Core promoter region (−35 and −10 element) shows poor sequence conservation even for promoters associated with primary category of TSSs (Supplementary Fig. S2) and becomes poorer for internal and antisense categories of promoters as compared with the primary category across the six different model organisms with varying genomic GC content. Interestingly no sequence motif signature was observed in their -35 region, while a secondary low stability peak was observed in the same region for primary category of TSSs across the organisms. The weak preference for AT rich tetramers and poor sequence motif conservation in core promoter elements (−35 and −10 elements) makes it difficult to predict these promoters by sequence motif based approaches.
Figure 2.
DNA sequence dependent structural properties: AFE, DNase I sensitivity, NPP and curvature profiles for 1001 nucleotides long internal (rows 1-6), antisense (rows 7-10) and Non-coding (rows 11-12) promoter sequences (extending from −500 to + 500 nucleotides with respect to TSS at 0). The black colour plots correspond to profiles of structural feature for shuffled sequences (upstream −500 to − 100, promoter region −80 to + 20 and downstream +100 to + 500 nucleotide position with respect to TSS at 0). The number of promoter sequences and whole genome GC% is mentioned along with the organism name, on the left of each row.
3.2 Predicting promoters associated with different categories of TSSs using structural features
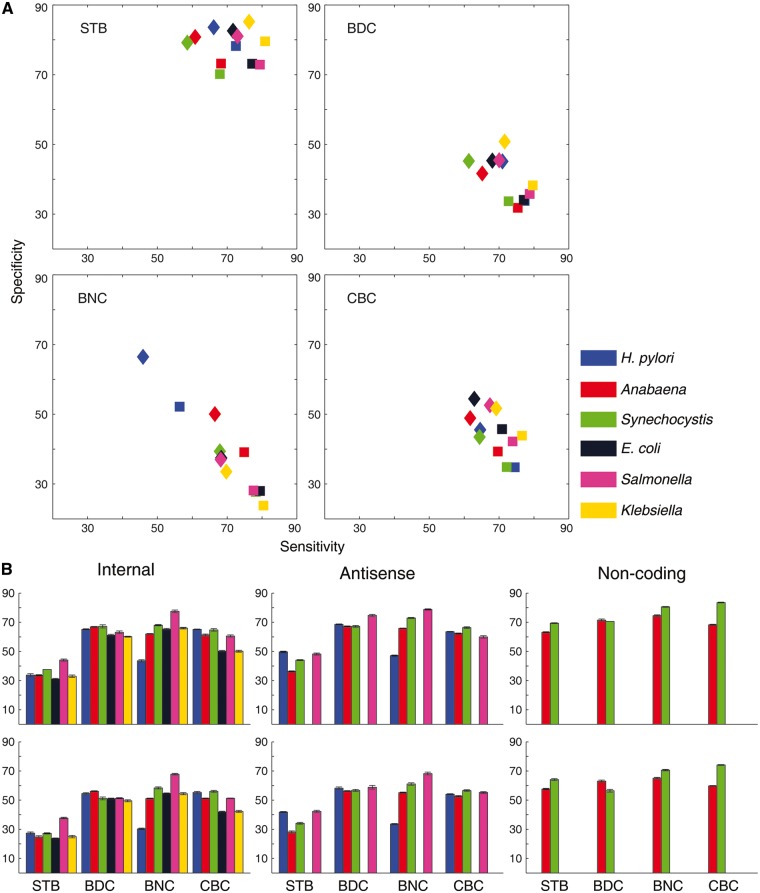
Promoters were predicted in the full set of 1001 nucleotides long sequences, flanking the annotated TSSs of all categories (primary, internal, antisense and non-coding). Two cycles of predictions were performed using 75 and 100 nucleotides window size (using two different sets of cut-off values derived using 75 and 100 nucleotides fragment length for cycle one and two, respectively). The sensitivity and specificity achieved in each cycle, using the four different properties are shown in Figure 3 for all six organisms. Higher specificity was achieved at the cost of sensitivity in the second cycle of prediction, which uses larger fragment length of DNA sequence and most importantly, it was found to be valid for all structural features in primary category of promoters. Bendability (DNase I sensitivity model) and curvature were found to be equally sensitive but less specific in nature as compared with stability in all six model organisms.
Figure 3.
Prediction statistics using sequence dependent structural properties (STB, stability; BDC, bendability using DNase I sensitivity; BNC, bendability using NPP model; CBC, curvature). (A) The performance of the classifier for primary promoters. Markers in Square and Diamond shape represent the first and second cycle of prediction, respectively. (B) Sensitivity values obtained for internal, antisense and Non-coding promoters, top and bottom row correspond to the first and second cycle of prediction respectively. Error bars represent the SD obtained from the 5-fold analysis.
Stability was found to be the most characteristic structural feature in all organisms and achieved higher precision values in the second cycle of prediction compared with cycle one (Table 2). Similarly, bendability (both models DNase I sensitivity and NPP) and curvature features were found to give high precision values in cycle two as compared with cycle one. Both prediction cycles were found to give an almost similar balanced-accuracy (BA), which is the mean of TP rate (sensitivity) and TN rate (specificity). Overall cycle one was found to be a better predictor, with better F-score (harmonic mean of sensitivity and precision) values across all the model organisms for all structural features.
Table 2.
Assessment of DNA structural features for their ability to differentiate between promoter and non-promoter sequences in six different model organisms
| Organism | Number of sequences | First cycle |
Second cycle |
Property | ||||
|---|---|---|---|---|---|---|---|---|
| Prec. | BA | F-score | Prec. | BA | F-score | |||
| H. pylori | 706 | 77 | 75 | 75 | 80 | 75 | 72 | STB |
| Anabaena | 2513 | 72 | 71 | 70 | 76 | 71 | 68 | |
| Synechocystis | 1639 | 69 | 69 | 69 | 74 | 69 | 65 | |
| E. coli | 1331 | 74 | 75 | 76 | 80 | 77 | 76 | |
| Salmonella | 981 | 75 | 76 | 77 | 79 | 77 | 76 | |
| Klebsiella | 1324 | 80 | 80 | 80 | 84 | 81 | 80 | |
| H. pylori | 706 | 54 | 56 | 63 | 56 | 58 | 63 | BDC |
| Anabaena | 2513 | 52 | 54 | 62 | 53 | 53 | 58 | |
| Synechocystis | 1639 | 52 | 53 | 61 | 53 | 53 | 57 | |
| E. coli | 1331 | 54 | 56 | 63 | 55 | 57 | 61 | |
| Salmonella | 981 | 55 | 57 | 65 | 56 | 58 | 62 | |
| Klebsiella | 1324 | 56 | 59 | 66 | 59 | 61 | 65 | |
| H. pylori | 706 | 54 | 54 | 55 | 58 | 56 | 51 | BNC |
| Anabaena | 2513 | 55 | 57 | 64 | 57 | 58 | 61 | |
| Synechocystis | 1639 | 52 | 53 | 62 | 53 | 54 | 59 | |
| E. coli | 1331 | 52 | 54 | 63 | 52 | 53 | 59 | |
| Salmonella | 981 | 52 | 53 | 62 | 52 | 53 | 59 | |
| Klebsiella | 1324 | 51 | 52 | 63 | 51 | 51 | 59 | |
| H. pylori | 706 | 53 | 55 | 62 | 54 | 55 | 59 | CBC |
| Anabaena | 2513 | 53 | 54 | 60 | 55 | 55 | 58 | |
| Synechocystis | 1639 | 53 | 54 | 61 | 53 | 54 | 58 | |
| E. coli | 1331 | 57 | 58 | 63 | 58 | 59 | 60 | |
| Salmonella | 981 | 56 | 58 | 64 | 59 | 60 | 63 | |
| Klebsiella | 1324 | 58 | 60 | 66 | 59 | 60 | 64 | |
Out of the total test dataset (promoters associated with the primary category of TSS), 1001 nucleotide long sequences with 30–65% GC content were considered. The sequences with promoter predictions falling in the 300 nucleotide region spanning −200 to +100 with respect to TSS at 0 were considered as TP while sequences with predictions falling in the coding region +200 to + 500 with respect to TSS were labelled as FP. Evaluation parameters precision (Prec.), BA and F-score were calculated using formulas explained in the ‘Material and methods’ section.
We categorized the predicted promoters based on the function performed by the associated gene, according to the Cluster of Orthologous Groups information obtained from NCBI (Supplementary Table S1). Promoters associated with genes involved in information storage and processing function (transcription, translation and replication), which are constitutively expressed in the cell are annotated well across the six different organisms. However, sensitivity values for an individual structural feature vary among the organisms, e.g. promoters associated with genes which have a role in replication and repair exhibit sensitivity values (using stability) ranging from a low of 38% for Synechocystis to a high of 72% for Klebsiella. Promoters of genes involved in cellular processing and cell signaling are relatively less well annotated. Genes involved in metabolism function show variation in the sensitivity values for different gene families. General function genes (which are based on prediction only) also get annotated reliably in all six prokaryotic organisms. Genes which are not characterized or with unknown function are found to be relatively less annotated in all organisms. Hence, prediction results suggest that structural features in the promoter regions are related to the function associated with the gene.
Internal, antisense and non-coding RNA promoters were also predicted using the cut-off values derived from primary promoter sequences. Higher sensitivity values were obtained in the first cycle of prediction as compared with the second cycle. Bendability (both by DNase I sensitivity and NPP model) and curvature were found to be more sensitive for these categories of promoters, as compared with stability. The reduction in the sensitivity values for stability feature for internal and antisense promoters was due to sharp and narrow low stability peak in the AFE profile (Fig. 2). The narrower peaks can be attributed to these categories of promoters being present within the coding region while primary promoters are mostly present in the relatively AT rich intergenic region upstream of TSS.
3.3 Promoter annotation using combination of structural features
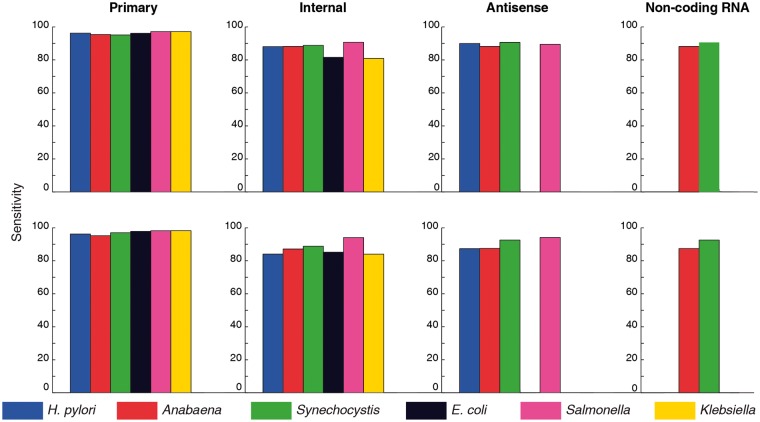
Although stability was found to be the most distinctive feature, differentiating the promoter from the flanking region for a given genomic sequence, it was able to achieve maximum 81% sensitivity for primary category of promoters in Klebsiella, in cycle one prediction (Fig. 3). In order to assess the number of promoter sequences that do not get identified by any structural feature, we applied a combinatorial approach. It was found that ≈95% primary category of promoter sequences get identified by at least one structural feature. Similar sensitivity values are obtained from a combinatorial approach using DNase I sensitivity and NPP models for bendability (as seen in Fig. 4). Likewise internal and antisense categories of promoters achieve ≈85 and ≈89% sensitivity, respectively. Further analysis of promoter prediction in the different categories of transcripts revealed several interesting facts. For example, ≈50% of the primary category of promoter sequences are characterized by all three structural features, viz. stability, bendability and curvature, while remaining promoter sequences lack either one or two structural features (Supplementary Fig. S3). Interestingly, majority of internal and antisense categories of promoters are characterized by bendability and curvature while promoters associated with non-coding RNA genes are similar to primary category of promoters (Supplementary Fig. S4). This suggests that the promoter sequences which occur in the coding region, such as internal and antisense promoters, are distinguishable by different structural features, when compared with the primary and non-coding RNA promoter sequences, which are present in the intergenic region.
Figure 4.
Bar plots showing sensitivity values obtained from cycle I predictions, using one or more structural features for different categories of TSSs. Top row correspond to stability, bendability using DNase I sensitivity and curvature while bottom row corresponds to stability, bendability using NPP and curvature.
3.4 DNA structural features of the promoter region are correlated with the gene expression
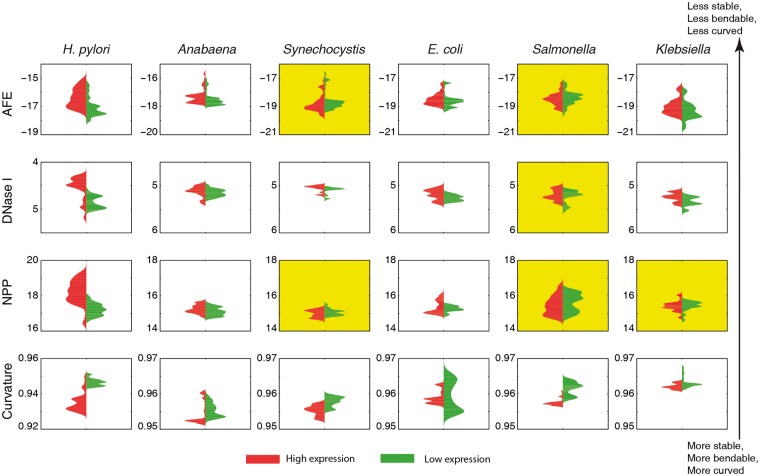
Sequence dependent structural properties of double stranded DNA are associated with the physiology of the promoter. For example DNA duplex has to be less stable to facilitate easier melting in the vicinity of the TSS during transcription initiation. Hence, in order to study the promoter architecture in terms of structural features, we characterized these features in two classes of promoters, viz. those associated with genes exhibiting high and low gene expression. Gene expression data was obtained from GEO, NCBI expression microarray profiling. The structural properties (stability, bendability and curvature) in the promoter regions associated with high gene expression are found to be more pronounced as compared with low gene expression. A violin plot drawn from the values of DNA structural features underscores the difference in the distribution of probability densities between the promoters associated with high and low expression categories of genes (Fig. 5). The promoters of highly expressed genes were less stable, less bendable and more curved as compared with promoters of lowly expressed genes. One-sided two sample KS test was performed to check if the difference in feature value between the two classes is significant enough for them to be compared (Supplementary Table S2). Interestingly curvature was found to be the only consistent property demarcating promoters from high and low gene expression classes, across all organisms, while other properties show one or more exceptions in case of Synechocystis, Salmonella and Klebsiella.
Figure 5.
Violin plot of DNA structural property values in the promoter regions (−100 to 0 nucleotide with respect to TSS at 0) associated with high and low gene expression. The x-axis shows the probability density while y-axis represents the DNA structural features value. Plots with shaded background indicate the cases which failed to reject the null hypothesis using two sample KS test at the level of significance of P = 0.01.
Trinucleotide preference analysis of the whole promoter region (UP elements and core promoter region) was performed in order to understand the underlying difference in terms of oligonucleotide composition. It showed that overall high expression genes favor rigid and minor groove facing histone preferring trimers, such as AAA/TTT and AAT/ATT while flexible or major groove facing histone preferring trinucleotides like GCC/GGC are less preferred (Supplementary Fig. S5). Preference for AAA/TTT reduces gradually as genomic GC% increases.
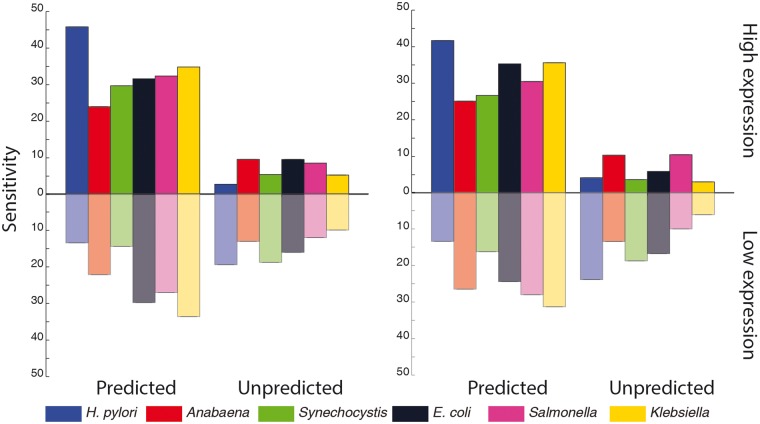
Promoter prediction using DNA structural features shows that sensitivity for promoters associated with high gene expression is higher as compared with the low gene expression (Fig. 6). Interestingly it was found that promoters associated with higher gene expression are better characterized (higher sensitivity) by a combination of all structural features (stability, bendability and curvature) as compared with those associated with low gene expression. Correspondingly, promoters for low gene expression category are less likely to be predicted as compared with the high gene expression category. Moreover, this trend is present across organisms with varying genomic GC content.
Figure 6.
Bar plot of sensitivity values obtained from cycle I predictions using all three structural features (predicted) or none (unpredicted) (stability, bendability using DNase I sensitivity model, bendability using NPP model and curvature). Left and right plots correspond to the DNase I sensitivity and NPP models of bendability.
4. Conclusion
DNA sequence dependent structural properties like duplex stability, protein induced bendability and intrinsic curvature are well studied and physiologically relevant properties for the promoter region. Since there is very weak sequence (tetramer) preference and poor sequence motif conservation in the internal and antisense promoters as compared with primary promoters, it is very difficult to identify them using sequence based approaches. Characteristic structural features, on the other hand, are present in the promoter regions of all categories of promoters identified in prokaryotic transcriptomes. Significantly, these properties are not organism specific in nature. However, promoters associated with primary, internal, antisense and non-coding RNA category of transcripts show differences in the profiles of their structural properties. In particular, internal and antisense categories of promoters show distinctly different structural feature profiles as compared with the primary category of promoters. Since a single feature does not predict all promoters, transcriptome can be better annotated using a combinatorial approach. Annotation and analysis of promoters corresponding to the different transcripts present in the transcriptome can provide a better understanding of the complexity involved in gene regulation process. Finally, the promoter DNA structural features show good correlation with the level of expression of the associated gene. Hence by examining structural features associated with promoter regions, one can possibly estimate the level of gene expression.
Supplementary Material
Acknowledgements
M.B. is the recipient of J. C. Bose National Fellowship of DST, India.
Conflict of interest
None declared.
Supplementary data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
References
- 1.MacQuarrie K. L., Fong A. P., Morse R. H., Tapscott S. J. 2011, Genome-wide transcription factor binding: beyond direct target regulation. Trends Genet., 27, 141–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Slattery M., Riley T., Liu P., et al. 2011, Cofactor binding evokes latent differences in DNA binding specificity between Hox proteins. Cell, 147, 1270–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gordan R., Shen N., Dror I., et al. 2013, Genomic regions flanking E-box binding sites influence DNA binding specificity of bHLH transcription factors through DNA shape. Cell Rep., 3, 1093–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levo M., Segal E. 2014, In pursuit of design principles of regulatory sequences. Nat. Rev. Genet., 15, 453–468. [DOI] [PubMed] [Google Scholar]
- 5.Meysman P., Marchal K., Engelen K. 2012, DNA structural properties in the classification of genomic transcription regulation elements. Bioinform. Biol. Insights, 6, 155–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maienschein-Cline M., Dinner A. R., Hlavacek W. S., Mu F. 2012, Improved predictions of transcription factor binding sites using physicochemical features of DNA. Nucleic Acids Res., 40, e175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou T., Shen N., Yang L., et al. 2015, Quantitative modeling of transcription factor binding specificities using DNA shape. Proc. Natl. Acad. Sci. USA, 112, 4654–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levo M., Zalckvar E., Sharon E., et al. 2015, Unraveling determinants of transcription factor binding outside the core binding site. Genome Res., 185033–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiu T. P., Yang L., Zhou T., et al. 2015, GBshape: a genome browser database for DNA shape annotations. Nucleic Acids Res., 43, D103–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khandelwal G., Lee R. A., Jayaram B., Beveridge D. L. 2014, A statistical thermodynamic model for investigating the stability of DNA sequences from oligonucleotides to genomes. Biophys J., 106, 2465–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khandelwal G., Jayaram B. 2012, DNA-water interactions distinguish messenger RNA genes from transfer RNA genes. J. Am. Chem. Soc., 134, 8814–6. [DOI] [PubMed] [Google Scholar]
- 12.Khandelwal G., Gupta J., Jayaram B. 2012, DNA-energetics-based analyses suggest additional genes in prokaryotes. J. Biosci., 37, 433–44. [DOI] [PubMed] [Google Scholar]
- 13.Florquin K., Saeys Y., Degroeve S., Rouze P., Van de Peer Y. 2005, Large-scale structural analysis of the core promoter in mammalian and plant genomes. Nucleic Acids Res., 33, 4255–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanhere A., Bansal M. 2005, Structural properties of promoters: similarities and differences between prokaryotes and eukaryotes. Nucleic Acids Res., 33, 3165–3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meysman P., Collado-Vides J., Morett E., Viola R., Engelen K., Laukens K. 2014, Structural properties of prokaryotic promoter regions correlate with functional features. PLoS One, 9, e88717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bansal M., Kumar A., Yella V. R. 2014, Role of DNA sequence based structural features of promoters in transcription initiation and gene expression. Curr. Opin. Struct. Biol., 25, 77–85. [DOI] [PubMed] [Google Scholar]
- 17.Rangannan V., Bansal M. 2009, Relative stability of DNA as a generic criterion for promoter prediction: whole genome annotation of microbial genomes with varying nucleotide base composition. Mol. Biosyst., 5, 1758–1769. [DOI] [PubMed] [Google Scholar]
- 18.Rangannan V., Bansal M. 2010, High-quality annotation of promoter regions for 913 bacterial genomes. Bioinformatics, 26, 3043–3050. [DOI] [PubMed] [Google Scholar]
- 19.Rangannan V., Bansal M. 2011, PromBase: a web resource for various genomic features and predicted promoters in prokaryotic genomes. BMC Res. Notes, 4, 257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar A., Bansal M. 2012, Characterization of structural and free energy properties of promoters associated with Primary and Operon TSS in Helicobacter pylori genome and their orthologs. J. Biosci., 37, 423–31. [DOI] [PubMed] [Google Scholar]
- 21.Du X., Wojtowicz D., Bowers A. A., Levens D., Benham C. J., Przytycka T. M. 2013, The genome-wide distribution of non-B DNA motifs is shaped by operon structure and suggests the transcriptional importance of non-B DNA structures in Escherichia coli. Nucleic Acids Res., 41, 5965–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Llorens-Rico V., Lluch-Senar M., Serrano L. 2015, Distinguishing between productive and abortive promoters using a random forest classifier in Mycoplasma pneumoniae. Nucleic Acids Res., 43, 3442–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wade J. T., Grainger D. C. 2014, Pervasive transcription: illuminating the dark matter of bacterial transcriptomes. Nat. Rev. Microbiol., 12, 647–653. [DOI] [PubMed] [Google Scholar]
- 24.Kopf M., Hess W. R. 2015, Regulatory RNAs in photosynthetic cyanobacteria. FEMS Microbiol. Rev., 39, 301–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shearwin K. E., Callen B. P., Egan J. B. 2005, Transcriptional interference–a crash course. Trends Genet., 21, 339–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brophy J. A., Voigt C. A. 2016, Antisense transcription as a tool to tune gene expression. Mol. Syst. Biol., 12, 854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fu X.-D. 2014, Non-coding RNA: a new frontier in regulatory biology. Natl. Sci. Rev., 1, 190–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weller K., Recknagel R. D. 1994, Promoter strength prediction based on occurrence frequencies of consensus patterns. J. Theor. Biol., 171, 355–9. [DOI] [PubMed] [Google Scholar]
- 29.Rhodius V. A., Mutalik V. K. 2010, Predicting strength and function for promoters of the Escherichia coli alternative sigma factor, sigmaE. Proc. Natl. Acad. Sci. USA, 107, 2854–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rhodius V. A., Mutalik V. K., Gross C. A. 2012, Predicting the strength of UP-elements and full-length E. coli sigmaE promoters. Nucleic Acids Res., 40, 2907–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sobetzko P., Travers A., Muskhelishvili G. 2012, Gene order and chromosome dynamics coordinate spatiotemporal gene expression during the bacterial growth cycle. Proc. Natl. Acad. Sci. USA, 109, E42–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sobetzko P., Glinkowska M., Travers A., Muskhelishvili G. 2013, DNA thermodynamic stability and supercoil dynamics determine the gene expression program during the bacterial growth cycle. Mol. Biosyst., 9, 1643–51. [DOI] [PubMed] [Google Scholar]
- 33.Sharma C. M., Hoffmann S., Darfeuille F., et al. 2010, The primary transcriptome of the major human pathogen Helicobacter pylori. Nature, 464, 250–5. [DOI] [PubMed] [Google Scholar]
- 34.Mitschke J., Vioque A., Haas F., Hess W. R., Muro-Pastor A. M. 2011, Dynamics of transcriptional start site selection during nitrogen stress-induced cell differentiation in Anabaena sp. PCC7120. Proc. Natl. Acad. Sci. USA, 108, 20130–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kopf M., Klahn S., Scholz I., Matthiessen J. K., Hess W. R., Voss B. 2014, Comparative analysis of the primary transcriptome of Synechocystis sp. PCC 6803. DNA Res., 21, 527–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim D., Hong J. S., Qiu Y., et al. 2012, Comparative analysis of regulatory elements between Escherichia coli and Klebsiella pneumoniae by genome-wide transcription start site profiling. PLoS Genet., 8, e1002867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kroger C., Dillon S. C., Cameron A. D., et al. 2012, The transcriptional landscape and small RNAs of Salmonella enterica serovar Typhimurium. Proc. Natl. Acad. Sci. USA, 109, E1277–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.SantaLucia J., Jr. 1998, A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl. Acad. Sci. USA, 95, 1460–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yella V. R., Kumar A., Bansal M. 2015, DNA Structure and Promoter Engineering In: Singh V., Dhar P. K. (eds), Systems and Synthetic Biology, Springer, Netherlands, pp. 241–4. [Google Scholar]
- 40.Brukner I., Sanchez R., Suck D., Pongor S. 1995, Trinucleotide models for DNA bending propensity: comparison of models based on DNaseI digestion and nucleosome packaging data. J. Biomol. Struct. Dyn., 13, 309–17. [DOI] [PubMed] [Google Scholar]
- 41.Satchwell S.C., Drew H.R., Travers A.A. 1986, Sequence periodicities in chicken nucleosome core DNA. J. Mol. Biol., 191, 659–75. [DOI] [PubMed] [Google Scholar]
- 42.Kanhere A., Bansal M. 2003, An assessment of three dinucleotide parameters to predict DNA curvature by quantitative comparison with experimental data. Nucleic Acids Res., 31, 2647–2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bolshoy A., McNamara P., Harrington R. E., Trifonov E. N. 1991, Curved DNA without A-A: experimental estimation of all 16 DNA wedge angles. Proc. Natl. Acad. Sci. USA, 88, 2312–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bansal M., Bhattacharyya D., Ravi B. 1995, NUPARM and NUCGEN: software for analysis and generation of sequence dependent nucleic acid structures. Comput. Appl. Biosci., 11, 281–7. [DOI] [PubMed] [Google Scholar]
- 45.Barrett T., Wilhite S. E., Ledoux P., et al. 2013, NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res., 41, D991–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anders S., Huber W. 2010, Differential expression analysis for sequence count data. Genome Biol., 11, R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.