Abstract
Herpes simplex virus type 1 (HSV-1) establishes a latent infection in sensory neurons from which the virus can periodically reactivate. Whilst latency establishment is thought to result from a failure to express immediate-early genes, we have previously shown that subpopulations of the latent neuronal reservoir have undergone lytic promoter activation prior to latency establishment. In the present study, we have investigated the biological properties of such latently infected neuronal subpopulations using Ai6 fluorescent reporter mice. Using this system we have determined that prior ICP0 or TK promoter activation does not correlate with increased latent virus DNA loads within individual cells and that neurons with evidence of historical lytic cycle promoter activity exhibit a comparable frequency of reactivation to that of the general latent cell population. Comparison of viral DNA content within cells harbouring latent HSV-1 genomes and those undergoing the earliest stages of reactivation has revealed that reactivation can initiate from cells harbouring a wide range of HSV-1 genome copies, but that exiting latency is biased towards cells bearing higher latent virus DNA loads.
Introduction
Herpes simplex virus type 1 (HSV-1) establishes lifelong latency within sensory neurons and can periodically reactivate to facilitate virus shedding at peripheral sites, often asymptomatically (Roizman & Whitley, 2013; Wagner & Bloom, 1997). Much has been learnt from small animal models of herpes simplex virus (HSV) latency both at the molecular and immunological level (Efstathiou & Preston, 2005; Nicoll et al., 2012b; Wagner & Bloom, 1997) but our understanding of latency within single neurons in vivo has been hampered by the lack of amenable systems to identify and isolate live cells from the infected host. For example, careful analyses of HSV-1 DNA copy numbers within individual human or mouse neurons using contextual analysis of DNA and laser-capture microdissection methodologies have shown that virus genome copies can vary over three orders of magnitude (Chen et al., 2002; Sawtell, 1997; Thompson & Sawtell, 2000; Wang et al., 2005), but studying the potential biological effect of this phenomenon has been limited to indirect examination (Sawtell et al., 1998). Reporter mice encoding conditionally expressed LacZ or yellow fluorescent protein (YFP) from the endogenous ROSA locus have proved useful for the stable visual marking of latently infected sensory neurons following infection with Cre recombinase-expressing HSV-1 recombinants, and have also revealed that lytic gene promoter activation and low level lytic gene transcription is compatible with latency in at least a subset of latently infected neurons (Ma et al., 2014; Nicoll et al., 2012a; Proença et al., 2008, 2011; Wakim et al., 2008). Again, while powerful, these reporter mouse systems have relied on tissue fixation for analysis, limiting further direct investigation of intact latently infected cells.
In this study, we have used ZsGreen Ai6 fluorescent reporter mice (Madisen et al., 2010) to develop a model of HSV-1 latency that allows for the direct visualization of live latently infected neurons. The strong fluorescence afforded by this reporter mouse strain also facilitates the isolation of these neurons, allowing for the characterization of neuronal cells that have entered latency with or without evidence of prior lytic cycle promoter activation.
We report here that HSV-1 expressing Cre recombinase under control of the human cytomegalovirus (HCMV) major immediate early promoter (MIEP) marks the latent reservoir with high efficiency in Ai6 fluorescent reporter mice, and that HSV-1 DNA copy numbers can vary by at least three orders of magnitude amongst infected neurons, consistent with previous studies using contextual analyses (Sawtell, 1997; Thompson & Sawtell, 2000).
Using this system we also report that latently infected neurons marked by immediate early (IE) and early (E) promoter activation prior to latency establishment do not harbour increased HSV-1 DNA loads relative to the entire infected cell reservoir. Furthermore, these neurons do not differ in their reactivation competence during ex vivo culture. Finally, we detail an ex vivo system for determining HSV-1 DNA copy number in isolated neurons capable of reactivating following heat-shock stimulation.
Results
Ai6 reporter mice facilitate the marking and visualization of sensory neurons latently infected with HSV
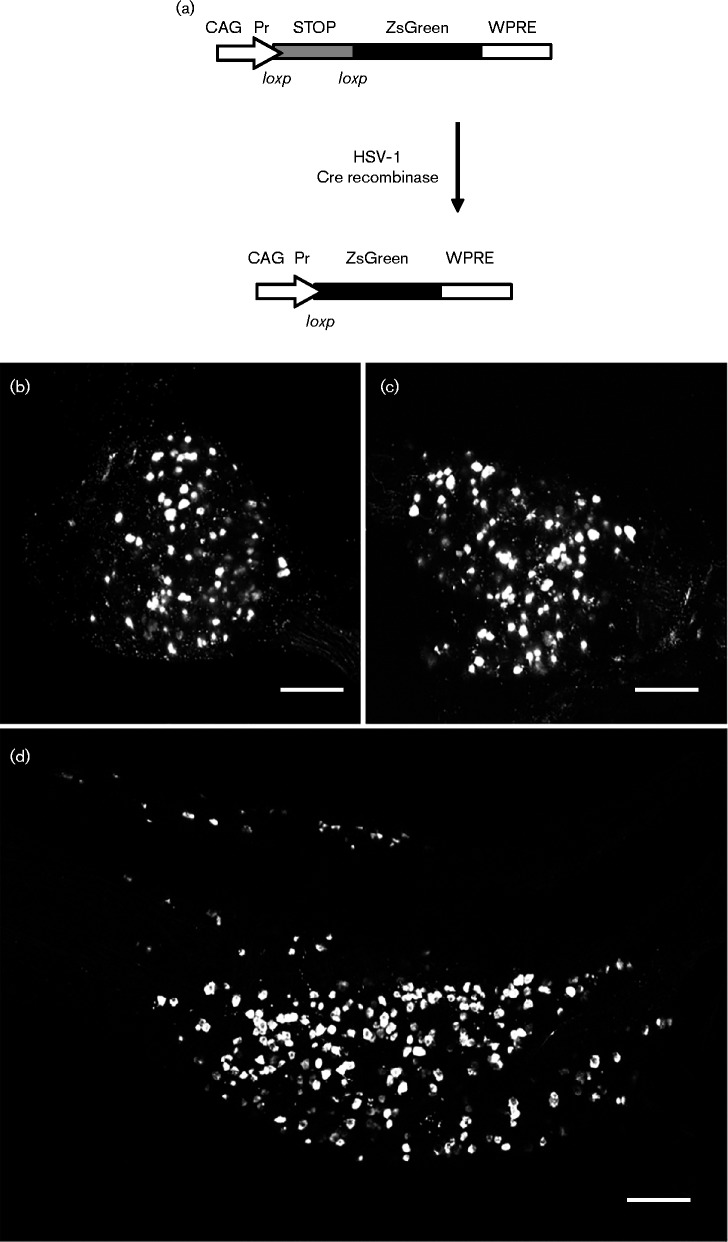
To detect live latently infected neurons we investigated the use of Ai6 reporter mice (Madisen et al., 2010), a transgenic strain that conditionally expresses ZsGreen fluorescent protein upon Cre recombinase excision of a stop cassette (Fig. 1a). Following peripheral infection with Cre-expressing HSV-1 recombinants, virions are transported to innervating sensory neurons. Surviving (and thus latently infected) sensory neurons in which Cre recombinase was expressed prior to virus genome silencing undergo a permanent genetic change that results in ZsGreen expression. Ai6 reporter mice were infected with 2 × 106 p.f.u. of HSV CMVCre (Proença et al., 2008) in the left ear or in both whisker pads. Animals were killed at 30 days post-infection (p.i.) and CII, CIII and CIV dorsal root ganglia (DRG) innervating the ear pinna or trigeminal ganglia (TG) innervating the whisker pad were dissected, fixed and imaged. Fig. 1(b, c, d) shows maximum intensity Z projections of confocal slices taken throughout the whole depth of the DRG or TG. Figs S1 and S2 (available in the online Supplementary Material) show 3D projections of these DRG and TG, respectively, and show the identification and spatial organization of latently infected neurons residing within these tissues. In both anatomical sites latently infected cells can be readily identified by direct fluorescent imaging.
Fig. 1.
Ai6 reporter mouse model. (a) Structure of the Rosa26 locus in the Ai6 reporter mouse before and after recombination with Cre recombinase. (b, c) Representative maximum intensity Z projections of confocal slices taken throughout the whole depth of a latently infected DRG. (d) Representative Z projections of confocal slices taken throughout the whole depth of a latently infected TG. Bars, 200 μm.
ZsGreen-positive cells harbour latent virus genomes
To rule out that cell marking was a consequence of either (1) abortive infection followed by subsequent genome loss or (2) bystander marking, the virus DNA content of individual fluorescent cells was determined (Fig. 2). Ai6 reporter mice were infected with HSV CMVCre on both whisker pads and latently infected TGs dissected at 33, 140 and 214 days p.i. Dissected TGs were dissociated into single cell suspensions and individual ZsGreen-positive cells were individually picked and subjected to quantitative (q)PCR for viral DNA load determination. At 33 days p.i., all analysed ZsGreen-positive cells (n = 36) were virus DNA positive, with a range of 1 to 702 HSV-1 genome copies per cell with a median of 41. Similar results were obtained following single cell analyses of latently infected TG sampled at 140 and 214 days p.i. Once again, at 140 days p.i. all analysed ZsGreen-positive cells (n = 54) contained viral DNA, and whilst possessing a higher median viral DNA copy number of 72, cells contained a similar range of HSV-1 DNA copies (3–590 copies per cell). At 214 days p.i. viral DNA copies per cell ranged from 4 to 365 with a median of 84 copies per cell (n = 61). Following removal of all ZsGreen-positive cells from the dissociated TG sampled at 140 days p.i., the remaining unmarked cells were pooled and analysed by qPCR for virus DNA. A total of 872 HSV-1 genome copies were detected in all remaining cells. Whilst we cannot determine the distribution of HSV-1 genomes within individual unmarked cells, a median of 72 copies may represent the equivalent of 12 ZsGreen-positive neurons. We conclude that marked cells are always associated with virus DNA and only a small proportion of viral DNA can be detected in pooled unmarked cells.
Fig. 2.
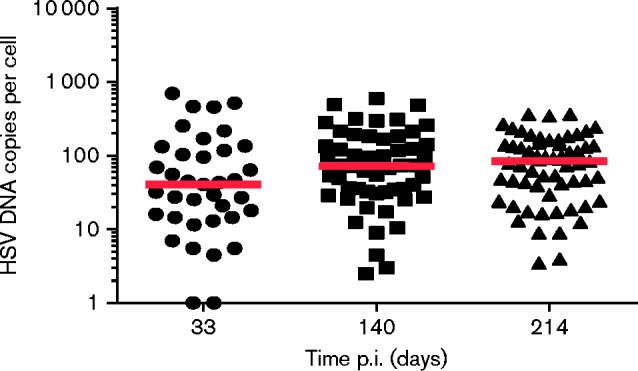
Single cell latent viral DNA loads. Dot plot representation of single cell latent DNA loads from three (1 per time point) mice killed at 33, 140 and 214 days p.i. Each dot represents DNA loads within a cell and the floating red bar represents the median of these data at each time point.
Neurons that experience immediate early or early promoter activity prior to latency establishment show no bias towards high virus DNA load
Using ROSA26R mice we have previously shown that HSV-1 ICP0 and ICP4 IE promoter activity was compatible with latency establishment in up to 35 % of latently infected neurons. Furthermore, evidence of E (TK) and late (L; gC) promoter activation could also be detected but only in a smaller subset of the latent population (up to 4 %) (Proença et al., 2008, 2011). The biological significance of these marked latent populations is currently unknown. Several studies have described a small population of latently infected cells containing >1000 HSV DNA copies (Chen et al., 2002; Ma et al., 2014; Sawtell, 1997; Thompson & Sawtell, 2000; Wang et al., 2005), and indeed our own data describe populations of neurons containing several hundred genome copies (Fig. 2). We thus tested for any correlation between latent populations containing high viral DNA loads and virus promoter activity experienced prior to latency establishment. Specifically, we sought to address two hypotheses: (1) cells marked by TK promoter activity may have experienced virus DNA amplification prior to latency establishment, thus contributing to an increased latent virus DNA load, (2) cells marked as a consequence of ICP0 promoter activity are restricted to neurons receiving a high genome input from the periphery. For either of these hypotheses to be correct, the distribution of HSV DNA loads within these marked populations should be biased towards higher copy numbers per cell than those from HSV CMVCre-marked cells, which marks the majority of the latent cells within the ganglion.
Groups of Ai6 mice were infected in parallel with HSV CMVCre, HSV ICP0Cre and HSV TKCre to examine single cell virus loads during latency. Mice were infected by scarification of the left ear (2 × 106 p.f.u.) or both whisker pads (5 × 106 p.f.u. per pad). During latency (30 days p.i.), animals were killed and single marked cells from either DRG or TG were isolated for viral genome quantification (Fig. 3). HSV CMVCre-marked cells (n = 59) isolated from DRG contained a median of 53 viral genomes per cell, which was larger than the median viral genome copy numbers detected following marking with HSV ICP0Cre and TKCre: 33 and 45 viral genomes, respectively (Fig. 3a). In TG, median viral genome copy numbers of 75, 63 and 34 were detected in HSV CMVCre-, HSV ICP0Cre- and HSV TKCre-infected cells, respectively (Fig. 3b). Irrespective of the promoter used to drive Cre expression or the sensory ganglion of origin, a similar distribution of genome copies per cell across each marked cell population was observed. Whilst no single TKCre-marked cell contained ≥ 1000 HSV-1 genomes (Fig. 3), no significantly different distribution of HSV-1 genomes was detected in cells marked by any virus group (Kruskal–Wallis analysis: P = 0.10 in DRG; P = 0.12 in TG). These data suggest that cell marking by the ICP0 promoter is not dependent on the input of multiple virus templates and that activation of the TK promoter in a manner compatible with latency is not linked to viral DNA amplification.
Fig. 3.
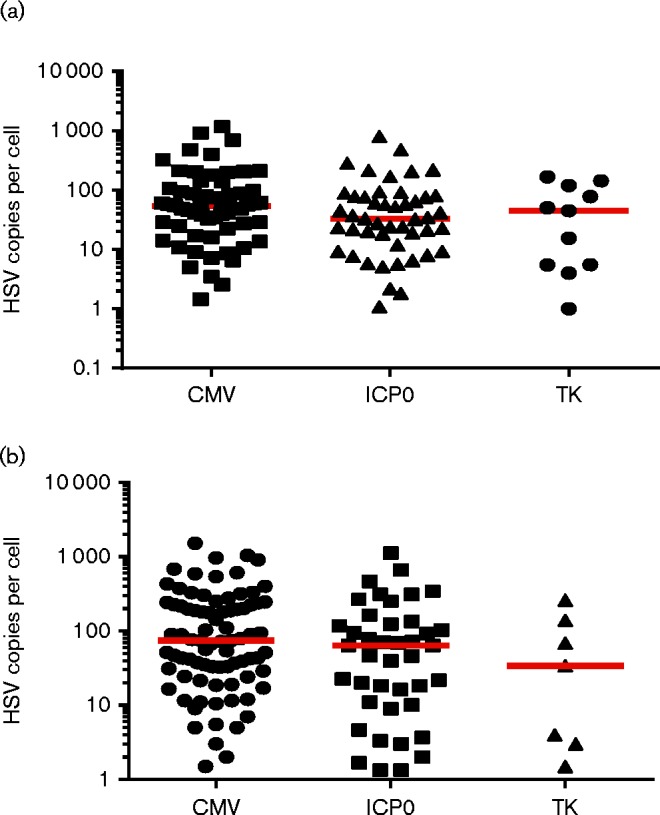
Single cell viral DNA loads in latent populations marked by CMV-, ICP0- and TK-promoter activity. Dot plot representation of single cell DNA loads in DRG (a) and TG (b). Each dot represents DNA loads within a cell and the floating red bar represents the median of these data for each promoter.
Reactivation of virus from marked neurons is not restricted to cells with prior evidence of lytic cycle promoter activity
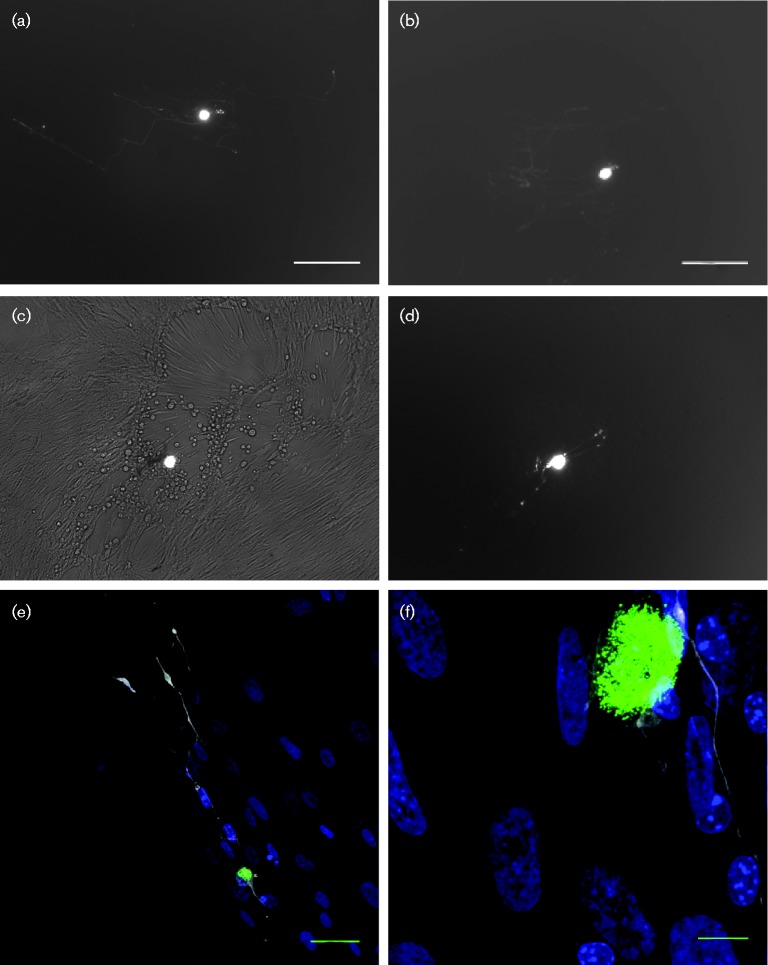
As neuronal subpopulations may differ in their permissiveness to HSV-1 lytic replication (Bertke et al., 2011), we hypothesized that cells undergoing limited lytic gene expression before ultimately establishing latency are the same cells which are later permissive for full virus reactivation. To initially examine reactivation from marked cell cultures, Ai6 mice were infected with HSV CMVCre by scarification of the left ear pinna. CII, CIII and CIV ganglia innervating the ear were dissected after 30 days p.i., dissociated into a single cell suspension and plated across a 24-well plate on to MRC5 cell monolayers to enhance the adherence of marked cells and allow for the identification of virus reactivation by cytopathic effect (CPE) in the monolayer. One hundred and sixty-two ZsGreen-positive cells were plated at a range of 2–15 positive cells per well. Within a day of plating, ZsGreen-positive neurites could be readily observed extending from marked neuronal cell bodies (Fig. 4a, b, d, e, f). Marked cells were examined daily for up to 7 days and MRC5 monolayers scored for virus-induced CPE (Fig. 4c). By 7 days post-explant, virus reactivation was observed in 4/24 wells. Heat-shock incubation of TG cultures at 43 °C for 2 h has been previously shown to induce HSV-1 reactivation (Halford et al., 1996), and indeed we were able to increase the frequency of reactivation in two subsequent experiments by heat-shocking neurons 1 day after plating (Table S1). To examine whether latent neurons marked by differing classes of lytic promoters differed in their reactivation competence, Ai6 mice were infected in parallel with 5 × 106 p.f.u. of HSV CMVCre, HSV ICP0Cre or HSV TKCre by scarification of both whisker pads. During latency (>30 days p.i.) TG were removed and dissociated to single cell suspensions. Single marked cells were isolated using glass micropipettes and plated one per well on MRC5 monolayers in 96-well plates. Once all remaining marked cells were removed from HSV ICP0Cre- and HSV TKCre-infected single cell suspensions, the remaining non-marked cells (enriched for, but not exclusively containing, neuronal cells) were plated in bulk across MRC5 monolayers to compare reactivation from marked and unmarked populations (the large number of cells marked by HSV CMVCre precluded a similar analysis for this recombinant). All plates were heat-shocked 1 day post-explant and CPE-positive wells enumerated 6 days later. Reactivated virus could be detected in wells containing cells marked by all three viruses (Table 1). Reactivation frequency from ZsGreen-positive cell populations was similar between all three viruses (HSV CMVCre vs HSV ICP0Cre: P = 0.196, HSV CMVCre vs HSV TKCre: P = 1.000, Fisher's Exact test). Whilst reactivated virus was detected in wells containing HSV ICP0Cre- and HSV TKCre-marked cells, the large majority of cells that were competent for reactivation were not in populations marked by these recombinants, as evidenced from the high rates of reactivation in wells containing bulk neuronal suspensions lacking ZsGreen-positive cells (Table 1). These data were corroborated in a second experiment with HSV ICP0Cre, in which reactivation occurred in all wells containing unmarked cells, and just 9.3–15 % of wells containing positive cells (Table 2). These results are consistent with only a small fraction of latent cells being marked by ICP0 or TK promoters (Proença et al., 2011). We conclude that cells competent for reactivation following explant dissociation and hyperthermic stress include, but are not restricted to, latent populations that experience either ICP0 or TK promoter activity prior to latency establishment.
Fig. 4.
Ex vivo culture of individual latently infected Ai6 mouse neurons on MRC5 cell monolayers. (a, b) Photomicrographs of two ZsGreen+ neurons. Bright cell bodies surrounded by faint axonal projections are clearly visible. Bars, 200 μm. (c) Reactivation from marked cells is clearly visible in MRC5 monolayer CPE local to the neuron. (d) Fluorescence-only photomicrograph of the neuron displayed in panel c. (e, f) Fluorescent photomicrographs of latently infected neurons in culture. Cultures were stained with anti-β-III-tubulin (white) and DAPI (blue). Endogenous ZsGreen is clearly visible in the neuron cell body. Bars, 50 μm (e); 10 μm (f).
Table 1. Analysis of reactivation frequency from ICP0- and TK promoter-marked cells following incubation at 43 °C for 2 h.
Latent ZsGreen-positive cells were plated at one positive cell per well. Remaining TG suspensions containing negative cells were plated separately. Reactivation was scored following the development of CPE within a well. Note: larger numbers of mouse TG were used to collect ICP0- and TK-marked cells to isolate these scarcer cell populations.
| Promoter | Total no. wells | Total no. wells reactivated | Reactivation (%) |
|---|---|---|---|
| CMVCre (1 mouse) | |||
| ZsGreen + | 72 | 9 | 12.5 |
| ICP0Cre (2 mice) | |||
| ZsGreen + | 48 | 2 | 4.2 |
| ZsGreen − | 48 | 30 | 62.5 |
| TKCre (4 mice) | |||
| ZsGreen + | 20 | 2 | 10 |
| ZsGreen − | 72 | 66 | 91.7 |
Table 2. Analysis of reactivation frequency from ICP0-promoter-marked cells following incubation at 43 °C for 2 h.
Latent ZsGreen-positive cells were plated at one positive cell per well. Remaining TG suspensions containing negative cells were plated separately. Reactivation was scored following the development of CPE within a well.
| Promoter | Total no. wells | Total no. wells reactivated | Reactivation (%) |
|---|---|---|---|
| ICP0Cre (1 mouse) | |||
| ZsGreen + | 13 | 2 | 15 |
| ZsGreen − | 11 | 11 | 100 |
| ICP0Cre (1 mouse) | |||
| ZsGreen + | 32 | 3 | 9.3 |
| ZsGreen − | 5 | 5 | 100 |
Explant reactivation is not restricted to cells harbouring a high viral load
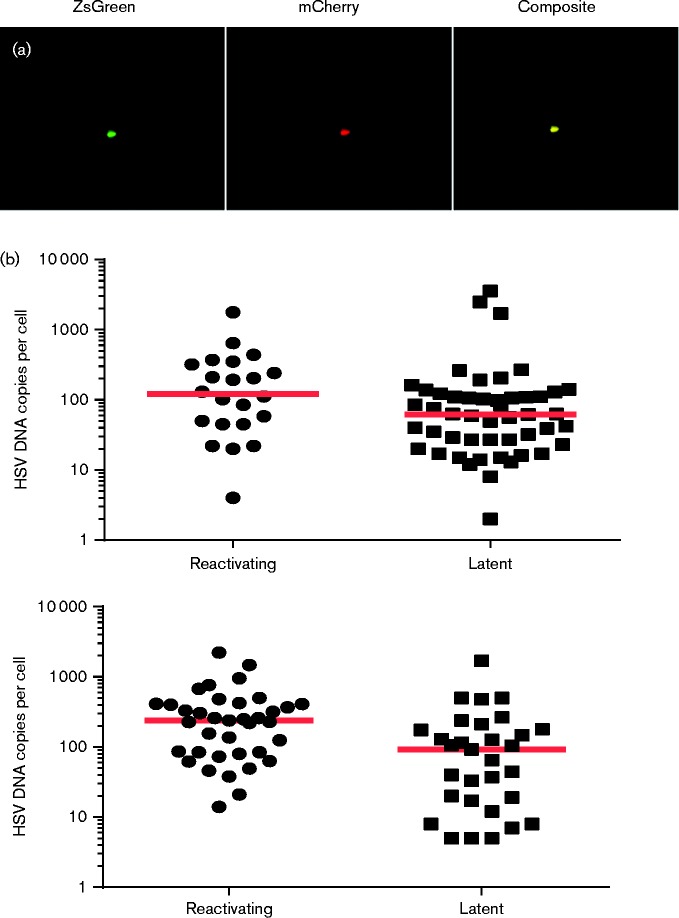
It is clear that not all latently infected cells reactivated in response to heat shock, and also that reactivation was not limited to cells that had experienced lytic promoter activation prior to latency establishment. From previous single cell PCR studies (Sawtell, 1997; Thompson & Sawtell, 2000) and our data (Figs 2 and 3), it is known that HSV-1 DNA copies within individual neurons range over three orders of magnitude. We therefore examined the hypothesis that reactivation-competent cells possess more HSV-1 genome copies than non-responsive cells. Ai6 reporter mice were infected with HSV-1 recombinant HSV CMVCreCherry, a reporter variant of HSV CMVCre which also expresses mCherry fluorescent protein from the CMV MIEP in the UL37/38 intergenic locus of HSV-1 (Fig. S3). Infection with HSV CMVCreCherry allows for the identification of both latently infected cells (ZsGreen-positive) and cells at early stages of reactivation (that are mCherry-positive as a consequence of de-repression of the latent virus genome). To quantify latent DNA loads in reactivating cells, viral DNA replication was blocked with phosphonoacetic acid (PAA). Latently infected neurons were purified from single cell TG suspension and plated on MRC5 cells in the presence of PAA at a concentration (100 μg ml− 1) that completely inhibited viral DNA amplification (data not shown). One day after plating, the cultures were heat shocked at 43 °C for 2 h and allowed to recover for 2 days. After this period, individual latent (ZsGreen-expressing) or reactivating (ZsGreen and mCherry-expressing) neurons were manually isolated from cultures. DNA was then extracted from single cells and qPCR performed. The qPCR data from two independent experiments is shown in Fig. 5. Viral DNA loads present in reactivating cells contained a similar range of HSV-1 genome copies as observed previously (Fig. 3). Furthermore, reactivating and latent marked cells harboured a similar range of HSV-1 DNA copies per cell, with latent and reactivating cells containing between 2–3565 and 4–2219 genomes per cell, respectively (Fig. 5). However, at a population level reactivation was biased towards cells containing more viral genomes, with a significant 2.5 × increase in the median HSV-1 DNA copy number per neuron observed in reactivating cells following heat shock stimulation (P = 0.036 and 0.006 in independent experiments; Mann–Whitney U-test) (Fig. 5). These data demonstrate that although there does not seem to be a minimal threshold viral load beneath which a neuron cannot reactivate following explantation, at a population level cells that contain high viral loads are more likely to do so.
Fig. 5.
Determining HSV-1 DNA loads in latent and reactivating neurons. (a) Fluorescence photomicrographs demonstrating endogenous ZsGreen and mCherry co-expression in reactivating neurons. (b) Dot plot representation of single cell DNA loads in heat-shocked neurons expressing ZsGreen and mCherry (‘reactivating’) and ZsGreen alone (‘latent’). Each graph displays an independent experiment. Each dot represents DNA loads within a cell and the floating red bar represents the median of these data at each time point.
Discussion
In this study we report on the characterization of an Ai6 ZsGreen reporter mouse model of HSV-1 latency. Upon infection of Ai6 transgenic mice (Madisen et al., 2010) with Cre recombinase-expressing HSV-1 recombinants, excision of a lox-stop-lox cassette within the mouse genome leads to constitutive expression of a strong ZsGreen fluorescence protein allowing for both the visual identification of infected cells and their isolation from suspensions made from infected tissues. Following infection with an HSV-1 recombinant that marks the vast majority of the cell reservoir (Proença et al., 2008), we have demonstrated that all marked cells contain virus DNA, ranging from < 10 to >1000 genomes per cell. These data are consistent with the range of HSV-1 DNA copy numbers reported from studies using contextual analysis of DNA (CXA-D) of mouse tissue (Sawtell, 1997) and laser-capture microdissection in human TG (Chen et al., 2002; Wang et al., 2005). However, we also detected a small amount of HSV-1 DNA within bulk unmarked tissues. As manual isolation of fluorescent cells was performed in our experiments, it is possible this DNA may have originated from a failure to remove all marked cells, the presence of infected cells in which Cre recombination failed, or undigested viral DNA in the medium originating from lysis of latent cells during isolation. A previous report using CXA-D determined that the majority of mouse neurons possess < 10 or 10–100 copies of HSV-1 (17syn+) DNA depending on input dose following corneal infection (Sawtell, 1997). Within our study, the majority of neurons (56 %) harboured 10–100 copies of HSV-1 DNA, suggesting cells possessing < 10 copies of DNA may be underrepresented in our fluorescent mouse model. If true, this may result from the different infection procedure, from virus-strain-specific differences in average HSV-1 copy number (Sawtell et al., 1998) and/or from a reduced success of Cre recombination following infection with < 10 HSV genomes.
Previously, we described a Cre-lox ROSA26R reporter mouse model of HSV-1 infection allowing for the identification and enumeration of latently infected cells in sensory ganglia (Proença et al., 2008). We also identified populations of latent cells undergoing lytic gene promoter activation prior to latency establishment (Proença et al., 2008), with roughly 24 % of all latent cells marked by ICP0 promoter activation in mouse TG, and 4 % of all latent cells marked by TK promoter activation in mouse DRG (Proença et al., 2011). These studies suggest a significant proportion of latent cells undergo lytic gene promoter activation prior to the establishment of latency, but the biological significance of these observations remains unclear. In the present study, we employed the Ai6 ZsGreen reporter mouse model to examine live latent cells marked with HSV-1 recombinants expressing Cre recombinase from ICP0 and TK promoters.
One of our key hypotheses was that cells marked by TK promoter activity may represent a population of neurons in which viral DNA replication could commence before latency establishment. If true, infected cells marked by infection with HSV TKCre may be expected to contain more HSV-1 DNA on average than the general latent cell reservoir. Whilst we could not analyse comparable numbers of marked cells between HSV CMVCre- and HSV TKCre-infected TG (as we could only recover ∼2 marked neurons per mouse), we found no evidence to support our hypothesis in cells analysed from both DRG and TG.
These data indicate that latent DNA copy number is not a result of viral gene expression, but rather most likely reflects virus seeding from the periphery and is in agreement with previous work (Thompson & Sawtell, 2000). Whilst a stricter examination of our hypothesis could be conducted using an HSV-1 reporter virus expressing Cre recombinase from a true-late promoter such as that of glycoprotein C (Proença et al., 2008), this virus marks half as many cells as TK promoter activity (Proença et al., 2011) and was not amenable to our investigation. We also addressed whether latent cells that experienced ICP0 promoter activity represent a population of cells infected with multiple virus templates. We hypothesized that high input load from the periphery (such as primary infection in the whisker pad) may overwhelm intracellular antiviral responses to virus DNA [such as sequestration and repression of HSV-1 DNA by nuclear ND-10 bodies and DNA repair machinery (Lilley et al., 2011; Lukashchuk & Everett, 2010)] and thus increase the likelihood of immediate-early gene expression before the cell can curtail infection. Our data do not show a bias towards high genome copies in ICP0-marked cells from either DRG or TG, suggesting that ICP0 promoter activation is not dependent on a high number of input virus templates. Indeed, careful analysis of alphaherpesvirus infection has determined gene expression is limited to < 7 genomes regardless of increasing m.o.i. in cell culture (Kobiler et al., 2010). Together these data suggest there is no inherent bias towards entry into lytic cycle in neurons receiving increasingly higher input viral genomes.
To further investigate the biological relevance of latent cell populations marked with ICP0 and TK promoter activity, we developed an ex vivo culture methodology to analyse reactivation from single cells following a heat-shock reactivation stimulus, which has been shown to stimulate reactivation both in vitro and in vivo (Halford et al., 1996; Sawtell & Thompson, 1992). Reactivation occurs at a low frequency in mouse models of infection. HSV-1 lytic antigen-positive cells can be detected in mouse ganglia at a frequency of 0.1 and 1–3 cells per ganglion during spontaneous (Feldman et al., 2002) and hyperthermia-induced reactivation (Sawtell & Thompson, 1992), respectively, using immunohistochemical approaches. We hypothesized that cells capable of reactivation may represent the minority population of neurons in which lytic promoter activity (and assumed gene expression) occurred before latency establishment. By culturing individual neurons isolated from TG infected with HSV CMVCre, which marks the vast majority of the latent cell reservoir (Proença et al., 2008), we observed reactivation in 12.5 % of cells following heat-shock stimulation (Table 1). Simultaneously, we assessed reactivation from cells marked by HSV ICP0Cre and TKCre, which comprise a minority of the infected cell population marked by HSV CMVCre (Proença et al., 2011). Reactivation of HSV ICP0Cre- and TKCre-marked cells occurred at a similar frequency to that in cells marked by HSV CMVCre, suggesting that reactivation competence was highly comparable between cells marked by all three recombinants (Tables 1 and 2). Whilst we cannot directly compare reactivation frequency between ZsGreen-positive cells (one individual marked neuron per well) and ZsGreen-negative cells (aliquots of bulk tissue which must comprise 76 % and over 96 % of the latent reservoir for HSV ICP0Cre and TKCre, respectively), we interpret these data to show that reactivation is not restricted to ICP0- or TKCre-marked cell populations. We conclude that reactivation competence of HSV-1 within a single neuron is independent of lytic promoter activation prior to latency establishment; however, it should be stressed that this conclusion relates to the use of heat shocking as an inducer of reactivation. The precise mechanism by which heat shocking derepresses the viral genome leading to full reactivation is not understood, and it would therefore be of interest to investigate alternative reactivation stimuli to fully understand the responsiveness of different subsets of neuronal populations.
Finally, we have demonstrated using a dual fluorescence HSV-1 latency model that at a population level reactivating neurons in culture contain 2–3 × more viral DNA copies than non-reactivating cells. As analysis of latent DNA loads required inhibition of de novo genome replication, full reactivation competence of mCherry-positive cells could not be determined. Nevertheless, our data demonstrate that genome de-repression occurs with greater frequency in populations of cells containing larger amounts of HSV-1 DNA. Furthermore, it is currently unknown whether a minimum number of HSV-1 genomes are required to successfully exit latency. Using mCherry expression from the HCMV promoter as a marker for cells harbouring reactivating virus, we found that HSV DNA copy numbers ranged from 4 to 2219 genomes, which is similar to the copy number range detected in non-reactivating cells (2–3565 genomes per cell). Our data demonstrate that although the exit of latency occurs with greater frequency in populations of cells with a higher median copy number than non-reactivating cells, there is no upper or lower threshold of viral DNA copy number per cell that is necessary for reactivation.
In summary, we have developed a fluorescent reporter mouse model of HSV-1 latency that allows for the isolation of single, live, latently infected cells. With this system we have determined that prior lytic promoter activation before latency establishment does not correlate with differing distributions in HSV-1 DNA copy number, nor the frequency with which reactivation occurs at the single cell level. We have additionally determined that HSV-1 reactivation can initiate within cells containing a range of virus DNA copies, suggesting that there is no threshold genome load required for reactivation.
Methods
Cells and viruses
All recombinants viruses are derived from HSV-1 strains SC16 (Hill et al., 1975) and 17syn+ (Brown et al., 1973). HSV CMVCre (Proença et al., 2008) is an SC16-based recombinant containing Cre recombinase under control of the human cytomegalovirus major immediate early promoter (HCMV MIEP) inserted in the non-essential Us5 gene.
Viruses were propagated and assayed using Baby Hamster Kidney-21 (BHK) cells. BHK cells were grown in Glasgow's modified Eagles medium (GIBCO) supplemented with 10 % FCS and 10 % tryptose phosphate broth (TPB). MRC-5 cells were maintained in Dulbecco's modified Eagle's medium (GIBCO) supplemented with 10 % FCS, 2 mM l-glutamine and 1 × non-essential amino acids. Neuronal explant cultures were maintained in MRC5 medium and where indicated supplemented with 0.1 mg PAA ml− 1.
Plasmids
pUL37/38CMVCherry is a plasmid designed to insert an mCherry expression cassette into the UL37/38 intergenic locus at position 84 252 bp of the HSV-1 genome (GenBank accession no. NC_001806; McGeoch et al., 1988). The plasmid contains the HSV-1 SnaBI fragment (nucleotides 82 644 to 85 780 bp). The expression cassette contains a HCMV MIEP, mCherry gene and nuclear localization sequence and was inserted in the AflII restriction site at position 84 252 bp of the HSV-1 genome.
Construction of recombinant viruses
HSV CMVCreCherry was generated by co-transfection of ScaI-linearized pUL37/38CMVCherry and HSV CMVCre-infected cell DNA resulting in the insertion of the mCherry expression cassette in the UL37/38 intergenic region. Three days post-transfection, infected BHK cell monolayers were harvested and recombinant progeny selected based on their ability to express the mCherry protein. Recombinant mCherry-expressing virus was purified by limiting dilution assay as previously described (Proença et al., 2008). Viral genomic structures were confirmed by restriction endonuclease digestion and Southern blot hybridization analyses (data not shown).
In vitro growth curves
In vitro assays were performed as described previously (Proença et al., 2008).
In vivo assays
In vivo replication assays were performed using seven- to eight-week-old female BALB/c or C57BL/6 mice as previously described (Proença et al., 2008).
Ai6 Rosa26ZsGreen reporter mice were used in cell marking studies. Groups of adult mice (>8 weeks of age) that differed in age by less than 12 days were anaesthetized by isoflurane inhalation before infection with the specified virus dose by scarification of the left ear or both whisker pads. Animal gender was matched per experiment or in the case of experiments including more than one virus, matched per time point. At various times p.i. mice were killed and CII, CIII and CIV DRG or both TG were harvested.
Confocal imaging
Whole DRG or TG were fixed in 4 % paraformaldehyde for 60 min or 90 min, respectively. TGs were cleared by incubation in ScaleU2 (Hama et al., 2011) in the dark at 4 °C for 21 days. TGs were imaged in 3.5 mm glass-bottom dishes through the whole depth of the tissue using a Leica SP5 confocal microscope. Images were acquired using the × 10 objective; when the whole tissue did not fit in the field of view several overlapping images were taken and assembled using FIJI's (Schindelin et al., 2012) stitching plugin, function 3d stitching (Preibisch et al., 2009).
Dissociation of sensory ganglia
Sensory ganglia were kept on ice in complete medium until dissociation could be performed. The ganglia were transferred to 0.2 % collagenase (CLS type 1, Worthington Biochemical Corporation), 0.04 mg DNase ml− 1, minced with spring scissors and incubated for 10 min at 37 °C. Following incubation, samples were gently pipetted up and down 50 times and incubated at 37 °C. This procedure was repeated every 5 min until a single cell suspension was observed. OptiPrep (Sigma) gradient enrichment of neurons (Bertke et al., 2011) was performed as follows. Cell suspensions were layered on top of 30, 25, 20, 15 and 10 % OptiPrep gradient in MRC5 medium. One millilitre of sample containing up to two TG or six DRG was loaded onto preformed gradients and centrifuged for 15 min at 800 g. Each gradient layer was harvested, transferred to a 6-well plate and its contents visualized using an inverted microscope. Gradient layers enriched in neurons (usually layers 15 % and over minus the cell pellet) were washed and resuspended in MRC5 medium.
Single cell purification
Single cell suspensions of two TG or six DRG were resuspended in 16 ml of MRC5 medium and distributed across eight 3.5 cm dishes. Single ZsGreen-expressing cells were picked by hand under a fluorescent inverted microscope using glass micropipettes (Subkhankulova et al., 2008), and cells were washed by transferring them to a second and third 3.5 cm tissue culture dish containing fresh MRC5 medium.
Single cell DNA extraction and real-time PCR
Single cells were transferred to a 200 μl PCR tube containing 10 μl of QuantiLyse optimized proteinase K extraction solution (Pierce et al., 2002) on ice. DNA extraction was performed in a PCR thermal cycler block by incubation at 50 °C for 150 min followed by inactivation of proteinase K at 95 °C for 15 min. Real-time PCR was performed in the same tube as the DNA extraction using a Rotor-Gene (Corbett Research) in triplicate for each reaction as described previously (Coleman et al., 2008). The primer sets and Taqman probes employed for amplification of the HSV-1 ICP0 and adenine phosphoribosyltransferase (APRT) promoters were as follows: ICP0: HSV-1 nt 2072–2090 forward primer, HSV-1 nt 2209–2139 reverse primer, probe HSV-1 nt 2115–2134; APRT: forward primer (nt 585–604), reverse primer (nt 706–688) (Dush et al., 1985) and Taqman probe (nt 667–640). Primers and HPLC-purified probes were manufactured by TIB-Molbiol. PCR products were quantified using a Rotor-Gene and associated software as the copy number per PCR, calculated from triplicate results from each PCR. A standard curve for each promoter region was generated using dilutions of appropriate plasmids.
Culture of latent neurons
Neurons to be cultured in vitro were purified as described above and plated in either 3.5 cm dishes, 24- or 96-well plates containing 90 % confluent MRC5 cells. MRC5 cell medium was used for short-term cultures in the presence of 0.1 mg PAA ml− 1 where specified.
Hyperthermia-induced reactivation
Hyperthermia-induced reactivation was performed 1 day post plating by incubation at 43 °C for 2 h followed by incubation at 37 °C. Positive reactivation of individually picked neurons was based on the appearance of CPE on MRC5 monolayers. In the case of bulk cultures of neurons derived from Ai6 mice latently infected with HSV CMVCreCherry, gradient-enriched neurons were cultured in the presence of PAA (100 μg ml− 1), and following hyperthermia treatment cells were incubated for 2 days at 37 °C prior to single cell picking of reactivating ‘red and green’ and non-reactivating ‘green’-only neurons.
Acknowledgements
This research was supported by the Medical Research Council (MRC) UK (MR/J00223). All animal experiments were approved by the University of Cambridge ethical review board and by the UK Home Office under the 1986 Animal (Scientific Procedures) Act as Project Licence number 80/2205.
Supplementary Data
Supplementary Data
References
- Bertke A. S., Swanson S. M., Chen J., Imai Y., Kinchington P. R., Margolis T. P. (2011). A5-positive primary sensory neurons are nonpermissive for productive infection with herpes simplex virus 1 in vitro J Virol 856669–6677 10.1128/JVI.00204-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S. M., Ritchie D. A., Subak-Sharpe J. H. (1973). Genetic studies with herpes simplex virus type 1. The isolation of temperature-sensitive mutants, their arrangement into complementation groups and recombination analysis leading to a linkage map J Gen Virol 18329–346 10.1099/0022-1317-18-3-329. [DOI] [PubMed] [Google Scholar]
- Chen X. P., Mata M., Kelley M., Glorioso J. C., Fink D. J. (2002). The relationship of herpes simplex virus latency associated transcript expression to genome copy number: a quantitative study using laser capture microdissection J Neurovirol 8204–210 10.1080/13550280290049642. [DOI] [PubMed] [Google Scholar]
- Coleman H. M., Connor V., Cheng Z. S., Grey F., Preston C. M., Efstathiou S. (2008). Histone modifications associated with herpes simplex virus type 1 genomes during quiescence and following ICP0-mediated de-repression J Gen Virol 8968–77 10.1099/vir.0.83272-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dush M. K., Sikela J. M., Khan S. A., Tischfield J. A., Stambrook P. J. (1985). Nucleotide sequence and organization of the mouse adenine phosphoribosyltransferase gene: presence of a coding region common to animal and bacterial phosphoribosyltransferases that has a variable intron/exon arrangement Proc Natl Acad Sci U S A 822731–2735 10.1073/pnas.82.9.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efstathiou S., Preston C. M. (2005). Towards an understanding of the molecular basis of herpes simplex virus latency Virus Res 111108–119 10.1016/j.virusres.2005.04.017. [DOI] [PubMed] [Google Scholar]
- Feldman L. T., Ellison A. R., Voytek C. C., Yang L., Krause P., Margolis T. P. (2002). Spontaneous molecular reactivation of herpes simplex virus type 1 latency in mice Proc Natl Acad Sci U S A 99978–983 10.1073/pnas.022301899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halford W. P., Gebhardt B. M., Carr D. J. (1996). Mechanisms of herpes simplex virus type 1 reactivation J Virol 705051–5060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hama H., Kurokawa H., Kawano H., Ando R., Shimogori T., Noda H., Fukami K., Sakaue-Sawano A., Miyawaki A. (2011). Scale: a chemical approach for fluorescence imaging and reconstruction of transparent mouse brain Nat Neurosci 141481–1488 10.1038/nn.2928. [DOI] [PubMed] [Google Scholar]
- Hill T. J., Field H. J., Blyth W. A. (1975). Acute and recurrent infection with herpes simplex virus in the mouse: a model for studying latency and recurrent disease J Gen Virol 28341–353 10.1099/0022-1317-28-3-341. [DOI] [PubMed] [Google Scholar]
- Kobiler O., Lipman Y., Therkelsen K., Daubechies I., Enquist L. W. (2010). Herpesviruses carrying a Brainbow cassette reveal replication and expression of limited numbers of incoming genomes Nat Commun 1146. 10.1038/ncomms1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley C. E., Chaurushiya M. S., Boutell C., Everett R. D., Weitzman M. D. (2011). The intrinsic antiviral defense to incoming HSV-1 genomes includes specific DNA repair proteins and is counteracted by the viral protein ICP0. PLoS Pathog 7e1002084. 10.1371/journal.ppat.1002084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukashchuk V., Everett R. D. (2010). Regulation of ICP0-null mutant herpes simplex virus type 1 infection by ND10 components ATRX and hDaxx J Virol 844026–4040 10.1128/JVI.02597-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J. Z., Russell T. A., Spelman T., Carbone F. R., Tscharke D. C. (2014). Lytic gene expression is frequent in HSV-1 latent infection and correlates with the engagement of a cell-intrinsic transcriptional response PLoS Pathog 10e1004237. 10.1371/journal.ppat.1004237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L., Zwingman T. A., Sunkin S. M., Oh S. W., Zariwala H. A., Gu H., Ng L. L., Palmiter R. D., Hawrylycz M. J. & other authors (2010). A robust and high-throughput Cre reporting and characterization system for the whole mouse brain Nat Neurosci 13133–140 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeoch D. J., Dalrymple M. A., Davison A. J., Dolan A., Frame M. C., McNab D., Perry L. J., Scott J. E., Taylor P. (1988). The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1 J Gen Virol 691531–1574 10.1099/0022-1317-69-7-1531. [DOI] [PubMed] [Google Scholar]
- Nicoll M. P., Proença J. T., Connor V., Efstathiou S. (2012a). Influence of herpes simplex virus 1 latency-associated transcripts on the establishment and maintenance of latency in the ROSA26R reporter mouse model J Virol 868848–8858 10.1128/JVI.00652-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicoll M. P., Proença J. T., Efstathiou S. (2012b). The molecular basis of herpes simplex virus latency FEMS Microbiol Rev 36684–705 10.1111/j.1574-6976.2011.00320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce K. E., Rice J. E., Sanchez J. A., Wangh L. J. (2019). QuantiLyse: reliable DNA amplification from single cells Biotechniques 321106–1111. [DOI] [PubMed] [Google Scholar]
- Preibisch S., Saalfeld S., Tomancak P. (2009). Globally optimal stitching of tiled 3D microscopic image acquisitions Bioinformatics 251463–1465 10.1093/bioinformatics/btp184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proença J. T., Coleman H. M., Connor V., Winton D. J., Efstathiou S. (2008). A historical analysis of herpes simplex virus promoter activation in vivo reveals distinct populations of latently infected neurones J Gen Virol 892965–2974 10.1099/vir.0.2008/005066-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proença J. T., Coleman H. M., Nicoll M. P., Connor V., Preston C. M., Arthur J., Efstathiou S. (2011). An investigation of herpes simplex virus promoter activity compatible with latency establishment reveals VP16-independent activation of immediate-early promoters in sensory neurones J Gen Virol 922575–2585 10.1099/vir.0.034728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roizman B., Whitley R. J. (2013). An inquiry into the molecular basis of HSV latency and reactivation Annu Rev Microbiol 67355–374 10.1146/annurev-micro-092412-155654. [DOI] [PubMed] [Google Scholar]
- Sawtell N. M. (1997). Comprehensive quantification of herpes simplex virus latency at the single-cell level J Virol 715423–5431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawtell N. M., Thompson R. L. (1992). Rapid in vivo reactivation of herpes simplex virus in latently infected murine ganglionic neurons after transient hyperthermia J Virol 662150–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawtell N. M., Poon D. K., Tansky C. S., Thompson R. L. (1998). The latent herpes simplex virus type 1 genome copy number in individual neurons is virus strain specific and correlates with reactivation J Virol 725343–5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S. & other authors (2012). Fiji: an open-source platform for biological-image analysis Nat Methods 9676–682 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subkhankulova T., Gilchrist M. J., Livesey F. J. (2008). Modelling and measuring single cell RNA expression levels find considerable transcriptional differences among phenotypically identical cells BMC Genomics 9268. 10.1186/1471-2164-9-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson R. L., Sawtell N. M. (2000). Replication of herpes simplex virus type 1 within trigeminal ganglia is required for high frequency but not high viral genome copy number latency J Virol 74965–974 10.1128/JVI.74.2.965-974.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner E. K., Bloom D. C. (1997). Experimental investigation of herpes simplex virus latency Clin Microbiol Rev 10419–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakim L. M., Jones C. M., Gebhardt T., Preston C. M., Carbone F. R. (2008). CD8(+) attenuation of cutaneous herpes simplex virus infection reduces the average viral copy number of the ensuing latent infection Immunol Cell Biol 86666–675 10.1038/icb.2008.47. [DOI] [PubMed] [Google Scholar]
- Wang K., Lau T. Y., Morales M., Mont E. K., Straus S. E. (2005). Laser-capture microdissection: refining estimates of the quantity and distribution of latent herpes simplex virus 1 and varicella-zoster virus DNA in human trigeminal Ganglia at the single-cell level J Virol 7914079–14087 10.1128/JVI.79.22.14079-14087.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data