Abstract
Methylenetetrahydrofolate reductase (MTHFR) is an important enzyme involved in folate metabolism and DNA synthesis. A number of studies have examined the association of MTHFR C677T and A1298C polymorphisms with non-Hodgkin lymphoma (NHL) susceptibility; however, the conclusions were contradictory. We searched available publications assessing the polymorphisms of MTHFR and NHL susceptibility from MEDLINE, EMBASE and CBM. Genotype-based mRNA expression analysis was performed using data from 270 individuals with three different ethnicities. Ultimately, a total of 7448 cases and 11146 controls from 25 studies were included for the C677T polymorphism, 6173 cases and 9725 controls from 19 studies for the A1298C polymorphism. Pooled results indicated that neither C677T nor A1298C polymorphism was associated with NHL susceptibility. However, C677T polymorphism showed a statistically significantly increased risk for Caucasians, but a decreased risk for Asians in the subgroup analysis by ethnicity. The same variants may confer increased susceptibility to develop follicular lymphoma (FL). Moreover, A1298C polymorphism was associated with increased NHL risk for Asians. This meta-analysis indicated that C677T polymorphism was associated with altered NHL susceptibility for Caucasians, Asians and FL. Increased NHL risk was also shown for A1298C among Asians. These findings warrant validation in large and well-designed prospective studies.
Cancer has been recognized as one of the most formidable public health challenges, with estimates of nearly 12.7 million cancer cases and 7.6 million cancer deaths having occurred in 2008. Non-Hodgkin lymphoma (NHL) is the eighth most frequently diagnosed tumor type among men and the tenth among women worldwide1. Noteworthily, according to GLOBOCAN 2008 estimates, 355900 new cases and 191400 deaths might have occurred in 2008. There are a variety of different subtypes of NHL. Generally NHL is categorized into two major groups: B cell lymphomas and T cell lymphomas, with B cell lymphomas making up majority of NHL cases (about 85%). Diffuse large B-cell lymphoma (DLBCL) and follicular lymphoma (FL) are the two major subtypes of B cell lymphomas2,3. North America, Australia/New Zealand, and Northern, Western, and Southern Europe with the highest incidence rates, and South-Central and Eastern Asia and the Caribbean with the lowest incidence rates1. Certain immunodefective conditions (e.g., immunosuppression, Epstein-Barr virus and human immunodeficiency virus infections) as well as occupational exposures to herbicides and chlorinated organic compounds are the main risk factors for NHL1,4. Moreover, deficiency of nutrients (e.g., folate) related to one-carbon metabolism is also a well-established risk factors for NHL5, which has been reported to likely lead to immune responses impaired6.
Folate is an important coenzyme in DNA synthesis. Methylenetetrahydrofolate reductase (MTHFR) is one of the most critical enzymes involved in folate metabolism and DNA synthesis, which catalyses the conversion of 5, 10-methylenetetrahydrofolate to 5-methyltetrahydrofolate irreversibly7. Reduced MTHFR activity may play an inhibitory role on the 5-methyltetrahydrofolate pathway, and may lead to the accumulation of 5-methylenetetrahydrofolate, and consequentially methylation of dUMP to dTMP is decreased. Uracil can misincorporate into DNA when the methylation of dUMP to dTMP is deficient, consequentially results in DNA double-strand breaks and other anomalies, if not repaired which may lead to carcinogenesis8,9,10.
The MTHFR gene is located at chromosome 1p36.3. Among all the identified single nucleotide polymorphisms (SNPs) in this gene, C677T (Ala222Val, rs1801133) and A1298C (Glu429Ala, rs1801131), likely associated with reduced enzyme activity, have been widely investigated in a variety of diseases, such as psychiatric disorders11, congenital anomalies12, colorectal cancer13, and so on. Numerous studies have focused on the relationship between these two polymorphisms and NHL risk14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39, but the conclusions remain controversial. The discrepancies among studies may be ascribed to the relatively small sample size in each investigation as well as ethnicity difference. Therefore, we performed this meta-analysis using genotype data from all eligible investigations to provide a more precise evaluation of the association of MTHFR C677T and A1298C polymorphisms with NHL susceptibility.
Results
Study characteristics
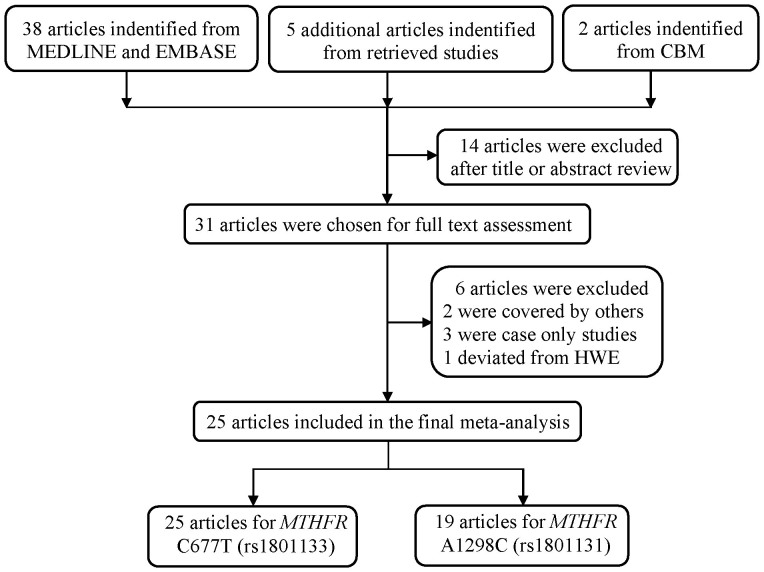
A total of 38 articles were initially indentified from MEDLINE and EMBASE, five more articles were indentified from the reference of retrieved studies, and ultimately, additional two articles were indentified from the CBM database (Figure 1). Of them, 14 articles were excluded after title and abstract assessment, while 31 articles met the crude inclusion criteria and were further evaluated. Among these remaining 31 publications, we further excluded two studies15,26 that were covered by other included investigations20,24, three case-only designed studies40,41,42, and one more study in which genotype frequency data in the controls for both of the C677T and A1298C polymorphisms were deviated from HWE (P = 0.009 for C677T and P = 0.040 for A1298C)38. Overall, 25 articles were included in the final meta-analysis. There were 25 articles with 7448 cases and 11146 controls for the C677T polymorphism, and 19 studies with 6173 cases and 9725 controls for the A1298C polymorphism (Table 1). Intriguingly, in the remaining studies, 12 and nine studies pertaining to C677T polymorphism provided detailed genotype frequency data for the DLBCL and FL subtype, respectively, while nine and eight studies regarding A1298C polymorphism provided detailed genotype data for the these two subtypes, respectively (Supplemental Table 1). Sample sizes of case in the incorporated studies ranged from 28 to 1103 for C677T polymorphism and from 31 to 1124 for A1298C polymorphism.
Figure 1. Flow diagram of selection of studies included in the current meta-analysis for the association between MTHFR gene polymorphisms and NHL susceptibility.
Table 1. Characteristics of studies included in the current meta-analysis.
| Surname | Year | Country | Ethnicity | Source | Genotype method | Case | Control | MAF | HWE | Score | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 11 | 12 | 22 | All | 11 | 12 | 22 | All | |||||||||
| C677T polymorphism | ||||||||||||||||
| Gonzalez Ordonez | 2000 | Spain | Caucasian | HB | PCR-RFLP | 21 | 21 | 5 | 47 | 92 | 88 | 20 | 200 | 0.32 | 0.876 | 6 |
| Lincz | 2003 | Australia | Caucasian | HB | PCR-RFLP | 73 | 58 | 17 | 148 | 145 | 133 | 21 | 299 | 0.29 | 0.198 | 7 |
| Toffoli | 2003 | Italy | Caucasian | PB | PCR-RFLP | 44 | 49 | 18 | 111 | 147 | 233 | 85 | 465 | 0.43 | 0.662 | 11 |
| Gemmati | 2004 | Italy | Caucasian | PB | PCR-RFLP | 60 | 101 | 39 | 200 | 78 | 128 | 51 | 257 | 0.45 | 0.908 | 10 |
| Linnebank | 2004 | German | Caucasian | PB | PCR-RFLP | 13 | 12 | 6 | 31 | 66 | 52 | 24 | 142 | 0.35 | 0.019 | 8 |
| Matsuo | 2004 | Japan | Asian | HB | PCR-RFLP | 165 | 122 | 63 | 350 | 182 | 230 | 88 | 500 | 0.41 | 0.301 | 8 |
| Rudd | 2004 | UK | Caucasian | HB | Taqman | 361 | 381 | 90 | 832 | 383 | 397 | 106 | 886 | 0.34 | 0.841 | 12 |
| Skibola | 2004 | USA | Caucasian | PB | Taqman | 122 | 160 | 52 | 334 | 288 | 350 | 84 | 722 | 0.36 | 0.149 | 14 |
| Lightfoot | 2005 | UK | Caucasian | PB | Taqman | 247 | 270 | 72 | 589 | 356 | 316 | 83 | 755 | 0.32 | 0.309 | 14 |
| Stanulla | 2005 | German | Caucasian | PB | PCR-RFLP | 207 | 216 | 64 | 487 | 184 | 152 | 43 | 379 | 0.31 | 0.179 | 9 |
| Chen | 2006 | China | Asian | HB | Taqman | 11 | 13 | 4 | 28 | 72 | 66 | 19 | 157 | 0.33 | 0.522 | 8 |
| Deligezer | 2006 | Turkey | Caucasian | HB | Taqman | 31 | 30 | 5 | 66 | 66 | 72 | 16 | 154 | 0.34 | 0.574 | 9 |
| Niclot | 2006 | France | Caucasian | PB | DHPLC | 66 | 86 | 20 | 172 | 92 | 88 | 24 | 204 | 0.33 | 0.674 | 8 |
| Timuragaoglu | 2006 | Turkey | Caucasian | PB | Realtime PCR | 31 | 22 | 5 | 58 | 36 | 36 | 10 | 82 | 0.34 | 0.829 | 9 |
| Lee | 2007 | Australia | Caucasian | PB | Taqman | 253 | 227 | 74 | 554 | 256 | 190 | 57 | 503 | 0.30 | 0.019 | 11 |
| Lim | 2007 | USA | Mixed | PB | Taqman | 499 | 477 | 127 | 1103 | 443 | 396 | 86 | 925 | 0.31 | 0.853 | 15 |
| Siraj | 2007 | Saudi Arabia | Caucasian | PB | PCR-RFLP | 109 | 45 | 6 | 160 | 372 | 126 | 13 | 511 | 0.15 | 0.553 | 10 |
| Gra | 2008 | Russia | Caucasian | HB | Hybridization | 39 | 28 | 9 | 76 | 85 | 79 | 13 | 177 | 0.30 | 0.354 | 8 |
| Kim | 2008 | Korea | Asian | PB | PCR-RFLP | 223 | 286 | 75 | 584 | 540 | 863 | 297 | 1700 | 0.43 | 0.133 | 12 |
| Berglund | 2009 | Sweden | Caucasian | PB | Illumina | 154 | 85 | 24 | 263 | 241 | 157 | 32 | 430 | 0.26 | 0.363 | 10 |
| Ismail | 2009 | Jordan | Caucasian | PB | PCR-RFLP | 34 | 19 | 2 | 55 | 94 | 66 | 10 | 170 | 0.25 | 0.722 | 10 |
| Wang | 2009 | Jamaica | Mixed | PB | Taqman | 329 | 58 | 5 | 392 | 204 | 57 | 5 | 266 | 0.13 | 0.664 | 14 |
| Kurzwelly | 2010 | German | Caucasian | PB | PCR-RFLP | 78 | 81 | 26 | 185 | 96 | 96 | 20 | 212 | 0.32 | 0.568 | 10 |
| Weiner | 2011 | Russia | Caucasian | PB | Taqman | 72 | 60 | 11 | 143 | 242 | 198 | 46 | 486 | 0.30 | 0.553 | 8 |
| Li | 2013 | USA | Mixed | PB | Taqman | 202 | 206 | 72 | 480 | 236 | 246 | 82 | 564 | 0.36 | 0.173 | 15 |
| A1298C polymorphism | ||||||||||||||||
| Lincz | 2003 | Australia | Caucasian | HB | PCR-RFLP | 64 | 68 | 13 | 145 | 124 | 139 | 31 | 294 | 0.34 | 0.385 | 7 |
| Toffoli | 2003 | Italy | Caucasian | PB | PCR-RFLP | 54 | 44 | 13 | 111 | 200 | 222 | 43 | 465 | 0.33 | 0.094 | 11 |
| Gemmati | 2004 | Italy | Caucasian | PB | PCR-RFLP | 96 | 90 | 14 | 200 | 126 | 110 | 21 | 257 | 0.30 | 0.659 | 10 |
| Linnebank | 2004 | German | Caucasian | PB | PCR-RFLP | 16 | 12 | 3 | 31 | 69 | 54 | 19 | 142 | 0.32 | 0.116 | 9 |
| Matsuo | 2004 | Japan | Asian | HB | PCR-RFLP | 209 | 122 | 19 | 350 | 327 | 150 | 23 | 500 | 0.20 | 0.282 | 8 |
| Rudd | 2004 | UK | Caucasian | HB | Taqman | 397 | 363 | 72 | 832 | 412 | 389 | 85 | 886 | 0.32 | 0.622 | 12 |
| Skibola | 2004 | USA | Caucasian | PB | Taqman | 178 | 128 | 27 | 333 | 341 | 310 | 71 | 722 | 0.31 | 0.964 | 14 |
| Lightfoot | 2005 | UK | Caucasian | PB | Taqman | 288 | 250 | 51 | 589 | 347 | 331 | 77 | 755 | 0.32 | 0.882 | 14 |
| Niclot | 2006 | France | Caucasian | PB | DHPLC | 79 | 76 | 17 | 172 | 102 | 81 | 15 | 198 | 0.28 | 0.844 | 8 |
| Lim | 2007 | USA | Mixed | PB | Taqman | 540 | 480 | 104 | 1124 | 461 | 393 | 81 | 935 | 0.30 | 0.831 | 15 |
| Siraj | 2007 | Saudi Arabia | Caucasian | PB | PCR-RFLP | 38 | 40 | 35 | 113 | 239 | 220 | 52 | 511 | 0.32 | 0.896 | 10 |
| Gra | 2008 | Russia | Caucasian | HB | Hybridization | 36 | 30 | 10 | 76 | 81 | 82 | 14 | 177 | 0.31 | 0.278 | 8 |
| Kim | 2008 | Korea | Asian | PB | Taqman | 372 | 182 | 29 | 583 | 1147 | 500 | 53 | 1700 | 0.18 | 0.868 | 12 |
| Berglund | 2009 | Sweden | Caucasian | PB | Illumina | 116 | 121 | 25 | 262 | 214 | 196 | 39 | 449 | 0.31 | 0.533 | 10 |
| Ismail | 2009 | Jordan | Caucasian | PB | PCR-RFLP | 20 | 23 | 12 | 55 | 76 | 81 | 13 | 170 | 0.31 | 0.172 | 10 |
| Wang | 2009 | Jamaica | Mixed | PB | Taqman | 277 | 98 | 15 | 390 | 201 | 65 | 9 | 275 | 0.15 | 0.198 | 14 |
| Kurzwelly | 2010 | German | Caucasian | PB | PCR-RFLP | 72 | 96 | 17 | 185 | 106 | 89 | 17 | 212 | 0.29 | 0.779 | 10 |
| Weiner | 2011 | Russia | Caucasian | PB | Taqman | 59 | 52 | 22 | 133 | 232 | 215 | 56 | 503 | 0.33 | 0.562 | 8 |
| Li | 2013 | USA | Mixed | PB | Taqman | 246 | 203 | 40 | 489 | 265 | 250 | 59 | 574 | 0.32 | 0.997 | 15 |
HB, Hospital based; PB, Population based; PCR-RFLP, Polymorphism chain reaction-restriction fragment length polymorphism; DHPLC, Denaturing high performance liquid chromatography; MAF, Minor allele frequency; HWE, Hardy-Weinberg equilibrium.
For the C677T polymorphism, there were 19 studies conducted in Caucasians, three studies in Asians, and three studies in mixed ethnic group. Of these studies, 18 were population based (PB) and seven were hospital based (HB) designed, respectively. Furthermore, 11 studies were considered as low quality (quality socre ≤ 9), and 14 (56%) were considered as high quality (quality score > 9). As to the A1298C polymorphism, there were 14 studies conducted in Caucasians, two studies in Asians, three studies in mixed ethnic group. While divided by the source of control, 15 studies were PB and four were HB. Among them, 6 were classified into low quality and 13 were classified into high quality. The mainly adopted genotyping methods were PCR-restriction fragment length polymorphism (12 and eight studies for C677T and A1298C, respectively) and Taqman (10 studies and eight studies for the C677T and A1298C, respectively).
Meta-analysis results
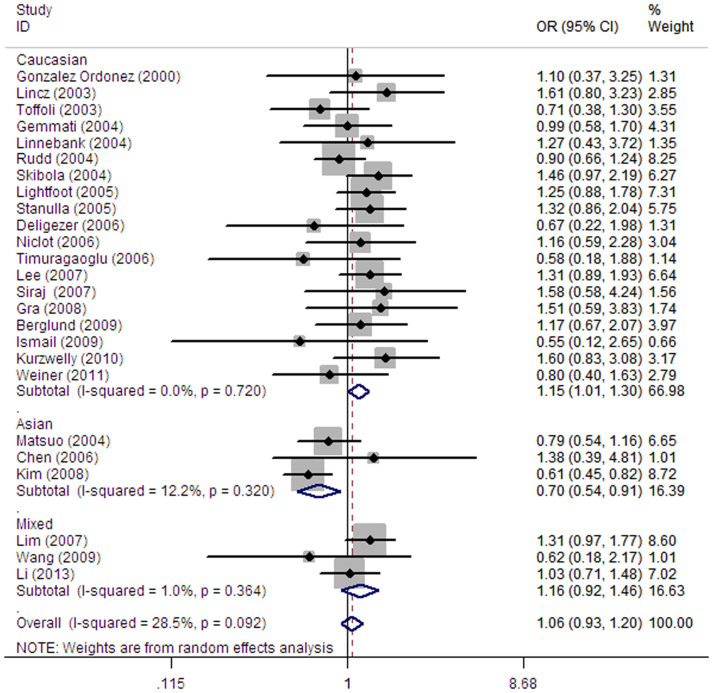
As shown in Table 2 and Figure 2, pooled analysis did not yield a significant association between MTHFR C677T polymorphism and overall NHL risk (homozygous: OR = 1.06, 95% CI = 0.93–1.20; heterozygous: OR = 0.97, 95% CI = 0.89–1.07; recessive: OR = 1.04, 95% CI = 0.95–1.15; dominant: OR = 0.99, 95% CI = 0.90–1.08 and allele comparing: OR = 1.01, 95% CI = 0.94–1.08). Next, we performed stratification analysis for the association between the C677T polymorphism variant genotypes by ethnicity, source of control, quality of studies, and tumor subtype. Stratification analysis by ethnicity revealed a statistically significantly increased NHL risk for Caucasians (homozygous: OR = 1.15, 95% CI = 1.01–1.30, and allele comparing: OR = 1.07, 95% CI = 1.01–1.13). In contrast, a significantly decreased risk of NHL was observed with the homozygous (OR = 0.70, 95% CI = 0.54–0.91) and dominant model (OR = 0.74, 95% CI = 0.59–0.91) genotypes for Asian group. Allele comparison further indicated that T variant allele is protective factor for NHL (T vs. C: OR = 0.81, 95% CI = 0.72–0.90). No significant association was found in the subgroup analysis by source of control and quality of studies. Moreover, stratification by tumor subtype demonstrated a significant increased risk of FL (homozygous: OR = 1.25, 95% CI = 1.01–1.53 and recessive: OR = 1.26, 95% CI = 1.04–1.53).
Table 2. Meta-analysis of the association between MTHFR C677T and A1298C polymorphisms and cancer risk.
| Variables | No. of study | Sample | Homozygous | Heterozygous | Recessive | Dominant | Allele Comparing | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Size | OR (95% CI) | P het | OR (95% CI) | P het | OR (95% CI) | P het | OR (95% CI) | P het | OR (95% CI) | P het | ||
| C677T (rs1801133) | TT vs. CC | CT vs. CC | TT vs. (CT + CC) | (CT + TT) vs. CC | T vs. C | |||||||
| All | 25 | 7448/11146 | 1.06 (0.93–1.20) | 0.092 | 0.97 (0.89–1.07) | 0.027 | 1.04 (0.95–1.15) | 0.465 | 0.99 (0.90–1.08) | 0.006 | 1.01 (0.94–1.08) | 0.006 |
| Ethnicity | ||||||||||||
| Caucasian | 19 | 4511/7034 | 1.15 (1.01–1.30) | 0.720 | 1.06 (0.97–1.15) | 0.639 | 1.11 (0.98–1.25) | 0.838 | 1.08 (0.99–1.16) | 0.546 | 1.07 (1.01–1.13) | 0.556 |
| Asian | 3 | 962/2357 | 0.70 (0.54–0.91) | 0.320 | 0.74 (0.55–1.01) | 0.108 | 0.81 (0.66–1.00) | 0.187 | 0.74 (0.59–0.91) | 0.238 | 0.81 (0.72–0.90) | 0.391 |
| Mixed | 3 | 1975/1755 | 1.16 (0.92–1.46) | 0.364 | 0.92 (0.71–1.19) | 0.067 | 1.15 (0.93–1.43) | 0.473 | 0.93 (0.71–1.22) | 0.034 | 0.96 (0.76–1.20) | 0.023 |
| Source of control | ||||||||||||
| PB | 18 | 5901/8773 | 1.08 (0.92–1.27) | 0.050 | 1.01 (0.92–1.12) | 0.086 | 1.05 (0.94–1.17) | 0.329 | 1.02 (0.92–1.13) | 0.010 | 1.02 (0.94–1.11) | 0.003 |
| HB | 7 | 1547/2373 | 0.95 (0.77–1.17) | 0.552 | 0.86 (0.69–1.06) | 0.120 | 1.02 (0.84–1.25) | 0.560 | 0.89 (0.75–1.05) | 0.244 | 0.95 (0.86–1.04) | 0.571 |
| Score | ||||||||||||
| Low | 11 | 1606/2780 | 1.04 (0.85–1.28) | 0.650 | 0.95 (0.78–1.16) | 0.048 | 1.09 (0.89–1.32) | 0.876 | 0.97 (0.81–1.17) | 0.078 | 1.00 (0.90–1.12) | 0.319 |
| High | 14 | 5842/8366 | 1.06 (0.89–1.27) | 0.018 | 0.99 (0.90–1.09) | 0.092 | 1.03 (0.93–1.15) | 0.139 | 1.00 (0.89–1.11) | 0.010 | 1.01 (0.92–1.11) | 0.001 |
| Subtype | ||||||||||||
| DLBCL | 12 | 1966/7271 | 1.03 (0.81–1.30) | 0.047 | 0.94 (0.78–1.13) | 0.002 | 1.02 (0.88–1.20) | 0.196 | 0.96 (0.80–1.14) | 0.002 | 1.00 (0.89–1.13) | 0.011 |
| FL | 9 | 1251/4508 | 1.25 (1.01–1.53) | 0.501 | 0.93 (0.75–1.14) | 0.034 | 1.26 (1.04–1.53) | 0.645 | 0.98 (0.81–1.19) | 0.049 | 1.05 (0.93–1.19) | 0.164 |
| A1298C (rs1801131) | CC vs. AA | AC vs. AA | CC vs. (AC + AA) | (AC + CC) vs. AA | C vs. A | |||||||
| All | 19 | 6173/9725 | 1.21 (0.97–1.50) | <0.001 | 1.02 (0.95–1.09) | 0.471 | 1.21 (0.98–1.49) | <0.001 | 1.05 (0.96–1.14) | 0.064 | 1.08 (0.98–1.18) | <0.001 |
| Ethnicity | ||||||||||||
| Caucasian | 14 | 3237/5741 | 1.24 (0.93–1.67) | <0.001 | 0.98 (0.89–1.08) | 0.490 | 1.25 (0.94–1.66) | <0.001 | 1.03 (0.92–1.17) | 0.075 | 1.09 (0.96–1.23) | <0.001 |
| Asian | 2 | 933/2200 | 1.54 (1.05–2.24) | 0.506 | 1.17 (0.99–1.39) | 0.494 | 1.46 (1.00–2.11) | 0.430 | 1.21 (1.03–1.42) | 0.646 | 1.20 (1.05–1.38) | 0.908 |
| Mixed | 3 | 2003/1784 | 0.96 (0.72–1.29) | 0.293 | 1.00 (0.87–1.14) | 0.471 | 0.98 (0.77–1.24) | 0.426 | 0.99 (0.85–1.15) | 0.291 | 0.99 (0.86–1.14) | 0.199 |
| Source of control | ||||||||||||
| PB | 15 | 4770/7868 | 1.26 (0.97–1.64) | <0.001 | 1.01 (0.93–1.10) | 0.400 | 1.25 (0.97–1.61) | <0.001 | 1.06 (0.95–1.18) | 0.039 | 1.09 (0.98–1.22) | <0.001 |
| HB | 4 | 1403/1857 | 0.98 (0.75–1.28) | 0.467 | 1.02 (0.88–1.19) | 0.381 | 0.99 (0.76–1.27) | 0.443 | 1.02 (0.88–1.19) | 0.356 | 1.02 (0.90–1.15) | 0.319 |
| Score | ||||||||||||
| Low | 6 | 907/1814 | 1.27 (0.94–1.71) | 0.669 | 1.08 (0.91–1.29) | 0.680 | 1.26 (0.94–1.68) | 0.612 | 1.11 (0.94–1.31) | 0.732 | 1.12 (0.98–1.27) | 0.673 |
| High | 13 | 5266/7911 | 1.21 (0.92–1.59) | <0.001 | 1.00 (0.93–1.08) | 0.299 | 1.21 (0.93–1.57) | <0.001 | 1.04 (0.93–1.16) | 0.019 | 1.08 (0.96–1.21) | <0.001 |
| Subtype | ||||||||||||
| DLBCL | 9 | 1624/6331 | 1.23 (0.84–1.79) | 0.002 | 1.02 (0.91–1.15) | 0.515 | 1.25 (0.86–1.81) | 0.001 | 1.06 (0.94–1.18) | 0.127 | 1.09 (0.92–1.28) | 0.001 |
| FL | 8 | 1039/4023 | 1.26 (0.88–1.79) | 0.081 | 1.04 (0.90–1.21) | 0.403 | 1.23 (0.90–1.67) | 0.153 | 1.07 (0.93–1.23) | 0.147 | 1.10 (0.93–1.30) | 0.036 |
HB, Hospital based; PB, Population based; DLBCL, diffuse large B-cell lymphoma; FL, follicular lymphoma.
Figure 2. Forest plots of effect estimates for MTHFR C677T polymorphism and NHL susceptibility (TT vs. CC).
For each study, the estimation of OR and its 95% CI are plotted with a box and a horizontal line. ◊, pooled ORs and its 95% CIs.
Similar to MTHFR C677T polymorphism, the presence of MTHFR A1298C polymorphism did not associate with an altered overall NHL risk (homozygous: OR = 1.21, 95% CI = 0.97–1.50; heterozygous: OR = 1.02, 95% CI = 0.95–1.09; recessive: OR = 1.21, 95% CI = 0.98–1.49; dominant: OR = 1.05, 95% CI = 0.96–1.14 and allele comparing: OR = 1.08, 95% CI = 0.98–1.18). Nevertheless, we observed a significantly increased risk of NHL for Asians (homozygous: OR = 1.54, 95% CI = 1.05–2.24; dominant: OR = 1.21, 95% CI = 1.03–1.42 and allele comparing: OR = 1.20, 95% CI = 1.05–1.38) but not other ethnic groups, when the analysis was stratified by ethnicity. No significant association was found in the remaining subgroup analyses by source of control, quality of studies and tumor subtype.
The correlation between the mRNA expression and genotypes
The correlation between MTHFR mRNA expressions levels by the genotypes were explored for three ethnic groups (i.e., CEU, YRI and Asian) and the whole group (Table 3). No significant alteration in the mRNA expression levels was found for the C677T polymorphism under all the genetic models.
Table 3. MTHFR mRNA expression by the genotypes of SNPs, using data from the HapMapa.
| Population | C667T (rs1801133) | A1298C (rs1801131) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| genotypes | No. | Mean ± SD | Pb | Ptrendc | genotypes | No. | Mean ± SD | Pb | Ptrendc | |
| CEU | CC | 48 | 6.14 ± 0.10 | 0.904 | AA | 41 | 6.12 ± 0.09 | 0.139 | ||
| CT | 37 | 6.14 ± 0.09 | 0.924 | AC | 36 | 6.15 ± 0.10 | 0.069 | |||
| TT | 5 | 6.12 ± 0.09 | 0.685 | CC | 13 | 6.16 ± 0.09 | 0.163 | |||
| Dominant | 42 | 6.14 ± 0.09 | 0.983 | Dominant | 49 | 6.15 ± 0.09 | 0.046 | |||
| YRId | CC | 70 | 6.20 ± 0.08 | 0.329 | AA | 71 | 6.20 ± 0.07 | 0.197 | ||
| CT | 19 | 6.19 ± 0.06 | 0.651 | AC | 17 | 6.19 ± 0.09 | 0.504 | |||
| TT | 1 | 6.09 | 0.171 | CC | 1 | 6.07 | 0.074 | |||
| Dominant | 20 | 6.19 ± 0.06 | 0.470 | Dominant | 18 | 6.18 ± 0.09 | 0.317 | |||
| Asiand | CC | 30 | 6.18 ± 0.08 | 0.116 | AA | 59 | 6.22 ± 0.10 | 0.074 | ||
| CT | 40 | 6.21 ± 0.09 | 0.120 | AC | 28 | 6.17 ± 0.09 | 0.024 | |||
| TT | 19 | 6.23 ± 0.11 | 0.052 | CC | 3 | 6.20 ± 0.06 | 0.814 | |||
| Dominant | 59 | 6.22 ± 0.10 | 0.057 | Dominant | 31 | 6.17 ± 0.08 | 0.028 | |||
| Alld | CC | 148 | 6.17 ± 0.09 | 0.360 | AA | 171 | 6.19 ± 0.09 | 0.186 | ||
| CT | 96 | 6.18 ± 0.09 | 0.732 | AC | 81 | 6.17 ± 0.09 | 0.107 | |||
| TT | 25 | 6.20 ± 0.12 | 0.156 | CC | 17 | 6.16 ± 0.09 | 0.261 | |||
| Dominant | 121 | 6.18 ± 0.10 | 0.422 | Dominant | 98 | 6.16 ± 0.09 | 0.069 | |||
aGenotyping data and mRNA expression levels for MTHFR by genotypes were obtained from the HapMap phase II release 23 data from EBV-transformed lymphoblastoid cell lines from 270 individuals.
bTwo-side Student's t test within the stratum.
cP values for the trend test of MTHFR mRNA expression among 3 genotypes for each SNP from a general linear model.
dThere were missing data because genotyping data were not available.
Interestingly, we found that C variant allele of MTHFR A1298C polymorphism significantly correlated with increased MTHFR mRNA expression levels among Caucasians (dominant model: P = 0.046), but decreased mRNA expression among Asians (heterozygous: P = 0.024 and dominant: P = 0.028).
Heterogeneity and sensitivity analyses
As shown in Table 2, substantial heterogeneities were observed among all investigations for the C677T polymorphism and NHL risk (homozygous: P = 0.092; heterozygous: P = 0.027; dominant model: P = 0.006 and allele comparing: P = 0.006), except for the recessive model (P = 0.465). We also observed considerable heterogeneities for the A1298C polymorphism (homozygous: P < 0.001; recessive model P < 0.001; dominant model: P = 0.064 and allele comparing: P < 0.001), except for the heterozygous model (P = 0.471). The meta-regression analysis did not yield any significant difference between subgroup analysis. Thus, leave-one-out sensitivity analyses indicated that no single study could alter the pooled ORs obviously (data not shown).
Publication bias
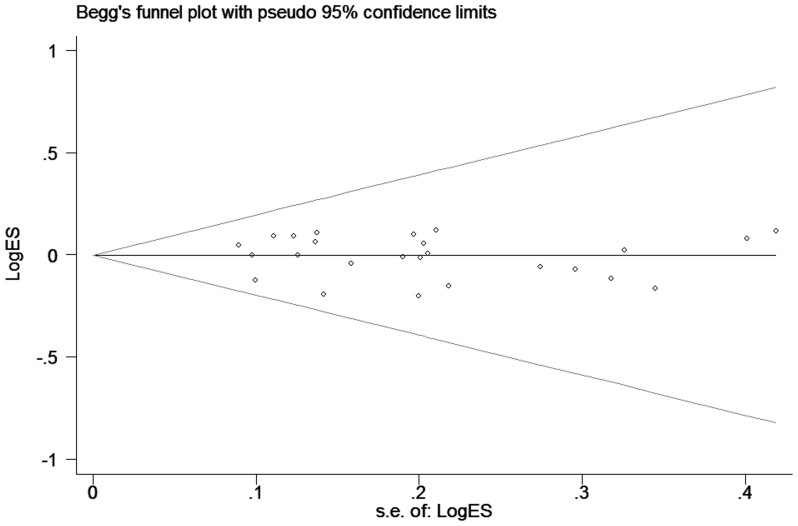
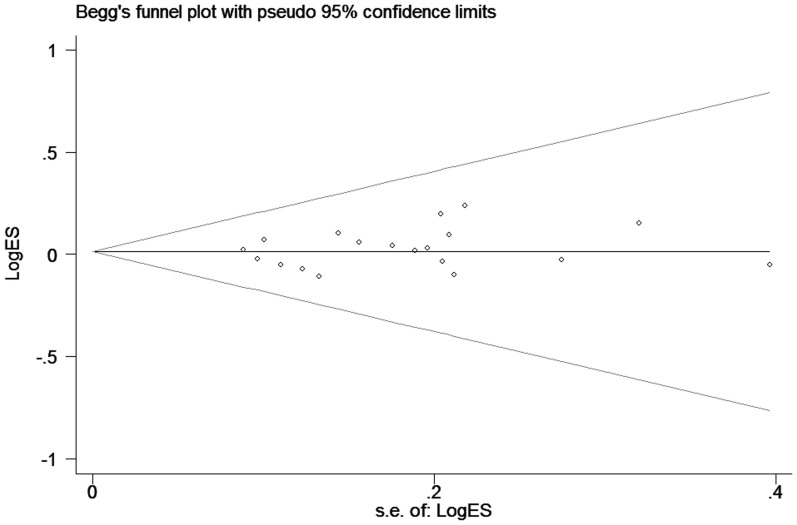
The shape of the funnel plots seemed asymmetry for the C677T and A1298C polymorphisms (Figure 3 and Figure 4), and we did not detect any significant publication bias by the Egger's test for C677T polymorphism (homozygous: P = 0.802; heterozygous: P = 0.462; recessive model: P = 0.667; dominant model: P = 0.568 and allele comparing: P = 0.761), and A1298C polymorphism (homozygous: P = 0.195; heterozygous: P = 0.767; recessive model: P = 0.274; dominant model: P = 0.312 and allele comparing: P = 0.152).
Figure 3. Funnel plot analysis to detect publication bias for C677T polymorphism by dominant model.
Each point represents a separate study for the indicated association.
Figure 4. Funnel plot analysis to detect publication bias for A1298C polymorphism by dominant model.
Each point represents a separate study for the indicated association.
Discussion
To our knowledge, the current meta-analysis is the largest one to investigate the association between MTHFR gene polymorphisms and NHL risk. Pooled analysis for the C677T polymorphism contained 25 studies with a total of 7448 NHL patients and 11146 controls; meanwhile, pooled analysis for the A1298C polymorphism encompassed 19 studies with 6173 NHL patients and 9725 controls. The meta-analysis observed no significant association between MTHFR C677T and A1298C polymorphisms and overall NHL risk. However, stratified analyses by ethnicity revealed that the C677T polymorphism increased NHL risk for Caucasians but decreased risk for Asians. The study of the same SNP also observed a significantly increased risk of FL, but not DLBCL, when the analysis was stratified by subtype of NHL. Moreover, the A1298C polymorphism was associated with increased risk for Asians, while no effect was observed for other ethnic groups. Interestingly, we found the MTHFR mRNA expression levels was slightly increased in the Asians carrying 677T alleles (P = 0.052), which was in accordance with our findings that C677T polymorphism was significantly associated with decreased NHL risk in the Asian group. Moreover, we also found the 1298C carriers showed significantly decreased MTHFR mRNA expression (P = 0.028), which corresponded to the evidence of association of 1298C polymorphism with increased NHL risk. Therefore, this results suggested that our findings from association studies for Asians may be biological plausible. MTHFR gene variants play an important role in the outcome of NHL patients. Examination of polymorphisms in the folate pathway genes might facilitate to reduce chemotherapy toxicity and improve survival by indicating when dose adjustments or alternative treatments are needed40.
Folate is a critical nutrient and coenzyme involved in DNA synthesis and methylation, and folate deficiency has been reported associated with numerous malignancies43. The product of MTHFR gene plays an important role in the methylation of homocysteine into methionine, sequentially leading to DNA methylation44. MTHFR C677T and A1298C polymorphisms were widely investigated in varieties of cancers. The former genetic variation is located in exon 4, and can lead to amino acid change from alanine to valine, which was first reported in 1995. This variation was reported to associate with reduced enzyme activity from a total of 40 subjects, with the CT and TT genotypes having ~60% and ~30% of the wild-type enzyme activity, respectively45. The A1298C polymorphism is located in exon 7. It has also been reported the CC genotype carriers having ~60% of wild-type enzyme activity46. Some of the previous studies failed to found this association21,47. Based on the largest meta-analysis to date, none of these two polymorphisms of interest was associated with overall NHL risk. Nonetheless, stratification analyses by ethnicity detected that analyzed SNPs significantly altered the risk of developing NHL in different ethnic groups. SNPexp online tool allows us to evaluate genotypes of MTHFR C677T and A1298C polymorphisms and their respective MTHFR transcript expression levels. With this in mind, we further investigated whether the biological results are in accordance with the observed association. We performed genotype-based mRNA expression analysis using the data from 270 individual with three ethnicities. We did not find a similar trend in the mRNA expression for the Caucasians but for the Asians, which may be due to the fact that the genotype counts for the homozygous variants is relatively small.
As so far, only two meta-analyses, which were nested in case-control studies, have investigated the association of MTHFR C677T and/or A1298C polymorphisms and NHL susceptibility. The study carried out by Lee et al.28 only studied the C677T polymorphism in Caucasians, consisting 13 studies with a total of 4245 cases and 5594 controls. The study observed increased NHL risk for T variant allele carrier in the Caucasians. Subjects with T alleles showed similarly increased risk of DLBCL and FL, in the subgroup analysis by NHL subtype. Another study included 4176 cases and 7585 controls for C677T, as well as 3648 cases and 6331 controls for A1298C polymorphism, in which no significant associations were observed for all subjects37. Some of the significant findings described above were not validated in our meta-analysis. For instance, the finding that the MTHFR C677T polymorphism was associated with elevated risk for DLBCL in homozygous model28 could not be duplicated in our study. Such associations were no longer significant in the current meta-analysis upon the inclusion of seven more studies. We also found some significant associations that were not observed in the previous studies, one example of which was that we found the C677T polymorphism decreased NHL risk for Asians, whereas the A1298C polymorphism conferred an increased risk to them. These new findings may be ascribed to the inclusion of more investigations with much large sample size in the current meta-analysis. It was noteworthy that we found the C677T polymorphism was associated with increased NHL risk for Caucasians while with decreased NHL susceptibility for Asians. The opposite findings in different ethnic groups may be resulted from ethnicity difference as well as the number of investigations. Earlier studies indicated that diet of Western contains high heterocyclic amines and polycyclic aromatic hydrocarbons48,49, which is relatively low in the diets of other ethnicities, which may contribute to the different effects of C677T polymorphism on cancer risk. Though we have included the latest investigations as well as publications written in Chinese, the current meta-analysis still has several limitations to be addressed. First, the sample size of cases from most eligible studies is relatively limited (<500), except for five studies21,23,28,29,32. Among the five studies, only one study has case more than one thousand29. The relative limited cases may have compromised statistical power. Second, we only included the studies written in English and Chinese, and might miss some investigations written in other languages. Third, heterogeneity was observed under some genetic models, so the results should be interpreted cautiously. Finally, our results were derived from unadjusted estimates due to lack of the original data, such as age, gender, smoking, drinking and dietary intaking habits, occupational exposures, as well as virus infections.
In summary, this study indicated that C677T and A1298C polymorphisms in the MTHFR gene may be associated with NHL susceptibility, especially for Caucasians, Asians and FL. Well-designed prospective studies with large sample size should be conducted to validate our findings.
Methods
Literature search strategy
We identified publications examined the association between MTHFR gene polymorphisms and NHL from MEDLINE and EMBASE using the following search items: “MTHFR or methylenetetrahydrofolate reductase”, “polymorphism or variant or variation” and “non-Hodgkin lymphoma or non-Hodgkin's lymphoma or NHL” (the last search updated was on January 8, 2014). We also identified related publications written in Chinese from Chinese Biomedical (CBM) database using the combinations terms of “MTHFR”, “NHL” and “polymorphism” in Chinese. Besides, we identified additional studies by searching of the references from retrieved studies manually. We only included the latest or the largest sample size studies in our final meta-analysis, if there exists more than one article published using the same subjects or overlapping data.
Inclusion and exclusion criteria
Studies included in the final meta-analysis had to meet the following criteria: (1) evaluate MTHFR C677T and/or A1298C polymorphisms and NHL risk; (2) be a case-control study, nested case-control study or a cohort study; (3) written in English or Chinese; (4) contain SNP genotype data; (5) independent from other studies; (6) provide sufficient data to calculate odds ratios (ORs) and their corresponding 95% confidence intervals (CIs).
The studies were excluded if genotype frequency data in the controls for MTHFR C677T and A1298C polymorphisms demonstrated departure from Hardy-Weinberg equilibrium (HWE) without further evidence showing that genotype distribution of other SNPs in controls followed HWE. In addition, case-only studies, case reports, conference abstract, reviews, meta-analyses and studies without detailed data were excluded.
Data extraction
Two investigators (JH and XL) independently extracted the following information from all eligible studies according to the inclusion and exclusion criteria: the first author's surname, year of publication, country of origin, ethnicity, cancer type and subtype (FL and DLBCL), control source (population based or hospital based), the total number of cases and controls, genotyping methods, minor allele frequency (MAF) for controls, P values for HWE for the control subjects and numbers of cases and controls with the CC, CT and TT genotypes for the C677T polymorphism and AA, AC and CC genotypes for the A1298C polymorphism. Any disagreement was resolved by discussion within our team numbers until consensus was reached.
Genotype based mRNA expression analysis
The genotypes data for MTHFR C677T and A1298C polymorphisms were available from HapMap (http://hapmap.ncbi.nlm.nih.gov/) for 270 subjects with three different ethnicities and their corresponding mRNA expression levels data were available from SNPexp (http://app3.titan.uio.no/biotools/tool.php?app=snpexp) as described previously50,51,52,53.
Quality assessment
Two investigators assessed the quality of each investigation using the quality assessment criteria (Supplemental Table 2), which was derived from previously published meta-analysis of molecular association studies54. Quality scores of studies ranged from 0 (lowest) to 15 (highest). Studies with scores ≤ 9 were categorized into low quality, while those with scores > 9 were considered as high quality. A third investigator (JZ) would be involved if there existed any disagreement.
Statistical methods
Crude ORs and their corresponding 95% CIs were used to evaluate the strength of associations between MTHFR gene polymorphisms and NHL risk. The pooled ORs were estimated for C677T polymorphism under the homozygous model (TT vs. CC), heterozygous model (CT vs. CC), recessive model (TT vs. CT + CC), dominant model (CT + TT vs. CC) and allele comparison (T vs. C). The same genetic models were also adopted for A1298C polymorphism as followed: homozygous model (CC vs. AA), heterozygous model (AC vs. AA), recessive model (CC vs. AC + AA), dominant model (AC + CC vs. AA) and allele comparison (C vs. A). Goodness-of-fit chi-square test was used to test deviation from HWE for the genotypes of controls. P < 0.05 was considered significant. The Chi-square based Q-test was performed to evaluate the heterogeneity across the studies. The random-effects model55 was chosen when significant heterogeneous exist (Pheterogeneity < 0.10); otherwise, fixed-effects model (the Mantel–Haenszel method)56 would be adopted. Stratification and meta-regression analyses were conducted to explore the potential source of heterogeneity across studies. Furthermore, stratification analyses were conducted by ethnicity (i.e., Asians, Caucasians, and Mixed that contained more than one ethnic group), control source (hospital-based and population-based), quality score of studies (low and high), and tumor subtype (FL and DLBCL). Sensitivity analysis was performed to assess the stability of the results by sequentially excluding one study at a time and recalculating the pooled ORs and their corresponding 95% CIs. Furthermore, both the Begg's funnel plot57 and the Egger's linear regression test58 were performed to assess the potential publication bias. The differences in mRNA expression levels for different genotypes were evaluated by one-way ANOVA, and the mRNA expression levels trend were evaluated by General linear model. All statistical tests were performed with STATA software (version 11.0; Stata Corporation, College Station, TX) and SAS software (version 9.1; SAS Institute, Cary, NC). All the statistics were two-sided, and P < 0.05 was considered as significant findings.
Supplementary Material
Dataset 1
Acknowledgments
This study was supported by grants from the National Science Fund for Distinguished Young Scholars (Grant No. 81325018) and the key project for International Cooperation and Exchange of the National Natural Science Foundation of China (Grant No. 81220108022).
Footnotes
The authors declare no competing financial interests.
Author Contributions All authors contributed significantly to this work. J.H., X.L., W.C. and S.H. performed the research study and collected the data; J.H., W.X. and G.S. analyzed the data; W.J. designed the research study; J.H., J.Z. and W.J. wrote the paper, and J.H. prepared Figures 1–4 and Supplemental Tables 1–2. All authors reviewed the manuscript. In addition, all authors approved the final draft.
References
- Jemal A. et al. Global cancer statistics. CA Cancer J Clin 61, 69–90 (2011). [DOI] [PubMed] [Google Scholar]
- Harris N. L. et al. The World Health Organization classification of neoplasms of the hematopoietic and lymphoid tissues: report of the Clinical Advisory Committee meeting--Airlie House, Virginia, November, 1997. Hematol J 1, 53–66 (2000). [DOI] [PubMed] [Google Scholar]
- Tan D. E. et al. Genome-wide association study of B cell non-Hodgkin lymphoma identifies 3q27 as a susceptibility locus in the Chinese population. Nat Genet 45, 804–7 (2013). [DOI] [PubMed] [Google Scholar]
- Muller A. M., Ihorst G., Mertelsmann R. & Engelhardt M. Epidemiology of non-Hodgkin's lymphoma (NHL): trends, geographic distribution, and etiology. Ann Hematol 84, 1–12 (2005). [DOI] [PubMed] [Google Scholar]
- Zheng T. et al. Epidemiology of non-Hodgkin lymphoma in Connecticut. 1935–1988. Cancer 70, 840–9 (1992). [DOI] [PubMed] [Google Scholar]
- Nauss K. M. & Newberne P. M. Effects of dietary folate, vitamin B12 and methionine/choline deficiency on immune function. Adv Exp Med Biol 135, 63–91 (1981). [DOI] [PubMed] [Google Scholar]
- Blount B. C. et al. Folate deficiency causes uracil misincorporation into human DNA and chromosome breakage: implications for cancer and neuronal damage. Proc Natl Acad Sci U S A 94, 3290–5 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skibola C. F. et al. Polymorphisms in the methylenetetrahydrofolate reductase gene are associated with susceptibility to acute leukemia in adults. Proc Natl Acad Sci U S A 96, 12810–5 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dianov G. L. et al. Repair of uracil residues closely spaced on the opposite strands of plasmid DNA results in double-strand break and deletion formation. Mol Gen Genet 225, 448–52 (1991). [DOI] [PubMed] [Google Scholar]
- Duthie S. J. & McMillan P. Uracil misincorporation in human DNA detected using single cell gel electrophoresis. Carcinogenesis 18, 1709–14 (1997). [DOI] [PubMed] [Google Scholar]
- Gilbody S., Lewis S. & Lightfoot T. Methylenetetrahydrofolate reductase (MTHFR) genetic polymorphisms and psychiatric disorders: a HuGE review. Am J Epidemiol 165, 1–13 (2007). [DOI] [PubMed] [Google Scholar]
- Botto L. D. & Yang Q. 5,10-Methylenetetrahydrofolate reductase gene variants and congenital anomalies: a HuGE review. Am J Epidemiol 151, 862–77 (2000). [DOI] [PubMed] [Google Scholar]
- Taioli E. et al. Meta- and pooled analyses of the methylenetetrahydrofolate reductase (MTHFR) C677T polymorphism and colorectal cancer: a HuGE-GSEC review. Am J Epidemiol 170, 1207–21 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez Ordonez A. J. et al. Normal frequencies of the C677T genotypes on the methylenetetrahydrofolate reductase (MTHFR) gene among lymphoproliferative disorders but not in multiple myeloma. Leuk Lymphoma 39, 607–12 (2000). [DOI] [PubMed] [Google Scholar]
- Matsuo K. et al. Association between polymorphisms of folate- and methionine-metabolizing enzymes and susceptibility to malignant lymphoma. Blood 97, 3205–9 (2001). [DOI] [PubMed] [Google Scholar]
- Lincz L. F. et al. Methionine synthase genetic polymorphism MS A2756G alters susceptibility to follicular but not diffuse large B-cell non-Hodgkin's lymphoma or multiple myeloma. Br J Haematol 120, 1051–4 (2003). [DOI] [PubMed] [Google Scholar]
- Toffoli G. et al. Methylenetetrahydrofolate reductase genotype in diffuse large B-cell lymphomas with and without hypermethylation of the DNA repair gene O6-methylguanine DNA methyltransferase. Int J Biol Markers 18, 218–21 (2003). [DOI] [PubMed] [Google Scholar]
- Gemmati D. et al. Common gene polymorphisms in the metabolic folate and methylation pathway and the risk of acute lymphoblastic leukemia and non-Hodgkin's lymphoma in adults. Cancer Epidemiol Biomarkers Prev 13, 787–94 (2004). [PubMed] [Google Scholar]
- Linnebank M. et al. The methionine synthase polymorphism D919G alters susceptibility to primary central nervous system lymphoma. Br J Cancer 90, 1969–71 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo K. et al. Methylenetetrahydrofolate reductase gene (MTHFR) polymorphisms and reduced risk of malignant lymphoma. Am J Hematol 77, 351–7 (2004). [DOI] [PubMed] [Google Scholar]
- Rudd M. F. et al. MTHFR polymorphisms and risk of chronic lymphocytic leukemia. Cancer Epidemiol Biomarkers Prev 13, 2268–70 (2004). [PubMed] [Google Scholar]
- Skibola C. F. et al. Polymorphisms and haplotypes in folate-metabolizing genes and risk of non-Hodgkin lymphoma. Blood 104, 2155–62 (2004). [DOI] [PubMed] [Google Scholar]
- Lightfoot T. J. et al. Risk of non-Hodgkin lymphoma associated with polymorphisms in folate-metabolizing genes. Cancer Epidemiol Biomarkers Prev 14, 2999–3003 (2005). [DOI] [PubMed] [Google Scholar]
- Stanulla M. et al. Methylenetetrahydrofolate reductase (MTHFR) 677C > T polymorphism and risk of pediatric non-Hodgkin lymphoma in a German study population. Blood 105, 906–7 (2005). [DOI] [PubMed] [Google Scholar]
- Niclot S. et al. Implication of the folate-methionine metabolism pathways in susceptibility to follicular lymphomas. Blood 108, 278–85 (2006). [DOI] [PubMed] [Google Scholar]
- Seidemann K. et al. MTHFR 677 (C --> T) polymorphism is not relevant for prognosis or therapy-associated toxicity in pediatric NHL: results from 484 patients of multicenter trial NHL-BFM 95. Ann Hematol 85, 291–300 (2006). [DOI] [PubMed] [Google Scholar]
- Timuragaoglu A., Dizlek S., Uysalgil N., Tosun O. & Yamac K. Methylenetetrahydrofolate reductase C677T polymorphism in adult patients with lymphoproliferative disorders and its effect on chemotherapy. Ann Hematol 85, 863–8 (2006). [DOI] [PubMed] [Google Scholar]
- Lee K. M. et al. One-carbon metabolism gene polymorphisms and risk of non-Hodgkin lymphoma in Australia. Hum Genet 122, 525–33 (2007). [DOI] [PubMed] [Google Scholar]
- Lim U. et al. Gene-nutrient interactions among determinants of folate and one-carbon metabolism on the risk of non-Hodgkin lymphoma: NCI-SEER case-control study. Blood 109, 3050–9 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siraj A. K. et al. Genetic polymorphisms of methylenetetrahydrofolate reductase and promoter methylation of MGMT and FHIT genes in diffuse large B cell lymphoma risk in Middle East. Ann Hematol 86, 887–95 (2007). [DOI] [PubMed] [Google Scholar]
- Gra O. A. et al. Polymorphisms in xenobiotic-metabolizing genes and the risk of chronic lymphocytic leukemia and non-Hodgkin's lymphoma in adult Russian patients. Am J Hematol 83, 279–87 (2008). [DOI] [PubMed] [Google Scholar]
- Kim H. N. et al. Association between folate-metabolizing pathway polymorphism and non-Hodgkin lymphoma. Br J Haematol 140, 287–94 (2008). [DOI] [PubMed] [Google Scholar]
- Berglund M., Enblad G., Turesson I., Edman V. & Thunberg U. Folate-metabolizing genes in lymphoma patients from Sweden. Scand J Immunol 70, 408–10 (2009). [DOI] [PubMed] [Google Scholar]
- Ismail S. I., Ababneh N. A., Khader Y., Abu-Khader A. A. & Awidi A. Methylenetetrahydrofolate reductase genotype association with the risk of follicular lymphoma. Cancer Genet Cytogenet 195, 120–4 (2009). [DOI] [PubMed] [Google Scholar]
- Wang S. S., Carreon J. D., Hanchard B., Chanock S. & Hisada M. Common genetic variants and risk for non-Hodgkin lymphoma and adult T-cell lymphoma/leukemia in Jamaica. Int J Cancer 125, 1479–82 (2009). [DOI] [PubMed] [Google Scholar]
- Kurzwelly D. et al. Genetic variants of folate and methionine metabolism and PCNSL incidence in a German patient population. J Neurooncol 100, 187–92 (2010). [DOI] [PubMed] [Google Scholar]
- Weiner A. S. et al. Polymorphisms in folate-metabolizing genes and risk of non-Hodgkin's lymphoma. Leuk Res 35, 508–15 (2011). [DOI] [PubMed] [Google Scholar]
- Nasr A. S., Sami R. M. & Ibrahim N. Y. Methylenetetrahydrofolate reductase gene polymorphisms (677C > T and 1298A > C) in Egyptian patients with non-Hodgkin lymphoma. J Cancer Res Ther 8, 355–60 (2012). [DOI] [PubMed] [Google Scholar]
- Li Q. et al. Role of one-carbon metabolizing pathway genes and gene-nutrient interaction in the risk of non-Hodgkin lymphoma. Cancer Causes Control 24, 1875–84 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemmati D. et al. Methylenetetrahydrofolate reductase C677T and A1298C gene variants in adult non-Hodgkin's lymphoma patients: association with toxicity and survival. Haematologica 92, 478–85 (2007). [DOI] [PubMed] [Google Scholar]
- Avivi I., Zuckerman T., Krivoy N. & Efrati E. Genetic polymorphisms predicting methotrexate blood levels and toxicity in adult non-Hodgkin lymphoma. Leuk Lymphoma 55, 565–70 (2014). [DOI] [PubMed] [Google Scholar]
- D'Angelo V. et al. Influence of methylenetetrahydrofolate reductase gene polymorphisms on the outcome of pediatric patients with non-Hodgkin lymphoma treated with high-dose methotrexate. Leuk Lymphoma 54, 2639–44 (2013). [DOI] [PubMed] [Google Scholar]
- Stolzenberg-Solomon R. Z. et al. Pancreatic cancer risk and nutrition-related methyl-group availability indicators in male smokers. J Natl Cancer Inst 91, 535–41 (1999). [DOI] [PubMed] [Google Scholar]
- Friso S. et al. A common mutation in the 5,10-methylenetetrahydrofolate reductase gene affects genomic DNA methylation through an interaction with folate status. Proc Natl Acad Sci U S A 99, 5606–11 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frosst P. et al. A candidate genetic risk factor for vascular disease: a common mutation in methylenetetrahydrofolate reductase. Nat Genet 10, 111–3 (1995). [DOI] [PubMed] [Google Scholar]
- van der Put N. M. et al. A second common mutation in the methylenetetrahydrofolate reductase gene: an additional risk factor for neural-tube defects? Am J Hum Genet 62, 1044–51 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crott J. W., Mashiyama S. T., Ames B. N. & Fenech M. F. Methylenetetrahydrofolate reductase C677T polymorphism does not alter folic acid deficiency-induced uracil incorporation into primary human lymphocyte DNA in vitro. Carcinogenesis 22, 1019–25 (2001). [DOI] [PubMed] [Google Scholar]
- Rohrmann S. et al. Cooking of meat and fish in Europe--results from the European Prospective Investigation into Cancer and Nutrition (EPIC). Eur J Clin Nutr 56, 1216–30 (2002). [DOI] [PubMed] [Google Scholar]
- Bogen K. T. & Keating G. A. U. S. dietary exposures to heterocyclic amines. J Expo Anal Environ Epidemiol 11, 155–68 (2001). [DOI] [PubMed] [Google Scholar]
- Holm K., Melum E., Franke A. & Karlsen T. H. SNPexp - A web tool for calculating and visualizing correlation between HapMap genotypes and gene expression levels. BMC Bioinformatics 11, 600 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger B. E. et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science 315, 848–53 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J. et al. Polymorphisms in the XPG gene and risk of gastric cancer in Chinese populations. Hum Genet 131, 1235–44 (2012). [DOI] [PubMed] [Google Scholar]
- He J. et al. Associations of Lys939Gln and Ala499Val polymorphisms of the XPC gene with cancer susceptibility: a meta-analysis. Int J Cancer 133, 1765–75 (2013). [DOI] [PubMed] [Google Scholar]
- Thakkinstian A. et al. Systematic review and meta-analysis of the association between complement component 3 and age-related macular degeneration: a HuGE review and meta-analysis. Am J Epidemiol 173, 1365–79 (2011). [DOI] [PubMed] [Google Scholar]
- DerSimonian R. & Laird N. Meta-analysis in clinical trials. Control Clin Trials 7, 177–88 (1986). [DOI] [PubMed] [Google Scholar]
- Mantel N. & Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 22, 719–48 (1959). [PubMed] [Google Scholar]
- Begg C. B. & Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics 50, 1088–101 (1994). [PubMed] [Google Scholar]
- Egger M., Davey Smith G., Schneider M. & Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ 315, 629–34 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Dataset 1