Abstract
Because of their biocompatibility and unique magnetic properties, superparamagnetic iron oxide nanoparticles NPs (SPIONs) are recognized as some of the most prominent agents for theranostic applications. Thus, understanding the interaction of SPIONs with biological systems is important for their safe design and efficient applications. In this study, SPIONs were coated with 2 different polymers: polyvinyl alcohol polymer (PVA) and dextran. The obtained NPs with different surface charges (positive, neutral, and negative) were used as a model study of the effect of surface charges and surface polymer materials on protein adsorption using a magnetic separator. We found that the PVA-coated SPIONs with negative and neutral surface charge adsorbed more serum proteins than the dextran-coated SPIONs, which resulted in higher blood circulation time for PVA-coated NPs than the dextran-coated ones. Highly abundant proteins such as serum albumin, serotransferrin, prothrombin, alpha-fetoprotein, and kininogen-1 were commonly found on both PVA- and dextran-coated SPIONs. By increasing the ionic strength, soft- and hard-corona proteins were observed on 3 types of PVA-SPIONs. However, the tightly bound proteins were observed only on negatively charged PVA-coated SPIONs after the strong protein elution.
Nanomedicine is an area of growing public interest. In recent years, nanoparticles (NPs) have become the most important components in nanotechnology. It is now well recognised that the surface of NPs is covered with various types of biomolecules (e.g., proteins) when they enter the biological fluids1,2,3,4,5. Therefore, the biomolecule-coated NPs (e.g., protein-“corona”-coated particles) can be considered as entirely new materials compared to manufactured products in laboratories. Depending on the physicochemical properties of the NPs (e.g., the size, the surface curvature, the composition, the charges, and the surface modifications) and the biological fluid that they contact, only a selective set of proteins with a high affinity for the NP surface will adhere and remain tightly bound for a long time1,2,6. Because the protein corona layer remains tightly bound to the NP surface for several hours, the living organisms (e.g., the cells and the tissues) most likely “feel” the NP protein corona instead of the pristine surface of the NPs7,8,9,10,11,12,13,14,15.
Among various types of NPs used in biomedical applications, superparamagnetic iron oxide NPs (SPIONs) are recognised as some of the most promising materials because of their multi-functional biomedical capabilities (e.g., targeted-imaging16, targeted delivery of drugs17, hyperthermia18, transfections19,20, and stem cell tracking21). Full information on the protein corona compositions at the surface of various types of SPIONs is required to thoroughly understand their toxicities and biological fate. Although there are extensive reports on the biological responses (e.g., the cellular uptakes, the cellular signalling pathways, and the biodistribution) of SPIONs with various physicochemical properties22,23,24,25,26,27,28,29,30,31, there are few reports on the compositions of the protein corona at their surfaces32.
Many methods have been developed to investigate the protein corona on the NP surface33. Among these methods, centrifugation has been recognised as the most reliable method to separate protein-NP complexes10. However, high-abundance proteins may be identified as being bound on the NP surface because of insufficient washing. The outcome of the centrifugation method is affected by the centrifugation force, the washing duration, the washing solution and the solution volumes on the detachment of the loosely attached proteins34. A magnetic separation technique and the magnetic properties of SPIONs are useful to overcome these problems. Moreover, this technique creates the precise gradient removal of the NP-attached protein coronas35,36. Thus, this study aims to probe the corona compositions of two important types of polymers (dextran and polyvinyl alcohol; PVA) with various surface charges using a magnetic separation method. In addition, we used different ionic strengths to separate the proteins from the NPs, washed the NPs with PBS to separate the loosely bound proteins and subsequently washed them with solutions of up to 2 molar of KCl to investigate the hard corona. The proteins that remained bound to the NPs were called “tightly bound” proteins.
Results
NP characterisation
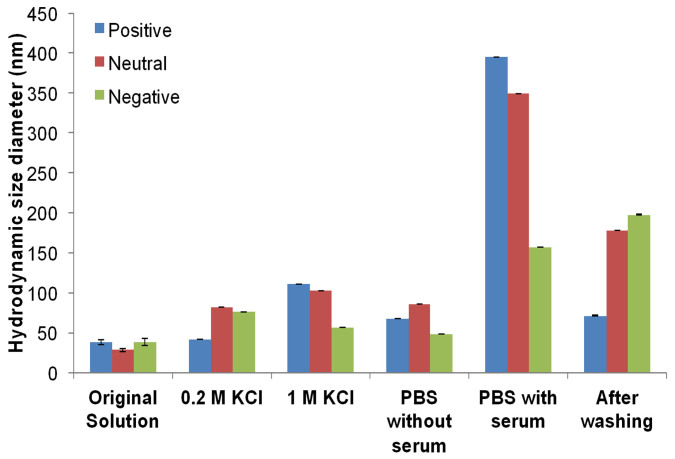
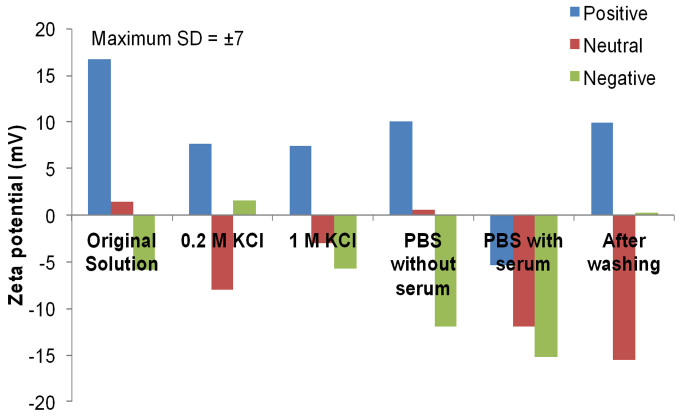
The NP size and charge of PVA- and dextran-coated SPIONs are listed in Table 1. In the absence of foetal bovine serum (FBS), the direct effects of different solutions on the size and charge of different PVA-coated SPIONs are shown in Figures 1 and 2. The increase of the NP size and the change in the zeta potential revealed the effect of salt on the NP stability. However, the zeta potentials of NPs in PBS are slightly changed. In addition to mimicking a physiological buffer, we chose PBS as a washing buffer in our work.
Table 1. Summary of PVA-coated SPIONs and dextran-coated SPIONs measured in DI water.
| Nanoparticle | Functional group | Hydrodynamic diameter (nm) | Zeta potential (mV) |
|---|---|---|---|
| PVA coated SPIONs | -NH2 | 37.9 ± 3 | +16.7 ± 1.5 |
| -OH | 28.3 ± 2.1 | 1.5 ± 2.2 | |
| -COOH | 38.1 ± 4.2 | −5.9 ± 0.9 | |
| Dextran coated SPIONs | -NH2 | 18.8 ± 0.2 | +36.5 ± 1.8 |
| -OH | 19.7 ± 0.4 | −17.9 ± 1.1 | |
| -COOH | 18.3 ± 0.2 | −26.7 ± 0.5 |
Figure 1. Size measurements of different surface-charged PVA-coated SPIONs in water (original solution), 0.2 M KCl, 1.0 M KCl, PBS without serum, PBS with serum and NPs after 30 column-volume washings with PBS.
Figure 2. Zeta potential measurements of different surface-charged PVA-coated SPIONs in water (original solution), 0.2 M KCl, 1.0 M KCl, PBS without serum, PBS with serum and NPs after 30 column-volume washings with PBS.
To determine the effect of the washing step on the protein adsorption on different surface-charge PVA-coated SPIONs, the size of the NPs before and after the interaction with serum also examined (Figures 1 and 2). The protein-NP interaction significantly increased the hydrodynamic size of the NPs. The increase of the NP hydrodynamic size is related to the protein diffusion layer on the NP surface. Severe washings of the fixed NPs reduced these hydrodynamic sizes; however, the tightly bound proteins (such as the attached proteins in the Stern layer) did not allow the particles to recover their initial sizes. According to Figure 2, after the interaction with serum, the net charge of each particle became negative. The zeta potential values of the SPIONs in PBS with and without serum were compared, which shows that the protein adsorption onto the NP surface resulted in the net-negative-charged layer on the NP surface. After washing with 30 column volumes (CV) of PBS, the obtained NP charges were remarkably different from those before washing because most of the proteins were removed from the NP surface. It is import to note that both, the positive charge and neutral (slightly positive charged SPIONs) passed the isoelectric point and therefore the possibility of agglomeration is heavily increased. This might be the reason why the particles show a larger hydrodynamic diameter before the washing step.
Optimisation of the washing procedure
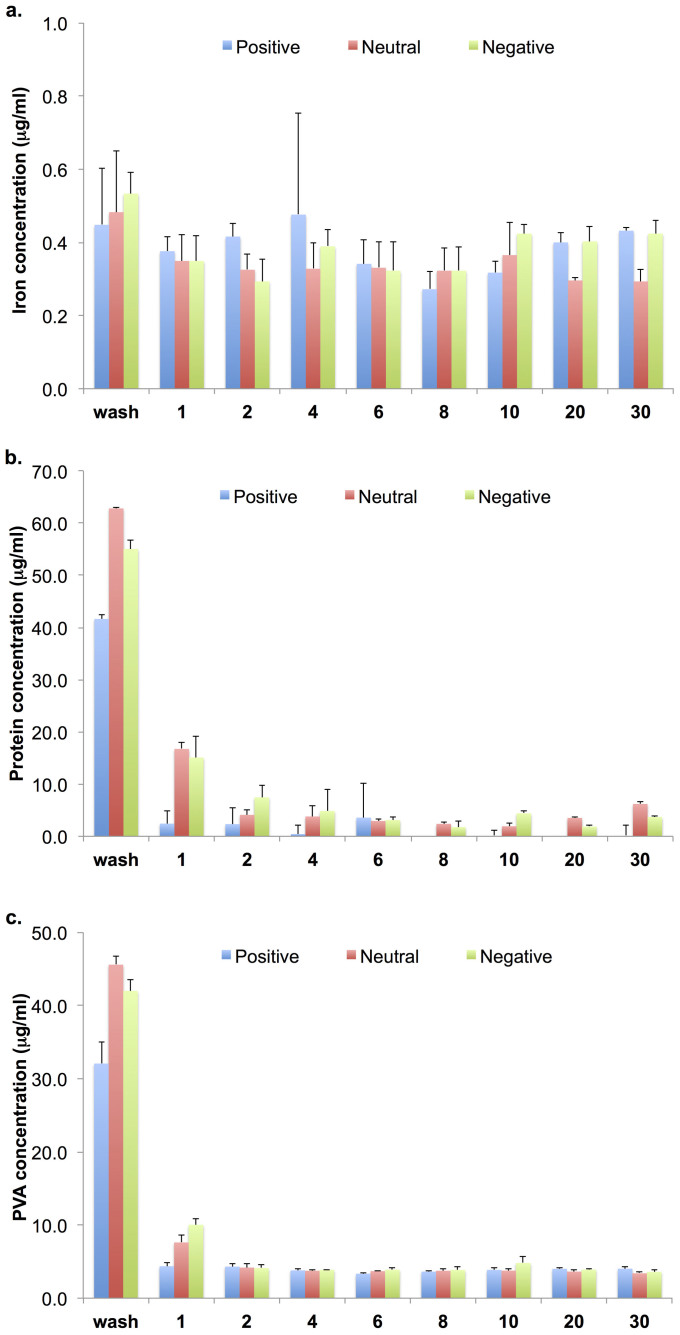
To understand the protein adsorption to the surface of the PVA-coated SPIONs, the washing procedure was optimised to remove the unbound proteins without the influence of the PVA that leave the NP surface and the trapped NPs. As shown in Figure 3, the minimum amount of the eluted unbound proteins from positively-, neutral and negative-NPs after washing with PBS are respectively 4, 8 and 8 CV, while the amount of PVA and SPION are stable at 2 CV. Thus, the optimum PBS washing column volume (to elute unbound proteins, to avoid the PVA leaving NP surface and to avoid the loosing of the trapped NPs during protein elution) are 4 for positively charged NPs and 8 for both neutral and negatively charged NPs.
Figure 3.
Iron (a), protein (b) and PVA (c) amounts in the solutions that were passed through the trapped NPs (PBS is the first collected fraction, and the numbers refer to the washing times).
Interaction of different surface-coated SPIONs with FBS proteins
In this study, we investigated the interaction of PVA- and dextran-coated SPIONs, which had various surface charges (positive, neutral, and negative), with a protein source (foetal bovine serum (FBS)). Note that the dextran-coated particles have notably low blood circulation time in the human body, and they are usually used to image the immune systems, the liver, and the spleen. PVA-coated SPIONs are also found in the liver and the spleen, but a significant amount of injected particles can remain longer in the blood circulation (Table 2, i.e., a half-life of several hours in mice instead of 18 min for Feridex®, which is a type of dextran-coated SPION37). Thus, they are notably suitable for targeting, early diagnosis, and specific protein-fishing applications3,17,21,38,39. Our unpublished data showed that different surface-charged PVA-coated SPIONs distributed differently in the organ parts of rats. Neutral and negatively charged NPs had longer half lives in the bloodstream than positively charged NPs: the amounts of neutral and negatively charged NPs in blood are 2 times higher than that of positive NPs 15 min after the injection. PVA of up to 195000 g/mol with a molecule size (Rh) of 13 nm are far above the critical cut-off of PVA in renal filtration (30,000 g/mol), and they can gradually excrete through the kidney for a long time without damaging the renal glomeruli40,41. The absence of the magnetic susceptibility of SPION in the urine of NP-injected rats indicated that only PVA, but not SPIONs, was eliminated through the kidneys into the urine. The positively charged PVA-coated SPIONs have been reported as a long-term (7 day) and non-toxic agent in in vivo and in vitro tests. Interestingly, from the observation of the positively charged PVA-coated NPs, no NPs and no PVA were observed in the urine after 7 days post-injection; however, the iron content in the liver was twice higher than after 15 min post-injection. The increased iron content in the liver indicated that within 15 min, the particles began to deposit in the liver without significant biological SPION degradation. Moreover, we found that the SPIONs were slowly metabolised in the liver afterwards. Nanoparticle uptake by immune cells may occur both in the bloodstream by monocytes, platelets, leukocytes, and dendritric cell (DC) and in tissues by the resident phagocytes (e.g., Kupffer cells in the liver, DC in the lymph nodes, and macrophages and B cells in the spleen). Nanoparticle uptake by immune cells may occur through various pathways and can be facilitated by the adsorption of opsonins (plasma proteins) at the particle surface38,42,43.
Table 2. Summary table of blood circulation of SPION 15 minutes after the injection.
| Type of SPION | Percentage of injected dose present in the blood after 15 min |
|---|---|
| Dextran coated | 50%37 |
| PVA-SPION positive | 45.4 ± 9.8% |
| PVA-SPION neutral | 87.1 ± 3.7% |
| PVA-SPION negative | 85.6 ± 3.2% |
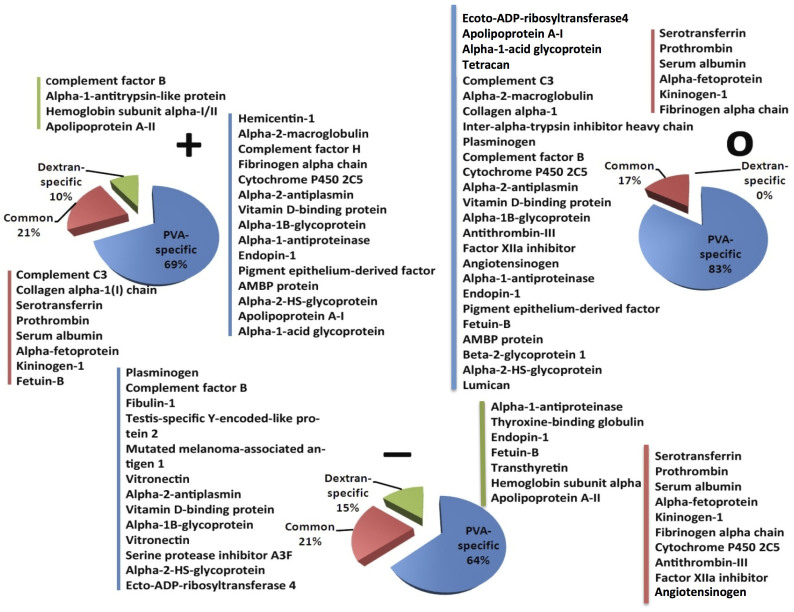
The changes in the temperature of a biological system could play an important role in the protein conformation and the decoration of the protein corona44; therefore, the temperature in this study was carried out at 37°C. As shown in Figure 4, the serum proteins adsorb more onto the PVA-coated SPIONs than onto the dextran-coated SPIONs. In addition, the chemistry of the coating and the charge can significantly affect the type of adsorbed proteins. Interestingly, all particles independently affected the chemistry and the charge of the surface layer, but only the following five detected proteins were adsorbed: serotransferrin, prothrombin, serum albumin, alpha-fetoprotein, and kininogen-1. Furthermore, depending on the NP charge, the proteins could adsorb onto both dextran- and PVA-coated SPIONs. Positively and negatively charged dextran-coated SPIONs showed the specific adsorption of proteins onto the NP surfaces, whereas the neutral dextran-coated SPIONs have no specific adsorbed proteins. All PVA-coated SPIONs show a notably specific adsorption of proteins, which implies that most proteins found on the PVA-coated NPs could not be detected on the dextran-coated ones. This result shows that the chemical composition and structure of the molecules must have a notably important impact on the composition of the soft and hard coronas. These differences in the composition of the protein corona could decrease the cellular uptake (e.g., by the RES system)45 of the PVA-coated SPIONs, which may be the main reason for significant variations in the blood circulation times of the dextran- and PVA-coated SPIONs.
Figure 4. Comparison and characterisation of the hard-corona compositions on PVA- and dextran-coated SPIONs with various surface charges (i.e., positive, neutral, and negative charges).
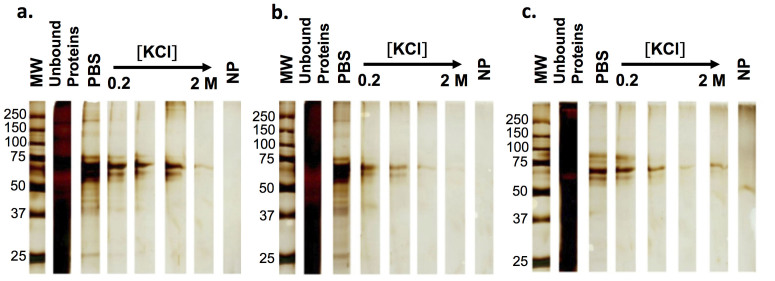
Figure 5 shows the silver-stained SDS-PAGE gel of corona proteins from the PVA-coated NPs. Different proteins were eluted at different KCl concentrations. The normalised SpC (NSpC) values for all proteins that were identified in the hard coronas of positive-, neutral- and negative-charge SPIONs in all washing steps were determined and presented in Table 3, Figures 6 and 7. First, only serotransferrin, prothrombin, serum albumin, alpha-fetoprotein, kininogen-1, fibrinogen alpha chain, cytochrome P450 2C5, alpha-2-antiplasmin, vitamin D-binding protein, alpha-1B-glycoprotein are found in the soft and hard (up to KCl concentration of 0.5 mol) protein corona of all three types of NPs. In a washing solution with higher ionic strengths, only serum albumin was detected on all 3 types of NPs. Note that the negatively charged PVA-coated SPIONs could maintain some specific proteins (cytochrome P450 2C5, alpha-2-antiplasmin, vitamin D-binding protein, and alpha-1B-glycoprotein) even after extensive washing, which shows that these proteins have the highest affinities to the surface of negative NPs among all proteins in the hard-corona composition. These 4 proteins were detected in the soft corona and sometimes in the hard protein corona of all types of SPIONs.
Figure 5. Silver-stained SDS-PAGE gels represent the proteins in each elution fraction of the positively (a), neutral (b), and negatively (c) charged PVA-coated SPIONs.
Table 3. Soft- and hard-corona proteins, which are dissociated from the positive-, neutral and negative-charge SPIONs after washing with various washing solutions, as identified by LC-MS/MS (the yellow-brownish underlined fields in the “protein identity” column denote that these proteins are detected at all 3 different charged PVA-coated SPIONs; the grey underlined fields in the “charge” column denote that these proteins are common with the dextran-coated SPIONs).
| Gel band Mw (Da) | Protein identity | Charge | NSpC | |||||
|---|---|---|---|---|---|---|---|---|
| PBS | 0.2 KCl | 0.5 KCl | 1 KCl | 2 KCl | NPs | |||
| Soft corona | Hard corona | Tightly bound | ||||||
| 621279 | Hemicentin-1 | positive | 1.3 | |||||
| 188377 | Complement C3 | neutral | 0.3 | |||||
| 188377 | Complement C3 | positive | 1.4 | 0.5 | ||||
| 164325 | Alpha-2-macroglobulin | neutral | 0.1 | |||||
| 164325 | Alpha-2-macroglobulin | positive | 0.4 | |||||
| 144053 | Complement factor H | positive | 0.1 | |||||
| 139681 | Collagen alpha-1 | neutral | 0.3 | |||||
| 139681 | Collagen alpha-1(I) chain | positive | 0.3 | 0.3 | ||||
| 106738 | Inter-alpha-trypsin inhibitor heavy chain | neutral | 0.6 | |||||
| 93364 | Plasminogen | negative | 2.0 | |||||
| 93364 | Plasminogen | neutral | 0.2 | |||||
| 86462 | Complement factor B | negative | 1.4 | |||||
| 86462 | Complement factor B | neutral | 0.8 | |||||
| 80521 | Fibulin-1 | negative | 1.2 | |||||
| 79571 | Testis-specific Y-encoded-like protein 2 | negative | 0.4 | 1.5 | ||||
| 79450 | Serotransferrin | negative | 13.3 | 8.9 | ||||
| 79450 | Serotransferrin | neutral | 9.6 | |||||
| 79450 | Serotransferrin | positive | 5.0 | 2.7 | 10.8 | 4.3 | ||
| 71610 | Prothrombin | negative | 1.7 | 0.8 | ||||
| 71610 | Prothrombin | neutral | 0.3 | 8.7 | ||||
| 71610 | Prothrombin | positive | 9.6 | |||||
| 71570 | Mutated melanoma-associated antigen 1 | negative | 0.9 | 2.5 | ||||
| 70858 | Serum albumin | negative | 39.1 | 46.4 | 61.5 | 100.0 | 71.2 | |
| 70858 | Serum albumin | neutral | 26.5 | 29.8 | 44.8 | 100.0 | 100.0 | |
| 70858 | Serum albumin | positive | 49.8 | 53.3 | 48.5 | 51.9 | 100.0 | |
| 70015 | Alpha-fetoprotein | negative | 9.3 | 9.2 | 25.6 | 28.8 | ||
| 70015 | Alpha-fetoprotein | neutral | 8.1 | 19.5 | 38.9 | |||
| 70015 | Alpha-fetoprotein | positive | 1.3 | 1.2 | 12.3 | 19.7 | ||
| 69720 | Kininogen-1 | negative | 3.6 | 2.5 | 12.9 | |||
| 69720 | Kininogen-1 | neutral | 1.3 | 14.2 | 16.3 | |||
| 69720 | Kininogen-1 | positive | 3.8 | 4.9 | 5.1 | 8.2 | ||
| 67385 | Fibrinogen alpha chain | negative | 3.5 | |||||
| 67385 | Fibrinogen alpha chain | neutral | 1.8 | 5.5 | ||||
| 67385 | Fibrinogen alpha chain | positive | 3.4 | 4.4 | 4.2 | |||
| 55791 | Cytochrome P450 2C5 | negative | 12.2 | |||||
| 55791 | Cytochrome P450 2C5 | neutral | 0.2 | |||||
| 55791 | Cytochrome P450 2C5 | positive | 1.3 | |||||
| 54915 | Vitronectin | negative | 2.3 | 2.1 | ||||
| 54906 | Alpha-2-antiplasmin | negative | 2.8 | 4.3 | 24.9 | |||
| 54906 | Alpha-2-antiplasmin | neutral | 0.9 | |||||
| 54906 | Alpha-2-antiplasmin | positive | 3.9 | |||||
| 54595 | Vitamin D-binding protein | negative | 8.0 | 11.8 | 37.5 | |||
| 54595 | Vitamin D-binding protein | neutral | 5.0 | |||||
| 54595 | Vitamin D-binding protein | positive | 7.3 | 9.3 | 2.6 | |||
| 53980 | Alpha-1B-glycoprotein | negative | 1.7 | 5.4 | 25.3 | |||
| 53980 | Alpha-1B-glycoprotein | neutral | 0.9 | |||||
| 53980 | Alpha-1B-glycoprotein | positive | 1.3 | |||||
| 53182 | Vitronectin | negative | 1.8 | 1.1 | ||||
| 52728 | Antithrombin-III | negative | 1.2 | |||||
| 52728 | Antithrombin-III | neutral | 0.6 | |||||
| 51875 | Factor XIIa inhibitor | negative | 1.2 | |||||
| 51875 | Factor XIIa inhibitor | neutral | 0.2 | |||||
| 51410 | Angiotensinogen | negative | 1.8 | |||||
| 51410 | Angiotensinogen | neutral | 0.2 | |||||
| 50458 | Serine protease inhibitor A3F | negative | 0.6 | |||||
| 46351 | Alpha-1-antiproteinase | neutral | 2.6 | |||||
| 46351 | Alpha-1-antiproteinase | positive | 4.1 | 2.7 | 3.1 | 5.0 | ||
| 46300 | Endopin-1 | neutral | 0.9 | |||||
| 46300 | Endopin-1 | positive | 2.0 | 1.8 | 3.1 | |||
| 46292 | Pigment epithelium-derived factor | neutral | 0.7 | |||||
| 46292 | Pigment epithelium-derived factor | negative | 3.3 | |||||
| 43280 | Fetuin-B | neutral | 0.5 | |||||
| 43280 | Fetuin-B | positive | 3.5 | 2.9 | ||||
| 39945 | AMBP protein | neutral | 3.1 | |||||
| 39945 | AMBP protein | positive | 2.8 | 2.1 | ||||
| 39284 | Beta-2-glycoprotein 1 | neutral | 1.0 | |||||
| 39038 | Alpha-2-HS-glycoprotein | negative | 4.8 | |||||
| 39038 | Alpha-2-HS-glycoprotein | neutral | 9.1 | 22.3 | ||||
| 39038 | Alpha-2-HS-glycoprotein | positive | 5.3 | 3.3 | ||||
| 39008 | Lumican | neutral | 0.3 | |||||
| 36130 | Ecto-ADP-ribosyltransferase 4 | negative | 0.9 | |||||
| 36130 | Ecto-ADP-ribosyltransferase 4 | neutral | 0.3 | |||||
| 30258 | Apolipoprotein A-I | neutral | 3.7 | |||||
| 30258 | Apolipoprotein A-I | positive | 1.9 | 4.2 | ||||
| 23398 | Alpha-1-acid glycoprotein | plain | 19.5 | |||||
| 23398 | Alpha-1-acid glycoprotein | positive | 8.1 | 2.8 | ||||
| 22498 | Tetranectin | plain | 0.5 | |||||
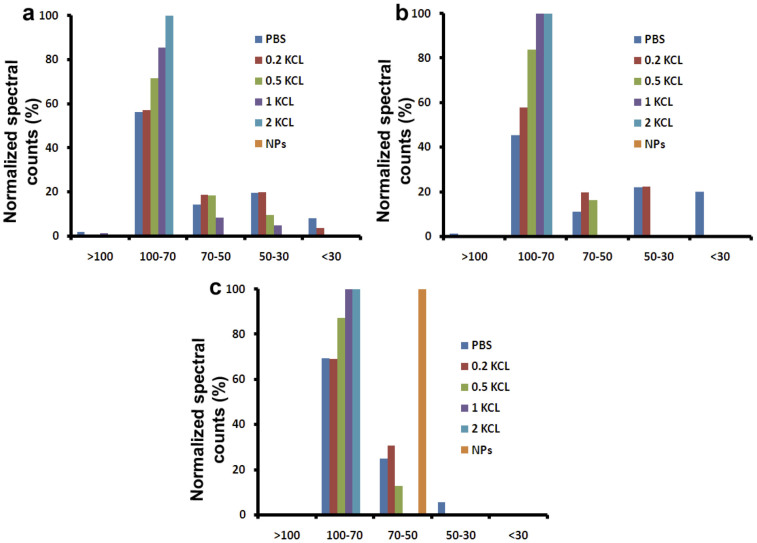
Figure 6. Normalised spectral counts (NSpC) of different molecular-weight (MW) range proteins in each elution fraction of the positively (a), neutral (b), and negatively (c) charged PVA-coated SPIONs.
Figure 7. Normalised spectral counts (NSpC) of the bound proteins in each elution fraction of the positively (a), neutral (b), and negatively (c) charged PVA-coated SPIONs (100% represents the total amount of adsorbed proteins in the protein corona).
Discussion
According to the results, one can conclude that most proteins that are adsorbed onto the surface of various PVA-coated SPIONs had relative large molecular weights (between 70–100 kDa), compared to the other defined range of molecular weights.
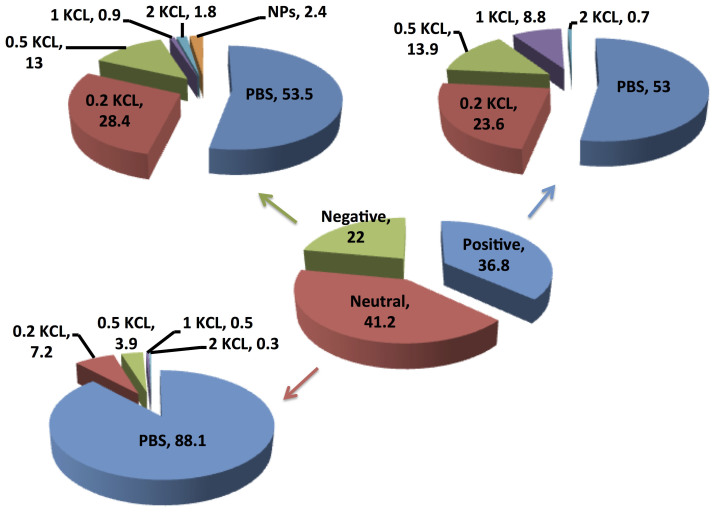
The total number of adsorbed proteins was represented as 100%. The serum proteins preferred to adsorb on the surface of positively charged and neutral PVA-coated SPIONs instead of the negative ones with a value of 36, 41 and 22%. However, 88% of the protein bound on neutral SPIONs was washed out in the PBS step, which indicates that the attached proteins on the surface of neutral SPIONs had a lower affinity than the positively and negatively charged SPIONs. A comparison between the adsorbed protein on negatively charged silica NP4 and the protein on the negatively charged PVA NPs showed that 9 proteins (serotransferrin, plasminogen, fibrinogen alpha chain, serum albumin, kininogen-1, vitamin D-binding protein alpha-2-antiplasmin, vitronectin, and alpha-2-HS-glycoprotein) could be found on both types of negatively charged NPs. A similar result was found when the negatively charged silica-coated SPIONs were compared with the negatively charged dextran-coated SPIONs. The 4 proteins that are found on all 3 types of negatively charged NPs (silica, PVA- and dextran-coated SPIONs) are as follows: serotransferrin, fibrinogen alpha chain, serum albumin, and kininogen-1. In addition, some proteins adsorbed at neutral and positively charged PVA- or dextran-coated SPIONs, which indicates a binding ability of these common and abundant proteins on different surface charges and surface materials.
The isoelectric point and therefore the overall charge of the proteins did not influence the adsorption behaviour of the proteins at the NP surface independently of the NP charge because there are several important additional factors (e.g., the inhomogeneous distribution of the surface charge on proteins, the hydrophobicity/hydrophilicity of the proteins and the NP surfaces, the London dispersion, the protein-protein interactions, and the hydrogen bonding) are active at the nano-bio interfaces46.
In summary, we have shown that the characteristics of the protein adsorption on two different surface coating using dextran- and PVA-coated SPIONs, strongly depend on the particle size, coating material, and the surface charge. Using a magnetic separation technique, we have shown that the protein components were eluted fraction by fraction when the salt concentration increased, which reveals the identification of a loosely and tightly bound protein profile. Comparing different surface materials, we found that the proteins preferred to bind on PVA-coated SPIONs instead of dextran-coated SPIONs. Serum albumin, serotransferrin, alpha-fetoprotein, and kininogen-1 are abundant proteins that were found on all types and surface charge of NPs. For the same charge with different materials, the complement factor B, alpha-1-antitrypsin like protein, haemoglobin subunit alpha-I/II, and apolipoprotein A-II specifically bind to the positively charged dextran-coated SPIONs but not the PVA-coated SPIONs. Alpha-1-antiproteinase, thyroxine-binding globulin, endopin-1, fetuin-B, transthyretin, haemoglobin subunit alpha, and apolipoprotein A-II were found specifically on the negatively charged dextran-coated SPIONs but not on the negatively charged PVA-coated SPIONs. However, there are no specific proteins bound on the neutral dextran-coated SPIONs. Interestingly, the proteins prefer to bind to the charged dextran-coated SPIONs and neutral PVA-coated SPIONs. With a semi-quantitative assessment using a spectral-counting method, we present the adsorption isotherms of different components in different adsorbed layers. For the PVA-coated SPIONs, the serum proteins preferred to bind to neutral NP with a loose binding. Intermediate-binding proteins were found on the positively charged NPs, as represented by many proteins that remained bound on the particles after an optimum wash of PBS but not on the NP fraction. Notably tightly bound proteins (cytochrome P450 2C5, alpha-2-antiplasmin, vitamin D-binding protein, and alpha-1B-glycoprotein) were observed on the negatively charged SPIONs.
The elution of NP-bound proteins with different salt concentration solutions reveals different binding affinities of each protein to the NP surfaces. Combining the results of intermediate-bound proteins and cell internalisation of positively charged PVA-coated SPIONs indicated that the tightly bound proteins and the intermediate-bound proteins are necessary for cell internalisation. This protein corona profile indicates the nature of the adsorbed proteins and the importance of protein affinity to specific surface charges and materials, which were related to both in vivo and in vitro biological responses e.g., the cell-NP interaction, the intracellular mechanism, the cell signalling, the immunisation, and the clearance mechanism.
Although there has been an overall lack of correlation between the NP properties such as the charge and the composition of the polymer coating and the total-protein corona, another recent study demonstrated the preferential adsorption of some classes of proteins based on the charge density47. However, little is known today about the specific interactions of NPs with the proteins, the cell surfaces and the intracellular organelles. The proteomic analysis, which uses state-of-the-art MS platforms, bioinformatics and system biology tools, must be included in such investigations to predict the behaviour of the engineered NPs in a biological environment48.
Methods
Preparation of dextran-coated NPs
SPIONs were synthesized and coated with various dextrans according to the previously reported protocols49,50,51.
PVA coatings
All chemicals were of the analytical reagent grade and used without further purification. SPIONs were synthesized with co-precipitation method52. Polyvinyl alcohol (PVA; Mowiol® 3-83) with an average molecular weight (MW) of 14,000 g/mol and a hydrolysis degree of 83% was supplied by CLARIANT. Vinyl alcohol/vinyl amine copolymer M12 with an average MW of 80,000–140,000 was supplied by ERKOL. PVA with randomly distributed carboxyl groups (KL-506) with an average molecular weight (MW) of 30,000–50,000 g/mol and a hydrolysis degree of 74–80% was supplied by the Kuraray Co., Ltd. The polymer solutions were prepared by dissolving the powders in water (2% M12, 10% Mowiol 3-83 and 6% KL-506) and rapidly heating the solutions for 15 min (Mowiol® 3-83 and KL-506) to 4 h (M12) at 90°C. Then, the polymer solution was filtered through a sterile filter (pore size: 200 nm). Ultrapure deionised water (Seralpur delta UV/UF setting, 0.055 μS/cm) was used in all synthesis steps. To obtain positively, neutral and negatively charged NPs, a mixture of polyvinyl alcohol (Mowiol®3-83) and vinyl alcohol/vinyl amine copolymer (M12, vinyl alcohol/vinyl alcohol copolymer) at a mass ratio of 45, 10% Mowiol 3-83 and 6% KL-506 solutions, respectively, were prepared by mixing with 10 mgFe/ml SPION suspension with a v/v ratio of 1:1. The mixture was preserved for at least 1 week at 4°C and maintained at that temperature until used. The particle suspension was adjusted to pH 7 for the protein-separation experiment.
Dynamic light scattering (DLS) for size and zeta measurement
The DLS measurements were performed using a Malvern PCS-4700 instrument for dextran-NPs, which was equipped with a 256-channel correlator. The 488.0 nm line of a Coherent Innova-70 Ar ion laser was used as the incident beam. The laser power was 250 mW. The scattering angles θ ranged between 40–140°. The temperature was maintained at 25°C with an external circulator. The zeta potential was determined using a Malvern Zetasizer 3000HSa. Each measurement was an average of 6 repetitions of one minute each and was repeated five times. The data were analysed according to the standard procedures and interpreted using a cumulant expansion of the field autocorrelation function to the second order. Moreover, to obtain the decay rate distribution, a constrained regularisation method, CONTIN, was used to invert the experimental data. PVA-SPIONs were measured at 25°C with Brookhaven Analyzer, ZetaPALS with CONTIN. The samples (after size and zeta measurement) were not used further for protein analysis.
NPs incubation
The interactions of positive-, neutral and negative-charge SPIONs with foetal bovine serum (FBS) were probed. Because the previous reports confirmed that the proteins could form a relatively stable corona over a period of 1 h6, we selected 1 h for the protein corona evaluation in our experiments. Note that in the media that contained serum, the serum-to-particle surface ratio was fixed at 2.8 ml per m2 to achieve good results10.
Optimisation of the washing procedure
To ensure that the NP-bound proteins were washed out without the interference of the PVA that left the NP surface, the effect of washing on the removal of PVA from NP surface was evaluated. The NPs were incubated with serum with the identical protein-to-particle surface ratio (2.8 ml per m2) for 1 h before loading and trapping in a magnetic separator. Then, the trapped NPs (NP-protein complex) were eluted with phosphate buffered saline (PBS) for 30 CV at different column volumes (1 CV is approximately 500 μl) at a flow rate of 0.5 ml/min. The washing fractions (the eluate) were collected at different column volumes. The particles were dispersed and diluted in PBS to measure the size and the zeta potential. In addition, the protein, the iron and the PVA were quantified using the Lowry assay, the iron Prussian blue assay and the PVA polymer assay, respectively.
Interaction of different surface-coated SPIONs with FBS proteins
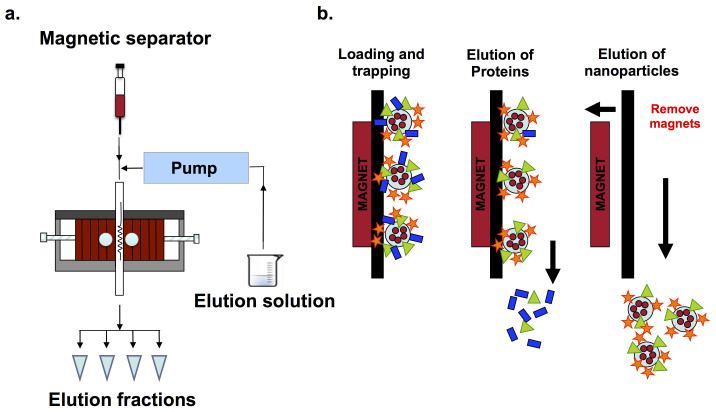
To obtain the SPION-“hard corona” composition, after the nanoparticle incubation, the NPs were trapped in a magnetic column. The setup and the protein-separation procedure using a magnetic separator are shown in Figure 8. The flow-through fractions were removed. The trapped NPs were washed with PBS according to the optimum condition, which was obtained from the optimisation procedure (4 column volumes (CV) for positively charged NPs and 8 CV for both neutral and negatively charged NPs). Then, the proteins were eluted from the NPs using 0.2, 0.5, 1 and 2 M KCl. Finally, the protein-NP complexes (NPs with tightly bound proteins or “hard-corona” composition) were removed from the column. The eluted proteins were assessed using SDS-PAGE and liquid chromatography mass spectrometry (LC-MS/MS).
Figure 8. Schematic representations of the magnetic separator setup for the protein separation (a) and the protein separation processes (b) including the trapping, protein elution and protein-NP complex elution.
Note that the washing process was designed to also remove the excess (unbound or loosely bound) proteins, and all stages were performed in low-protein-attachment Eppendorf tubes.
Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE)
The eluted proteins from all collection fractions and NP-protein complexes were resuspended in Laemmli sample buffer and boiled for 5 min at 100°C. An equal-volume sample was loaded and separated on a 12% polyacrylamide gel. Gel electrophoresis was performed at 120 V. The gels were stained using the silver staining method. All bands were cut and analysed using the LC-MS/MS technique.
Animal experiments
Two-month-old female Lewis rats (approximately 200 g, purchased from Javier France) were used for blood circulation study of SPION. Then either 1.4 mL of 150 mM NaCl or 7 mg of Fe of SPIONs (1.4 mL of suspension of positive, neutral and negative PVA-SPION) was injected intravenously through the tail. The rats were anesthetised with 5% isoflurane and sacrificed 15 min after the injection. The whole blood was collected and the amount of SPION still in circulation was measured using a MS3 magneto-susceptometer (Bartington®) with a MS2G sensor53 The results obtained in mgFe are given in percentage of the injected dose following the equation: percentage of injected dose = mgFe measured in blood/7 mgFe. The experiments were done in triplicate for each condition.
Relative protein quantification
To obtain the total number of the LC-MS/MS spectra for all peptides that are attributed to a matched protein, the protein amounts were semi-quantitatively assessed by applying the spectral-counting method (SpC) to obtain the percentage of participating proteins in the hard-corona composition4,15. The normalised SpC amounts of each protein, which were identified in the LC-MS/MS study of smooth and jagged surfaces, were calculated by applying the following equation:
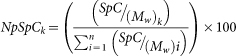 |
where NpSpCk is the normalised percentage of the spectral count for protein k, SpC is the identified spectral count, and MW is the molecular weight (in kDa) of the protein k.
Supplementary Material
In vivo evaluations
Acknowledgments
This work has been supported by the NanoDiaRA project, European Commission (FP7-NMP-2008-LARGE) under grant agreement no. 228929, and the Swiss National Science Foundation (SNSF) fund number 205321-120161/1. The authors would like to acknowledge A. Gramoun, Department of Radiology, University of Geneva and Geneva University Hospital, Geneva, Switzerland for helping us with the animal tests.
Footnotes
The authors declare no competing financial interests.
Author Contributions U.S., M.M., L.M., J.S. and H.H. designed the project. U.S., M.M., L.M. and J.S. performed the experiments and analysed the data. U.S., M.M. and H.H. wrote the manuscript.
References
- Cedervall T. et al. Detailed identification of plasma proteins adsorbed on copolymer nanoparticles. Angew. Chem. Int. Ed. Engl. 46, 5754–5756 (2007). [DOI] [PubMed] [Google Scholar]
- Cedervall T. et al. Understanding the nanoparticle–protein corona using methods to quantify exchange rates and affinities of proteins for nanoparticles. PNAS 104, 2050–2055 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoudi M., Hofmann H., Rothen-Rutishauser B. & Petri-Fink A. Assessing the In Vitro and In Vivo Toxicity of Superparamagnetic Iron Oxide Nanoparticles. Chem. Rev. 112, 2323–2338 (2012). [DOI] [PubMed] [Google Scholar]
- Monopoli M. P. et al. Physical−Chemical Aspects of Protein Corona: Relevance to in Vitro and in Vivo Biological Impacts of Nanoparticles. J. Am. Chem. Soc. 133, 2525–2534 (2011). [DOI] [PubMed] [Google Scholar]
- Mahmoudi M. et al. Protein-nanoparticle interactions: opportunities and challenges. Chem. Rev. 111, 5610–5637 (2011). [DOI] [PubMed] [Google Scholar]
- Walczyk D., Bombelli F. B., Monopoli M. P., Lynch I. & Dawson K. A. What the Cell ‘Sees' in Bionanoscience. J. Am. Chem. Soc. 132, 5761–5768 (2010). [DOI] [PubMed] [Google Scholar]
- Hellstrand E. et al. Complete high-density lipoproteins in nanoparticle corona. FEBS Journal 276, 3372–3381 (2009). [DOI] [PubMed] [Google Scholar]
- Lindman S. et al. Systematic Investigation of the Thermodynamics of HSA Adsorption to N-iso-Propylacrylamide/N-tert-Butylacrylamide Copolymer Nanoparticles. Effects of Particle Size and Hydrophobicity. Nano Lett. 7, 914–920 (2007). [DOI] [PubMed] [Google Scholar]
- Linse S. et al. Nucleation of protein fibrillation by nanoparticles. PNAS 104, 8691–8696 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundqvist M. et al. Nanoparticle size and surface properties determine the protein corona with possible implications for biological impacts. PNAS 105, 14265–14270 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch I. & Dawson K. A. Protein-nanoparticle interactions. Nano Today 3, 40–47 (2008). [Google Scholar]
- Lynch I., Dawson K. A. & Linse S. Detecting Cryptic Epitopes Created by Nanoparticles. Sci. STKE 2006, pe14 (2006). [DOI] [PubMed] [Google Scholar]
- Lynch I. et al. The nanoparticle–protein complex as a biological entity; a complex fluids and surface science challenge for the 21st century. Adv. Colloid Interface Sci. 134–135, 167–174 (2007). [DOI] [PubMed] [Google Scholar]
- Dawson K. A., Salvati A. & Lynch I. Nanotoxicology: Nanoparticles reconstruct lipids. Nat. Nanotechnol. 4, 84–85 (2009). [DOI] [PubMed] [Google Scholar]
- Monopoli M. P., Bombelli F. B. & Dawson K. A. Nanobiotechnology: nanoparticle coronas take shape. Nat. Nanotechnol. 6, 11–12 (2011). [DOI] [PubMed] [Google Scholar]
- Rosen J. E., Chan L., Shieh D.-B. & Gu F. X. Iron oxide nanoparticles for targeted cancer imaging and diagnostics. Nanomedicine: Nanotechnology, Biology Med. 8, 275–290 (2012). [DOI] [PubMed] [Google Scholar]
- Mahmoudi M., Sant S., Wang B., Laurent S. & Sen T. Superparamagnetic iron oxide nanoparticles (SPIONs): Development, surface modification and applications in chemotherapy. Adv. Drug Delivery Rev. 63, 24–46 (2011). [DOI] [PubMed] [Google Scholar]
- Laurent S., Dutz S., Häfeli U. O. & Mahmoudi M. Magnetic fluid hyperthermia: Focus on superparamagnetic iron oxide nanoparticles. Adv. Colloid Interface Sci. 166, 8–23 (2011). [DOI] [PubMed] [Google Scholar]
- Ang D., Tay C. Y., Tan L. P., Preiser P. R. & Ramanujan R. V. In vitro studies of magnetically enhanced transfection in COS-7 cells. Mat. Sci. Eng. C 31, 1445–1457 (2011). [Google Scholar]
- Soenen S. J., De Smedt S. C. & Braeckmans K. Limitations and caveats of magnetic cell labeling using transfection agent complexed iron oxide nanoparticles. Contrast Med. Mol. Imag. 7, 140–152 (2012). [DOI] [PubMed] [Google Scholar]
- Mahmoudi M. et al. Magnetic Resonance Imaging Tracking of Stem Cells in Vivo Using Iron Oxide Nanoparticles as a Tool for the Advancement of Clinical Regenerative Medicine. Chem. Rev. 111, 253–280 (2011). [DOI] [PubMed] [Google Scholar]
- Mahmoudi M., Simchi A., Milani A. S. & Stroeve P. Cell toxicity of superparamagnetic iron oxide nanoparticles. J. Colloid Interface Sci. 336, 510–518 (2009). [DOI] [PubMed] [Google Scholar]
- Neuwelt E. A. et al. Delivery of virus-sized iron oxide particles to rodent CNS neurons. Neurosurgery 34, 777–784 (1994). [DOI] [PubMed] [Google Scholar]
- Müller R. H., Maaβen S., Weyhers H., Specht F. & Lucks J. S. Cytotoxicity of magnetite-loaded polylactide, polylactide/glycolide particles and solid lipid nanoparticles. Int. J. Pharm. 138, 85–94 (1996). [Google Scholar]
- Berry C. C., Wells S., Charles S., Aitchison G. & Curtis A. S. G. Cell response to dextran-derivatised iron oxide nanoparticles post internalisation. Biomaterials 25, 5405–5413 (2004). [DOI] [PubMed] [Google Scholar]
- Berry C. C., Wells S., Charles S. & Curtis A. S. G. Dextran and albumin derivatised iron oxide nanoparticles: influence on fibroblasts in vitro. Biomaterials 24, 4551–4557 (2003). [DOI] [PubMed] [Google Scholar]
- Gupta A. K., Berry C., Gupta M. & Curtis A. Receptor-mediated targeting of magnetic nanoparticles using insulin as a surface ligand to prevent endocytosis. IEEE Trans Nanobiosci. 2, 255–261 (2003). [DOI] [PubMed] [Google Scholar]
- Gupta A. K. & Curtis A. S. Lactoferrin and ceruloplasmin derivatized superparamagnetic iron oxide nanoparticles for targeting cell surface receptors. Biomaterials 25, 3029–3040 (2004). [DOI] [PubMed] [Google Scholar]
- Gupta A. K., Naregalkar R. R., Vaidya V. D. & Gupta M. Recent advances on surface engineering of magnetic iron oxide nanoparticles and their biomedical applications. Nanomedicine 2, 23–39 (2007). [DOI] [PubMed] [Google Scholar]
- Gupta A. K. & Wells S. Surface-modified superparamagnetic nanoparticles for drug delivery: preparation, characterization, and cytotoxicity studies. IEEE Trans Nanobiosci. 3, 66–73 (2004). [DOI] [PubMed] [Google Scholar]
- van den Bos E. J. et al. Improved Efficacy of Stem Cell Labeling for Magnetic Resonance Imaging Studies by the Use of Cationic Liposomes. Cell Transplant. 12, 743–756 (2003). [DOI] [PubMed] [Google Scholar]
- Laurent S., Burtea C., Thirifays C., Rezaee F. & Mahmoudi M. Significance of cell ‘observer' and protein source in nanobiosciences. J. Colloid Interface Sci. 392, 431–445 (2013). [DOI] [PubMed] [Google Scholar]
- Maiorano G. et al. Effects of Cell Culture Media on the Dynamic Formation of Protein−Nanoparticle Complexes and Influence on the Cellular Response. ACS Nano 4, 7481–7491 (2010). [DOI] [PubMed] [Google Scholar]
- Mahmoudi M. et al. Irreversible changes in protein conformation due to interaction with superparamagnetic iron oxide nanoparticles. Nanoscale 3, 1127–1138 (2011). [DOI] [PubMed] [Google Scholar]
- Steitz B. et al. Fixed Bed Reactor for Solid-Phase Surface Derivatization of Superparamagnetic Nanoparticles. Bioconjugate Chem. 18, 1684–1690 (2007). [DOI] [PubMed] [Google Scholar]
- Salaklang J. et al. Superparamagnetic Nanoparticles as a Powerful Systems Biology Characterization Tool in the Physiological Context. Angew. Chem. Int. Ed. 47, 7857–7860 (2008). [DOI] [PubMed] [Google Scholar]
- Prigent P. et al. in Molecular and Cellular MR Imaging (Modo M., & Bulte J.) 59–83 (CRC Press, 2007). [Google Scholar]
- Reinert M. et al. Metabolic pathway and distribution of superparamagnetic iron oxide nanoparticles: in vivo study. Int. J. Nanomedicine 1793 (2011). 10.2147/IJN.S23638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann-Amtenbrink M., Hofmann H. & Montet X. Superparamagnetic nanoparticles - a tool for early diagnostics. Swiss Med Wkly 140, w13081 (2010). [DOI] [PubMed] [Google Scholar]
- Jiang Y. et al. In-vivo studies on intraperitoneally administrated poly(vinyl alcohol). J. Biomed. Mater. Res. Part B Appl. Biomater. 93, 275–284 (2010). [DOI] [PubMed] [Google Scholar]
- Venturoli D. & Rippe B. Ficoll and dextran vs. globular proteins as probes for testing glomerular permselectivity: effects of molecular size, shape, charge, and deformability. Am. J. Physiol. Renal. Physiol. 288, F605–F613 (2005). [DOI] [PubMed] [Google Scholar]
- Dobrovolskaia M. A. & McNeil S. E. Immunological properties of engineered nanomaterials. Nat. Nanotechnol. 2, 469–478 (2007). [DOI] [PubMed] [Google Scholar]
- Levy M. et al. Long term in vivo biotransformation of iron oxide nanoparticles. Biomaterials 32, 3988–3999 (2011). [DOI] [PubMed] [Google Scholar]
- Mahmoudi M., Shokrgozar M. A. & Behzadi S. Slight temperature changes affect protein affinity and cellular uptake/toxicity of nanoparticles. Nanoscale 5, 3240–3244 (2013). [DOI] [PubMed] [Google Scholar]
- Lesniak A. et al. Effects of the Presence or Absence of a Protein Corona on Silica Nanoparticle Uptake and Impact on Cells. ACS Nano 6, 5845–5857 (2012). [DOI] [PubMed] [Google Scholar]
- Puzyn T. et al. Using nano-QSAR to predict the cytotoxicity of metal oxide nanoparticles. Nat. Nanotechnol. 6, 175–178 (2011). [DOI] [PubMed] [Google Scholar]
- Capriotti A. L. et al. Do plasma proteins distinguish between liposomes of varying charge density? J. Proteomics 75, 1924–1932 (2012). [DOI] [PubMed] [Google Scholar]
- Lai Z. W., Yan Y., Caruso F. & Nice E. C. Emerging Techniques in Proteomics for Probing Nano–Bio Interactions. ACS Nano 6, 10438–10448 (2012). [DOI] [PubMed] [Google Scholar]
- Rauch J., Kolch W. & Mahmoudi M. Cell Type-Specific Activation of AKT and ERK Signaling Pathways by Small Negatively-Charged Magnetic Nanoparticles. Sci. Rep. 2, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. et al. Ultra-large-scale syntheses of monodisperse nanocrystals. Nat. Mater. 3, 891–895 (2004). [DOI] [PubMed] [Google Scholar]
- Mahmoudi M. & Serpooshan V. Silver-Coated Engineered Magnetic Nanoparticles Are Promising for the Success in the Fight against Antibacterial Resistance Threat. ACS Nano 6, 2656–2664 (2012). [DOI] [PubMed] [Google Scholar]
- Chastellain M., Petri A. & Hofmann H. Particle size investigations of a multistep synthesis of PVA coated superparamagnetic nanoparticles. J. Colloid Interface Sci. 278, 353–360 (2004). [DOI] [PubMed] [Google Scholar]
- Maurizi L., Sakulkhu U., Gramoun A., Vallee J.-P. & Hofmann H. A fast and reproducible method to quantify magnetic nanoparticle biodistribution. Analyst 139, 1184 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
In vivo evaluations