Abstract
Hibiscus syriacus (L.) (rose of Sharon) is one of the most widespread garden shrubs in the world. We report a draft of the H. syriacus genome comprised of a 1.75 Gb assembly that covers 92% of the genome with only 1.7% (33 Mb) gap sequences. Predicted gene modeling detected 87,603 genes, mostly supported by deep RNA sequencing data. To define gene family distribution among relatives of H. syriacus, orthologous gene sets containing 164,660 genes in 21,472 clusters were identified by OrthoMCL analysis of five plant species, including H. syriacus, Arabidopsis thaliana, Gossypium raimondii, Theobroma cacao and Amborella trichopoda. We inferred their evolutionary relationships based on divergence times among Malvaceae plant genes and found that gene families involved in flowering regulation and disease resistance were more highly divergent and expanded in H. syriacus than in its close relatives, G. raimondii (DD) and T. cacao. Clustered gene families and gene collinearity analysis revealed that two recent rounds of whole-genome duplication were followed by diploidization of the H. syriacus genome after speciation. Copy number variation and phylogenetic divergence indicates that WGDs and subsequent diploidization led to unequal duplication and deletion of flowering-related genes in H. syriacus and may affect its unique floral morphology.
Keywords: Hibiscus syriacus, Whole Genome Duplication, Diploidization, Multivoltinism, Homeolog
1. Introduction
Hibiscus syriacus (L.) (rose of Sharon) is a fast-growing deciduous shrub of the Malvaceae family, which includes species such as Gossypium raimondii and Theobroma cacao. Although its’ name indicates this species was first identified in Syria, H. syriacus likely originated from the Korean peninsula and southern China and has since spread to Western countries. In temperate zones, H. syriacus is a commonly grown ornamental species with attractive white, pink, red, lavender, or purple flowers displayed over a long blooming period, though individual flowers last only a day. Its Korean name, Mugunghwa, literally means ‘flowering forever’. In addition to its ornamental value, H. syriacus acts as an ozone bioindicator,1 and its dried flowers and root bark are used in Oriental herbal medicines. Specifically, a novel cyclic peptide (Hibispeptin A) and three naphthalene compounds (syriacusins A-C) isolated from the plant’s root bark have been used as anti-pyretic, anti-helminthic and anti-fungal agents.2,3
Polyploidy is a well-established influence on plant genome evolution but is now recognized as a common phenomenon in diverse eukaryotes,4,5 as signs of whole-genome duplication (WGD) have been detected in many sequenced genomes. Recent genome analysis demonstrated that most eudicot plants descended from an ancient hexaploid ancestor and followed lineage-specific polyploidization6 and that two rounds of WGD occurred in ancestral vertebrates.7 In general, changes in ploidy are expected to be deleterious and an ‘evolutionary dead end’ for many species.8 However, polyploidization of plants mediated their survival during the Cretaceous-Tertiary extinction event by increasing their genetic diversity.9 Each round of polyploidization was followed by many gene deletions (homeolog gene loss), interchromosomal rearrangements, neofunctionalization, and subfunctionalization.4,5 In Malvaceae plants, Gossypium includes five tetraploid taxa (AD1 to AD5, 2n = 4x) and 45 diploid taxa (2n = 2x).10 Among them, G. raimondii10 (DD, D-genome), G. arboreum11 (AA, A-genome) and G. hirsutum12 (AtDt) genomes were reported. Hibiscus also includes many polyploid species, such as H. syriacus (2n = 4x = 80), H. aspera (2n = 8x = 72), and H. rosa-sinensis (2n = 16x = 144) and diploid species [H. pedunculatus (2n = 2x = 30) and H. phoeniceus (2n = 2x = 22)].13
Here, we report the genome sequence of H. syriacus and the possible correlation between polyploidization and its phenotypes. Comparative genomic analysis of Malvaceae species, including H. syriacus, T. cacao, and G. raimondii (DD), provides clues of the recent polyploidization in H. syriacus by WGDs and unequal regulation of gene dosage by subsequent paleopolyploidy. Our investigation of copy number variations of floral regulators in Malvaceae plants also offers insight into the evolution of flowering phenotypes in H. syriacus. Moreover, the reference genome of H. syriacus is an important resource for identifying relationships between polyploidization and gene diversity. To our knowledge, this is the first report on whole genome sequence analysis of polyploidy woody plants and the effects of WGD on their unique phenotypes.
2. Materials and methods
2.1. Plant materials and whole genome sequencing
Leaves of H. syriacus plants >100-years-old and nominated as National Monument of Korea trees (serial number 520) were harvested and frozen immediately in liquid nitrogen. Genomic DNA for Paired-end (PE) and Mate-pair (MP) libraries was extracted, and libraries for next-generation sequencing were constructed according to the manufacturer’s instructions (Illumina, San Diego, CA, USA). The quality of each library was validated using the KAPA SYBR FAST Universal 2× qPCR Master Mix (Kapa Biosystems, Boston, MA, USA). Each library was sequenced with the Illumina HiSeq 2000 platform.
2.2. Genome assembly, scaffolding and gap-closing
Genome assembly was performed using both Platanus v1.2.114 and SSPACE v2.0.15 To generate longer initial contigs, single reads merged using FLASH v1.2.216 and reads from the PE libraries were assembled using Platanus with parameters to resolve heterozygosity in the H. syriacus genome (-u 0.2 -c 5 -d 0.3 -m 460). The scaffolding process was performed with Platanus and SSPACE. We first determined mapping seed length for scaffolding and then generated longer scaffolds using optimized Platanus parameters (-l 5 -s 41 -u 0.3). To extend scaffold length, SSPACE fulfilled serial scaffolding with hash parameters for the scaffolds generated by Platanus. Lastly, remaining gaps were filled with Platanus and GapCloser version 1.10 (http://soap.genomics.org.cn/down/GapCloser_release_2011.tar.gz) using reads from the PE and MP libraries.
2.3. Genome annotation
Annotation of the H. syriacus genome was performed using the KOBIC annotation pipeline (modified PGA pipeline17) consisting of repeat masking, mapping of different protein sequence sets, and ab initio prediction performed by AUGUSTUS v3.0.3.18 The protein sequences of A. thaliana (TAIR10, http://www.arabidopsis.org), T. cacao19 and G. raimondii10 were mapped using GeneWise v2.120 to generate protein-based gene models for consensus modeling. AUGUSTUS was used for gene prediction in H. syriacus. Then predicted gene models from AUGUSTUS were validated using BLASTp with protein sequences from the three genomes (T. cacao, G. raimondii and A. thaliana) as queries and erratic gene models were filtered with a BLASTp cut-off value of query coverage ≥ 0.3. The predicted gene models from GeneWise were also filtered using query coverage ≥ 0.3. Remaining gene models of GeneWise depicted as GeneWise format were reformatted as GFF3 data and used to determine the consensus gene model via EVidenceModeler (EVM),21 which combines ab initio gene predictions with protein alignments into weighted consensus gene structures (ab initio predictions = 1, protein alignment = 5, transcript alignment assemblies = 7). Biological functions of the final gene models were assigned using InterPro,22 plant protein sequences in the RefSeq23 and UniProt databases,24 which includes SWISS-PROT and TrEMBL data as described in previous study.17 For functional annotation, three quality criteria were concerned: (i) bit score of the BLAST result is >50 and e-value is <e−10; (ii) subject coverage of the BLAST result is >60%; and (iii) top token score from lexical analysis is >0.5. To infer function for the protein-coding genes, we used InterProScan25 version 5.4 to scan protein sequences against the protein signatures from InterPro.
2.4. RNA sequencing and de novo transcriptome assembly
Total RNA was extracted from plant leaves, petals, ovaries, and roots using TRIzol reagent (Invitrogen, CA, USA) following the manufacturer’s instructions. RNA-Seq libraries were generated using purified total RNA and sequenced using an Illumina HiSeq 2000 system. Thirty-six gigabases of raw reads were generated and preprocessed using DynamicTrim and LengthSort in SolexaQA.26 The preprocessed raw reads were then used for transcriptome assembly and DEG analysis. Velvet v1.2.07 was used to assess k-mer sizes and assembled contigs, which were then merged using Oases v0.2.08. Assembled transcripts were validated using BLASTx (e-value < e−10, best hit) against 1,917,424 protein sequences from 39 plant genomes selected from each family including Arabidopsis thaliana, Brassica rapa, Solanum lycopersicum, Solanum phureja, T. cacao, G. raimondii, Oryza sativa, Zea mays, Cucumis sativus, Vitis vinifera.
2.5. Evaluation of genome assembly
For validation of the assembled genome sequence, CEGMA (Core Eukaryotic Genes Mapping Apporoach) v2.527 and BUSCO (Benchmarking Universal Single-Copy Orthologs) v1.2228 were used in H. syriacus genome using default parameters. The CEGMA mapped a gene structures to new genomic sequence using a set of highly conserved protein family in eukaryotes by Hidden Markov Model. We evaluated 248 core eukaryotic genes defied by CEGMA to our genome sequence. The BUSCO provides completeness assessment of assembled genome based on orthologous group with single copy from OrthoDB (http://www.orthodb.org) using hidden Markov model for profile of amino acid alignments. For BUSCO assessments, we used 429 gene sets of conserved orthologs in eukaryotes.
2.6. Detection of gene families in the H. syriacus genome
OrthoMCL v2.0.229 was used to identify gene family clusters in H. syriacus and the other four sequenced genomes which are G. raimondii, T. cacao, A. trichopoda and A. thaliana (In the first step, a set of high quality of gene models was obtained by rejecting low-quality sequences based on default parameters in OrthoMCL. The default parameters of rejecting low quality protein sequences were (i) shorter than 10 amino acids (ii) >20% stop-codons (iii) >20% non-standard amino acids. Pairwise sequence similarities between all input protein sequences were calculated by all-by-all BLASTp with an e-value cut-off of 1e−05 and a minimum match length of 50%. To define ortholog cluster structure, a Markov clustering algorithm was applied with an inflation value (−I) of 1.5 (default value in OrthoMCL). Putative splice variants were removed from the data set; longest protein sequences were kept and subsequently filtered for premature stop codons and incompatible sequences.
2.7. Detection of collinearity blocks in Malvaceae plants
MCScanX30 was used to construct synteny and collinearity blocks between H. syriacus and G. raimondii against T. cacao.19 First, homologous gene pairs were identified using protein sequences from the three genomes and then scanned inter- and intra-species by BLASTp (options with −e 1e−10 –b 5 –v 5). The BLASTp output was used with merged GFFs of three species to perform MCscanX with default parameters. We generated gene synteny and collinearity data to align proteins of the two species against reference chromosome of T. cacao. Collinearity blocks containing fewer than five proteins were excluded. To search a candidate of duplicated regions, we made the groups of collinear block from multiple collinear blocks which have similar protein members (>80%), and the same chromosome in T. cacao. Then, each block in H. syriacus and G. raimondii was counted by overlap against the cluster blocks of T. cacao. The duplicated regions in H. syriacus and G. raimondii were identified, if the number of blocks was more than two.
2.8. Estimation of speciation time in Malvaceae plants
To construct a phylogenetic tree of the five species (A. trichopoda, A. thaliana, G. raimondii, T. cacao and H. syriacus), we extracted 941 single-copy gene sets from all genomes in the OrthoMCL clusters. We performed multiple alignments of the CDSs of each gene set using Prank (−f = nexus -codon).31 The alignment file was used to construct a phylogenetic tree based on calculations of divergence time for the five species.32 For accurate tree construction, we assigned taxon sets based on previously calculated speciation of A. thaliana, G. raimondii and T. cacao. The Bayesian software package BEAST32 (v1.8.2) was used to estimate divergence times and construct the final tree. The Markov chain Monte Carlo (MCMC) analyses in BEAST was conducted for 10 million generations with samples every 1,000 steps and the effective sampling size was over 150 for all of parameters. We used SRD0633 as a substitution model and the Yule process34 as a traditional speciation model.
2.9. Identification of TF candidates
We identified TF candidates as previously described.17 Briefly, predicted proteins containing TF domains were screened by InterProScan22 search against Pfam35 databases. The TF candidates were classified based on rules as indicated at PlnTFDB (http://plntfdb.bio.uni-potsdam.de/v3.0/rules.php, Rules for the classification of TFs).36 In the case of TF gene families that don’t have Pfam ID, domain alignments as Clustal format were downloaded from PlnTFDB and Hidden Markov Model profiles were built and screened using HMMER.37 The assigned TF candidates were confirmed by BLASTp against plant TF protein sequences downloaded from PlnTFDB (http://plntfdb.bio.uni-potsdam.de/v3.0/downloads.php).
2.10. Identification of genes encoding nucleotide-binding site proteins
To identify nucleotide-binding site (NBS)-encoding genes, representative genes from each plant genome were screened using the raw Hidden Markov Model (HMMER3.0)38 to search for the Pfam NBS family PF00931 domain (e-value cut-off of 1.0). All putative NBS protein sequences were analysed and manually curated by a BLASTp search against known R gene sequences in GenBank. To further identify TIR homologs and sequences encoding CC and LRR motifs, candidate NBS-LRR protein sequences were characterized using SMART,39 the Pfam database40 and the COILS programme41 with a threshold of 0.9 to detect CC domain specifically.
3. Results
3.1. Genome sequencing and assembly
H. syriacus plants over 100-years-old were selected for genome sequencing. Illumina whole-genome shotgun sequencing generated 233.3 Gb (122.8× coverage) of genomic sequences (Supplementary Table S1). PE libraries (250–500 bp) were generated, and 2 kb and 5 kb MP libraries were sequenced with a read length of 101 or 151 bp. Pre-processing analysis of raw sequences was performed to remove extraneous sequences for accurate genome assembly as described in the previous report.17 After filtering, 156.6 Gb (82.4× coverage) of H. syriacus genome sequences were used for further analysis (Supplementary Table S2). K-mer distribution analysis, which provides information related to low frequencies, sequencing depth, level of heterozygosity, and genome size42 was then applied using Jellyfish (Supplementary Fig. S1). The estimated genome size of 1,901 Mb was calculated by dividing the total volume by the peak of distribution as described previously.17
Validation of the assembled genome was performed using 128,888 representative transcripts derived from de novo assembly of a combined transcriptome from all libraries (Supplementary Table S3). To confirm sequence alignment between transcriptome assembly and scaffolds, we performed BLAST comparisons for the transcriptome assembly and scaffolds as queries and subjects, respectively. We found that 117,431 (91.1%) assembled transcripts as query sequences matched to scaffolds based with 98% identity. In addition, 93,688 (79.8%) transcripts matched to genome sequence with query coverage over 80%, and 82,394 (70.2%) transcripts matched over 90% coverage by the assembled scaffolds (Supplementary Fig. 2). The quality of the assembly was also evaluating using CEGMA27 (Core Eukaryotic Genes Mapping Approach) and BUSCO28 (Benchmarking Universal Single-Copy Orthologs). These analyses showed 92.74% of completeness (230 of 248 CEGs) from CEGMA and 92% of complete BUSCOs. These results suggested that H. syriacus genome assembly was high quality. As a result, 1,748 Mb (91.9% of 1,901 Mb) of genomic sequences were assembled into 77,492 scaffolds. The assembled genome was comprised of 33 Mb (1.9%) gap sequences and 1,715 Mb of contigs with N50 = 30 kb (Table 1 and Supplementary Table S4).
Table 1.
Summary of H. syriacus genome assembly
| Number of scaffolds | 77,492 |
| Total length of scaffolds | 1,748 Mb |
| N50 of scaffolds | 140 kb |
| Longest (shortest) length of scaffolds | 1.54 Mb (500 bp) |
| Number of contigs | 172,672 |
| Total length of contigs | 1,715 Mb |
| N50 of contigs | 30.0 kb |
| Longest (shortest) length of contigs | 643 kb (87 bp) |
| Number of gap sequences | 33 Mb (1.9%) |
| GC content | 34.04% |
| Total size of TEs | 1,095 Mb (57.6%) |
3.2. Genome annotation
Annotation of the H. syriacus genome was performed using the KOBIC Genome Annotation pipeline (Supplementary Fig. S3), including masking repetitive sequences, transcriptome mapping, reference protein mapping using GeneWise, ab initio gene prediction, and determination of consensus gene models using EVM.
Before masking repetitive sequences, repeat annotation was performed by RepeatModeler and RepeatMasker (http://www.repeatmasker.org) for the assembled genome. Due to a lack of repeat sequence information for this genome, we constructed a de novo repeat library using RepeatModeler, and RepeatMasker was applied for annotation of the constructed repeat library. Repeat sequences, except for unknown transposable elements (TEs), were masked so we could identify essential gene families, such as those that encode receptor-like kinases and nucleotide-binding proteins. TEs comprised 1,095.8 Mb (57.6%) of the genome (Supplementary Table S5) and mostly included long terminal repeats (LTRs), which accounted for approximately 30% of total TEs. Gypsy and Copia retrotransposons were the most common LTRs detected.
Transcripts mapping was performed using TopHat and Cufflinks, and protein alignment was performed by GeneWise. The protein sequences of A. thaliana (TAIR10), T. cacao and G. raimondii were mapped to generate protein-based gene models. For annotation of duplicated genes or gene families, mapping regions of a reference protein in the H. syriacus genome were determined from tBLASTn (default e-value 10) results using custom Perl scripts. These steps prevented mis-annotation of duplicated genes due to lack of mapping data for reference proteins from parsing single best-matched regions in the H. syriacus genome. We annotated 87,603 genes using KOBIC annotation prediction with an average CDS length of 1,188 bp, similar to that for G. raimondii (Table 2). Consensus gene models were evaluated using 88.4 Gb of Illumina-derived RNA-Seq data. Overall, 91.76% of the predicted coding sequences were supported by Illumina data, demonstrating the high accuracy of KOBIC annotation prediction. The H. syriacus genome contains two times more genes than G. raimondii and three times more genes than T. cacao (Table 2), suggesting a polyploid genome as first indicated in a previous report.13
Table 2.
Statistics of H. syriacus gene models
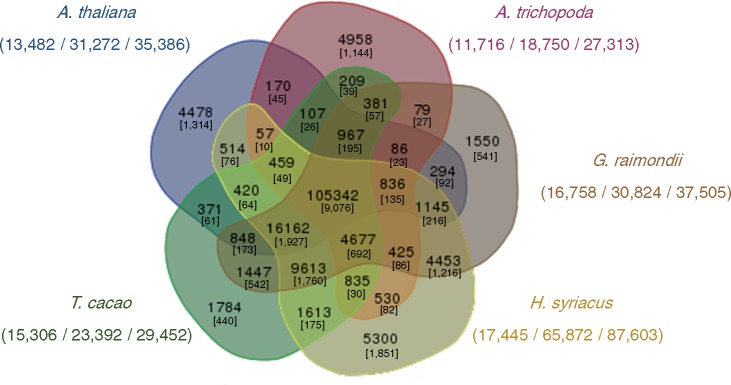
We also performed OrthoMCL analysis to detect orthologous genes among Malvaceae plants, A. thaliana, and Amborella trichopoda. We identified 21,472 orthologous gene sets containing 164,660 genes, 5,300 of which were H. syriacus-specific (Fig. 1). Interestingly, these genes were the number of gene was three times larger than those of G. raimondii and T. cacao, further indicating H. syriacus’ polyploidy. In addition, relatively large numbers of singletons in the genomes of A. thaliana and A. trichopoda suggest that the Malvaceae lineage diverged long ago and now shows a high degree of evolutionary distance from other eudicots. For further analysis, estimation of speciation time and comparison of genome structures among Malvaceae plants were performed using paired gene sets.
Figure 1.
Distribution of orthologous gene families of H. syriacus, G. raimondii, T. cacao, A. trichopoda and A. thaliana, from which 169,570 sequences were clustered into 9,076 groups. The number of clustered groups and genes in each species are shown on the left and center, and total gene numbers are shown on the right.
3.3. Genome structure and polyploidization of H. syriacus
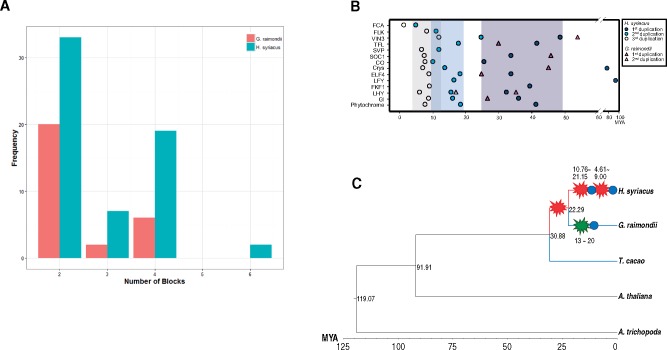
To compare genome structures among Malvaceae plants, collinearity blocks were detected using MCScanX.30 Two WGDs or a triplication event have occurred in G. raimondii (DD),43,44 while none have occurred in T. cacao. Therefore, the T. cacao genome was used as a template to detect collinearity blocks in G. raimondii and H. syriacus (Fig. 2A). We detected T. cacao collinearity blocks in G. raimondii and H. syriacus with frequencies ranging from 2 to 7. The H. syriacus genome contains four times as many collinearity blocks than G. raimondii and blocks two times larger, indicating WGD events in H. syriacus. Duplication patterns were identified using phylogenetic analyses, which revealed single-copy flowering regulator genes in the diploid genomes of A. thaliana, A. trichopoda and T. cacao (Fig. 2B and Supplementary Fig. S4). Duplication of the GIGANTEA (GI) gene indicated WGDs occurred three times in H. syriacus but that many descendant genes since the first WGD have been lost (Fig. 2B), such as the CONSTANS and SOC1 genes (Supplementary Fig. S4). Thus, diploidization and homeolog loss in H. syriacus, first proposed in previous studies,5 included duplication of distinct, individual gene families stemming from random homeolog gene loss after each WGD. Paleohexaploidy has occurred in the G. raimondii genome,10,43,44 and duplication patterns we observed were consistent with these previous results.
Figure 2.
Collinearity block detection and calculation of gene duplication times. (A) Collinearity blocks of the T. cacao genome were detected in G. raimondii and H. syriacus. (B) Calculation of divergence times of individual gene families. Circles and triangles indicate H. syriacus and G. raimondii, respectively, and shade boxes indicate each WGD. (C) Divergence time of Malvales plants. H. syriacus diverged from the H. syriacus-G. raimondii common ancestor 22.28 MYA. Red (H. syriacus) and green (G. raimondii) stars indicate WGD and blue circles indicate diploidization events.
To estimate divergence time among Malvaceae plants, we calculated synonymous substitution rates (Ks) and constructed phylogenetic trees via the BEAST package using single-copy genes in OrthoMCL clusters. The trees revealed that the Malvaceae family diverged from a Brassicae-Malvaceae common ancestor approximately 91.91 MYA (Fig. 2C) and that H. syriacus, G. raimondii and T. cacao belong to a common subclade that diverged from a common ancestor approximately 30.88 MYA, which corroborates earlier studies.10 Occurrence of duplications in G. raimondii genes ranged from 24.46 to 45.46 MYA (Fig. 2B), while H. syriacus individual gene duplications before speciation and WGD events ranged from 25.23 to 48.23 MYA and from 4.61 to 21.15 MYA, respectively (Fig. 2B and C). These results suggest that one WGD occurred in H. syriacus before speciation and two WGDs occurred after speciation.
Previous reports indicate transcriptional factors (TFs) were retained as duplicated genes, while other genes remained singletons.5 We investigated the duplication status of TFs in H. syriacus, and other Malvaceae plants and identified 9,642 TFs and transcriptional regulators in 81 families in the H. syriacus genome. Eighteen H. syriacus TF gene families, including AP2-ERF, AUX/IAA, and FAR1, contained more genes than those in diploid genomes (Supplementary Table S6). In particular, the H. syriacus genome contains 10 times more FAR1 family genes than the other genomes we analysed, although 19 TF genes showed convergent evolution patterns, and the proportions of other major TF family genes were similar across species. Thus, complex WGD events followed by diploidization led to unequal regulation of gene dosage and caused gene family copy number variations in H. syriacus.
3.4. Flowering-time and disease-resistance genes in H. syriacus
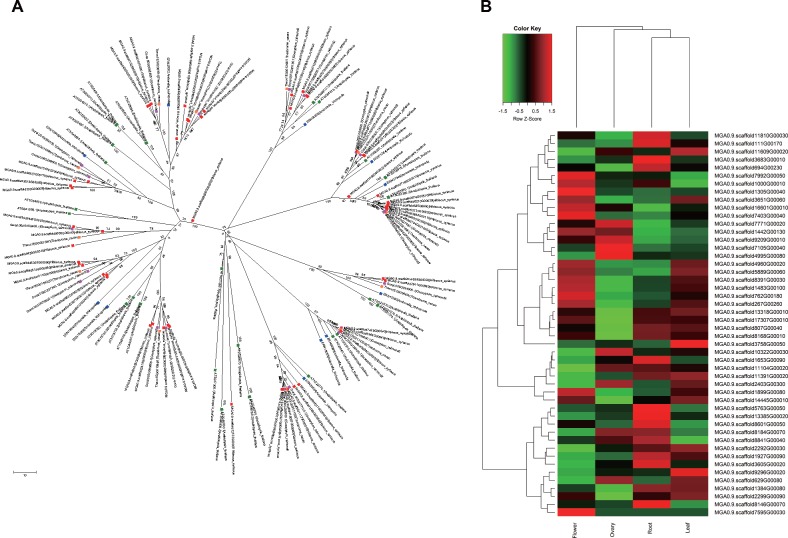
Genetic and molecular mechanisms of floral development in different plant species is highly conserved45 and include four major flowering pathways (photoperiod, autonomous, vernalization and gibberellin) well-characterized in A. thaliana. Main flowering signals are regulated by the FLOWERING LOCUS T (FT) in the photoperiod pathway, while the vernalization pathway acts via removal of an FT repressor after exposure to certain stimuli. H. syriacus is a long-day flowering plant with a long blooming period and can express a multivoltinism phenotype with 20–30 blossoms per day. However, the flowers of H. syriacus open daily and last for only one day. To uncover the genetic mechanisms controlling these phenotypes, we investigated genes involved in the four major flowering pathways of A. trichopoda, A. thaliana, T. cacao, G. raimondii and H. syriacus and their expression pattern in H. syriacus tissues (flowers, ovaries, roots and leaves) (Fig. 3). Phylogenetic analysis of flowering-time genes identified H. syriacus-specific clusters (Fig. 3B). Because flowering time is frequently dependent on gene copy number,46 we used reference genes from Arabidopsis species to determine copy number variation of these genes among diverse plant genomes (Table 3) and found that copy numbers of H. syriacus were two to seven times greater than those in the other four genomes analysed. Among the flowering-time regulatory genes examined in our study, numbers of genes involved in circadian rhythm regulation (CO, ELF4, FKF1, GI, LHY, PHYs) and flower initiation (FCA, FLK, FT, LFY, VIN3, SOC1, TFL, SVP) were significantly higher in H. syriacus than in the other genomes. Moreover, the copy number of FAR1 family genes that modulate phytochrome A signaling showed high copy numbers in H. syriacus compared to other genomes (Supplementary Table S6). Plants with spike inflorescence, such as barley, rice, and wheat, also contain high copy numbers of FAR1 genes; thus, high copy numbers of FAR1 may also affect the flowering phenotype of H. syriacus.
Figure 3.
Phylogenetic tree of photoperiod/circadian clock genes. (A) The evolutionary history of these genes was inferred using the minimum evolution method. Blue (A. trichopoda), green (A. thaliana), pink (G. raimondii), orange (T. cacao) and red (H. syriacus) indicate genes from each species. (B) Expression patterns of photoperiod/circadian clock genes in petals, ovaries, roots, and leaves.
Table 3.
Comparison of flowering-time gene copy numbers
| Regulators | Arabidopsis locus | Copy number |
|||
|---|---|---|---|---|---|
| H. syriacus | T. cacao | G. raimondii | A. trichopoda | ||
| CO | AT5G15840 | 6 | 1 | 1 | 0 |
| ELF4 | AT2G40080 | 7 | 1 | 4 | 1 |
| FCA | AT4G16280 | 2 | 1 | 1 | 1 |
| FKF1 | AT1G68050 | 4 | 1 | 2 | 1 |
| FLK | AT3G04610 | 3 | 1 | 2 | 1 |
| FT | AT1G65480 | 2 | 1 | 1 | 1 |
| GI | AT1G22770 | 5 | 1 | 2 | 0 |
| LFY | AT5G61850 | 4 | 1 | 1 | 1 |
| LHY | AT1G01060 | 7 | 1 | 3 | 1 |
| VIN3 | AT5G57380 | 7 | 1 | 2 | 1 |
| SOC1 | AT2G45660 | 4 | 1 | 2 | 1 |
| TFL | AT5G03840 | 4 | 2 | 2 | 0 |
| SVP | AT1G24260 | 3 | 2 | 2 | 0 |
| PHYA | AT1G09570 | 4 | 1 | 1 | 1 |
| PHYB | AT2G18790 | 4 | 0 | 1 | 1 |
| PHYC | AT5G35840 | 2 | 1 | 1 | 1 |
| PHYE | AT4G18130 | 4 | 1 | 0 | 0 |
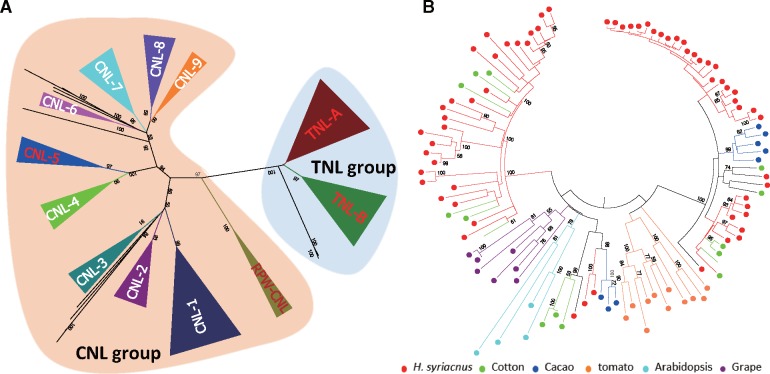
Most disease-resistance (R) family genes encode intracellular proteins with a NBS and leucine-rich repeats (LRR).47–51 The NBS-encoding R gene family is one of the largest in the H. syriacus genome, with 472 genes, approximately three times greater than A. trichopoda and A. thaliana. These genes are divided into two clades based on presence of the distinct toll interleukin receptor (TIR) domain.50 TIR genes in H. syriacus (n = 76, 17%) are markedly over-represented compared to those of S. lycopersicum (25 genes, 9%), G. raimondii (27 genes, 9%) and T. cacao (17 genes, 6%) (Table 4 and Fig. 4). More than 70% of NBS-encoding genes in Malvaceae plants (H. syriacus, T. cacao, and G. raimondii) are shared among 26 subclasses, indicating that most R genes are derived from a common ancestor (Supplementary Table S7). In addition, 125 NBS-encoding genes in H. syriacus from four subclasses are expanded approximately five time more than other Malvaceae and 18 NBS-encoding genes from seven subclasses are unique to H. syriacus (Supplementary Table S8). Notably, genes in TIR and RPW8 motif-encoding subclasses (NBS cluster 11 and NBS cluster 20, respectively) exhibited extensive expansion in the H. syriacus genome, underwent unequal duplication events, and displayed great diversity among plant genomes (Fig. 4, Supplementary Fig. S5 and Supplementary Table S8). The different R gene repertoires in the H. syriacus genome suggest that expansion and diversity of clustered R genes might involve lineage-specific gene duplication events, eventually leading to divergent evolution in close relatives. These results provide useful preliminary information to support further comparative analysis of flowering-time and disease-resistance genes in other perennial plant species.
Table 4.
Comparative NBS-LRR gene family numbers
| Predicted domain | Class | H. syriacus | G. raimondii | T. cacao | S. lycopersicum | A. thaliana | V. vinifera | O. sativa | A. trichopoda |
|---|---|---|---|---|---|---|---|---|---|
| TIR group | |||||||||
| TIR-NBS-LRR | TNL | 68 | 26 | 14 | 19 | 87 | 19 | 0 | 9 |
| TIR-NBS | TN | 9 | 1 | 3 | 6 | 17 | 4 | 2 | 2 |
| % on NBS genes | 17 | 9 | 6 | 9 | 61 | 7 | 0.4 | 10 | |
| Non-TIR group | |||||||||
| CC-NBS- LRR | CNL | 183 | 220 | 202 | 116 | 52 | 138 | 337 | 27 |
| CC-NBS | CN | 77 | 24 | 25 | 37 | 3 | 19 | 104 | 27 |
| NBS-LRR | NL | 81 | 28 | 34 | 39 | 8 | 110 | 70 | 18 |
| NBS | N | 54 | 4 | 9 | 50 | 3 | 32 | 14 | 29 |
| % on NBS genes | 84 | 91 | 94 | 91 | 39 | 93 | 99.6 | 90 | |
| Total NBS genes | 472 | 303 | 287 | 267 | 170 | 322 | 527 | 112 | |
| % on total genes | 0.53 | 0.81 | 0.97 | 0.77 | 0.63 | 1.22 | 1.35 | 0.41 | |
| Total no. of genes | 87,603 | 37,505 | 29,452 | 34,727 | 27,206 | 26,346 | 39,049 | 26,846 |
Figure 4.
Phylogenetic relationships of NBS-LRR genes with >80% bootstrap values. (A) Phylogenetic relationships of predicted NBS-LRR genes in H. syriacus. Red (TNL-A, TNL-B, CNL-5 and RPW-CNL subgroups) indicates expanded subgroups of the H. syriacus genome compared to other plant genomes (Supplementary Table S8). (B) Detailed phylogenetic relationships of expanded TNL subgroups are shown. Intact NB-ARC domains of H. syriacus (red), G. raimondii (green), T. cacao (blue), S. lycopersicum (orange), A. thaliana (light blue) and V. vinifera (purple) were used in the phylogenetic construction.
4. Discussion
Polyploidy is an important mechanism of plant speciation that occurs in many angiosperms5 and leads to increased genetic diversity compared to their diploid progenitors. Initial polyploidy events were followed by successive paleopolyploidy or diploidization events to stabilize the newly expanded genomes. Paleopolyploidy or diploidization returns a polyploidy genome to a diploid-like state and is characterized by loss of duplicated genes, chromosomes, and repetitive DNA, gene silencing, and altered chromosome pairings.4,5 The newly formed polyploids may experience rapid homeolog gene loss, genome reconstruction post-polyploidization, and altered patterns of gene expression.5 Retained diploid genes are less often duplicated due to cumulative losses of the homeologous copy in a duplicated gene pair. Consequently, some genes are consistently returned to singleton status, while others, such as those encoding TFs, are retained in duplicate.5,52 Duplicated TF genes were commonly found in the H. syriacus genome, although 25% of these genes showed evidence of convergent evolution, and their copy numbers varied greatly. Our phylogenetic analysis of individual genes in H. syriacus also indicated that homeolog gene loss events and diploidization occurred after WGD. Recent studies have suggested that one duplicate gene may be more susceptible to loss than others,4 which could account for unequal gene dosage and corresponding phenotypic changes in H. syriacus.
The flowering phenotype of H. syriacus is characterized by multivoltinism, a long blooming period, and high blossom turnover. We found that the copy numbers of most flowering-related genes, such as GIGANTEA, CONSTANS, and ELF4 (but not FT), were higher in H. syriacus than in the diploid genomes of T. cacao, A. trichopoda, and A. thaliana. In addition, FAR1 genes, which modulate phytochrome A signaling by directly activating transcription of FHY1 and FHL and lead to accumulation of nuclear phytochrome A, were significantly increased in H. syriacus. FAR1 regulates the circadian clock, and its high copy number could directly affect the flowering phenotype of H. syriacus as seen in plants with spike inflorescence.
The investigation of duplication event timing in H. syriacus genome showed that two recent WGDs occurred after speciation. In the past 50 million years, the average global temperature has been <20°C,53 which is far below the optimal flowering temperature for H. syriacus. Lower temperatures, especially between 5 and 20 MYA, could have been an environmental suppressor of H. syriacus pollination that prompted polyploidization to overcome these unfavourable conditions and unreduced gamete formation.5 Furthermore, low temperatures could have exerted selective pressure on H. syriacus to extend its blooming period for increased chance of pollination.
Perennial plants are prone to invasion by pathogens before reproduction, and many fungal and bacterial diseases often threaten the life cycle of H. syriacus. Aside from primary defenses, such as thickened cell walls and secondary metabolites, plants have numerous disease resistance (R) genes that confer protection against various pathogens. In H. syriacus, NBS-containing R genes account for ∼0.53% of its total predicted genes, which is lower than other plant genomes studies, whose R gene proportions ranged from 0.63 to 1.35%. However, subsets of these genes, including those with TIR- and RPW8-encoding motifs, are markedly over-represented in the H. syriacus genome compared to those of other plants. Genes in the RPW8-NBS-LRR subclass provide broad-spectrum resistance against powdery mildew pathogens in Arabidopsis,54 and genes in TIR-NBS subclasses are conserved in basal angiosperms and eudicots, such as A. trichopoda (Supplementary Table S8 and Supplementary Fig. S5), but are absent in most monocots.55,56 Their greater dominance in the H. syriacus genome indicates divergent evolution of TIR- and RPW-containing NBS genes from an ancestral origin may have led to more extensive expansion of this gene family. Moreover, the long life cycle of woody plants makes it difficult for them to adapt to pathogens undergoing more rapid evolution, thus favouring R gene maintenance and expansion for the plants’ survival.
Polyploidization in plants is a common mechanism for their adaptation to environmental change. After divergence from the H. syriacus-G. raimondii common ancestor, two WGDs and subsequent diploidization occurred in the H. syriacus genome to promote the plants’ survival in unfavourable environments. During the diploidization events, low temperatures may have selected for the maintenance of duplicate flowering-related genes whose high copy numbers led to the multivoltinism and long blooming period phenotypes expressed by H. syriacus. Further analyses H. syriacus, T. cacao and G. raimondii (DD) genomes with another diploid genome, G. arboretum (AA)11 and allotetraploid genome, G. hirsutum (AtDt)12 will provide more information of evolution of Malvaceae plants.
Availability
This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession MBGJ00000000. The version described in this article is version MBGJ01000000. The raw sequence reads has been deposited at DDBJ/ENA/GenBank under accession SRP087036 (PRJNA341314). In addition, the genome data of H. syriacus are accessible at https://hibiscus.kobic.re.kr/hibiscus.en.
Conflict of interest
None declared.
Supplementary data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
Funding
This work was supported by the Korean Research Institute of Bioscience and Biotechnology initiative programme and by a grant from the Agricultural Genome Center of the Next Generation Biogreen 21 Programme (Project No. PJ011275, PJ011088 and PJ011100) of the Rural Development Administration, Republic of Korea.
Genome data including genome assembly, annotation and transcriptome data were also provided through webpage (https://hibiscus.kobic.re.kr/hibiscus.en).
Supplementary Material
References
- 1.Paoletti E., Ferrara A. M., Calatayud V., et al. 2009, Deciduous shrubs for ozone bioindication: Hibiscus syriacus as an example. Environ. Pollut., 157, 865–70. [DOI] [PubMed] [Google Scholar]
- 2.Yun B.-S., Ryoo I.-J., Lee I.-K., Yoo I.-D. 1998, Hibispeptin A, a novel cyclic peptide from Hibiscus syriacus. Tetrahedron Lett., 39, 993–6. [Google Scholar]
- 3.Yoo I.-D., Yun B.-S., Lee I.-K., Ryoo I.-J., Choung D.-H., Han K.-H. 1998, Three naphthalenes from root bark of Hibiscus syriacus. Phytochemistry, 47, 799–802. [DOI] [PubMed] [Google Scholar]
- 4.Semon M., Wolfe K. H. 2007, Consequences of genome duplication. Curr. Opin. Genet. Dev., 17, 505–12. [DOI] [PubMed] [Google Scholar]
- 5.Soltis P. S., Marchant D. B., Van de Peer Y., Soltis D. E. 2015, Polyploidy and genome evolution in plants. Curr. Opin. Genet. Dev., 35, 119–25. [DOI] [PubMed] [Google Scholar]
- 6.Jaillon O., Aury J. M., Noel B., et al. 2007, The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature, 449, 463–7. [DOI] [PubMed] [Google Scholar]
- 7.Dehal P., Boore J. L. 2005, Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol., 3, e314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Otto S. P., Whitton J. 2000, Polyploid incidence and evolution. Annu. Rev. Genet., 34, 401–37. [DOI] [PubMed] [Google Scholar]
- 9.Fawcett J. A., Maere S., Van de Peer Y. 2009, Plants with double genomes might have had a better chance to survive the Cretaceous-Tertiary extinction event. Proc. Natl. Acad. Sci. U S A, 106, 5737–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang K., Wang Z., Li F., et al. 2012, The draft genome of a diploid cotton Gossypium raimondii. Nat. Genet., 44, 1098–03. [DOI] [PubMed] [Google Scholar]
- 11.Li F., Fan G., Wang K., et al. 2014, Genome sequence of the cultivated cotton Gossypium arboreum. Nat. Genet., 46, 567–72. [DOI] [PubMed] [Google Scholar]
- 12.Li F., Fan G., Lu C., et al. 2015, Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat. Biotechnol., 33, 524–30. [DOI] [PubMed] [Google Scholar]
- 13.Darlington C. D., Wylie A. P. 1955, Chromosome Atlas of Flowering Plants. Allen & Unwin: London. [Google Scholar]
- 14.Kajitani R., Toshimoto K., Noguchi H., et al. 2014, Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res., 24, 1384–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boetzer M., Henkel C. V., Jansen H. J., Butler D., Pirovano W. 2011, Scaffolding pre-assembled contigs using SSPACE. Bioinformatics, 27, 578–9. [DOI] [PubMed] [Google Scholar]
- 16.Magoc T., Salzberg S. L. 2011, FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics, 27, 2957–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim S., Park M., Yeom S. I., et al. 2014, Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat. Genet., 46, 270–8. [DOI] [PubMed] [Google Scholar]
- 18.Stanke M., Keller O., Gunduz I., Hayes A., Waack S., Morgenstern B. 2006, AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res., 34, W435–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Argout X., Salse J., Aury J. M., et al. 2011, The genome of Theobroma cacao. Nat. Genet., 43, 101–8. [DOI] [PubMed] [Google Scholar]
- 20.Birney E., Clamp M., Durbin R. 2004, GeneWise and Genomewise. Genome Res., 14, 988–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haas B. J., Salzberg S. L., Zhu W., et al. 2008, Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol., 9, R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones P., Binns D., Chang H. Y., et al. 2014, InterProScan 5: genome-scale protein function classification. Bioinformatics, 30, 1236–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pruitt K. D., Tatusova T., Brown G. R., Maglott D. R. 2012, NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res., 40, D130-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.UniProt, C. 2013, Update on activities at the Universal Protein Resource (UniProt) in 2013. Nucleic Acids Res., 41, D43–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zdobnov E. M., Apweiler R. 2001, InterProScan–an integration platform for the signature-recognition methods in InterPro. Bioinformatics, 17, 847–8. [DOI] [PubMed] [Google Scholar]
- 26.Cox M. P., Peterson D. A., Biggs P. J. 2010, SolexaQA: At-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinformatics, 11, 485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parra G., Bradnam K., Korf I. 2007, CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics, 23, 1061–7. [DOI] [PubMed] [Google Scholar]
- 28.Simao F. A., Waterhouse R. M., Ioannidis P., Kriventseva E. V., Zdobnov E. M. 2015, BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics, 31, 3210–2. [DOI] [PubMed] [Google Scholar]
- 29.Li L., Stoeckert C. J., Jr., Roos D. S. 2003, OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res., 13, 2178–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y., Tang H., Debarry J. D., et al. 2012, MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res., 40, e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loytynoja A. 2014, Phylogeny-aware alignment with PRANK. Methods Mol. Biol., 1079, 155–70. [DOI] [PubMed] [Google Scholar]
- 32.Drummond A. J., Suchard M. A., Xie D., Rambaut A. 2012, Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol. Biol. Evol., 29, 1969–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shapiro B., Rambaut A., Drummond A. J. 2006, Choosing appropriate substitution models for the phylogenetic analysis of protein-coding sequences. Mol. Biol. Evol., 23, 7–9. [DOI] [PubMed] [Google Scholar]
- 34.Gernhard T. 2008, The conditioned reconstructed process. J. Theor. Biol., 253, 769–78. [DOI] [PubMed] [Google Scholar]
- 35.Punta M., Coggill P. C., Eberhardt R. Y., et al. 2012, The Pfam protein families database. Nucleic Acids Res, 40, D290–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perez-Rodriguez P., Riano-Pachon D. M., Correa L. G., Rensing S. A., Kersten B., Mueller-Roeber B. 2010, PlnTFDB: updated content and new features of the plant transcription factor database. Nucleic Acids Res., 38, D822–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Finn R. D., Clements J., Eddy S. R. 2011, HMMER web server: interactive sequence similarity searching. Nucleic Acids Res., 39, W29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marchin M., Kelly P. T., Fang J. 2005, Tracker: continuous HMMER and BLAST searching. Bioinformatics, 21, 388–9. [DOI] [PubMed] [Google Scholar]
- 39.Schultz J., Milpetz F., Bork P., Ponting C. P. 1998, SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. U S A, 95, 5857–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Finn R. D., Bateman A., Clements J., et al. 2014, Pfam: the protein families database. Nucleic Acids Res., 42, D222–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lupas A., Van Dyke M., Stock J. 1991, Predicting coiled coils from protein sequences. Science, 252, 1162–4. [DOI] [PubMed] [Google Scholar]
- 42.Potato Genome Sequencing C., Xu X., Pan S., et al. 2011, Genome sequence and analysis of the tuber crop potato. Nature, 475, 189–95. [DOI] [PubMed] [Google Scholar]
- 43.Muhlhausen S., Kollmar M. 2013, Whole genome duplication events in plant evolution reconstructed and predicted using myosin motor proteins. BMC Evol. Biol., 13, 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen J., Zhang Y., Liu J., Xia M., Wang W., Shen F. 2014, Genome-wide analysis of the RNA helicase gene family in Gossypium raimondii. Int. J. Mol. Sci., 15, 4635–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schiessl S., Samans B., Hüttel B., Reinhardt R., Snowdon R. J. 2014, Capturing sequence variation among flowering-time regulatory gene homologues in the allopolyploid crop species Brassica napus. Front. Plant Sci., 5, 404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grover C. E., Gallagher J. P., Wendel J. F. 2015, Candidate gene identification of flowering time genes in cotton. Plant Genome, 8. [DOI] [PubMed] [Google Scholar]
- 47.Dangl J. L., Jones J. D. 2001, Plant pathogens and integrated defence responses to infection. Nature, 411, 826–33. [DOI] [PubMed] [Google Scholar]
- 48.McDowell J. M., Woffenden B. J. 2003, Plant disease resistance genes: recent insights and potential applications. Trends Biotechnol., 21, 178–83. [DOI] [PubMed] [Google Scholar]
- 49.Meyers B. C., Kozik A., Griego A., Kuang H., Michelmore R. W. 2003, Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell, 15, 809–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee H. A., Yeom S. I. 2015, Plant NB-LRR proteins: tightly regulated sensors in a complex manner. Brief. Funct. Genomics, 14, 233–42. [DOI] [PubMed] [Google Scholar]
- 51.Seo E., Kim S., Yeom S.-I., Choi D. 2016, Genome-wide comparative analyses reveal the dynamic evolution of nucleotide-binding leucine-rich repeat gene family among Solanaceae plants. Front. Plant Sci., 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Freeling M., Scanlon M. J., Fowler J. E. 2015, Fractionation and subfunctionalization following genome duplications: mechanisms that drive gene content and their consequences. Curr. Opin. Genet. Dev., 35, 110–8. [DOI] [PubMed] [Google Scholar]
- 53.Zachos J., Pagani M., Sloan L., Thomas E., Billups K. 2001, Trends, rhythms, and aberrations in global climate 65 Ma to present. Science, 292, 686–93. [DOI] [PubMed] [Google Scholar]
- 54.Xiao S., Ellwood S., Calis O., et al. 2001, Broad-spectrum mildew resistance in Arabidopsis thaliana mediated by RPW8. Science, 291, 118–20. [DOI] [PubMed] [Google Scholar]
- 55.Pan Q., Wendel J., Fluhr R. 2000, Divergent evolution of plant NBS-LRR resistance gene homologues in dicot and cereal genomes. J. Mol. Evol., 50, 203–13. [DOI] [PubMed] [Google Scholar]
- 56.Tarr D. E., Alexander H. M. 2009, TIR-NBS-LRR genes are rare in monocots: evidence from diverse monocot orders. BMC Res. Notes, 2, 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.