Abstract
Seed metabolites are critically important both for plant development and human nutrition; however, the natural variation in their levels remains poorly characterized. Here we profiled 121 metabolites in mature seeds of a wide panel Oryza sativa japonica and indica cultivars, revealing correlations between the metabolic phenotype and geographic origin of the rice seeds. Moreover, japonica and indica subspecies differed significantly not only in the relative abundances of metabolites but also in their corresponding metabolic association networks. These findings provide important insights into metabolic adaptation in rice subgroups, bridging the gap between genome and phenome, and facilitating the identification of genetic control of metabolic properties that can serve as a basis for the future improvement of rice quality via metabolic engineering.
Rice (Oryza sativa L.) is a major staple food feeding over 50% of the global population. Cultivated rice was domesticated from the common ancestor of wild rice (Oryza rufipogon) under both natural and human selective pressures, and displays large genetic diversity across thousands of varieties1,2. Rice cultivars are mainly grouped into two subspecies japonica and indica with marked differences in plant architecture, agronomic and physiological features (e.g., stress resistance, cold tolerance, and seed quality)1. Although japonica and indica cultivars exhibit clear variation in genome sequences as well as in the morphological and physiological features2,3, the gap between DNA sequence information and end phenotypes, particularly chemical composition and its effect on plant development and adaption, remains largely obscure.
Seed quality is of immense agronomical importance, and is ultimately determined by chemical composition4,5,6. Metabolites in seeds function not only as energy components for the seed but also as nutrients for humans and livestocks. As metabolism is strongly influenced by heritable factors, the genetic basis underlying metabolic traits has recently become of major research interest7,8. Recent evidence suggests that there is considerable metabolic diversity in seeds from different rice cultivars. Kusano et al (2007) identified 10 metabolites with variations across 62 rice varieties using one- and two-dimensional gas chromatography-time-of-flight–mass spectrometry (GC-GC-TOF-MS)9. Heuberger et al (2010) detected 3,097 signals in 10 rice varieties using ultra performance liquid chromatography (UPLC-MS)10. Similarly, using GC-TOF-MS, Lou et al (2011) identified 41 metabolites showing a wide range of variations in 48 distinct rice germplasms11, whilst Kim et al (2012) identified 52 metabolites in seven cultivars by GC-TOF-MS12. Matsuda et al (2012) conducted metabolic quantitative trait loci (mQTL) analysis in rice grains using inbred lines, and determined few loci affecting levels of metabolites13. Despite the insights provided by these studies, metabolomic analysis of seeds with the goal of comparing and contrasting japonica and indica rice subspecies using a large collection of different inbred lines has not yet been reported.
Here we performed a large-scale non-targeted metabolomic analysis in seeds of 100 japonica and indica cultivars with a broad genetic diversity, and observed that japonica and indica had diversified metabolomes, reflecting their unique metabolic properties which can be regarded as a local adaptive response. Network-based analysis8,14 of the resulting metabolic profiles and other morphological traits identified further differences in the coordinated change of metabolite abundances in the two types of geographically dispersed cultivars.
Results
Metabolic profiles of rice seeds
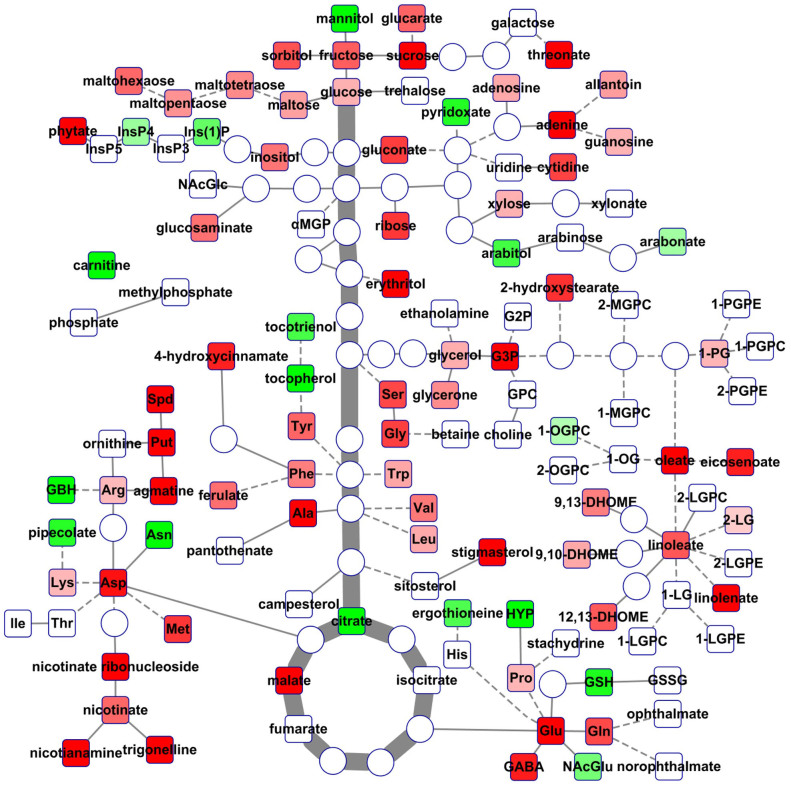
To evaluate metabolite composition in seeds across different rice cultivars, we analyzed a rice core collection representative of most japonica and indica cultivars. We identified a total of 121 metabolites with known structures, including: amino acids and their derivatives, carbohydrates, lipids, CPGECs (cofactors, prosthetic groups and electron carriers) as well as nucleotides and secondary metabolites. In comparison with previous reports by Lou et al. (2011) and Matsuda et al. (2012), our study extended both the numbers of rice genotypes and metabolites identified11,13. The identified metabolites were mapped onto nine super-metabolic pathways and their underlying 31 sub-pathways (Fig. 1 and Supplementary Table S1) as defined by the Plant Metabolic Net (PMN) and Kyoto Encyclopedia of Genes and Genomes (KEGG). These metabolites cover most of central metabolism and reflect the physiological state and nutritional value of rice seeds. Notably, the abundance of each metabolite differs remarkably among all tested inbred lines (Supplementary Table S1). Therefore, these lines may represent useful rice germplasm for breeders, which can pursue hybrids with high levels of desirable nutrients via hybridization and selection. The absence of glycolytic intermediates in mature rice seeds supported the fact that the physiological activity of desiccated seed is quite low.
Figure 1. Rice seed metabolome.
The identified 121 metabolites are mapped on a simplified metabolic network. Squares denote metabolites detected in this study, while circles represent undetected metabolites. Red (green) squares denote the metabolites with higher (lower) mean levels in japonica in comparison to indica, determined by Nested ANOVA. The darker the color is, the more significant the difference is. For full metabolite names, refer to Supplementary Table S1.
Japonica and indica have distinctive seed metabolomes
Although japonica and indica sub-species have been reported to have the same origin1,2,15, they have been domesticated under widely different environmental conditions. For example, japonica cultivars are mainly cultivated in temperate environment at high latitudes or altitudes with cool climate conditions, while indica cultivars are usually grown in tropical and subtropical regions at low latitudes or altitudes16. To assess differences in chemical composition between japonica and indica sub-species, we compared their mature seed metabolomes. Principle component analysis (PCA) revealed that the first three components can separate japonica and indica cultivars although there were some overlaps (Fig. 2), supported by the hierarchical clustering (Supplementary Fig. S1). The separation between japonica and indica at the metabolic level suggests that the two subspecies may employ different metabolic strategies to adapt to the growth environments.
Figure 2. Principal component (PC) analysis of the rice seed metabolomes.
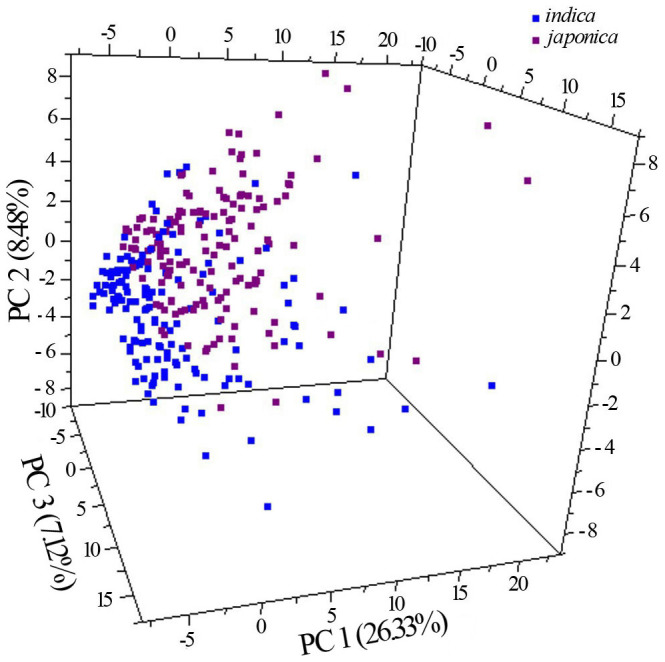
The first two PCs explain 34.81% of variance separating japonica from indica cultivars.
To further reveal the difference between japonica and indica cultivars in seed metabolome, we performed nested ANOVA and observed that the relative abundance of 92 metabolites including 28 amino acids and their derivatives, 23 carbohydrates, 22 lipids, 12 CPGECs, five nucleotides and two secondary metabolites exhibited statistically significant differences between japonica and indica cultivars with 66 metabolites higher in japonica and 26 higher in indica (Fig. 1 and Supplementary Table S1). To reveal the metabolites that can discriminate japonica from indica, we performed Random Forest ranking analysis, the accuracy of which was 94% in this study. We observed that particular metabolites including 14 amino acids can be used to differentiate between japonica and indica subspecies, with asparagine ranked the highest (Table 1). The order of the top 30 metabolites ranked according to the Random Forest analysis was, furthermore, remarkably similar to the ranking based on statistical significance from the ANOVA, supporting the reliability of possible application of these metabolites as biomarkers for the discrimination of japonica from indica subspecies.
Table 1. The top 30 metabolites contributing significantly to the correct classification of japonica and indica samples generated by Random Forest ranking.
| Biochemical Name | Mean Decrease Accuracy | Japonica/indica | P-value | FDR-value | Super Pathway |
|---|---|---|---|---|---|
| asparagine | 9.4445 | 0.68 | 3.62E-09 | 6.71E-09 | Amino acid |
| 4-guanidinobutanoate | 9.1961 | 0.68 | 1.00E-14 | 8.50E-14 | Amino acid |
| alanine | 9.0173 | 1.73 | 1.00E-15 | 1.50E-14 | Amino acid |
| trigonelline | 8.435 | 2.63 | 1.00E-15 | 1.90E-14 | CPGEC |
| gamma-tocopherol | 8.2808 | 0.50 | 8.90E-14 | 4.93E-13 | CPGEC |
| glutamate | 8.2736 | 1.38 | 1.20E-14 | 8.50E-14 | Amino acid |
| phytate | 7.7472 | 1.54 | 1.26E-06 | 1.52E-06 | Carbohydrate |
| 13-HODE-9-HODE | 6.263 | 1.88 | 4.36E-11 | 1.16E-10 | Lipids |
| agmatine | 6.2329 | 2.10 | 8.06E-13 | 3.20E-12 | Amino acid related compound |
| putrescine | 6.1999 | 2.07 | 1.16E-11 | 3.58E-11 | Amine |
| gamma-aminobutyrate | 6.1925 | 2.29 | 1.04E-10 | 2.40E-10 | Amino acid |
| glycine | 6.0299 | 1.35 | 4.95E-06 | 4.92E-06 | Amino acid |
| adenine | 5.8866 | 1.75 | 9.07E-12 | 3.15E-11 | Nucleotide |
| trans-4-hydroxyproline | 5.5906 | 0.60 | 0.0002 | 0.0001 | Amino acid |
| serine | 5.4119 | 1.43 | 2.17E-08 | 3.41E-08 | Amino acid |
| gluconate | 4.9611 | 2.89 | 5.96E-13 | 2.76E-12 | Carbohydrate |
| carnitine | 4.8728 | 0.69 | 3.91E-07 | 5.44E-07 | CPGEC |
| sucrose | 4.826 | 1.19 | 1.07E-09 | 2.28E-09 | Carbohydrate |
| inositol-1-phosphate | 4.8148 | 0.83 | 0.0006 | 0.0004 | Carbohydrate |
| nicotianamine | 4.5049 | 1.22 | 7.41E-07 | 9.37E-07 | CPGEC |
| citrate | 4.4552 | 0.80 | 0.0002 | 0.0001 | Carbohydrate |
| 1,3-dihydroxyacetone | 4.114 | 1.43 | 2.21E-08 | 3.41E-08 | Carbohydrate |
| tyrosine | 4.0544 | 1.29 | 6.31E-06 | 6.02E-06 | Amino acid |
| pipecolate | 4.054 | 0.78 | 9.28E-05 | 6.98E-05 | Amino acid |
| arginine | 3.921 | 1.15 | 0.0097 | 0.0042 | Amino acid |
| pyridoxate | 3.9206 | 0.84 | 0.0004 | 0.0002 | CPGEC |
| guanosine | 3.9175 | 1.10 | 0.0163 | 0.0067 | Nucleotide |
| stigmasterol | 3.8728 | 1.18 | 2.31E-09 | 4.59E-09 | Lipids |
| mannitol | 3.8254 | 0.70 | 0.0064 | 0.003 | Carbohydrate |
| spermidine | 3.8041 | 1.47 | 1.94E-06 | 2.16E-06 | Amine |
Higher values of mean decrease accuracy correspond to a larger importance of the metabolite in classifying japonica from indica cultivars. The column of japonica/indica shows the ratios of relative metabolite levels between japonica and indica. Red and green shaded cells indicate that the mean values are significantly higher in japonica and indica, respectively. P-value and FDR-value indicate the significance and false discovery rate of difference of the relative metabolite levels between japonica and indica, respectively. CPGEC: Cofactors, Prosthetic Groups, and Electron Carriers.
The metabolites of differential behavior between japonica and indica subspecies were mainly associated with three metabolic functions, namely, nitrogen metabolism, stress responses and inorganic nutrition storage and translocation. In general japonica appeared to have higher levels of nitrogen containing compounds, such as: gamma-aminobutyrate, serine, alanine, glutamate, glycine, glutamine, and agmatine, and polyamines (putrescine and spermidine) than indica (Supplementary Table S1). By contrast, japonica cultivars had lower levels of reduced glutathione, trans-4-hydroxyproline, asparagine and 4-guanidinobutanoate etc., compared to indica cultivars (Supplementary Table S1). In vegetative tissues nitrogen containing molecules, such as: glutamate, glutamine, and asparagine, play key roles in nitrogen assimilation, recycling, translocation and storage17,18. Previous studies demonstrated that japonica and indica display different nitrogen uptake efficiency during the vegetative growth stage19,20, whilst our data support previous claims that they likely have different strategies for nitrogen remobilization for yield and quality in seed during grain filling and maturation stage21. This difference in nitrogen-containing metabolites may result from the adaption to their original growth conditions and the nitrogen availability20, although it should be stressed that the plants studied here were grown ex situ. Moreover, metabolites associated with stress responses such as γ-aminobutyric acid (GABA), alanine, linolenate, 13-HODE-9-HODE (9,13-DHOME), 12,13-hydroxyoctadec-9(Z)-enoate (12,13-DHOME,), 9,10-hydroxyoctadec-12(Z)-enoic acid (9,10-DHOME,) and gluconate accumulated to higher levels in japonica cultivars. However, lower levels of anti-oxidative metabolites, such as reduced glutathione (GSH), γ-tocopherol, γ-tocotrienol and pyridoxate were observed in japonica. This result suggests that japonica seeds exhibit lower capacity for oxidative remediation than indica, which is consistent with observations that japonica cultivars are more susceptible than indica to irradiation and oxidative stress in seeds and seedlings22,23. Moreover, metabolites related to inorganic nutrition storage and translocation, such as phytate, gluconate and nicotianamine, were higher in japonica than indica cultivars, indicating a different ability of translocation and storage of those inorganic nutrients. It is known that negatively charged phosphate in phytate makes it an efficient chelating agent of positively charged mineral cations, such as K, Mg, Fe, Ca and Zn24. In addition, gluconate is also a strong chelating agent25, while nicotianamine is responsible for the translocation of Fe and Mn from leaf to the developing seeds26. Previous studies reported that the accumulations of Cu, Fe, Zn, and Mn in roots and shoots of japonica are higher than those of indica27, the metabolomic data in this study may imply higher amounts of certain inorganic nutrients in japonica seeds, as compared with those in indica.
Metabolite-metabolite correlation analysis
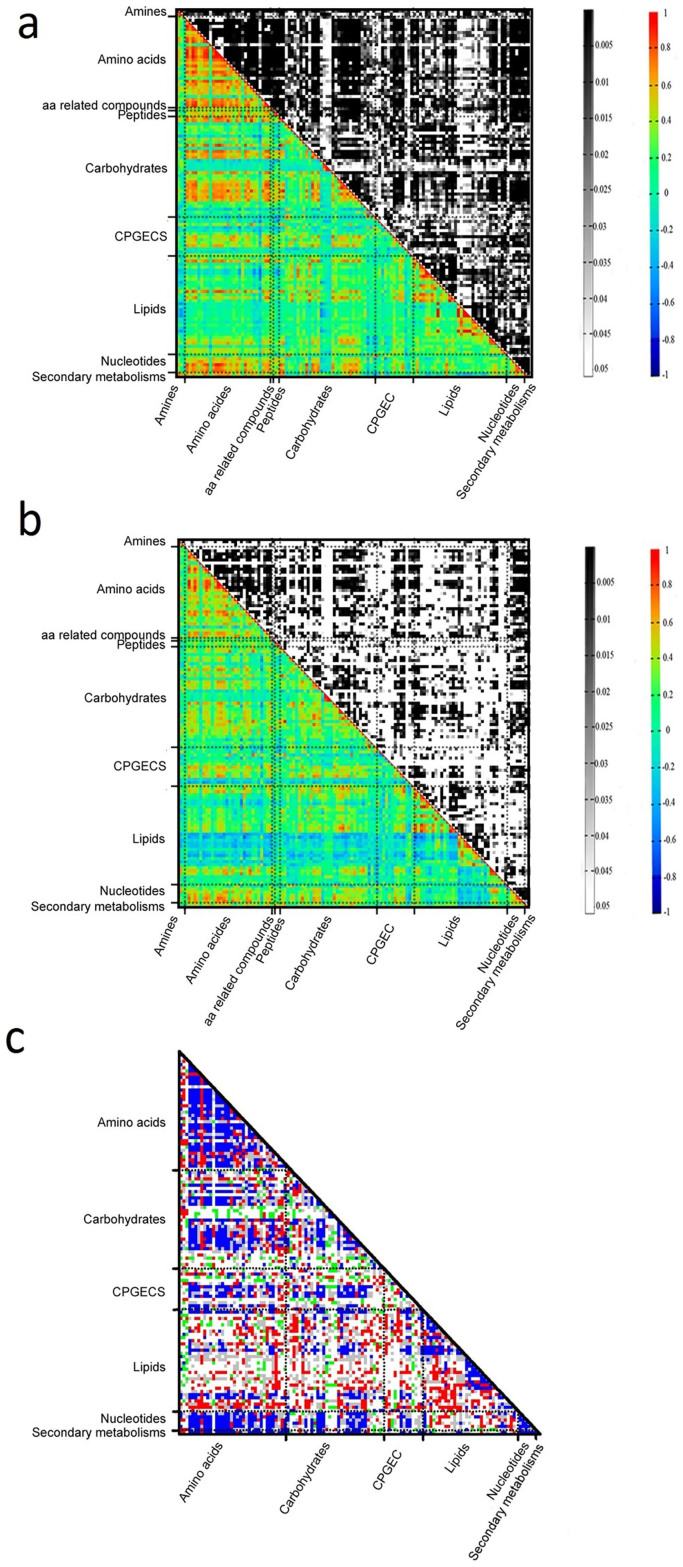
Correlation analysis can be used to reveal relationships among metabolites13,14. When applied to our dataset, this analysis revealed both positive and negative correlations among metabolites in both subspecies (Fig. 3a, 3b and Supplementary Data 1). At a threshold of correlation value greater than 0.50 (r-value ≥ 0.5), there were 868 and 1448 pairs of positive correlations, and 34 and 13 pairs of negative correlations, in japonica and indica, respectively. Generally, metabolites with high correlations observed in both indica and japonica were either amongst amino acids or between amino acids and carbohydrates or nucleotides. Additionally, some correlations among lipids, particularly phospholipids, were quite high: 12,13-hydroxyoctadec-9(Z)-enoate (12,13-DHOME) and 9,10-hydroxyoctadec-12(Z)- enoic acid (9,10-DHOME), which share the same common substrates and enzymes in their biosynthetic pathways, displayed the highest positive association in japonica (r-value = 0.99, p-value < 1.00E-16), while 1-oleoylglycerol (1-OG) and 2-oleoylglycerophosphocholine (2-OGPC), 1-oleoylglycerophosphocholine (1-OGPC) and 2-OGPC, 1-palmitoylglycerophosphocholine (1-PGPC) and 2-OGPC, 1-OG and 1-OGPC, 1-linoleoylglycerophosphocholine (1-LGPC) and 1-myristoylglycerophosphocholine (1-MGPC), and 1-OGPC and 1-PGPC accounted for the six highest positive correlations in indica (r-value > 0.97 and p-value < 1.00E-16). Remarkably, the strongest negative associations in japonica and indica were 1-linoleoylglycerophosphoethanolamine (1-LGPE) with choline (r = −0.77, p-value = 5.14E-11) and reduced glutathione (GSH) with linoleate (r = −0.62, p-value = 1.89E-06), respectively. Altogether, this analysis and other reports8 uncovered a conserved and highly coordinated interplay of amino acids, and of amino acids and carbohydrates in the crop seed metabolic network and a unique concerted interaction of lipids in rice seeds.
Figure 3. Comparison of metabolite-metabolite correlation in japonica and indica rice seeds.
(a) Heatmap of metabolite-metabolite correlation and significance in inidca. In the colored area, rectangles represent Pearson correlation coefficient (r) values of metabolite pairs (see correlation color key). In the black and white area, rectangles represent the respective p-values (see significance color key). (b) Heatmap of metabolite-metabolite correlation and significance in japonica. (c) Fisher's z-transformation analysis of differential metabolite-metabolite correlations between japonica and indica subspecies. Red rectangles indicate r-values of indica that are significantly bigger than those of japonica. Green rectangles indicate r-values of indica that are significantly smaller than those of japonica. Blue rectangles indicate r-values that are significant in both indica and japonica, but not significantly different between indica and japonica. Grey rectangles indicate r-values that are at least significant in one subspecies, but not significant between indica and japonica. White rectangles indicate r-values that are significant neither in indica nor in japonica, and not significantly different between indica and japonica.
Fisher's z-transformation analysis was employed to assess differential metabolite-metabolite correlations between japonica and indica subspecies. A total of 286 pair-wise associations were significantly different at False Discovery Rate at least 0.05 (FDR-value ≤ 0.05) between japonica and indica (Supplementary Data 1). The vast majority of the differential correlations were among different classes of metabolites such as carbohydrates, amino acids and lipids, with few differential correlations within metabolites of the same classes (Figure 3c and Supplementary Fig. S2). However, 11 metabolite-metabolite pairs associated with phospholipids displayed the most contrasting correlation trends between japonica and indica subspecies (Supplementary Data 1). Our results indicated that the correlations between metabolites of the same class are relatively conserved whilst those between metabolites of the different classes are rather diverse between japonica and indica subspecies. Therefore, our analysis provides insight that these two rice subspecies evolved distinct regulatory strategies for certain sets of metabolites, facilitating their adaptation to their specific growth conditions. Future detailed investigations into the role of these metabolites will allow elucidation of the network of key metabolic regulators in rice seeds.
Metabolite-morphological trait correlation analysis
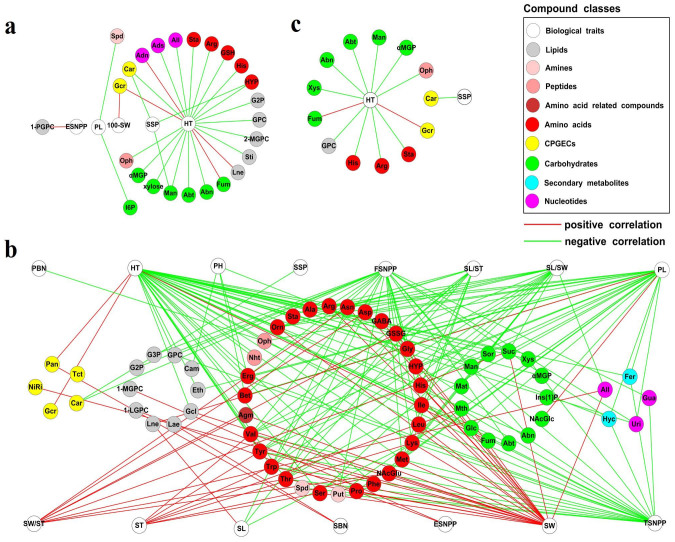
It has previously been reported that levels of certain metabolites are associated with other morphological traits7,14,28. To examine whether such associations are present in rice, we performed correlation analysis between the 121 seed metabolite traits (metabolites) and the 17 morphological traits we measured. This analysis revealed that most of the metabolites were negatively correlated with the measured morphological traits (Fig. 4a and 4b and Supplementary Data 1), which is similar to previous observations in tomato7,14,28. Generally, japonica cultivars displayed less metabolite-morphological trait associations than indica did (Fig. 4a and 4b), i.e. six positive and 23 negative associations in japonica (Fig. 4a), while the number of those associations in indica was 49 and 142, respectively (Fig. 4b). Furthermore, japonica and indica shared 13 metabolite-morphological trait associations (Fig. 4c), for example heading time (HT), an important and complex trait controlling the adaptation of rice cultivars to their growth environment, was negatively correlated with three amino acids (such as histidine and arginine), five carbohydrates (such as arabitol and mannitol), one lipid (glycerophosphorylcholine, GPC), and one peptide (ophthalmate), whilst being positively correlated with fumarate and glucarate.
Figure 4. Correlations between metabolite levels and morphological traits.
Full names of the abbreviation of metabolites and morphological traits refer to Supplementary Table S1 and Methods, respectively. Details about the associations are listed in Supplementary Data 1. Positive and negative correlations are represented by red and green edges, respectively. Each color denotes a compound class as shown in the top right legend. (a) Six positive and 23 negative correlations observed in japonica cultivars. (b) 49 positive and 142 negative correlations determined in indica cultivars. (c) 13 correlations shared between japonica and indica cultivars.
Fisher's z-transformation analysis was also employed to assess differential metabolite-morphological trait correlations between japonica and indica subspecies. Only five pair-wise associations were significantly different (FDR-value ≤ 0.05) between these two subspecies (Supplementary Data 1). They were seed width (SW) with oxidized glutathione and tyrosine, and panicle length with phytate, linolenate and fumarate.
Network-based analyses
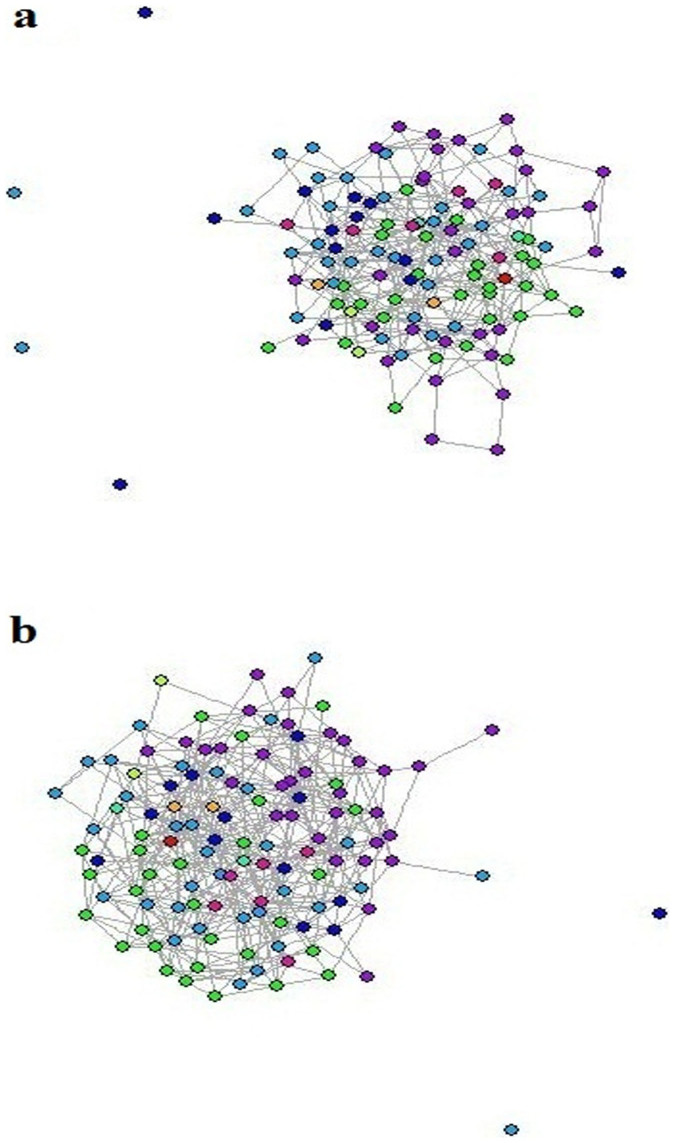
In order to explore which structural properties of the reconstructed networks reflect the partition of metabolites into differential and non-differential groups between two subspecies, japonica and indica networks were created separately by graphical LASSO. The indica network contained 339 edges, while the japonica network was denser and included 419 edges (Fig. 5a). The Indica network had five connected components, of which the largest one had 117 nodes (i.e. metabolites), and the remaining four were isolated nodes (i.e., glucosaminate, trehalose, gamma-tocopherol, and gamma-tocotrienol). The Japonica network exhibited three connected components, of which the largest one contained 119 nodes, and the other two were isolated nodes (i.e., glucosaminate, and gamma-tocopherol) (Fig. 5b). Of the four isolated nodes in the two networks, only gamma-tocotrienol and gamma-tocopherol showed differential behavior between the subspecies.
Figure 5. Delaunay triangulation networks of japonica and indica.
Different colors of nodes represent different types of metabolites: green, amino acid; light green, secondary metabolite; dark pink, nucleotide; orange, amine; cambridge blue, carbohydrate; blue, cofactors and prosthetic groups; red, amino acid related metabolite; purple, lipid; turquoise, peptide. (a) The japonica network consists of five connected components, the largest of which includes 117 nodes and 419 edges, and the remaining four are isolated nodes. (b) The indica network consists of three connected components, the largest of which includes 119 nodes and 339 edges, and the remaining two are isolated nodes.
The intersection shared by indica and japonica networks contained 121 nodes, with 93 edges and 34 connected components, while the symmetric difference contained 572 edges falling into three connected components, of which two were isolated nodes and one contained the remaining 119 metabolites. In the network intersection, 21 of the connected components were isolated nodes, and the remaining 13 connected components which may correspond to conserved associations due to similar underlying metabolic processes between the two rice subspecies. The decrease in the number of edges and the increase in the number of connected components between the intersection and each individual network imply that the japonica and indica networks have only few edges in common connecting a small number of nodes. Five out of these 13 connected components were observed to be enriched on the basis of all three ontologies used (Supplementary Fig. S3 and Supplementary Table S2). For example, connected component 1 was enriched in fatty acids (such as phospholipids and glycerophospholipids), which was in agreement with the metabolite-metabolite correlation analysis result (Fig. 3 and Supplementary Data 1).
Altogether, the edges in the network intersection were distributed among 100 nodes (metabolites), which indicate that 21 metabolites do not share edges between the japonica and indica networks. Nevertheless, 17 of these 21 metabolites were not isolated in the indica network, including: tryptophan, lysine, pipecolate, glutamine, N-acetylglutamate, betaine, N-acetylglucosamine, 1,3-dihydroxyacetone, sorbitol, glucarate (saccharate), nicotianamine, nicotinate ribonucleoside, trigonelline (N-methylnicotinate), glycerol, 13-HODE-9-HODE, beta-sitosterol, and adenosine. In the japonica network, additional two metabolites, i.e., gamma-tocotrienol and trehalose, were not isolated. These metabolites are involved in creating connected cultivar-specific subnetworks, which, in the indica network, include 13 metabolites on nine edges, and in the japonica network, consist of 12 metabolites and 10 edges (Supplementary Fig. S4).
To analyze the position of the differential metabolites within the network, the following node properties: degree, eccentricity, closeness, betweenness, eigencentriality and coreness were examined and the hypothesis that the average of each property for the differential metabolites is smaller/greater than the average of the non-differential metabolites was tested. Surprisingly, none of the 12 hypotheses (i.e., six properties and smaller/greater relationship) could be validated in the symmetric difference between the two networks. However, in the intersection, the differential metabolites have smaller degree, coreness, and closeness than the non-differential metabolites (Supplementary Table S3). This implied that a metabolite showing differential behavior is expected on average to be of larger eccentricity than a non-differential metabolite. Altogether, these findings indicated that differential metabolites are on the periphery of the network intersection, pulling the conserved processes in the cultivar-specific direction.
We further analyzed the metabolite community using three different approaches (i.e. fast greedy community detection, edge betweenness approach and leading eigenvector approach) (Supplementary Data 2). Since the fast greedy community detection resulted in the largest modularity in both networks, the resulting communities were then used to test for enrichment of terms based on the three previously used ontologies. The findings are presented in Supplementary Data 2 for the indica and japonica networks, respectively. Interestingly, the negligible overlap between the network communities, arising due to structural differences and quantified by the adjusted Rand index, corresponded to the observed differences in the enriched terms. While the modular structure of the indica network suggested orchestration of carbohydrate and amino acid metabolism, in the japonica network this seemed to be the case rather for lipid and amino acid metabolism.
Metabolomics profiles support isolation-by-distance model
To determine if the metabolic phenotype forms a random or structured spatial pattern, we investigated the relationship between the geographic proximity of the rice cultivars and their metabolic phenotypes. The geographic variability analysis29,30 showed positive associations between metabolic phenotype and geographic origin in rice seeds, revealing a robust pattern of isolation-by-distance. These structured patterns were observed for different classes of metabolites, and were supported by significant ranges for Moran's I (0.57–0.59), Geary's C (0.40–0.42), and Global G (0.022) (Supplementary Table S4).
Discussion
During their evolutionary process, plants, as sessile organisms, developed an array of molecular mechanisms to adapt to the varying environments, resulting in diversified molecular phenotypes and morphological traits (e.g., flowering time, yield and organ size)31. Understanding of the molecular factors determining plant adaptation is of general significance in plant research, and of particular importance in staple crops, which depends mainly on the quantification of the relationship between phenotypic characteristics and growth habitats or genotypic characteristics. In this study, using a standardized non-targeted UHPLC/MS/MS and GC-MS based metabolic profiling approach combined with robust statistical analysis, we investigated the relationship between biochemical characteristics and geographic origins, genotypic characteristics, and morphological traits in the mature seeds of japonica and indica rice.
Seed development can be divided into three stages: morphogenesis, maturation, and desiccation. During these processes, various metabolites such as amino acids, sugar alcohols (e.g. erythritol, arabitol, sorbitol and mannitol), betaines (e.g. trigonelline) and oligosaccharides (such as trehalose) accumulate in seeds32. Amino acids not only play an important role in maintaining the osmotic potential during seed desiccation but also serve as precursors for protein synthesis during germination. Asparagine is an ideal nitrogen storage and transport molecule17, and it is the major form for transport from the root to the leaves though the xylem and from the leaves to the developing seeds through the phloem in plants, and thereafter rapidly mobilized into glutamate and other amino acids during germination18. Alanine is a significant portion of the free amino acid pool under anaerobic and other stress conditions such as desiccation33. Metabolic profiling in mature rice seed therefore mainly reflects the metabolome of seed desiccation, the final stage of seed development, which seemingly exhibits highly convergent local adaptation. The opposite abundance of asparagine and alanine between japonica and indica cultivars indicates that they harbor different strategies for nitrogen utilization in seeds, probably via the regulation of the expression of transaminases or asparaginases. Another line of evidence suggestive of a different amino acid regulation is provided by the apparent difference in the metabolic fluxes between Arg and GBH vs Arg and polyamines in the two subspecies (Fig. 1).
The metabolic differences described here may be, at least partially, the result of mechanisms which sense or regulate stress responses. For example reactive oxygen species (ROS) generated during the seed desiccation process has an important role in cellular signaling for seed development34,35. Moreover, GABA and alanine are the two main amino acids accumulating under hypoxic stress conditions where they have been suggested to represent an important adaptive strategy to store carbon and nitrogen in preparation for the return to normal oxygen condition such as germination33,36. Furthermore in this vein, oxylipins are a large family of lipids-derived metabolites, which play significant roles in plant development and defense37. These compounds can be produced by non-enzymatic oxidation of polyunsaturated fatty acids, mainly linolenate (18:3) and linoleate (18:2)38. The lipid peroxidation products (9,13-DHOME, 12,13-DHOME and 9,10-DHOME) of the oxylipin pathways are consistent markers of higher oxidative states39, as are oxidized sugars40, such as gluconate. In contrast, reduced glutathione (GSH), tocopherols and pyridoxate directly act as protective agents against oxidative stress41,42,43. Compared with indica, japonica cultivars contain higher levels of oxidized metabolites (incuding GABA, alanine, gluconate and oxylipins) and concomitant lower levels of reduced metabolites (such as GSH, γ-tocopherol, γ-tocotrienol and pyridoxate), which suggest lower capacity for oxidative remediation in these cultivars. This is consistent with the previous findings that the overall susceptibility to oxidative damage is less in indica rice seeds than that in japonica22.
By investigating the interactions between metabolites, network-based analysis can help interpret complex datasets through the identification of key network components. Metabolite-metabolite correlation analysis generated a large number of significant correlations, the vast majority of which are positive, in both japonica and indica. The highly coordinated regulation of metabolic abundance in seeds has previously been also observed in other species, such as tomato, strawberry and Arabidopsis8,44,45. The highly synchronized patterns of change in metabolite levels of different species suggest that the regulation of metabolic process in seeds is conserved and tightly regulated during the evolutional history. On the other hand, japonica and indica also display significant differences in some metabolite-metabolite correlations. Firstly, there are less positive correlations but more negative correlations in japonica than in indica. Secondly, the magnitude of most significant correlations is different between the two subspecies. This phenomenon may be caused by variant feedback regulation and different preference of metabolic pathways between japonica and indica, which have not been proved in this study. Future efforts should be paid to elucidate the mechanism of different regulation of seed metabolome using a combination of molecular and biochemical methods.
Metabolite-morphological trait correlation analysis revealed more negative correlations in both indica and japonica cultivars, indicating an opposite change of specific rice phenotypic traits with the particular metabolite levels across the population, which is consistent with the findings in the pericarp and seed of tomato8,28. We propose that certain morphological traits, such as HT, TSNPP, PL and plant height (PH) affect rice seed metabolites. Negative correlations between seed metabolites and HT are mainly for amino acids and their derivatives. Similarly, in tomato the negative correlations between seed metabolites and harvest index are mainly for amino acids and N-containing compounds8. Therefore, a conserved metabolic regulatory mechanism may operate in plants, regulating N partitioning and seed development, which finally modulates plant yield. The genetic and molecular mechanism underlying this phenomenon merits further investigation. In addition, four seed metabolites (stachydrine, mannitol, arabinol, and arabitol) negatively correlate with filled seed number per panicle (FSNPP) but two metabolites (1-MGPC and 1-OGPC) positively correlate with ESNPP, revealing the competition for nutrition among the seeds in the same plant. The similar competitive phenomenon was observed in other species including badamu46, apple47, citrus48, papaya49, and tomato50. Furthermore, two seed metabolites (GPC and mannitol) negatively correlate with plant height. Altogether, metabolite and morphological trait correlation analysis uncovers common regulatory nodes for seed metabolic pathways in both rice subspecies.
Additionally, although there were only five significantly different metabolite and morphological trait correlations between japonica and indica subgroups, they tend to be subspecies-specific (Fig. 4), which may facilitate the dissection of the mechanisms underlying the metabolomic difference between these two subspecies. Furthermore, the same morphological trait in different subspecies highly correlate with different metabolites, for example, HT in japonica and indica correlates with different metabolites (Supplementary Data 1).
Further network-based analysis reveals that the network structural properties between japonica and indica vary significantly. Japonica network has more edges which largely differ from that of indica network. This is manifested in the finding that japonica has a distinct metabolite community from indica, in which carbohydrates and lipids metabolism instead of carbohydrates and amino acids exhibit orchestrated changes. On the other hand, our analysis also reveals a conserved sub-network between these two subspecies, which is validated by three additional ontology enrichment analyses, indicating both cultivar-specific and conserved metabolic processes between japonica and indica rice seeds.
To dissect the genetic basis of natural variation for complex metabolic traits in plants, association mapping including quantitative trait locus (QTL) analysis and genome-wide association study (GWAS) analysis has been increasingly applied51,52,53. By metabolite profiling and genetic analysis of 41 Arabidopsis accessions and two recombinant inbred lines, Trontin et al (2011) framed the range of natural variation of the major seed flavonoids and identified three genes co-localized with the considered QTLs and many other unknown genes may responsible for the these metabolic changes51. By mapping a QTL for natural variation in maize resistance to Rhopalosiphum maidis, Meihls et al (2013) identified a DIMBOA-Glc methyltransferase, whose activity was found to be associated with the formation of plant defense chemicals54. With the improvement and decline in cost of high-throughput genotyping and phenotyping technologies, GWAS studies are a preferred approach for dissecting complex QTLs. In Arabidopsis, GWAS analysis associated several lignin precursors with cinnamoyl-CoA, a target for improving the quality of lignocellulosic biomass by genetic engineering55, identified two major polymorphic loci controlling glucosinolate variation in natural populations related to plant defense56, and verified that allelic variation at BCAT2 is responsible for the natural variation of seed branched-chain amino acid levels57. In maize, GWASs have been reported for starch, oil, protein58 and vitamin E59,60, uncovering beneficial alleles can be identified and deployed for improvement of grain quality. In soybean, GWAS not only identified most known QTLs controlling seed protein and oil, but also narrowed genomic regions that facilitates more precise marker-assisted allele selection and positional cloning of the causal genes61. Unfortunately, due to the lack of genomic data of these cultivars, we could not perform either QTL mapping or GWAS analysis; therefore, the mechanistic architecture underlying the metabolic variation between japonica and indica subgroup cultivars remains to be elucidated.
In summary, this study provides a comprehensive study of natural variation in the rice seed metabolome and the correlation within and between metabolites and rice morphological properties. Although the observed metabolic variation between japonica and indica is exceedingly likely to be associated with the genetic variation between these two subspecies, the exact molecular mechanisms underlying the metabolic variation has not been studied here due to sampling limitation. However, this certainly merits further investigation using either novel approaches such as GWAS or specific materials resulting from bi-parental crosses (e.g., chromosomal segment substitution lines). Nevertheless, the reconstructed metabolic networks already indicate which processes are important in the two types of cultivars, thus, facilitating the identification of genetic regulators for future rice breeding and metabolic engineering strategies.
Methods
Materials
The study is based on our core collection of rice cultivars containing 51 japonica and 49 indica with a broad genetic and ecological diversity (Supplementary Table S5). All plants were planted in a paddy field in Minhang (31.03°N, 121.45°E), Shanghai, during the summer season in 2011. The experimental design was a randomized complete block design (including two rows of each inbred line and ten plants each row). Hundreds of individual mature rice seeds from four panicles of two individual plants were collected, frozen with liquid nitrogen and stored at −80°C until metabolomics analysis. Meanwhile, 17 morphological traits including: heading time (HT), plant height (PH), tiller number (TN), panicle length (PL), 100-seed weight (100-SW), seed length (SL), seed width (SW), seed thickness (ST), the ratio of seed length to seed width (SL/SW), the ratio of seed length to seed thickness (SL/ST), the ratio of seed width to seed thickness (SW/ST), filled seed number per panicle (FSNPP), empty seed number per panicle (ESNPP), total seed number per panicle (TSNPP), seed set percent (SSP), primary branch number per panicle (PBN), second branch number per panicle (SBN) were measured.
Metabolite profiling
Samples were ground into fine powder, and methanol extracts from 40 mg sample were then analyzed using a well established analytical platform: ultra-HPLC (UHPLC)-tandem mass spectrometry (MS/MS) and gas chromatography (GC)-MS. The detailed information about these platforms has been published elsewhere62,63,64. For LC platform, chromatographic separation and full scan mass spectra were performed to record retention time, mass-to-charge ratio, and MS/MS2 of all detectable ions. For GC platform, the samples were derivatized with BSTFA (bistrimethyl-silyl-triflouroacetamide), and the retention time and mass-to-charge ratio for all detectable ions were measured. The ion features of the samples were matched in an automated way against in-house built reference libraries of chemical standard entries for identification of metabolites.
Data analysis
Integrated peak ion counts were used to compare relative abundances of a metabolite in each sample. The missing values for a given metabolite were imputed with the observed minimum detected value for statistical analysis, assuming that they were below the limits of instrument detection sensitivity. Metabolic differences between japonica and indica seeds were determined using nested ANOVA in the R package. In addition, the metabolite profiles were subject to principal component (PC), and network-based analyses, the latter based on metabolite-metabolite and metabolite-morphological trait correlations employing the mean profile values8. To assess differential metabolite-metabolite and metabolite-morphological trait correlations between japonica and indica subspecies, Fisher's z-transformation analysis was employed. An edge was established between two metabolites if their 5-fold cross-validated partial correlation was different from zero, estimated from the sparse precision matrix based on LASSO65. Proximity network analysis were carried out as previously reported29,30. All statistical and network-based analyses were performed in R statistical environment and SIMPCA P software. Metabolic pathway and the graphical presentation of metabolite-morphological trait correlation were composed with Cytoscape version 2.8.3. The heatmaps of metabolite-metabolite correlation were visualized with MultiExperiment Viewer (MeV) version 4.8. Tests are deemed significant at level of 5%.
Supplementary Material
Supplementary Information
Acknowledgments
We thank Professor Qianqian from China National Rice Research Institute and Professor Lijun Luo from Shanghai Agrobiological Gene Center for providing the rice seeds used in present study. We also thank Mr. Jie Zong and Dai Chen from Novel Bioinformatics Ltd., Co. for their technical assistance in bioinformatics analysis, and Mr. Zhijing Luo and Ms. Mingjiao Chen for growing rice materials. This work was supported by the National Transgenic Plant Special Fund (2011ZX08012-002 and 2013ZX08012-002), China Innovative Research Team, Ministry of Education, 111 Project (B14016).
Footnotes
The authors declare no competing financial interests.
Author Contributions C.H., X.Z. and B.C. performed rice seed sampling and morphological trait observation. C.R., S.Q. and Q.L. carried out the metabolic profiling. D.A. and L.G. performed the annotation of the metabolites. C.H., J.S., H.L., J.W., X.C. and J.R. performed the data analysis. T.T. accomplished the classification of the identified chemicals, and S.K. and Z.N. carried out the network-based analysis and geographic origin analysis. C.H., J.S., L.G., Z.N., A.R.F. and D.Z. wrote the manuscript. J.S. and D.Z. designed the experiments and supervised the research.
References
- Khush G. S. Origin, dispersal, cultivation and variation of rice. Plant Mol Biol. 35, 25–34 (1997). [PubMed] [Google Scholar]
- Huang X. et al. A map of rice genome variation reveals the origin of cultivated rice. Nature 490, 497–501 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X. et al. Genome-wide association studies of 14 agronomic traits in rice landraces. Nat Genet. 42, 961–967 (2010). [DOI] [PubMed] [Google Scholar]
- Fitzgerald M. A., McCouch S. R. & Hall R. D. Not just a grain of rice: the quest for quality. Trends Plant Sci. 14, 133–139 (2009). [DOI] [PubMed] [Google Scholar]
- Kovach M. J., Calingacion M. N., Fitzgerald M. A. & McCouch S. R. The origin and evolution of fragrance in rice (Oryza sativa L.). Proc Natl Acad Sci.USA. 106, 14444–14449 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ufaz S. & Galili G. Improving the content of essential amino acids in crop plants: goals and opportunities. Plant Physiol. 147, 954–961 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer N. et al. Mode of inheritance of primary metabolic traits in tomato. Plant Cell 20, 509–523 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toubiana D. et al. Metabolic profiling of a mapping population exposes new insights in the regulation of seed metabolism and seed, fruit, and plant relations. PLoS Genet. 8, e1002612 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusano M. et al. Application of a metabolomic method combining one-dimensional and two-dimensional gas chromatography-time-of-flight/mass spectrometry to metabolic phenotyping of natural variants in rice. J Chromatogr B. 855, 71–79 (2007). [DOI] [PubMed] [Google Scholar]
- Heuberger A. L. et al. Metabolomic and functional genomic analyses reveal varietal differences in bioactive compounds of cooked rice. PLoS One 5, e12915 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou Q. et al. Profiling and Association Mapping of Grain Metabolites in a Subset of the Core Collection of Chinese Rice Germplasm (Oryza sativa L.). Journal of agricultural and food chemistry. 59, 9257–9264 (2011). [DOI] [PubMed]
- Kim J. K., Park S. Y., Lim S. H., Yeo Y., Cho H. S. & Ha S. H. Comparative metabolic profiling of pigmented rice (Oryza sativa L.) cultivars reveals primary metabolites are correlated with secondary metabolites. J Cereal Sci 57, 14–20 (2012). [Google Scholar]
- Matsuda F. et al. Dissection of genotype–phenotype associations in rice grains using metabolome quantitative trait loci analysis. Plant J. 70, 624–636 (2012). [DOI] [PubMed] [Google Scholar]
- Toubiana D., Fernie A. R., Nikoloski Z. & Fait A. Network analysis: tackling complex data to study plant metabolism. Trends Biotechnol. 31, 29–36 (2012). [DOI] [PubMed] [Google Scholar]
- Molina J. et al. Molecular evidence for a single evolutionary origin of domesticated rice. Proc. Natl. Acad. Sci. USA. 108, 8351–8356 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Z. et al. Differentiation and distribution of indica and japonica rice varieties along the altitude gradients in Yunnan Province of China as revealed by InDel molecular markers. Genet. Resour. Crop Ev. 57, 891–902 (2010). [Google Scholar]
- Hernández-Sebastià C. et al. Free amino acid profiles suggest a possible role for asparagine in the control of storage-product accumulation in developing seeds of low-and high-protein soybean lines. J. Exp. Bot 56, 1951–1963 (2005). [DOI] [PubMed] [Google Scholar]
- Gaufichon L., Reisdorf-Cren M., Rothstein S. J., Chardon F. & Suzuki A. Biological functions of asparagine synthetase in plants. Plant Sci. 179, 141–153 (2010). [Google Scholar]
- Britto D. T. & Kronzucker H. J. Bioengineering nitrogen acquisition in rice: can novel initiatives in rice genomics and physiology contribute to global food security? BioEssays 26, 683–692 (2004). [DOI] [PubMed] [Google Scholar]
- Fan X. et al. Comparing nitrate storage and remobilization in two rice cultivars that differ in their nitrogen use efficiency. J. Exp. Bot. 58, 1729–1740 (2007). [DOI] [PubMed] [Google Scholar]
- Koutroubas S. & Ntanos D. Genotypic differences for grain yield and nitrogen utilization in Indica and Japonica rice under Mediterranean conditions. Field Crop. Res. 83, 251–260 (2003). [Google Scholar]
- Ramarathnam N., Osawa T., Namiki M. & Kawakishi S. Studies on changes in fatty acid composition and content of endogenous antioxidants during γ irradiation of rice seeds. J. Amer. Oil Chem. Soc. 66, 105–108 (1989). [Google Scholar]
- Liu F. et al. Gene expression profiles deciphering rice phenotypic variation between Nipponbare (Japonica) and 93-11 (Indica) during oxidative stress. PLoS One 5, e8632 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson R. S., Bailey K. B., Gibbs M. & Ferguson E. L. A review of phytate, iron, zinc, and calcium concentrations in plant-based complementary foods used in low-income countries and implications for bioavailability. Food Nutr. Bull. 31, 134–146 (2010). [DOI] [PubMed] [Google Scholar]
- Gao F., Lu Q. & Komarneni S. Gluconate controls one-dimensional growth of tellurium nanostructures. J. Mater. Res. 21, 343–348 (2006). [Google Scholar]
- Lee S. et al. Iron fortification of rice seeds through activation of the nicotianamine synthase gene. Proc. Natl. Acad. Sci. USA 106, 22014–22019 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihara T., Goto F., Shoji K. & Kohno Y. Cross relationships of Cu, Fe, Zn, Mn, and Cd accumulations in common japonica and indica rice cultivars in Japan. Environ. Exp. Bot. 68, 180–187 (2010). [Google Scholar]
- Schauer N. et al. Comprehensive metabolic profiling and phenotyping of interspecific introgression lines for tomato improvement. Nat. Biotechnol. 24, 447–454 (2006). [DOI] [PubMed] [Google Scholar]
- Kleessen S. et al. Structured patterns in geographic variability of metabolic phenotypes in Arabidopsis thaliana. Nat. Commun. 3, 1319 (2012). [DOI] [PubMed] [Google Scholar]
- Kleessen S., Klie S. & Nikoloski Z. Data Integration through Proximity-Based Networks Provides Biological Principles of Organization across Scales. Plant Cell 25, 1917–1927 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genga A. et al. Plant Metabolomics: A characterisation of plant responses to abiotic stresses. In: Shanker, A. (Ed.). Abiotic Stress in Plants-Mechanisms and Adaptations. InTech Europe, Rijeka, Croatia(2011).
- Obendorf R. L. Oligosaccharides and galactosyl cyclitols in seed desiccation tolerance. Seed Sci. Res. 7, 63–74 (1997). [Google Scholar]
- De Sousa C. & Sodek L. Alanine metabolism and alanine aminotransferase activity in soybean (Glycine max) during hypoxia of the root system and subsequent return to normoxia. Environ. Exp. Bot 50, 1–8 (2003). [Google Scholar]
- Hendry G. A. F. et al. Free radical processes and loss of seed viability during desiccation in the recalcitrant species Quercus robur L. New Phytol. 122, 273–279 (2006). [DOI] [PubMed] [Google Scholar]
- Deng Z. Y., Gong C. Y. & Wang T. Use of proteomics to understand seed development in rice. Proteomics (2013). [DOI] [PubMed] [Google Scholar]
- Miyashita Y. & Good A. G. Contribution of the GABA shunt to hypoxia-induced alanine accumulation in roots of Arabidopsis thaliana. Plant Cell Physiol. 49, 92–102 (2008). [DOI] [PubMed] [Google Scholar]
- Vellosillo T. et al. Oxylipins Produced by the 9-Lipoxygenase Pathway in Arabidopsis Regulate Lateral Root Development and Defense Response through a Specific Signaling Cascade. Plant Cell, 831–846 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller M. J. & Berger S. Reactive electrophilic oxylipins: pattern recognition and signalling. Phytochemistry 70, 1511–1521 (2009). [DOI] [PubMed] [Google Scholar]
- Delaplace P., Rojas-Beltran J., Frettinger P., Du Jardin P. & Fauconnier M. L. Oxylipin profile and antioxidant status of potato tubers during extended storage at room temperature. Plant Physiol. Bioch. 46, 1077–1084 (2008). [DOI] [PubMed] [Google Scholar]
- Moreira P. et al. Detection and localization of markers of oxidative stress by in situ methods: application in the study of Alzheimer disease. Methods in molecular biology (Clifton, NJ) 610, 419 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbasi A. R., Hajirezaei M., Hofius D., Sonnewald U. & Voll L. M. Specific roles of α-and γ-tocopherol in abiotic stress responses of transgenic tobacco. Plant Physiol. 143, 1720–1738 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney S., Leuendorf J. E., Hendrickson C. & Hellmann H. Vitamin B6: a long known compound of surprising complexity. Molecules 14, 329–351 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer C. H. & Noctor G. Ascorbate and glutathione: the heart of the redox hub. Plant Physiol. 155, 2–18 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fait A. et al. Reconfiguration of the achene and receptacle metabolic networks during strawberry fruit development. Plant Physiol. 148, 730–750 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelovici R. et al. Deciphering transcriptional and metabolic networks associated with lysine metabolism during Arabidopsis seed development. Plant Physiol. 151, 2058–2072 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morandi B., Corelli Grappadelli L., Rieger M. & Lo Bianco R. Carbohydrate availability affects growth and metabolism in peach fruit. Physiol. Plantarum 133, 229–241 (2008). [DOI] [PubMed] [Google Scholar]
- Link H. Significance of flower and fruit thinning on fruit quality. Plant Growth Regul. 31, 17–26 (2000). [Google Scholar]
- Kubo T., Hohjo I. & Hiratsuka S. Sucrose accumulation and its related enzyme activities in the juice sacs of satsuma mandarin fruit from trees with different crop loads. Sci. Hortic-Amsterdam 91, 215–225 (2001). [Google Scholar]
- Zhou L., Christopher D. A. & Paull R. E. Defoliation and fruit removal effects on papaya fruit production, sugar accumulation, and sucrose metabolism. J. Amer. Soc. Hortic. Sci. 125, 644–652 (2000). [Google Scholar]
- Abe Y. et al. The SMALL AND ROUND SEED1 (SRS1/DEP2) gene is involved in the regulation of seed size in rice. Genes Genet Syst. 85, 327–339 (2010). [DOI] [PubMed] [Google Scholar]
- Trontin C., Tisné S., Bach L. & Loudet O. What does Arabidopsis natural variation teach us (and does not teach us) about adaptation in plants? Curr Opin Plant Biol. 14, 225–231 (2011). [DOI] [PubMed] [Google Scholar]
- Routaboul J. M. et al. Metabolite profiling and quantitative genetics of natural variation for flavonoids in Arabidopsis. J Exp Bot. 63, 3749–3764 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan E. K., Rowe H. C., Corwin J. A., Joseph B. & Kliebenstein D. J. Combining genome-wide association mapping and transcriptional networks to identify novel genes controlling glucosinolates in Arabidopsis thaliana. PLoS Biol. 9, e1001125 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meihls L. N. et al. Natural variation in maize aphid resistance is associated with 2, 4-dihydroxy-7-methoxy-1, 4-benzoxazin-3-one glucoside methyltransferase activity. Plant Cell 25, 2341–2355 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedelsheimer C. et al. Genome-wide association mapping of leaf metabolic profiles for dissecting complex traits in maize. Proc Natl Acad Sci 109, 8872–8877 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan E. K., Rowe H. C. & Kliebenstein D. J. Understanding the evolution of defense metabolites in Arabidopsis thaliana using genome-wide association mapping. Genetics 185, 991–1007 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelovici R. et al. Genome-wide analysis of branched-chain amino acid levels in Arabidopsis seeds. Plant Cell 25, 4827–4843 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook J. P. et al. Genetic architecture of maize kernel composition in the nested association mapping and inbred association panels. Plant Physiol 158, 824–834 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. et al. Genome-wide association study dissects the genetic architecture of oil biosynthesis in maize kernels. Nat Genet. 45, 43–50 (2013). [DOI] [PubMed] [Google Scholar]
- Lipka A. E. et al. Genome-wide association study and pathway level analysis of tocochromanol levels in maize grain. G3 Genes Genomes Genetics 3, 1287–1299 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang E. Y. et al. A genome-wide association study of seed protein and oil content in soybean. BMC Genom. 15, 1 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T. et al. Untargeted metabolomic profiling as an evaluative tool of fenofibrate-induced toxicology in Fischer 344 male rats. Toxicologic Patholo 37, 521–535 (2009). [DOI] [PubMed] [Google Scholar]
- Evans A. M., DeHaven C. D., Barrett T., Mitchell M. & Milgram E. Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal Chem. 81, 6656–6667 (2009). [DOI] [PubMed] [Google Scholar]
- Clarke J. D. et al. Assessment of genetically modified soybean in relation to natural variation in the soybean seed metabolome. Sci Rep 3, 3082 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibshirani R. The lasso method for variable selection in the Cox model. Statistics in Medicine 16, 385–395 (1997). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information