Abstract
All extant lamprey karyotypes are characterized by almost all dot-shaped microchromosomes. To understand the molecular basis of chromosome structure in lampreys, we performed chromosome C-banding and silver staining and chromosome mapping of the 18S–28S and 5S ribosomal RNA (rRNA) genes and telomeric TTAGGG repeats in the Arctic lamprey (Lethenteron camtschaticum). In addition, we cloned chromosome site-specific repetitive DNA sequences and characterized them by nucleotide sequencing, chromosome in situ hybridization, and filter hybridization. Three types of repetitive sequences were detected; a 200-bp AT-rich repetitive sequence, LCA-EcoRIa that co-localized with the 18S–28S rRNA gene clusters of 3 chromosomal pairs; a 364-bp AT-rich LCA-EcoRIb sequence that showed homology to the EcoRI sequence family from the sea lamprey (Petromyzon marinus), which contains short repeats as centromeric motifs; and a GC-rich 702-bp LCA-ApaI sequence that was distributed on nearly all chromosomes and showed significant homology with the integrase-coding region of a Ty3/Gypsy family long terminal repeat (LTR) retrotransposon. All three repetitive sequences are highly conserved within the Petromyzontidae or within Petromyzontidae and Mordaciidae. Molecular cytogenetic characterization of these site-specific repeats showed that they may be correlated with programed genome rearrangement (LCA-EcoRIa), centromere structure and function (LCA-EcoRIb), and site-specific amplification of LTR retroelements through homogenization between non-homologous chromosomes (LCA-ApaI).
Keywords: lamprey, chromosome evolution, repetitive sequence, satellite DNA, retrotransposon
1. Introduction
Lampreys (Petromyzontiformes) and hagfishes (Myxiniformes), which are known as cyclostomes or agnathans, occupy a basal position in the phylogeny of extant vertebrates and possess anatomical and developmental features that are quite different from those of gnathostomes (jawed vertebrates). Therefore, these animals are excellent models for understanding the evolution of vertebrates in a wide range of scientific fields, including genomics, immunology, endocrinology, neurobiology, and developmental biology. Molecular phylogenetic analyses have shown that extant lampreys and hagfishes diverged from the common ancestors of jawless vertebrates ∼480 million years ago (MYA).1–3 All extant lamprey species are classified into a single order, Petromyzontiformes, which is composed of three families, Geotriidae, Mordaciidae, and Petromyzontidae.4 Geotriidae and Mordaciidae are distributed in the southern hemisphere, whereas Petromyzontidae are distributed in the Northern Hemisphere.5,6 Their karyotypes are very different from those of other vertebrates and are characterized by almost all dot-shaped chromosomes (regarded as microchromosomes). Species of Geotriidae and Petromyzontidae have 142–184 chromosomes, whereas Mordaciidae species have 76 chromosomes.7–14 However, the chromosome number in hagfishes is much smaller and variable (2n = 14, 28, 34, or 36).11,13 The GC-content of 4-fold degenerate sites (GC4) in the protein-coding regions also differ greatly between lampreys and hagfishes; GC4 is much higher in lampreys than in hagfishes.2 These results suggested that drastic chromosomal and genomic rearrangements occurred in either or both of the lineages after divergence from the common ancestor of cyclostomes.13
Highly repetitive DNA sequences are classified into two categories based on their genomic organization and chromosomal distribution.15 The first category consists of interspersed repetitive sequences such as short interspersed nuclear elements (SINEs) and long interspersed nuclear elements (LINEs), which are distributed throughout the genome. The second category is chromosome site-specific and highly repetitive sequences such as satellite DNA, which are associated with constitutive or centromeric heterochromatin. Chromosome site-specific repetitive sequences generally show a high rate of nucleotide substitution and are divergent between even closely related species.16,17 Thus, identification and comparison of site-specific repetitive sequences aids in tracing the evolutionary process of speciation. An EcoRI family of satellite DNA sequences identified in the sea lamprey Petromyzon marinus (Petromyzontidae) was the first reported in lampreys18 and was shown to be distributed in the centromeric regions of 12 or 34 chromosomes.14,18 This repetitive sequence has high AT content and contains short repeat units. Sequences from the same family of EcoRI satellite DNA were later identified in two other Petromyzontidae species Lampetra planeri and Lampetra zanandreai, and these DNA sequences were localized to only two chromosomes in La. zanandreai.12 In lampreys, molecular and cytogenetic characterization of chromosome site-specific repetitive sequences is limited to this EcoRI satellite DNA.
The Arctic lamprey (Lethenteron camtschaticum, Petromyzontidae) is the most common and widespread lamprey in the arctic region from Lapland in the north to Kamchatka in the east and Japan and Korea in the south. To our knowledge, there has been only one study on the karyotype of this species, whose chromosome number has been reported to range from 144 to 162.19 In this study, we aimed to characterize the molecular basis of its chromosome structure through conventional and molecular cytogenetic approaches and to clone chromosome site-specific repetitive sequences associated with the heterochromatin of Le. camtschaticum. We performed conventional Giemsa staining of chromosomes, chromosome banding (C-banding and silver staining), and chromosome mapping of the 18S–28S and 5S ribosomal RNA (rRNA) genes and telomeric TTAGGG repeats. Next, we isolated three different chromosome site-specific repetitive sequences, determined their nucleotide sequences and chromosomal distribution, and examined the conservation of these nucleotide sequences in other lamprey species. Based on the results, we discuss the process of chromosomal evolution in lampreys.
2. Materials and methods
2.1. Animals
The Arctic lamprey (Lethenteron camtschaticum; synonym Le. japonicum, Petromyzontidae) was used for chromosome preparation and molecular cloning of repetitive sequences. After decapitation of two male and three female Le. camtschaticum, 5 ml of whole blood was collected from each individual in a test tube containing 20 µl of sodium heparin (final concentration: 10 IU/ml) (Mochida Pharmaceutical Co. Ltd., Tokyo, Japan) from each individual using a syringe and was used for cell culture and extraction of genomic DNA. All experimental procedures using animals were performed in accordance with the guidelines established by the Animal Care Committee of Hokkaido University.
2.2. Lymphocyte culture
Heparinized blood of Le. camtschaticum was added to Medium 199 (Thermo Fisher Scientific-Gibco, Carlsbad, CA, USA) containing 60 µg/ml kanamycin sulfate (Sigma-Aldrich, St. Louis, MO, USA) at a ratio of 1:5. After centrifugation at 300×g for 5 min, lymphocytes were obtained with a sterile Pasteur pipette from the buffy coat layer that is situated between the plasma and erythrocyte layers. The lymphocytes were suspended in culture medium [Medium 199 supplemented with 10% FBS, 100 µg/ml lipopolysaccharide (LPS, Sigma-Aldrich), 18 µg/ml phytohemagglutinin, M form (Thermo Fisher Scientific-Gibco), 60 µg/ml kanamycin sulfate, and 1% of Antibiotic–Antimycotic liquid (PSA, Thermo Fisher Scientific-Gibco)] and cultured in plastic bottles at 20 °C under CO2-free condition.
2.3. Chromosome preparation and banding
Lymphocytes were cultured for 6 days and exposed to 500 ng/ml colcemid for 2.5 h before harvest. The cells were collected by centrifugation (300×g, 7 min) and treated with 0.075 M KCl for 20 min at room temperature. The suspension was then fixed in methanol/acetic acid (3:1) and spread on clean glass slides using an air-drying method. For karyotyping, the slides were stained with 3% Giemsa solution (pH 6.8) for 10 min. The chromosomes were counted on printed pictures using a colony counter. C-bands were obtained by the barium hydroxide/saline/Giemsa (BSG) method.20 C-banding and Ag-NOR staining21 were performed on slides previously used for fluorescence in situ hybridization (FISH) analysis.
2.4. Molecular cloning of repetitive sequence
Extraction of genomic DNA and molecular cloning of repetitive sequences were performed as previously described.22 Genomic DNA was digested with restriction enzymes, electrophoresed on a 1.2% agarose gel, and DNA bands containing repetitive sequences were isolated from the gel and cloned into vectors. After the nucleotide sequences of the cloned repetitive sequences were obtained, their chromosome distribution, genomic organization, and sequence conservation in other lamprey species were examined. For detailed information, see Supplementary data.
2.5. Molecular phylogeny
Phylogenetic trees of the repetitive DNA sequences were constructed using the neighbor-joining (NJ) method as implemented in MEGA 6.0 software.23 We used the Kimura-2-parameter model for nucleotide sequences and the Poisson model for amino acid sequences with Gamma distribution parameter α = 4 for rates and pattern settings. The bootstrap branch-supporting statistic was calculated for 1,000 replicates.
2.6. FISH
To examine the chromosomal distribution of the repetitive sequences isolated in this study, 18S–28S and 5S rRNA genes, and telomeric TTAGGG repeats, FISH analysis was performed as previously described.24 For detailed information, see Supplementary data.
2.7. Hybridization analysis of DNA blots
The genomic organization of repetitive sequences and their sequence conservation were examined by Southern blot hybridization and slot-blot hybridization, respectively. For detailed methods, see Supplementary data.
3. Results
3.1. Karyotype of Le. camtschaticum and the chromosomal location of the major rRNA genes and telomeric repeat sequences
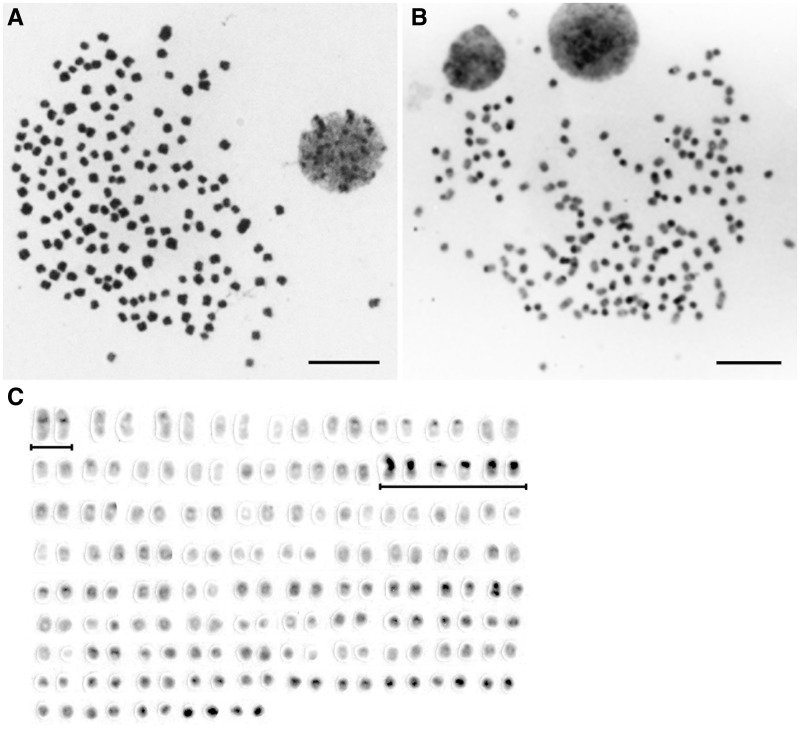
Figure 1A shows a Giemsa-stained metaphase spread of Le. camtschaticum, which consists of mostly small acrocentric or dot-shaped chromosomes and several pairs of small submetacentric chromosomes. The modal diploid chromosome number for 81 metaphase spreads was 168 (Supplementary Table S1). We collected chromosome samples from five individuals (two males and three females), and no differences in chromosome number and size were found between males and females. C-banding detected distinctive C-positive heterochromatin blocks on ∼30 pairs of chromosomes (Fig. 1B and C). Large C-bands were localized to the centromeric regions of the largest pair of submetacentric chromosomes and the centromeric regions of three other pairs of chromosomes. Distinctive C-bands were also located in other many small acrocentric and dot-shaped chromosomes.
Figure 1.
Giemsa-stained and C-banded chromosomes of Lethenteron camtschaticum. (A) Giemsa-stained metaphase spread. (B) C-banded metaphase spread. (C) C-banded karyotype. Scale bars represent 10 µm. The largest pair of submetacentric chromosomes and three pairs of chromosomes that have large C-bands are underlined.
FISH signals produced by the fluorescent probe for the 18S–28S rRNA genes were localized to six chromosomes; two signals were located in the pericentromeric regions of submetacentric chromosomes, and the others were in the terminal regions of four acrocentric chromosomes, which corresponded to C-band-positive heterochromatin regions (Supplementary Fig. S1A). To identify the active sites of the 18S–28S rRNA genes, we performed silver staining on the same chromosome slides used for FISH analysis of the 18S–28S rRNA genes.25 Ag-NORs were observed in the pericentromeric region of one submetacentric chromosome pair, but not on the other four acrocentric chromosomes bearing 18S–28S rRNA signals (Supplementary Fig. S1B). Only slight variations in the size of the Ag-NOR signals were observed among the individuals examined (Supplementary Fig. S1C). The 5S rDNA signals were detected on a single pair of small chromosomes (Supplementary Fig. S1D), which was different from the chromosomes on which the 18S–28S rRNA signals and C-positive bands were localized (data not shown). FISH signals for TTAGGG repeat sequences were detected on the terminal ends of all chromosomes, and there were large differences in the size of signals between chromosomes (Supplementary Fig. S1E).
3.2. Nucleotide sequences of the repetitive sequence families in the Le. camtschaticum genome
We isolated repetitive sequences of the Le. camtschaticum genome by agarose gel electrophoresis of genomic DNA digested with restriction endonucleases (Supplementary Fig. S2). Nine clones obtained from a 200-bp EcoRI band were grouped into the same repetitive sequence family, named LCA-EcoRIa (Supplementary Fig. S3A). The lengths of the nine fragments (LC149791–LC149799) were 200–202 bp, and the sequence similarity between the fragments ranged from 93.7 to 100.0%. The average GC content of the fragments was 47.5% (range 47.0–48.0%). Another EcoRI sequence family, LCA-EcoRIb, was isolated from a 360-bp EcoRI band (Supplementary Fig. S3B). The length of all 11 isolated fragments (LC149800–LC149810) was 364 bp, and the sequence similarity between fragments ranged from 96.1 to 99.7%. The sequences were AT-rich, and the average GC content was 41.5% (range 40.4–42.0%). Seven clones of the LCA-ApaI sequence family were isolated from a 700-bp ApaI band. The lengths of the seven fragments (LC149811–LC149817) ranged from 700 to 702 bp, and the consensus length was 702 bp (Supplementary Fig. S3C). The sequence similarity between the fragments ranged from 91.3 to 99.4%. In contrast to the LCA-EcoRIa and LCA-EcoRIb sequences, the LCA-ApaI sequences were GC-rich, and the average GC content was 57.2% (range 56.7–57.8%).
3.3. Similarity of the repetitive sequence families to other nucleotide sequences
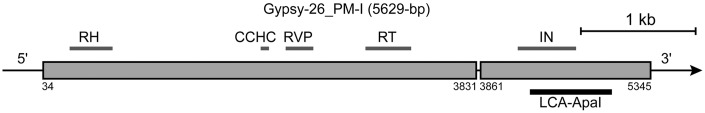
Homology searches using the three repetitive sequence families were performed using the NCBI non-redundant sequence database (http://blast.ncbi.nlm.nih.gov) and Repbase (http://www.girinst.org/repbase/).26 The 200-bp LCA-EcoRIa consensus sequence showed high sequence identity with the nucleotide sequence of a BAC clone (KF318012) isolated from this species, with 94.5% identity at positions 6–205 and 97.6% identity from 206 to 371 in KF318012. The search also identified two partial sequences in a tandem repeat element, rpt200, in Germ1 (GQ215662) of P. marinus, which is a genomic DNA sequence that is eliminated from germ cells by a programmed genome rearrangement.27 The sequences showed 76.8% identity at positions 5,133–5,283 (named as the rpt200_1 fragment) and 80.9% identity at positions 7,489–7,556 (rpt200_2) in the rpt200 element (Fig. 2A). The LCA-EcoRIb consensus sequence showed 84.8, 89.4, and 89.1% identity to the EcoRI family of satellite DNA sequences in P. marinus (X92515), La. planeri, and La. zanandreai, respectively (Fig. 2B).12,18 Short repeats, such as centromeric motifs, that appear in the centromeric sequences of several eukaryotic species,28–31 were detected in the consensus sequences of LCA-EcoRIb as well as the EcoRI satellite DNA sequences of the three lamprey species12,18 (Fig. 2B). A search using the LCA-ApaI sequence identified a partial sequence of the P. marinus long terminal repeat (LTR) retrotransposon Gypsy-26_PM-I in Repbase (Fig. 2C). A search using the InterProScan tool (http://www.ebi.ac.uk/interpro/) revealed that LCA-ApaI shared 70.7% identity with a partial region of the integrase domain at position 4,252–4,954 of Gypsy-26_PM-I (Fig. 3).
Figure 2.
Comparison of the nucleotide sequences of the LCA-EcoRIa, LCA-EcoRIb, and LCA-ApaI sequences with homologous sequences. (A) Alignment of the LCA-EcoRIa consensus sequence with two partial sequences of the tandem repeat sequence rpt200 (rpt200_1 at positions 5,133–5,283 and rpt200_2 at positions 7,489–7,556 of the Germ1 sequence (GQ215662) of P. marinus. (B) Alignment of the LCA-EcoRIb consensus sequence with the EcoRI satellite DNA sequence families from La. planeri (LPL),12 La. zanandreai (LZA),12 and P. marinus (PMA; X92515).18 The 6-bp motif (A/T)(G/C)AAA(T/C) and its complementary sequence, which are homologous to centromeric motifs observed in other vertebrates,28–31 are shown in squares on the consensus sequence of LCA-EcoRIb. (C) Alignment of the LCA-ApaI consensus sequence with a partial sequence of the P. marinus Ty3/Gypsy family of LTR retrotransposons (Gypsy-26_PM-I at positions 4,252–4,954). Dots indicate identity with the LCA-EcoRIa, LCA-EcoRIb, or LCA-ApaI consensus sequence. The numbers at the end of lines are the sequence lengths.
Figure 3.
Schematic representation of the Ty3/Gypsy family of LTR retrotransposons based on the Gypsy-26_PM-I sequence of P. marinus. Gypsy-26_PM-I contains sequences encoding a reverse transcriptase (RT), retroviral aspartyl protease (RVP), ribonuclease H (RH), and integrase (IN). The region at positions 4,252–4,954, which is homologous to the LCA-ApaI sequence, is shown with a bold underline. Bold lines above the ORF indicate the conserved domains.
3.4. Chromosomal distribution of the repetitive sequences
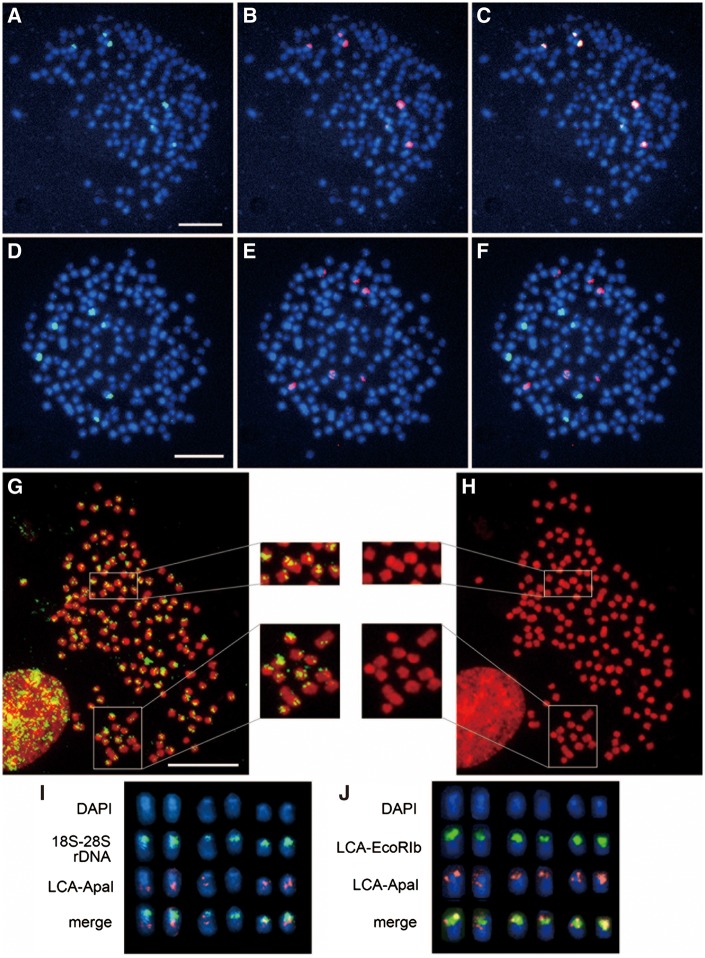
LCA-EcoRIa 2 (LC149792), LCA-EcoRIb 4 (LC149803), and LCA-ApaI 1 (LC149811) fragments were used for FISH analysis as representative probes of each repetitive sequence family. The results showed that the LCA-EcoRIa sequences were localized to the pericentromeric regions of six chromosomes (Fig. 4A), and the hybridization signals completely overlapped the signals for the 18S–28S rRNA genes (Fig. 4B and C). Hybridization signals for the LCA-EcoRIb sequence were observed in the centromeric regions of six chromosomes (Fig. 4D), which differed from the chromosomes bearing the 18S–28S rRNA genes (Fig. 4E and F). The LCA-ApaI sequence was localized to the centromeric and/or pericentromeric regions of almost all chromosomes. C-band positive bands were also located in the centromeric regions of many small acrocentric and dot-shaped chromosomes (Fig. 1B and C), indicating that the C-positive heterochromatin regions overlapped or partially overlapped the signals of LCA-ApaI sequence. However, the size of signals differed among the chromosomes (Fig. 4G and H). The signals were generally located in the centromeric regions on PI-positive blocks and/or in pericentromeric regions. Approximately 10 chromosomes showed large signals, and several large submetacentric chromosomes had more than 1 hybridization site. Two-color FISH analysis of LCA-ApaI and the 18S–28S rRNA genes showed that the LCA-ApaI sequences were located adjacent to the 18S–28S rRNA gene cluster but hardly overlapped it (Fig. 4I). However, the LCA-ApaI signals partially overlapped the LCA-EcoRIb signals (Fig. 4J).
Figure 4.
Chromosomal distribution of the LCA-EcoRI and LCA-ApaI sequences. (A) DAPI-stained metaphase spread hybridized with biotin-labeled LCA-EcoRIa (green). (B) The same metaphase spread in (A) hybridized with a DIG-labeled 18S–28S rRNA gene probe (red). (C) Merged image of the signals for biotin-labeled LCA-EcoRIa and DIG-labeled 18S–28S rDNA. (D–F) Hybridization patterns of biotin-labeled LCA-EcoRIb (green) (D) and DIG-labeled 18S–28S rDNA (red) (E) on the same DAPI-stained metaphase spread and their merge (F). (G, H) Hybridization patterns of biotin-labeled LCA-ApaI (green) on a PI-stained metaphase spread (G) and the PI-stained metaphase spread (H). Enlarged photographs of chromosomes hybridized to the LCA-ApaI probe in two parts of the metaphase spread are shown. Squares with white outlines on the metaphase spreads indicate the magnified areas. (I) Comparison of the hybridization patterns of biotin-labeled 18S–28S rDNA (green) and DIG-labeled LCA-ApaI (red) on six DAPI-stained chromosomes. (J) Comparison of the chromosome distribution patterns of biotin-labeled LCA-EcoRIb (green) and DIG-labeled LCA-ApaI (red). In the merged images (I, J), the regions of overlap between the two probes are shown in yellow. Scale bars represent 10 µm. LCA-EcoRIb 4 (LC149803) and LCA-ApaI 1 (LC149811) fragments were used as probes.
3.5. Molecular phylogeny of the LCA-EcoRIb and LCA-ApaI sequences
We performed phylogenetic analyses to examine the evolutionary relationships between the LCA-EcoRIb sequence and the EcoRI satellite sequences of other lamprey species as well as the LCA-ApaI sequence and Ty3/Gypsy LTR retrotransposons of other species (Supplementary Fig. S4). No sequences homologous to LCA-EcoRIa were found other than the rpt200 sequence of P. marinus; therefore, we did not perform molecular phylogenetic analysis of this sequence. NJ tree analysis of the EcoRI satellite repeats of lampreys revealed that all fragments of LCA-EcoRIb formed a clade supported by a high bootstrap value (100%), which was distinct from the clade containing the other three lamprey species (Supplementary Fig. S4A). Phylogenetic analysis of the LCA-ApaI sequences was performed using sequences encoding the integrase domain of the LTR retrotransposons obtained from GenBank (http://www.ncbi.nlm.nih.gov; Supplementary Table S2), most of which were used in a previous phylogenetic study.32 A close phylogenetic relationship between the LCA-ApaI sequences and Gypsy-26_PM-I of P. marinus was supported by a high bootstrap value (100%; Supplementary Fig. S4B).
3.6. Genomic organization of the repetitive sequences
FISH data revealed that LCA-EcoRIa is located in the spacer region of the 18S–28S rRNA gene cluster and tandem duplicated with the 18S–28S rDNA; therefore, we did not study the genomic organization of this repeated sequence. To examine the organization of the LCA-EcoRIb and LCA-ApaI sequences in the Le. camtschaticum genome, we performed Southern blot hybridizations using genomic DNA digested with AluI, ApaI, EcoRI, HpaII, MspI, and PstI (Supplementary Fig. S5). In a blot probed with LCA-EcoRIb, polymeric ladder bands of the 364-bp monomer units were detected in the EcoRI and AluI digests, which are characteristic of tandem repeat satellite DNA sequences (Supplementary Fig. S5A). Additional hybridization bands were observed interstitially in the ladder bands of the 364-bp monomer units in the AluI digests, which were derived from internal AluI cleavage sites in the monomer unit (Supplementary Fig. S5A). In contrast to the EcoRI and AluI digests, intense hybridization signals were observed at higher molecular weights in the ApaI, PstI, and MspI (HpaII) digests, indicating that these restriction sites are not highly conserved. Genomic DNA digested with the methylation-sensitive HpaII and methylation-insensitive MspI enzymes produced the same hybridization patterns, which indicates that their cleavage sites are not methylated.
Hybridization with LCA-ApaI also resulted in a ladder of signals consisting of multiples of the 702-bp monomer in the ApaI digest (Supplementary Fig. S5B). The intensity of ladder bands increased for higher-order repeating structures, indicating that the ApaI site was less conserved in the tandem arrays. In contrast, AluI and MspI sites in the tandem arrays were highly conserved because no multimeric bands were detected in the AluI and MspI digests. The hybridization bands at lower molecular weight than the 702-bp band in the AluI and MspI digests were derived from the internal cleavage sites contained in the monomer unit (Supplementary Fig. 3C). The interstitial band positioned between the monomeric and dimeric bands in the ApaI digest suggested that the ApaI site was highly conserved in another additional unit of the LCA-ApaI sequence. The hybridization bands smaller than the monomer unit were observed in the MspI digest, whereas higher molecular weight bands were observed in the HpaII digest blot, indicating that the LCA-ApaI sequence was highly methylated.
3.7. Sequence conservation of the repetitive sequences
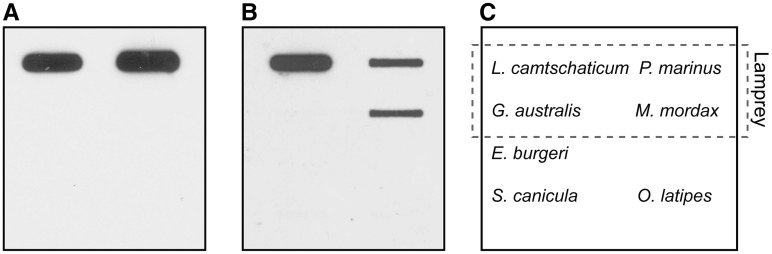
Conservation of the LCA-EcoRIb and LCA-ApaI sequences was examined by slot-blot hybridization using genomic DNA from four Petromyzontiformes species [Le. camtschaticum and P. marinus (Petromyzontidae), Geotria australis (Geotriidae), and Mordacia mordax (Mordaciidae)], inshore hagfish (Eptatretus burgeri, Myxinidae, Myxiniformes), lesser spotted catshark (Scyliorhinus canicula, Scyliorhinidae, Carcharhiniformes), and medaka (Oryzias latipes, Adrianichthyidae, Beloniformes) (Fig. 5). Using the LCA-EcoRIb probe, intense chemiluminescent hybridization signals were detected in P. marinus, which is in the same family (Petromyzontidae) as Le. camtschaticum. No signals were detected in the other species (Fig. 5A). Using the LCA-ApaI probe, an intense hybridization signal was detected in Le. camtschaticum, and less intense signals were observed in P. marinus and M. mordax (Fig. 5B).
Figure 5.
Slot-blot hybridization of the LCA-EcoRIb (A) and LCA-ApaI (B) sequences in four lamprey species (Lethenteron camtschaticum, Petromyzon marinus, Geotria australis, and Mordacia mordax), the inshore hagfish (Eptatretus burgeri), the lesser spotted catshark (Scyliorhinus canicula), and the medaka (Oryzias latipes). (C) A schematic representation of the blotted genomic DNAs is shown. Each lane contained 100 ng of genomic DNA. LCA-EcoRIb 4 (LC149803) and LCA-ApaI 1 (LC149811) fragments were used as probes.
4. Discussion
Here, we determined the chromosome number and C-banded karyotype of Le. camtschaticum in somatic cells obtained from lymphocyte cultures. The diploid chromosome number was 168, which was not consistent with that reported in a previous study (2n = 144–162).19 This discrepancy may be due to technical artifacts owing to the extremely small size and high number of tiny chromosomes in lampreys. The chromosome number of Le. camtschaticum determined in this study was similar to those reported in somatic cells of Lampetra (La. fluviatilis [2n = 164], La. lamottenii [2n = 166], La. planeri [2n = 164], La. zanandreai [2n = 164]),8,12 and Petromyzon marinus (2n = 164–168).7,10,14 C-positive bands were distributed in the centromeric regions of ∼30 pairs of chromosomes, which was more than the 20 pairs reported in La. zanandreai.12 In this study, the 18S–28S rRNA genes were localized to six chromosomes; however, the gene copies were only active in a single pair of submetacentric chromosomes. In other species, the 18S–28S rRNA genes were localized to the pericentromeric regions of four and two chromosomes in somatic cells of La. zanandreai12 and P marinus,14 respectively. These results suggest that the chromosomal location of the 18S–28S rRNA genes in lampreys is variable. However, the chromosomal location of the 5S rRNA genes has not been reported for any other lampreys than Le. camtschaticum, in which, these genes are localized to two chromosomes. The 5S rRNA genes are generally localized to a single pair of chromosomes in fishes, amphibians, and mammals,33 suggesting that chromosomal distribution of the 5S rRNA genes is conserved in vertebrates.
We isolated three families of repetitive sequences from Le. camtschaticum, two AT-rich sequences (LCA-EcoRIa and LCA-EcoRIb) and one GC-rich sequence (LCA-ApaI). The LCA-EcoRIa sequence, which co-localized with the 18S–28S rDNA, showed significant homology with the tandem rpt200 repeat of P. marinus. This sequence is located in the spacer regions between the intact 18S rDNA and fragmented 28S rDNA sequences in P. marinus, which are found in the Germ1 sequence (GQ215662) that is eliminated from germ cells by a programmed genome rearrangement.27 In the primary spermatocytes of P. marinus (2n = 198) at meiotic prophase I, FISH signals for rpt200 and 28S rDNA were distributed on about 20/99 and 10/99 chromosome pairs, respectively, and the signals co-localized on some chromosomes.14 However, in somatic cells of P. marinus (2n = 164–168), the rpt200 signals were localized to ∼14 pairs of chromosomes, and 28S rDNA signals were observed on only one pair of chromosomes, to which the rpt200 signal co-localized.14 This result indicates that most 18S–28S rDNA copies and several rpt200 copies are eliminated in somatic cells.14,27 The germline DNA loss in somatic cells has also been reported in the Korean lamprey (Lampetra morii, Petromyzontidae) recently.34 Therefore, the presence and co-localization of the rpt200-related LCA-EcoRIa sequence and 18S–28S rDNA in a small number of chromosome pairs in somatic cells of Le. camtschaticum might be related to a programmed genome rearrangement in this species.
The LCA-EcoRIb sequence is a homolog of an EcoRI satellite DNA sequence that has been isolated from three lamprey species (La. planeri, La. zanandreai, and P. marinus).12,18 These AT-rich EcoRI sequence families, including LCA-EcoRIb, range from 362 to 370 bp, and the sequence divergence between LCA-EcoRIb and the EcoRI satellite DNAs is relatively low (10.6–15.2%). These sequences contain short repeats, such as centromeric motifs, which are also observed in the centromeric sequences of several eukaryotic species such as bream, trout, mouse, and human,28–31 suggesting that these satellite DNAs and the LCA-EcoRIb sequence could be important for centromere structure and function in lampreys. A zoo-blot hybridization revealed that the EcoRIb sequence is conserved within Petromyzontidae, and it is distributed on six chromosomes in the somatic cells of Le. camtschaticum, on two chromosomes in La. zanandreai,12 and on 34 chromosomes in P. marinus.14 These results suggest that the EcoRI satellite sequence might have been acquired in the Petromyzontidae lineage and amplified on many more chromosomes in Petromyzon than in Lampetra and Lethenteron through homogenization between non-homologous chromosomes, called concerted evolution (reviewed in Elder and Turner35 and Plohl et al.36).
A search of RepBase revealed that the LCA-ApaI sequence showed significant homology with a partial integrase-coding region of the Ty3/Gypsy retrotransposon of P. marinus (Gypsy-26_PM-I), indicating that the LCA-ApaI sequence might be a LTR retrotransposon-derived repeat. LTR retrotransposons of the Ty3/Gypsy superfamily have been isolated from fungi, plants, and animals.37–39 The results of the slot-blot analysis indicate that the LCA-ApaI sequence is well conserved in Petromyzontidae and Mordaciidae, but not in Geotriidae. Mordaciidae, which is distributed in the southern hemisphere, has a lower chromosome number (2n = 76) than Petromyzontidae (2n = 142–174) in the northern hemisphere and Geotriidae (∼180 chromosomes) in the southern hemisphere, suggesting that extensive centric fusions occurred in Mordaciidae.9,13 Molecular phylogenetic analyses suggested that Mordaciidae is in a separate clade from Petromyzontidae and Geotriidae,40–43 which is reflected by the difference in chromosome numbers between Petromyzontidae/Geotriidae and Mordaciidae. The phylogenetic relationship among these three families strongly suggests that the LCA-ApaI sequence appeared in the common ancestor of Petromyzontiformes and amplified in Petromyzontidae and Mordaciidae but not in Geotriidae, in which no hybridization signal was detected by slot blot, although it is also possible that the LCA-ApaI sequence disappeared in the Geotriidae lineage.
A draft genome assembly of P. marinus has been reported, and 7,752 distinct families of repetitive sequences were identified, accounting for 34.7% of the genome.44 The genome sequence of P. marinus will facilitate cloning of repetitive sequences from another lamprey species and their comparison, which is needed to provide further insights into the molecular basis of chromosome structure and evolution in lampreys.
Accession numbers
Conflict of interest
None declared.
Supplementary data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
Funding
This work was supported by a Grant-in-Aid for Scientific Research on Innovative Areas (No. 23113004) and a Grant-in-Aid for Scientific Research (B) (No. 22370081) from the Ministry of Education, Culture, Sports, Science and Technology of Japan. Our gratitude extends to suppliers of genomic DNA or tissues of cyclostome and chondrichthyan species, namely, H. Kawauchi, R. Kusakabe, S. Moriyama, M. Nozaki, K.G. Ota, J. Joss, S. Kuratani, and A. Takahashi.
Supplementary Material
References
- 1.Stock D. W., Whitt G. S. 1992, Evidence from 18S ribosomal RNA sequences that lampreys and hagfishes form a natural group. Science, 257, 787–89. [DOI] [PubMed] [Google Scholar]
- 2.Kuraku S., Kuratani S. 2006, Time scale for cyclostome evolution inferred with a phylogenetic diagnosis of hagfish and lamprey cDNA sequences. Zool. Sci., 23, 1053–64. [DOI] [PubMed] [Google Scholar]
- 3.Kuraku S., Ota K.G., Kuratani S. 2009, Cyclostomata In: Hedges S.B., Kumar S. eds. Timetree of Life. New York: Oxford University Press, pp. 317–9. [Google Scholar]
- 4.Potter I.C., Gill H.S., Renaud C.B. 2014, Petromyzontidae: lampreys In: Burr B.M., Warren M.L., Jr., eds.North American Freshwater Fishes: Natural History, Ecology, Behavior, and Conservation. Baltimore: Johns Hopkins University Press, pp. 106–39. [Google Scholar]
- 5.Potter I.C., Strahan R. 1968, The taxonomy of the lampreys Geotria and Mordacia and their distribution in Australia. Proc. Linn. Soc. Lond., 179, 229–40. [Google Scholar]
- 6.Gill H.S., Renaud C.B., Chapleau F., Mayden R.L., Potter I.C., Douglas M.E. 2003, Phylogeny of living parasitic lampreys (Petromyzontiformes) based on morphological data. Copeia, 2003, 687–703. [Google Scholar]
- 7.Potter I.C., Rothwell B. 1970, The mitotic chromosomes of the lamprey, Petromyzon marinus L. Experientia, 26, 429–30. [DOI] [PubMed] [Google Scholar]
- 8.Robinson E.S., Potter I.C., Webb C.J. 1974, Homogeneity of holarctic lamprey karyotypes. Caryologia, 27, 443–54. [Google Scholar]
- 9.Robinson E.S., Potter I.C. 1981, The chromosomes of the southern hemispheric lamprey, Geotria australis Gray. Experientia, 37, 239–40. [Google Scholar]
- 10.Smith J.J., Stuart A.B., Sauka-Spengler T., Clifton S.W., Amemiya C.T. 2010, Development and analysis of a germline BAC resource for the sea lamprey, a vertebrate that undergoes substantial chromatin diminution. Chromosoma, 119, 381–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arai R. 2011, Fish Karyotypes: A Check List. Heidelberg: Springer. [Google Scholar]
- 12.Caputo V., Giovannotti M., Cerioni P.N., Splendiani A., Tagliavini J., Olmo E. 2011, Chromosomal study of a lamprey (Lampetra zanandreai Vladykov, 1955) (Petromyzonida: Petromyzontiformes): conventional and FISH analysis. Chromosome Res., 19, 481–91. [DOI] [PubMed] [Google Scholar]
- 13.Caputo Barucchi V., Giovannotti M., Nisi Cerioni P., Splendiani A. 2013, Genome duplication in early vertebrates: insights from agnathan cytogenetics. Cytogenet. Genome Res., 141, 80–9. [DOI] [PubMed] [Google Scholar]
- 14.Covelo-Soto L., Morán P., Pasantes J.J., Pérez-García C. 2014, Cytogenetic evidences of genome rearrangement and differential epigenetic chromatin modification in the sea lamprey (Petromyzon marinus). Genetica, 142, 545–54. [DOI] [PubMed] [Google Scholar]
- 15.Singer M.F. 1982, Highly repeated sequences in mammalian genomes. Int. Rev. Cytol., 76, 67–112. [DOI] [PubMed] [Google Scholar]
- 16.Murphy T.D., Karpen G.H. 1998, Centromeres take flight: alpha satellite and the quest for the human centromere. Cell, 93, 317–20. [DOI] [PubMed] [Google Scholar]
- 17.Henikoff S., Ahmad K., Malik H.S. 2001, The centromere paradox: stable inheritance with rapidly evolving DNA. Science, 293, 1098–102. [DOI] [PubMed] [Google Scholar]
- 18.Boán F., Viñas A., Rodríguez J.M., Sánchez L., Gómez-Márquez J. 1996, A new EcoRI family of satellite DNA in lampreys. FEBS Lett., 394, 187–90. [DOI] [PubMed] [Google Scholar]
- 19.Kitada J., Tagawa M. 1975, Somatic chromosomes of three species of Cyclostomata. Chrom. Inform. Serv., 18, 10–2. [Google Scholar]
- 20.Sumner A.T. 1972, A simple technique for demonstrating centromeric heterochromatin. Exp. Cell Res., 75, 304–6. [DOI] [PubMed] [Google Scholar]
- 21.Howell W.M., Black D.A. 1980, Controlled silver-staining of nucleolus organizer regions with a protective colloidal developer: a 1-step method. Experientia, 36, 1014–5. [DOI] [PubMed] [Google Scholar]
- 22.Uno Y., Asada Y., Nishida C., Takehana Y., Sakaizumi M., Matsuda Y. 2013, Divergence of repetitive DNA sequences in the heterochromatin of medaka fishes: molecular cytogenetic characterization of constitutive heterochromatin in two medaka species: Oryzias hubbsi and O. celebensis (Adrianichthyidae, Beloniformes). Cytogenet. Genome Res., 141, 212–26. [DOI] [PubMed] [Google Scholar]
- 23.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. 2013, MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol., 30, 2725–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsuda Y., Chapman V.M. 1995, Application of fluorescence in situ hybridization in genome analysis of the mouse. Electrophoresis, 16, 261–72. [DOI] [PubMed] [Google Scholar]
- 25.Howell W.M., Hsu T.C., Block B.M. 1977, Visualization of centriole-bodies using silver stain. Chromosoma, 65, 9–20. [DOI] [PubMed] [Google Scholar]
- 26.Kohany O., Gentles A.J., Hankus L., Jurka J. 2006, Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinformatics, 25, 474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith J.J., Antonacci F., Eichler E.E., Amemiya C.T. 2009, Programmed loss of millions of base pairs from a vertebrate genome. Proc. Natl. Acad. Sci. U.S.A., 106, 11212–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wong A. K., Rattner J. B. 1988, Sequence organization and cytological localization of the minor satellite of mouse. Nucleic Acids Res., 16, 11645–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vissel B., Nagy A., Choo K. H. 1992, A satellite III sequence shared by human chromosomes 13, 14, and 21 that is contiguous with alpha satellite DNA. Cytogenet. Cell Genet., 61, 81–6. [DOI] [PubMed] [Google Scholar]
- 30.Garrido-Ramos M. A., Jamilena M., Lozano R., Ruiz Rejón C., Ruiz Rejón M. 1995, The EcoRI centromeric satellite DNA of the Sparidae family (Pisces, Perciformes) contains a sequence motive common to other vertebrate centromeric satellite DNAs. Cytogenet. Cell Genet., 71, 345–51. [DOI] [PubMed] [Google Scholar]
- 31.Caputo V., Giovannotti M., Nisi Cerioni P., Splendiani A., Olmo E. 2009, Chromosomal study of native and hatchery trouts from Italy (Salmo trutta complex, Salmonidae): conventional and FISH analysis. Cytogenet. Genome Res., 124, 51–62. [DOI] [PubMed] [Google Scholar]
- 32.Malik H.S., Eickbush T.H. 1999, Modular evolution of the integrase domain in the Ty3/Gypsy class of LTR retrotransposons. J. Virol., 73, 5186–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martins C., Wasko A.P. 2004, Organization and evolution of 5S ribosomal DNA in the fish genome In: Williams C.R., ed. Focus on Genome Research. Hauppauge: Nova Science Publishers; pp. 289–318. [Google Scholar]
- 34.Yan X., Meng W., Wu F., Xu A., Chen S., Huang S. 2016, The nuclear DNA content and genetic diversity of Lampetra morii. PLoS ONE, 11, e0157494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elder J. F., Turner B. J. 1995, Concerted evolution of repetitive DNA sequences in eukaryotes. Q Rev Biol, 70, 297–320. [DOI] [PubMed] [Google Scholar]
- 36.Plohl M., Luchetti A., Meštrović N., Mantovani B. 2008, Satellite DNAs between selfishness and functionality: structure, genomics and evolution of tandem repeats in centromeric (hetero)chromatin. Gene, 409, 72–82. [DOI] [PubMed] [Google Scholar]
- 37.Miller K., Lynch C., Martin J., Herniou E., Tristem M. 1999, Identification of multiple Gypsy LTR-retrotransposon lineages in vertebrate genomes. J. Mol. Evol., 49, 358–66. [DOI] [PubMed] [Google Scholar]
- 38.Llorens C., Muñoz-Pomer A., Bernad L., Botella H., Moya A. 2009, Network dynamics of eukaryotic LTR retroelements beyond phylogenetic trees. Biol. Direct, 4, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Piskurek O., Nishihara H., Okada N. 2009, The evolution of two partner LINE/SINE families and a full-length chromodomain-containing Ty3/Gypsy LTR element in the first reptilian genome of Anolis carolinensis. Gene, 441, 111–8. [DOI] [PubMed] [Google Scholar]
- 40.Baldwin J., Mortimer K., Patak A. 1988, Evolutionary relationships among lamprey families: Amino acid composition analysis of lactate dehydrogenase. Biochem. Syst. Ecol., 16, 351–3. [Google Scholar]
- 41.Silver M.R., Kawauchi H., Nozaki M., Sower S.A. 2004, Cloning and analysis of the lamprey GnRH-III cDNA from eight species of lamprey representing the three families of Petromyzoniformes. Gen. Comp. Endocrinol., 139, 85–94. [DOI] [PubMed] [Google Scholar]
- 42.Takahashi A., Nakata O., Moriyama S., et al. 2006, Occurrence of two functionally distinct proopiomelanocortin genes in all modern lampreys. Gen. Comp. Endocrinol., 148, 72–8. [DOI] [PubMed] [Google Scholar]
- 43.Kuraku S. 2013, Impact of asymmetric gene repertoire between cyclostomes and gnathostomes. Semin. Cell Dev. Biol., 24, 119–27. [DOI] [PubMed] [Google Scholar]
- 44.Smith J.J., Kuraku S., Holt C., et al. 2013, Sequencing of the sea lamprey (Petromyzon marinus) genome provides insights into vertebrate evolution. Nat. Genet., 45, 415–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.