Abstract
Repression of the cellular CIITA gene is part of the immune evasion strategy of the γherpes virus Epstein–Barr virus (EBV) during its lytic replication cycle in B-cells. In part, this is mediated through downregulation of MHC class II gene expression via the targeted repression of CIITA, the cellular master regulator of MHC class II gene expression. This repression is achieved through a reduction in CIITA promoter activity, initiated by the EBV transcription and replication factor, Zta (BZLF1, EB1, ZEBRA). Zta is the earliest gene expressed during the lytic replication cycle. Zta interacts with sequence-specific elements in promoters, enhancers and the replication origin (ZREs), and also modulates gene expression through interaction with cellular transcription factors and co-activators. Here, we explore the requirements for Zta-mediated repression of the CIITA promoter. We find that repression by Zta is specific for the CIITA promoter and can be achieved in the absence of other EBV genes. Surprisingly, we find that the dimerization region of Zta is not required to mediate repression. This contrasts with an obligate requirement of this region to correctly orientate the DNA contact regions of Zta to mediate activation of gene expression through ZREs. Additional support for the model that Zta represses the CIITA promoter without direct DNA binding comes from promoter mapping that shows that repression does not require the presence of a ZRE in the CIITA promoter.
Introduction
Epstein–Barr virus infects people and has a life-long association with them, occasionally causing diseases including infectious mononucleosis, Burkitt's lymphoma, Hodgkin's lymphoma and nasopharygeal carcinoma (Magrath, 2012; Molyneux et al., 2012; Saha & Robertson, 2011). Epstein–Barr virus infects human B-lymphocytes and epithelial cells and establishes long-term latency in memory B-lymphocytes (Babcock et al., 1998). These cells are largely protected from immune attack by the silencing of viral gene expression. The virus is sporadically reactivated following B-cell activation and differentiation into plasma cells (Crawford & Ando, 1986; Laichalk et al., 2002; Laichalk & Thorley-Lawson, 2005). As EBV enters the lytic replication cycle, it expresses around 90 viral genes that are required for the regulation of viral gene expression, replication of the viral genome, assembly, packaging and egress of the virion (Farrell, 2005). Many viral genes expressed during viral lytic replication are excellent targets for immune recognition (Adhikary et al., 2006; Long et al., 2011). Attack by the immune system during viral replication would threaten cell survival and thus the successful generation of virions, but EBV has evolved several strategies to evade immune responses during viral lytic replication (Zuo & Rowe, 2012).
An important regulator of EBV lytic replication, termed Zta (BZLF1, ZEBRA, EB1), is a transcription factor, a replication factor and it disrupts several signal transduction pathways (Kenney, 2007). Routes by which Zta activates gene expression have been documented for both viral and host promoters. Many promoters are targeted by the interaction of the sequence-specific DNA-binding domain of Zta with sequence-specific 7 nt DNA elements, termed ZREs (Adamson & Kenney, 1999; Bergbauer et al., 2010; Bhende et al., 2004, 2005; Broderick et al., 2009; Dickerson et al., 2009; Flower et al., 2011; Holley-Guthrie et al., 1990; Kalla et al., 2012, 2010; Karlsson et al., 2008; Kenney et al., 1989; Ramasubramanyan et al., 2012a, b; Sinclair, 2003; Sinclair et al., 1991; Woellmer et al., 2012). At least 32 distinct ZRE sequence variants are specifically recognized by Zta (Flower et al., 2011).
Downregulation of gene expression by Zta has been documented for the TNFR1 gene, through the cellular C/EBP genes (Bristol et al., 2010). Additionally, post-translational modifications of Zta have been shown to reduce the ability of Zta to regulate gene expression, specifically phosphorylation at residue S209 (Asai et al., 2009) and sumoylation through residue K12 (Hagemeier et al., 2010; Murata et al., 2010).
Zta has been shown to downregulate the expression of the master regulator of MHC class II gene expression, CIITA, in an EBV-positive B-cell line, with both protein and RNA levels decreasing following induction of EBV lytic cycle activation (Li et al., 2009). The product of CIITA is a non-DNA-binding cellular transcriptional co-activator, which acts through interaction with DNA-bound proteins that lack integral activation domains. CIITA activates the expression of MHC class II genes (Chang et al., 2002) and the reduced expression of CIITA observed in B-cells undergoing lytic cycle correlates with the reduced expression of MHC class II observed at the cell surface (Li et al., 2009). Repression of CIITA gene expression is also driven by the related γherpesvirus, KSHV (Cai et al., 2013). Here, we investigate the route by which Zta represses CIITA expression.
Results
CIITA promoter is specifically repressed by the EBV Zta protein
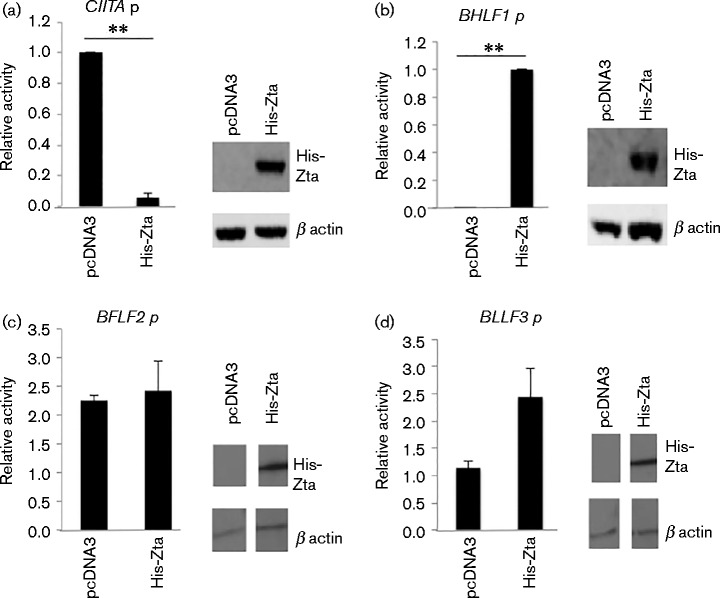
The effect of Zta expression on the activity of the CIITA promoter and the viral BHLF1 promoter were compared in EBV-positive Raji cells following co-transfection of reporter constructs with an expression vector for a polyhistidine-tagged version of Zta (Bailey et al., 2009). The impact of Zta expression was expressed relative to the maximal activity for each promoter (Fig. 1a, b). Expression of Zta repressed the CIITA promoter, whilst in the same experiment it dramatically activated expression of a viral promoter containing ZREs (BHLF1). This provided confirmation of the results of Li et al. showing that a short region of the CIITA promoter is sufficient to mediate repression following Zta expression (Li et al., 2009). The repression of CIITA promoter activity could result from the overexpression of a transactivator domain that non-specifically sequesters basal transcription factors or co-activators, thereby inhibiting all RNA polymerase II-dependent transcription. To address whether this was the case, we undertook experiments to explicitly determine whether Zta repressed other promoters. We generated promoter-reporter gene constructs for two viral promoters, BFLF2 and BLLF3. The impact of His-Zta expression on each promoter was assessed in Raji cells (Fig. 1c, d). This showed that neither BLLF3 nor BFLF2 promoters were repressed by Zta expression. We further investigated the repression of CIITA in BL cells by following two downstream targets of CIITA expression, HLA-DOA and HLA-DBM. Both were downregulated at the RNA level following Zta expression in BL cells (Fig. S1, available in the online Supplementary Material).
Fig. 1.
Repression of CIITA promoter by Zta is specific. (a–d) The CIITA ( − 286 to +54) (a), BHLF1 (b), BFLF2 (c) and BLLF3 (d) promoter-luciferase plasmids and the indicated expression vectors were introduced into cells by electroporation, and 48 h later cells were harvested and the luciferase activity determined. Promoter activity in Raji cells relative to the maximal activity of CIITA promoter (transfected with control plasmid) with the standard deviation from six assays (three replicate samples from each of two separate experiments) is shown. For comparisons +/ − Zta, ** represents p of significant difference < 0.01. The expression of His-Zta and endogenous protein were analysed by Western blot of proteins from the transfected cells.
As Raji cells contain an EBV genome, changes in viral gene expression may occur as a consequence of activating a partial lytic replication cycle through the expression of Zta (Kallin & Klein, 1983). In order to determine whether Zta relies on additional viral components to repress CIITA expression, we introduced the CIITA promoter-reporter gene into an EBV-negative subclone of Akata Burkitt's lymphoma cells (AK31) (Jenkins et al., 2000). In this cell background we saw that co-expression of Zta drove repression of the CIITA promoter-reporter gene around fivefold (Fig. 2). This clearly demonstrates that Zta-mediated repression of CIITA does not depend on additional EBV genes.
Fig. 2.
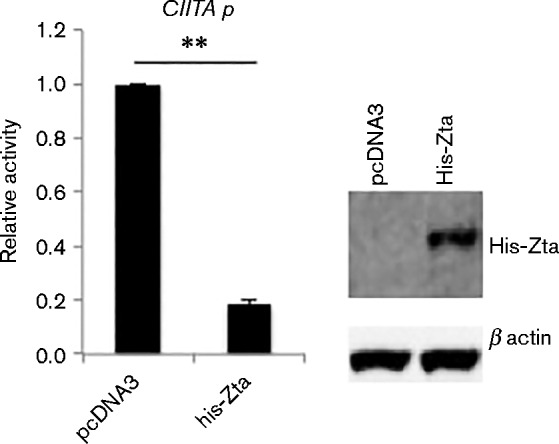
Repression of CIITA promoter by Zta is independent of other viral proteins. The CIITA ( − 286 to +54) promoter-luciferase plasmid and the indicated expression vectors were introduced into an EBV-negative subclone of the Akata BL cells (AK31) by electroporation, and 48 h later cells were harvested and the luciferase activity determined. Promoter activity is expressed relative to the maximal activity of CIITA promoter (transfected with control plasmid) with the standard deviation from six assays (three replicate samples from each of two separate experiments). For comparisons +/ − Zta, ** represents p of significant difference < 0.01. The expression of His-Zta and endogenous protein were analysed by Western blot of proteins from the transfected cells.
To explore the relevance of post-translational modifications of Zta to the Zta-mediated repression of the CIITA promoter, we generated mutants of Zta at amino acid residues K12 and S209 to prevent either sumoylation or phosphorylation. Following transfection, we found that neither post-translational modification was required for Zta to repress the CIITA promoter (Table 1).
Table 1. Impact of mutation of K12 and S209 on Zta-mediated repression.
| His-Zta | His-Zta K12R | His-Zta S209A | His-Zta S209D | |
|---|---|---|---|---|
| CIITA promoter activity | 1.00 | 1.00 | 1.00 | 1.00 |
| Relative promoter activity following His-Zta expression | 0.16 | 0.07 | 0.07 | 0.04 |
| Standard deviation | 0.15 | 0.08 | 0.01 | 0.00 |
Domains of Zta mediating repression of CIITA promoter
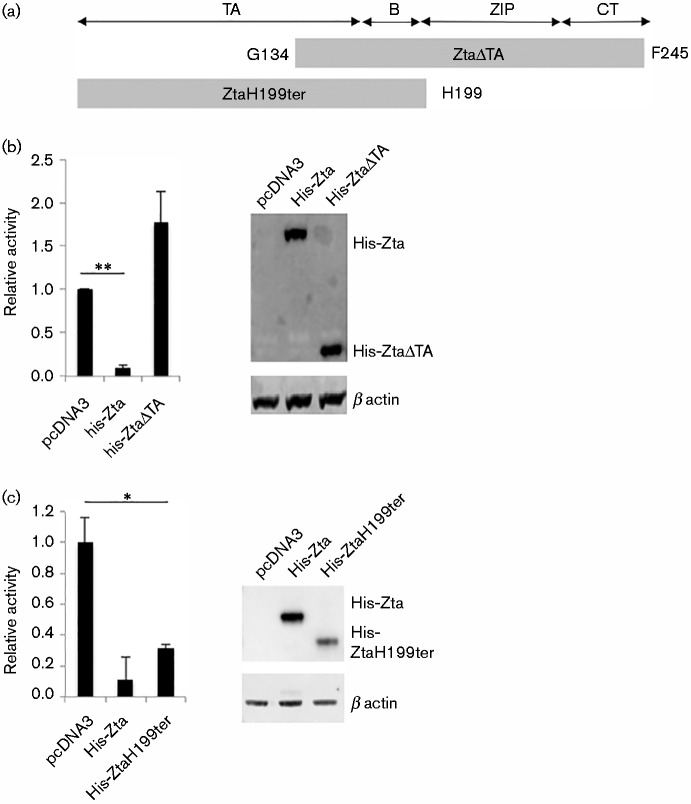
We then explored which domains of Zta protein mediate the repression of CIITA. Two versions of Zta were generated, both retaining the nuclear localization signal (Mikaélian et al., 1993). One mutant omits the N-terminal transactivation domain (ZtaΔTA); this protein was previously shown to be able to bind to DNA but not to transactivate a reporter construct (Packham et al., 1990). The second omits the dimerization and C-terminal region (ZtaH199ter) and has been shown previously to be unable to bind DNA (Hicks et al., 2003) or to form dimers (Schelcher et al., 2005) (Fig. 3a). Following transfection into Raji cells, we find that deletion of the transactivation domain ablates the ability of Zta to repress the CIITA promoter, despite the proteins being expressed at equivalent levels (Fig. 3b). In contrast, deletion of the dimerization and C-terminal regions of Zta only resulted in a small reduction in the repression of the CIITA promoter (Fig. 3c). The slightly lower level of repression observed with ZtaH199ter might result from the reduced abundance of this form of the protein. Taken together, these data show that a major component of Zta-mediated repression of the CIITA promoter occurs independently of a need for Zta to form dimers.
Fig. 3.
Zta repression of CIITA promoter requires the transactivation domain. (a) Schematic of the Zta protein and the two mutant versions that were evaluated. TA, transactivation domain; B, basic DNA contact region; ZIP, dimerization bZIP domain; CT, carboxy terminal region (required for dimerization and replication). (b, c) The CIITA promoter-luciferase plasmids ( − 286 to +54) and either control, His-Zta or His-Zta mutant expression vectors were introduced into Raji BL cells by electroporation. 48 h later, cells were harvested and the luciferase activity and protein concentrations determined. For comparisons +/ − Zta, ** represents p of significant difference < 0.01; * represents p of significant difference < 0.05. (b) Promoter activity of − 286 to +54 promoter with His-ZtaΔTA with the standard deviation from six assays (three replicate samples from each of two separate experiments), together with a Western blot. (c) Promoter activity of − 286 to +54 promoter with His-Zta199ter with the standard deviation from six assays (three replicate samples from each of two separate experiments), together with a Western blot.
It has been shown previously that a Zta binding site within the CIITA promoter allows repression by Zta (Li et al., 2009). Our data showed that dimerization is not an obligate requirement for repression, implying that DNA binding is not required. To explore this further, we assessed the promoter for potential ZREs (Flower et al., 2011) and found only one, which was shown previously to be a Zta binding site (Li et al., 2009). Using chromatin precipitation coupled with next-generation DNA sequencing we confirmed that Zta bound to the promoter region of CIITA (Fig. S2), but noted that this does not distinguish between direct and indirect binding. To evaluate the relevance of the potential ZRE, we generated a promoter reporter construct in which the region containing a ZRE was deleted (Fig. 4a). Both of these promoters were expressed at equivalent levels (Table 2). Both full-length Zta and the dimerization-deficient mutant Zta-H199ter repressed the promoter missing the ZRE (Fig. 4b). This supports our contention that the ability of Zta to repress the expression of CIITA does not rely on direct DNA binding.
Fig. 4.
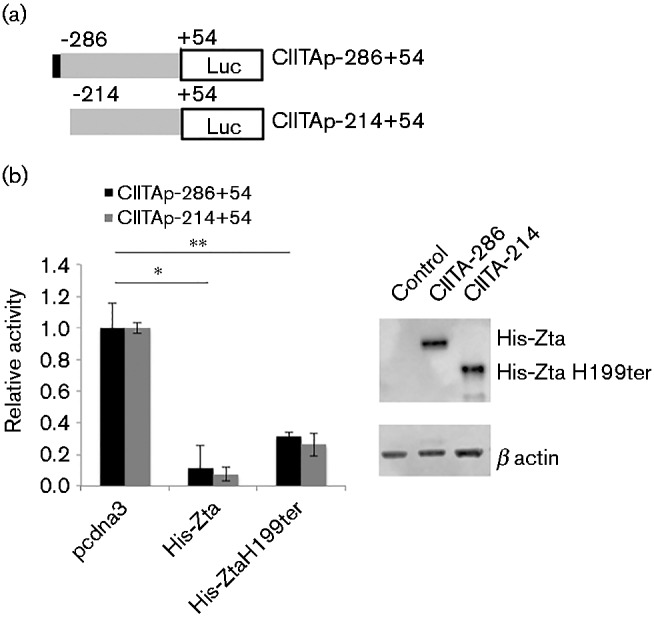
Zta repression of CIITA promoter occurs without binding to the ZRE. (a) Schematic of the CIITA mutant promoters used in these experiments. The location of the ZRE is indicated by a filled box. (b) The CIITA promoter-luciferase plasmids (either − 286 to +54 or − 214 to +54) and either control, His-Zta or His-Zta mutant expression vectors were introduced into Raji BL cells by electroporation. 48 h later, cells were harvested and the luciferase activity and protein concentrations determined. The CIITA basal promoter activity is shown (grey bars) together with the His-Zta mediated activity (black bars), with the standard deviation from six assays (three replicate samples from each of two separate experiments). For comparisons +/ − Zta, ** represents p of significant difference < 0.01; * represents p of significant difference < 0.05. For both His-Zta and His-ZtaH199ter, the significance is equal for each of the different promoters. Western blot analysis of protein expression in the transfected cells is also shown.
Table 2. Impact of Zta expression on − 286 and − 214 CIITA promoters.
| Average luciferase units CIITA ( − 286/+54) | Average luciferase units CIITA ( − 214/+54) | |
|---|---|---|
| Control | 440050+/ − 7113 | 472832+/ − 15031 |
| His-Zta | 18448+/ − 464 | 66785+/ − 1917 |
From these data we devised a model to account for Zta-mediated activation and repression of gene expression. In cells expressing MHC class II, CIITA expression is driven by the interaction of cellular factors (RNA polymerase II and cellular co-activators) (Fig. 5a). Once Zta is expressed it interferes with the activation machinery operating at the CIITA promoter, without the need to dimerize or bind to the promoter (Fig. 5b).
Fig. 5.
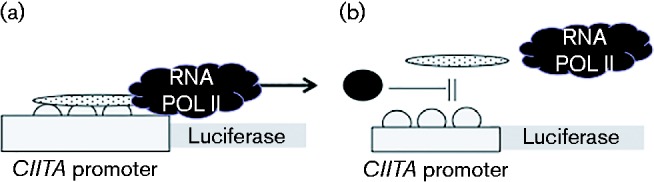
Proposed model to explain Zta-mediated gene repression of the CIITA promoter. (a) The active CIITA promoter is shown. Basal transcription factors are represented as white spheres and co-activators by the stippled oval. RNA polymerase II is represented by the black cloud, with transcription indicated by an arrow. (b) The ability of a non-DNA-binding form of Zta (filled oval) to repress expression of the CIITA promoter suggests that some repression can occur without direct DNA contact. The simplest model to account for this has the N-terminal part of Zta blocking the interaction of the basal transcription machinery.
Discussion
The EBV protein Zta is often described as the master regulator of EBV lytic cycle replication. Indeed, the ability of Zta to regulate viral gene expression is crucial to the success of viral lytic replication, as mutation of the BZLF1 gene in recombinant EBV demonstrates (Feederle et al., 2000). The activation of viral gene expression is considered to occur through the interaction of Zta with sequence-specific ZREs in the promoters of viral genes, and the attraction of co-activator proteins such as p300, TFIID and other RNA polymerase II components to the promoters (Lieberman & Berk, 1991, 1994) (Fig. 5 and Fig. S3). Recent genome-wide analyses have shown that Zta has extensive interactions across the EBV genome and a specific role in the transcriptional activation of many viral promoters (Bergbauer et al., 2010; Ramasubramanyan et al., 2012a).
Li discovered that Zta-mediated repression of CIITA expression occurs in EBV-positive Raji cells, but did not investigate whether other viral genes are required for the repression (Li et al., 2009). We confirm this and furthermore we show that the expression of two CIITA-dependent genes is also downregulated. As Zta expression in EBV-positive BL cells is sufficient to initiate the viral lytic replication cycle, many downstream changes in gene expression are expected, and it is important to determine whether repression requires Zta action alone or whether it acts in concert with additional viral proteins. Our demonstration that Zta is able to repress the CIITA promoter in an EBV-negative BL cell line unequivocally demonstrates that Zta-mediated repression does not require other viral gene products.
The relevance of two forms of post-translational modification of Zta that have been described as transcriptionally repressive was explored. The involvement of phosphorylation at S209 by the viral protein kinase BGLF4 (Asai et al., 2009) was investigated using the phospho-mimetic mutant version of Zta, S209D, and the phosphorylation dead mutant version, Zta S209A. Covalent addition of SUMO at K12 (Hagemeier et al., 2010; Murata et al., 2010) was assessed using the non-sumoylatable mutant version, Zta K12R. Both of these post-translational modifications have been described as transcriptionally repressive (Asai et al., 2009; Hagemeier et al., 2010; Murata et al., 2010). As none of these Zta mutants compromised the ability of Zta to repress the CIITA promoter, we conclude that neither post-translational modification is likely to be responsible for the observed repression of the CIITA promoter by Zta.
Zta also regulates gene expression by disrupting transcriptional activation by NFκB and p53 (Morrison & Kenney, 2004; Zhang et al., 1994). This occurs through physical interactions between Zta and the p65 component of NFκB and between Zta and p53 protein (Morrison & Kenney, 2004; Zhang et al., 1994). However, it is unlikely that either NFκB or p53 play a role in Zta-mediated CIITA repression, as both require the dimerization region of Zta, which is not necessary for repression of CIITA. In addition, mutation of the NFκB interaction site in the CIITA promoter does not alter either basal expression or Zta mediated repression (N. Balan & A. J. Sinclair, unpublished data). It is intriguing that Zta has been shown previously to modulate expression of a viral promoter (Zp) without the need to bind directly to DNA (Flemington et al., 1994).
A previous study suggests that Zta repression of the CIITA promoter is driven through the interaction of Zta with a single ZRE within the promoter (Li et al., 2009). This is supported by the impact of mutations of the ZRE within the promoter and by the inability of Zta to repress the CIITA promoter when the basic region is lost. This study places emphasis on a need of Zta to bind directly to DNA to effect repression. Our experiments support a different conclusion in which Zta represses CIITA expression without binding directly to DNA. We rationalize the need for the basic region of the Zta protein based on a requirement for the nuclear localization domain, which is contained therein (Mikaélian et al., 1993). Without entry to the nucleus, Zta would not be able to repress the CIITA promoter through either direct or indirect DNA binding.
In summary, we show that Zta-mediated repression of the CIITA promoter can occur without Zta contacting DNA directly; this is supported by the retention of repression when (i) the ZRE is deleted and (ii) by a version of Zta that is defective for dimerization and therefore defective for DNA binding. This discovery leads us to propose a mechanism to describe gene repression by Zta. In this model the amino terminal region of Zta is able to impede the function of an essential component of the transcriptionally active CIITA promoter, for example a DNA-bound transcription factor or a transcription factor-associated co-activator, thereby preventing its productive association with RNA Pol II and its accessory proteins (Fig. 5).
Methods
Plasmid constructs
The CIITA promoter ( − 286 to +54) was cloned with a KpnI restriction enzyme site included at the 5′ end and a HindIII site at the 3′ end of the sequence. The promoter was subcloned into the pGL3 enhancer plasmid, which contains a luciferase reporter construct downstream from a multi-cloning site and which includes a distal SV40 enhancer (Promega). A 5′ deletion version of the promoter was generated ( − 214 to +54); the location of the 5′ end of this promoter is immediately 3′ from the ZRE.
The BHLF1, BFLF2 and BLLF3 promoters were cloned with BamHI restriction enzyme sites added at the 5′ end and HindIII sites at the 3′ end. The DNA sequence between co-ordinates 40 472 and 40 818, 45 793 and 44 746, and 76 186 and 77 231 of the EBV genome (Human herpesvirus 4 complete wild-type genome Accession: NC_007605.1) was synthesized for the promoter regions for the BHLF1, BFLF2 and BLLF3 genes, respectively. The promoters were subcloned into the pCpGL plasmid (Klug & Rehli, 2006), which is based on pGL3 basic and contains a luciferase reporter construct downstream from a multi-cloning site.
A plasmid driving the expression of hexa-histidine tagged Zta (His-Zta) (Bailey et al., 2009) was used to express His-Zta, compared to the vector control pcDNA3 (Invitrogen).
Expression vectors for His-Zta K12R, His-Zta S209A and His-Zta S209D were generated by site-directed mutagenesis of His-Zta using the primers shown in Table 3. An expression vector for His Zta-199ter, which introduces a termination codon at the amino acid 199 of the Zta coding sequence, and His Zta-ΔTA, which deletes amino acids 1–133 of Zta, were generated by gene synthesis (Invitrogen).
Table 3. Oligonucleotides used to generate mutations.
| His-Zta S209A amino acid serine 209 mutated to alanine | GGCTGCTGCCAAATCAGCTGAAAATGACAGGCTGCGCC; GGCGCAGCCTGTCATTTTCAGCTGATTTGGCAGCAGCC |
| His-Zta S209D amino acid serine 209 mutated to glutamic acid | GGCTGCTGCCAAATCAGATGAAAATGACAGGCTGCGCC; GGCGCAGCCTGTCATTTTCATCTGATTTGGCAGCAGCC |
| His-Zta K12R amino acid lysine 12 mutated to arginine | CtCGACttCtGAAGAtGtAAgAtttACACCtGACCCAtACC; GGTATGGGTCAGGTGTAAATCTTACATCTTCAGAAGTCGAG |
Cell culture
Plasmids were introduced into EBV-positive Raji cells (Pulvertaft, 1965) or EBV-negative Akata cells (Jenkins et al., 2000) by electroporation. 1 × 107 cells in 0.25 ml medium were incubated with 10 μg plasmid DNA and pulsed with 250V at a capacitance of 975 μF in a Gene Pulser II electroporator (Bio-Rad).
Luciferase assays
post-transfection, cells were harvested into 250 μl of Passive Lysis Buffer (Promega) and incubated at room temperature for 15 min. The lysed cells were then centrifuged for 10 min at 13 300 g in a Thermo Scientific Heraeus Fresco centrifuge and the supernatant was used to determine luciferase activity. 10 μl aliquots of each lysate sample were pipetted into a 96-well white luminescence plate and analysed using luciferase detection kit reagents with a Glomax detection system (Promega). A protein concentration assay was undertaken (Bio-Rad) and promoter activity was expressed as luciferase RLU μg− 1 protein. Significance of different promoter activity was assessed using a Student's paired t-test with two-tail distribution.
Protein analysis
Proteins were extracted from cells by boiling in 2 × laemmli sample buffer and fractionated on Novex protein gels (Invitrogen). Following transfer to nitrocellulose membranes, the blots were incubated with the Zta-specific antibody sc-17503 (Santa Cruz), which recognizes the amino-terminal region of Zta, BZ1, which recognizes the basic and dimerization regions of Zta (Young et al., 1991) or a rabbit polyclonal beta actin antibody (Sigma), followed by detection with HRP-linked secondary antibodies and ECL (Ramasubramanyan et al., 2012b).
Acknowledgements
This work was supported by a grant from the Medical Research Council UK (MR/J001708/1). We thank Professor Takada for Akata cells, Professor Farrell for AK31 cells and Professor Rowe for BZ1 monoclonal antibody.
Supplementary Data
Supplementary Data
References
- Adamson A. L., Kenney S. (1999). Epstein-Barr virus BZLF1 protein interacts physically and functionally with the histone acetylase CREB-binding protein J Virol 736551–6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhikary D., Behrends U., Moosmann A., Witter K., Bornkamm G. W., Mautner J. (2006). Control of Epstein-Barr virus infection in vitro by T helper cells specific for virion glycoproteins J Exp Med 203995–1006 10.1084/jem.20051287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai R., Kato A., Kawaguchi Y. (2009). Epstein-Barr virus protein kinase BGLF4 interacts with viral transactivator BZLF1 and regulates its transactivation activity J Gen Virol 901575–1581 10.1099/vir.0.010462-0. [DOI] [PubMed] [Google Scholar]
- Babcock G. J., Decker L. L., Volk M., Thorley-Lawson D. A. (1998). EBV persistence in memory B cells in vivo Immunity 9395–404 10.1016/S1074-7613(00)80622-6. [DOI] [PubMed] [Google Scholar]
- Bailey S. G., Verrall E., Schelcher C., Rhie A., Doherty A. J., Sinclair A. J. (2009). Functional interaction between Epstein-Barr virus replication protein Zta and host DNA damage response protein 53BP1 J Virol 8311116–11122 10.1128/JVI.00512-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergbauer M., Kalla M., Schmeinck A., Göbel C., Rothbauer U., Eck S., Benet-Pagès A., Strom T. M., Hammerschmidt W. (2010). CpG-methylation regulates a class of Epstein-Barr virus promoters PLoS Pathog 6e1001114. 10.1371/journal.ppat.1001114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhende P. M., Seaman W. T., Delecluse H. J., Kenney S. C. (2004). The EBV lytic switch protein, Z, preferentially binds to and activates the methylated viral genome Nat Genet 361099–1104 10.1038/ng1424. [DOI] [PubMed] [Google Scholar]
- Bhende P. M., Seaman W. T., Delecluse H. J., Kenney S. C. (2005). BZLF1 activation of the methylated form of the BRLF1 immediate-early promoter is regulated by BZLF1 residue 186 J Virol 797338–7348 10.1128/JVI.79.12.7338-7348.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bristol J. A., Robinson A. R., Barlow E. A., Kenney S. C. (2010). The Epstein-Barr virus BZLF1 protein inhibits tumor necrosis factor receptor 1 expression through effects on cellular C/EBP proteins J Virol 8412362–12374 10.1128/JVI.00712-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broderick P., Hubank M., Sinclair A. J. (2009). Effects of Epstein-Barr virus on host gene expression in Burkitt's lymphoma cell lines Chinese journal of cancer 28813–821. [DOI] [PubMed] [Google Scholar]
- Cai Q., Banerjee S., Cervini A., Lu J., Hislop A. D., Dzeng R., Robertson E. S. (2013). IRF-4-mediated CIITA transcription is blocked by KSHV encoded LANA to inhibit MHC II presentation PLoS Pathog 9e1003751. 10.1371/journal.ppat.1003751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C. H., Gourley T. S., Sisk T. J. (2002). Function and regulation of class II transactivator in the immune system Immunol Res 25131–142 10.1385/IR:25:2:131. [DOI] [PubMed] [Google Scholar]
- Crawford D. H., Ando I. (1986). EB virus induction is associated with B-cell maturation Immunology 59405–409. [PMC free article] [PubMed] [Google Scholar]
- Dickerson S. J., Xing Y., Robinson A. R., Seaman W. T., Gruffat H., Kenney S. C. (2009). Methylation-dependent binding of the epstein-barr virus BZLF1 protein to viral promoters PLoS Pathog 5e1000356. 10.1371/journal.ppat.1000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell P. J. (2005). Epstein-Barr virus genome. In Epstein-Barr virus, pp. 263–288. Edited by Robertson E. S.Wymondham: Caister. [Google Scholar]
- Feederle R., Kost M., Baumann M., Janz A., Drouet E., Hammerschmidt W., Delecluse H. J. (2000). The Epstein-Barr virus lytic program is controlled by the co-operative functions of two transactivators EMBO J 193080–3089 10.1093/emboj/19.12.3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemington E. K., Lytle J. P., Cayrol C., Borras A. M., Speck S. H. (1994). DNA-binding-defective mutants of the Epstein-Barr virus lytic switch activator Zta transactivate with altered specificities Mol Cell Biol 143041–3052 10.1128/MCB.14.5.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flower K., Thomas D., Heather J., Ramasubramanyan S., Jones S., Sinclair A. J. (2011). Epigenetic control of viral life-cycle by a DNA-methylation dependent transcription factor PLoS One 6e25922. 10.1371/journal.pone.0025922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemeier S. R., Dickerson S. J., Meng Q., Yu X., Mertz J. E., Kenney S. C. (2010). Sumoylation of the Epstein-Barr virus BZLF1 protein inhibits its transcriptional activity and is regulated by the virus-encoded protein kinase J Virol 844383–4394 10.1128/JVI.02369-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks M. R., Al-Mehairi S. S., Sinclair A. J. (2003). The zipper region of Epstein-Barr virus bZIP transcription factor Zta is necessary but not sufficient to direct DNA binding J Virol 778173–8177 10.1128/JVI.77.14.8173-8177.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holley-Guthrie E. A., Quinlivan E. B., Mar E. C., Kenney S. (1990). The Epstein-Barr virus (EBV) BMRF1 promoter for early antigen (EA-D) is regulated by the EBV transactivators, BRLF1 and BZLF1, in a cell-specific manner J Virol 643753–3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins P. J., Binné U. K., Farrell P. J. (2000). Histone acetylation and reactivation of Epstein-Barr virus from latency J Virol 74710–720 10.1128/JVI.74.2.710-720.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalla M., Schmeinck A., Bergbauer M., Pich D., Hammerschmidt W. (2010). AP-1 homolog BZLF1 of Epstein-Barr virus has two essential functions dependent on the epigenetic state of the viral genome Proc Natl Acad Sci U S A 107850–855 10.1073/pnas.0911948107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalla M., Göbel C., Hammerschmidt W. (2012). The lytic phase of epstein-barr virus requires a viral genome with 5-methylcytosine residues in CpG sites J Virol 86447–458 10.1128/JVI.06314-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallin B., Klein G. (1983). Epstein-Barr virus carried by Raji cells: a mutant in early functions? Intervirology 1947–51 10.1159/000149336. [DOI] [PubMed] [Google Scholar]
- Karlsson Q. H., Schelcher C., Verrall E., Petosa C., Sinclair A. J. (2008). Methylated DNA recognition during the reversal of epigenetic silencing is regulated by cysteine and serine residues in the Epstein-Barr virus lytic switch protein PLoS Pathog 4e1000005. 10.1371/journal.ppat.1000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney S. C. (2007). Reactivation and lytic replication of EBV. In Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis. Edited by Arvin A., other editorsCambridge: Cambridge University Press; http://www.ncbi.nlm.nih.gov/books/NBK47442/. [Google Scholar]
- Kenney S., Holley-Guthrie E., Mar E. C., Smith M. (1989). The Epstein-Barr virus BMLF1 promoter contains an enhancer element that is responsive to the BZLF1 and BRLF1 transactivators J Virol 633878–3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug M., Rehli M. (2006). Functional analysis of promoter CpG methylation using a CpG-free luciferase reporter vector Epigenetics: official journal of the DNA Methylation Society 1127–130. [DOI] [PubMed] [Google Scholar]
- Laichalk L. L., Thorley-Lawson D. A. (2005). Terminal differentiation into plasma cells initiates the replicative cycle of Epstein-Barr virus in vivo J Virol 791296–1307 10.1128/JVI.79.2.1296-1307.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laichalk L. L., Hochberg D., Babcock G. J., Freeman R. B., Thorley-Lawson D. A. (2002). The dispersal of mucosal memory B cells: evidence from persistent EBV infection Immunity 16745–754 10.1016/S1074-7613(02)00318-7. [DOI] [PubMed] [Google Scholar]
- Li D., Qian L., Chen C., Shi M., Yu M., Hu M., Song L., Shen B., Guo N. (2009). Down-regulation of MHC class II expression through inhibition of CIITA transcription by lytic transactivator Zta during Epstein-Barr virus reactivation J Immunol 1821799–1809 10.4049/jimmunol.0802686. [DOI] [PubMed] [Google Scholar]
- Lieberman P. M., Berk A. J. (1991). The Zta trans-activator protein stabilizes TFIID association with promoter DNA by direct protein-protein interaction Genes Dev 5 (12B), 2441–2454 10.1101/gad.5.12b.2441. [DOI] [PubMed] [Google Scholar]
- Lieberman P. M., Berk A. J. (1994). A mechanism for TAFs in transcriptional activation: activation domain enhancement of TFIID-TFIIA–promoter DNA complex formation Genes Dev 8995–1006 10.1101/gad.8.9.995. [DOI] [PubMed] [Google Scholar]
- Long H. M., Leese A. M., Chagoury O. L., Connerty S. R., Quarcoopome J., Quinn L. L., Shannon-Lowe C., Rickinson A. B. (2011). Cytotoxic CD4+ T cell responses to EBV contrast with CD8 responses in breadth of lytic cycle antigen choice and in lytic cycle recognition J Immunol 18792–101 10.4049/jimmunol.1100590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magrath I. (2012). Towards Curative Therapy in Burkitt Lymphoma: The Role of Early African Studies in Demonstrating the Value of Combination Therapy and CNS Prophylaxis Adv Hematol 2012130680. 10.1155/2012/130680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikaélian I., Drouet E., Marechal V., Denoyel G., Nicolas J. C., Sergeant A. (1993). The DNA-binding domain of two bZIP transcription factors, the Epstein-Barr virus switch gene product EB1 and Jun, is a bipartite nuclear targeting sequence J Virol 67734–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneux E. M., Rochford R., Griffin B., Newton R., Jackson G., Menon G., Harrison C. J., Israels T., Bailey S. (2012). Burkitt's lymphoma Lancet 3791234–1244 10.1016/S0140-6736(11)61177-X. [DOI] [PubMed] [Google Scholar]
- Morrison T. E., Kenney S. C. (2004). BZLF1, an Epstein-Barr virus immediate-early protein, induces p65 nuclear translocation while inhibiting p65 transcriptional function Virology 328219–232 10.1016/j.virol.2004.07.020. [DOI] [PubMed] [Google Scholar]
- Murata T., Hotta N., Toyama S., Nakayama S., Chiba S., Isomura H., Ohshima T., Kanda T., Tsurumi T. (2010). Transcriptional repression by sumoylation of Epstein-Barr virus BZLF1 protein correlates with association of histone deacetylase J Biol Chem 28523925–23935 10.1074/jbc.M109.095356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packham G., Economou A., Rooney C. M., Rowe D. T., Farrell P. J. (1990). Structure and function of the Epstein-Barr virus BZLF1 protein J Virol 642110–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulvertaft J. V. (1965). A Study of Malignant Tumours in Nigeria by Short-Term Tissue Culture J Clin Pathol 18261–273 10.1136/jcp.18.3.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramasubramanyan S., Kanhere A., Osborn K., Flower K., Jenner R. G., Sinclair A. J. (2012a). Genome-wide analyses of Zta binding to the Epstein-Barr virus genome reveals interactions in both early and late lytic cycles and an epigenetic switch leading to an altered binding profile J Virol 8612494–12502 10.1128/JVI.01705-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramasubramanyan S., Osborn K., Flower K., Sinclair A. J. (2012b). Dynamic chromatin environment of key lytic cycle regulatory regions of the Epstein-Barr virus genome J Virol 861809–1819 10.1128/JVI.06334-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha A., Robertson E. S. (2011). Epstein-Barr virus-associated B-cell lymphomas: pathogenesis and clinical outcomes Clin Cancer Res 173056–3063 10.1158/1078-0432.CCR-10-2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schelcher C., Valencia S., Delecluse H. J., Hicks M., Sinclair A. J. (2005). Mutation of a single amino acid residue in the basic region of the Epstein-Barr virus (EBV) lytic cycle switch protein Zta (BZLF1) prevents reactivation of EBV from latency J Virol 7913822–13828 10.1128/JVI.79.21.13822-13828.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair A. J. (2003). bZIP proteins of human gammaherpesviruses J Gen Virol 841941–1949 10.1099/vir.0.19112-0. [DOI] [PubMed] [Google Scholar]
- Sinclair A. J., Brimmell M., Shanahan F., Farrell P. J. (1991). Pathways of activation of the Epstein-Barr virus productive cycle J Virol 652237–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woellmer A., Arteaga-Salas J. M., Hammerschmidt W. (2012). BZLF1 governs CpG-methylated chromatin of Epstein-Barr Virus reversing epigenetic repression PLoS Pathog 8e1002902. 10.1371/journal.ppat.1002902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young L. S., Lau R., Rowe M., Niedobitek G., Packham G., Shanahan F., Rowe D. T., Greenspan D., Greenspan J. S., other authors (1991). Differentiation-associated expression of the Epstein-Barr virus BZLF1 transactivator protein in oral hairy leukoplakia J Virol 652868–2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Gutsch D., Kenney S. (1994). Functional and physical interaction between p53 and BZLF1: implications for Epstein-Barr virus latency Mol Cell Biol 141929–1938 10.1128/MCB.14.3.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J., Rowe M. (2012). Herpesviruses placating the unwilling host: manipulation of the MHC class II antigen presentation pathway Viruses 41335–1353 10.3390/v4081335. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data