Abstract
X-chromosome inactivation (XCI) equalizes gene expression between the sexes by inactivating one of the two X chromosomes in female mammals. Xist has been considered as a major cis-acting factor that inactivates the paternally derived X chromosome (Xp) in preimplantation mouse embryos (imprinted XCI). Ftx has been proposed as a positive regulator of Xist. However, the physiological role of Ftx in female animals has never been studied. We recently reported that Ftx is located in the cis-acting regulatory region of the imprinted XCI and expressed from the inactive Xp, suggesting a role in the imprinted XCI mechanism. Here we examined the effects on imprinted XCI using targeted deletion of Ftx. Disruption of Ftx did not affect the survival of female embryos or expression of Xist and other X-linked genes in the preimplantation female embryos. Our results indicate that Ftx is dispensable for imprinted XCI in preimplantation embryos.
Female mammals have a unique mechanism of gene dosage compensation called X-chromosome inactivation (XCI), which is an essential epigenetic process for development. In mice, inactivation of the paternally derived X chromosome (Xp), but not of the maternally derived X chromosome (Xm), is initiated in the preimplantation embryos (imprinted XCI)1,2,3. This imprinted inactivation of the Xp is maintained in post-implantation extra-embryonic tissues. At the blastocyst stage, reactivation of the Xp occurs in the inner cell mass that will later form the embryo. Thus, the imprinted XCI is erased once, subsequently, random XCI of either the Xp or Xm chromosomes is initiated in the post-implantation embryo by a cis-acting region on the X chromosome termed the X-inactivation centre (Xic), which is essential as a genomic element for XCI4,5. Xic encodes a long non-coding (lnc)RNA Xist, which is expressed from the inactive X chromosome and represents the trigger for chromosome-wide silencing after implantation. Female mice with disrupted Xist alleles inherited from the father show severe growth retardation and die during post-implantation embryogenesis because of the failure of the imprinted XCI in extra-embryonic tissues6. Although Xist was also reported to play a major role in imprinted XCI before implantation7, the precise mechanism of imprinted XCI and how Xist expression is controlled remain unknown.
We recently identified a number of imprinted genes expressed predominantly from the inactive Xp at the preimplantation stage8, including three lncRNAs: Xist, Ftx, Jpx; two micro (mi) RNAs, miR-374-5p and miR-421-3p; and a protein-coding gene, Zcchc13. Combined with previous reports showing that Tsix was imprinted and expressed from the Xm9,10, these newly identified imprinted genes form a large cluster named the “Tsix-Zcchc13 imprinted gene cluster”. Furthermore Okamoto et al. indicated that part of this cluster, from Tsix to Ftx excluding Zcchc13, is included in the 210 kb region that contains sufficient factors to trigger imprinted XCI11,12. They reported that a 210 kb single-copy transgene (Tg80) region containing Tsix to Ftx induced imprinted cis-inactivation in autosomes in preimplantation embryos when it was derived paternally. Among the four imprinted genes located in this transgene, namely Tsix, Xist, Jpx, and Ftx, Ftx is the most abundant transcript at the preimplantation embryo stage except for Xist. Furthermore, Chureau et al. proposed that Ftx is a positive regulator of Xist, as deletion of the Ftx promoter leads to decreased Xist expression in male embryonic stem (ES) cells13. However, its role in XCI remains to be elucidated for female embryos.
Taking these reports into consideration, we focused on the function of imprinted Ftx at the preimplantation stage. Using Ftx-deficient female mice, we analysed the effects of deletion of Ftx on female viability and imprinted XCI during preimplantation development.
Results
Generation of Ftx-deficient mice
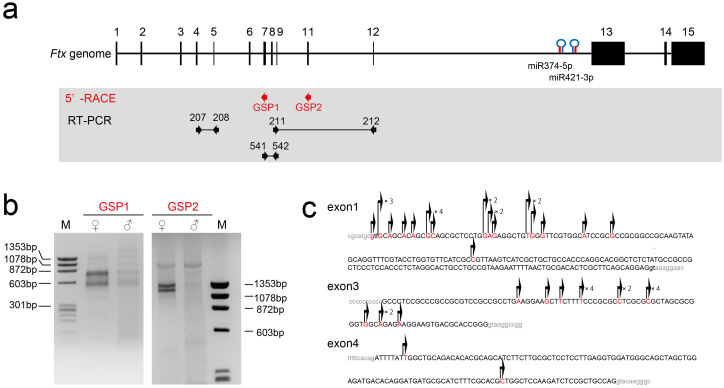
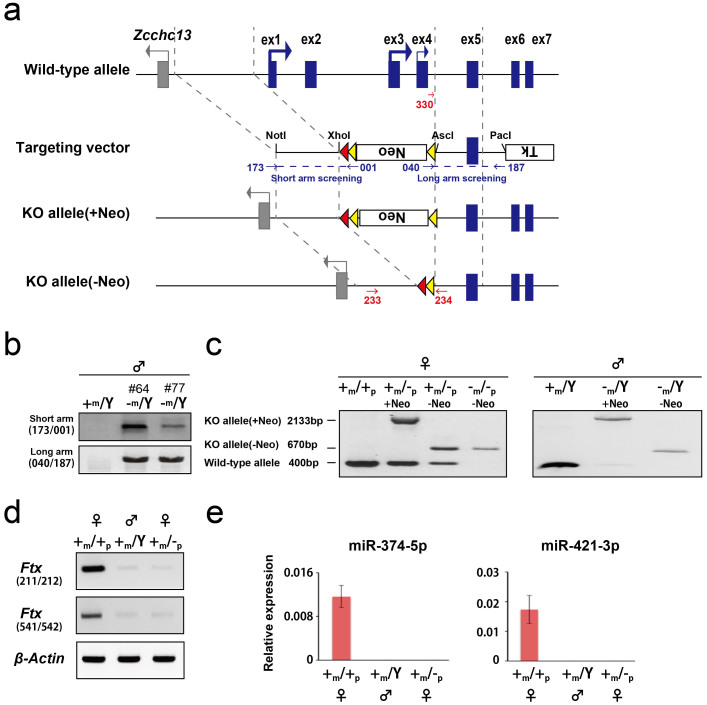
To generate Ftx-deficient mice, we determined the transcription start site of Ftx using 5′-rapid amplification of cDNA ends (RACE). We designed two gene-specific primers (GSPs) at exon 7 (GSP1) and exon 11 (GSP2) of Ftx based on information from mouse genomic sequences (UCSC mm9; http://genome.ucsc.edu/) (Figure 1a). We recently reported that Ftx is imprinted and exhibits predominant expression in female blastocysts8. Consistent with this, we detected two major bands in female blastocysts using both primers (Figure 1b), indicating that these transcription start sites were indeed imprinted. Sequencing of these two major bands revealed that the transcripts were transcribed from exons 1 and 3 (24 and 17 clones, respectively, Figure 1c). In addition to these two major start sites, a few clones were also identified from exon 4 as start sites (two clones, Figure 1c). Therefore, we disrupted Ftx by homologous recombination (Figure 2a). The targeting vector replaced exons 1–4 with a neomycin (Neo) cassette in the mutated allele, resulting in complete disruption of imprinted Ftx lncRNA expression. Correct targeting of the Ftx allele in ES cell clones by homologous recombination was confirmed by polymerase chain reaction (PCR) amplification (Figure 2b). Two independent Ftx-deficient ES cell lines, #64 and #77, were injected into blastocysts to create germline chimeras. The mutated allele derived from recombinant ES cells was transmitted to female offspring from chimeric males, and the Neo cassette was then removed by mating with CAG-FLPe transgenic mice (Figure 2c). We confirmed complete loss of Ftx expression in Ftx-deficient blastocysts by reverse transcription (RT)–PCR analysis (Figure 2d, RT-PCR primers (211/212) amplified exon 9–12, and primers (541/542) amplified exon 7–9 as shown in Figure 1a).
Figure 1. Identification of the 5′ ends of Ftx by 5′-RACE.
(a) Ftx genomic structure in the mouse genome is shown. Exons are indicated by rectangles (black). The two miRNAs, miR-374-5p and miR-421-3p, are clustered in the intron of Ftx. The locations of the gene-specific primers (GSP1 and GSP2) used for 5′-RACE analysis are shown as red arrows. The RT–PCR primer positions used to assess the Ftx expression levels are shown as black arrows. (b) Ethidium bromide-stained gel of 5′-RACE products: ♀, female blastocysts; ♂, male blastocysts; M, molecular-weight markers (PhaiX174 HaeIII digest). (c) Summary of 5′-RACE sequences. Transcriptonal start sites of each clone are indicated in red and with arrows. The numbers of clones of the same squence are shown to the right of the arrows. The starts sites tended to distribute within each exon. Reported exon sequences are indicated by capital letters.
Figure 2. Targeted disruption of the Ftx gene.
(a) Scheme for generating the Ftx knockout (KO) allele. Ftx exons are shown as blue boxes. Two major start sites and one minor one are indicated by heavy and light arrows, respectively. FRT sites, denoted as yellow triangles, flank the neomycin-resistance gene (Neo) as a positive gene selection marker, and loxP sites are shown as red triangles. Primers used for validating gene recombination in ES cells are shown as blue arrows. Primers used for genotyping are shown as red arrows. (b) Validation of recombination on the chromosomal long and short arm target sequences of ES cells (cell lines #64 and #77). (c) Genotyping results for wild-type females (+m/+p, where m is the maternally derived X chromosome and p is the paternally derived X chromosome), heterozygous females (+m/−p), homozygous females (−m/−p), wild-type males (+m/Y, where Y is the Y chromosome) and hemizygous males (−m/Y). The PCR primers (233, 234, and330) used for genotyping are depicted in Fig2-a. (d) Expression of Ftx in Ftx-deficient blastocysts. PCR primers used for RT–PCR are shown in parentheses. Uncropped images of the full-length gels are presented in Supplementary Figure S1. (e) The expression levels of miR-374-5p and miR-421-3p were quantified by q-PCR in wild-type female (+m/+p), male (+m/Y) and heterozygous female (+m/−p) blastocysts. The ratios of the number of cDNA copies for each miRNA to that of cDNA copies for miR-295 are indicated as expression levels. The bar graphs and lines show the mean ± standard deviation (SD) (n = 3).
Expression of miRNAs in Ftx-deficient blastocysts
Two miRNAs, miR-374-5p and miR-421-3p, are located within the Ftx intron in the same orientation as the Ftx transcript (Figure 1a). We confirmed that these miRNAs were not expressed in female blastocysts (+m/−p) when the Ftx-targeted allele was derived from the father, whereas these miRNAs were predominantly expressed in wild-type female blastocysts (+m/+p) (Figure 2e). This finding indicated that the Ftx-deficient mice failed to express miR-374-5p and miR-421-3p simultaneously, and suggested that both miRNAs were transcribed from the same promoter as Ftx.
Mating of Ftx-deficient mice
Paternal transmission of the Xist-deficient allele produces female-specific embryonic death because of failure of imprinted XCI in extra-embryonic cell lineages6. We examined whether paternal transmission of Ftx would affect female development, as with Xist. Mating Ftx-deficient mice (−/Y) with wild-type Mus mus domesticus (C57BL/6N) mice (+/+) generally produced offspring at the expected Mendelian ratios (Table 1, chi-square = 0.52). A similar result was observed in another mouse line from recombinant ES cell line #77 (Supplementary Table S1, chi-square = 0.52). Moreover, other crosses, namely +/− × +/Y, +/− × −/Y, and −/− × −/Y, also produced offspring at the expected Mendelian ratios (chi-square >0.1). All offspring of Ftx-deficient mice were viable and fertile which indicated that deleting Ftx had no serious effects on female viability.
Table 1. Mating of Ftx-deficient mice (line #64).
| Cross | Male pups | Female pups | Total pups | Mean litter size | |
|---|---|---|---|---|---|
| (♀ × ♂) | +/Y | +/− | +/O* | ||
| +/+ × −/Y | 40 | 45 | 1 | ||
| 40 | 46 | 86 | 7.8 | ||
*: XO female.
Expression of Xist in Ftx-deficient blastocysts
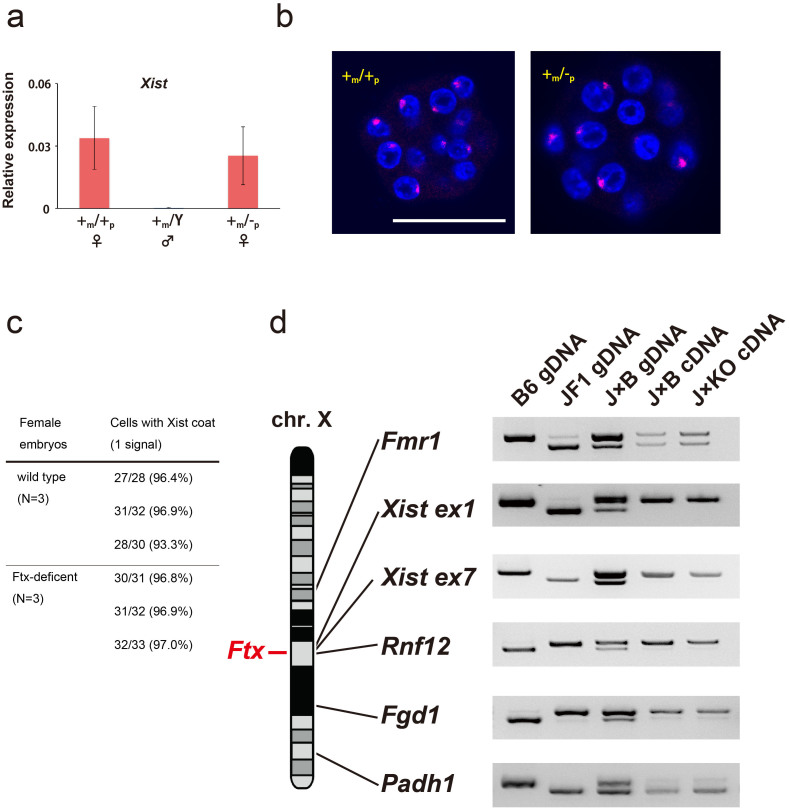
Xist was reported to be upregulated by Ftx in male ES cells13. Based on this report, we examined whether disruption of Ftx would affect Xist expression in preimplantation embryos. First, we carried out quantitative (q)-PCR, and found that the expression level of Xist in the Ftx-deficient blastocysts was not affected significantly; it was the same as in wild-type blastocysts (Figure 3a). Next, we analysed Xist expression by RNA-fluorescence in situ hybridisation (FISH) in the Ftx-deficient blastocysts. A single Xist RNA accumulation signal was detected in a Ftx-deficent as well as a wild-type blastomere at the blastocyst stage, indicating that the localisation of Xist had not been disturbed (Figure 3b). Figure 3c shows summary of Xist RNA FISH analysis. There was no significant difference of the number of the cells with Xist RNA signal between Ftx-deficient and wild-type embryos. These data indicate that deleting Ftx had no apparent effect on Xist expression at the preimplantation stage.
Figure 3. Expression of Xist in Ftx-deficient blastocysts.
(a) q-PCR analysis of Xist RNA in Ftx-deficient blastocysts. The ratios of the number of cDNA copies for Xist to that of cDNA copies for a housekeeping gene, β-actin, are indicated as expression levels. The bar graphs and lines show the mean ± standard deviation (SD) (n = 3). (b) RNA FISH analysis for Xist (red) expression in Ftx-deficient blastocysts. The nuclei of blastocysts were stained with DAPI (blue). Scale bar, 50 μm. (c) Summary of Xist RNA FISH analysis. Three wild-type (+m/+p) and Three Ftx-deficient (+m/−p) blastocysts were examined. Cells with 2 Xist signals were not observed. (d) Analysis of X-linked gene expression levels in Ftx-deficient blastocysts. No differences in the allelic expression patterns of X-linked genes between JF1 and C57BL/6N embryos were detected using RFLP. Uncropped images of the full-length gels are presented in Supplementary Figure S1.
Effects of Ftx disruption on imprinted XCI in preimplantation embryos
Our results with crossing experiments among Ftx-deficient mice and normal Xist expression in the Ftx-deficient blastocysts indicated that disruption of Ftx has no major role on imprinted XCI. However, the effect of Ftx-disruption on each X-linked gene was still uncertain. We next performed restriction enzyme fragment polymorphism (RFLP) analysis to examine the allelic expression patterns of Xist and four X-linked genes, Fmr1, Rnf12, Fgd1, and Padh1, which are all subject to imprinted XCI. We took advantage of a single nucleotide polymorphism found in each gene between C57BL/6N and the Japanese wild mouse M. m. molossinus (JF1), and examined the expression patterns of all five genes in Ftx-deficient blastocysts. The expression patterns of all genes were similar to those of wild-type embryos (Figure 3d). Taken together, the results indicate that disruption of Ftx expression produced no obvious effects on imprinted XCI during preimplantation development in mice.
Discussion
In this study, we examined whether Ftx might be involved in the mechanisms of imprinted XCI at the preimplantation stage. Although Xist deletion results in female-specific embryonic death when it is derived from the father because of the lack of XCI in extra-embryonic tissues6, disruption of Ftx did not affect survival rates or litter sizes in both sexes, demonstrating that this ncRNA is not necessary for the survival of female mice. Further studies indicated that Ftx disruption did not cause major deficiency in the expression of Xist or the expression of X-linked genes in preimplantation embryos. These results are inconsistent with the hypothesis that Ftx is a positive regulator of Xist in XCI, as proposed by Chureau et al.13. Because they analysed the expression of Xist using Ftx-deficient XY male ES cells that had one active X chromosome and lacked XCI, their results might not reflect the situation for female cells in vivo. Chureau also reported that, in addition to Xist, Ftx deletion decreased the expression levels of the adjacent imprinted genes, Jpx and, Zcchc1313. It would be of interest to examine whether these adjacent genes are also affected in female cells. We were also interested in whether Ftx is involved in the onset of the imprinted Xist upregulation during early preimplantation stages because previous studies have shown that Xist RNA upregulation is detected from the two- to four-cell stage onward14,15. We used q-PCR to examine the Ftx expression at preimplantation stages and found that the expression begins at the blastocyst stage (Figure S1). This suggests that Ftx is not involved in the onset of this Xist upregulation. As far as we examined, Ftx disruption had no apparent effects on imprinted XCI at the preimplantation stage. This indicates that Ftx is dispensable for imprinted XCI, and that the remaining genes included in the 210 kb regulatory region of imprinted XCI—Tsix and/or Jpx—might play essential roles in the regulation of imprinted Xist.
Tsix, an antisense transcript to Xist, is another lncRNA that functions as a repressor of Xist during the initiation of random XCI in post-implantation embryos9,10. It inhibits the expression of Xist on one female X chromosome to maintain the active state of the randomly chosen one. It is imprinted and expressed from maternally derived X chromosomes at the preimplantation stage10. However, whether Tsix is also required for the imprinted expression of Xist in preimplantation embryos remains unclear. Tsix expression was reported to start later than Xist expression from Xp alleles, implying that there were mechanisms other than Tsix that repress Xist expression from Xm alleles16. Further study is required to reveal the role of Tsix in imprinted XCI at the preimplantation stage.
Jpx is located just downstream of the Ftx locus. Its deletion in XX female ES cells results in failure of accumulation of Xist on the inactive X chromosome and XCI in differentiated ES cells, indicating that Jpx can upregulate Xist expression in random XCI17. However, how Jpx regulates Xist expression in vivo is not known. In the case of imprinted XCI, there are only three paternally expressed imprinted genes, Xist, Ftx, and Jpx, located in the 210 kb transgene. Apart from Ftx, Jpx is the only candidate gene that potentially upregulates Xist. Based on its proposed role in ES cells17, Studies in Jpx-targeted mice will be of interest to delineate the necessity of this imprinted ncRNA in imprinted XCI.
In this study, we focused on imprinted XCI, but not on random XCI. Because the Tsix–Zcchc13 cluster is included within the Xic region that plays a major role in random XCI, it is necessary to elucidate whether Ftx is involved in random XCI in post-implantation embryos. Further study is required to elucidate the mechanism of XCI, but the results presented in this paper will help clarify the answer.
Methods
Animals
All animal experiments were performed according to the guidelines of the Committee on the Use of Live Animals in Teaching and Research of Tokyo Medical and Dental University, and animal protocols were reviewed and approved by the animal welfare committee of the Tokyo Medical and Dental University (Permit Number: 0130228A).
Embryo collection
For blastocyst collection, 8-week-old C57BL/6N female mice (Crea Inc., Tokyo, Japan) were superovulated with an intraperitoneal (i.p.) injection of 5 IU of pregnant mare serum gonadotropin (Aska Pharmaceutical Co. Ltd., Tokyo, Japan) followed by an i.p. injection of 5 IU of human chorionic gonadotropin (hCG; Aska Pharmaceutical Co. Ltd.) 48 h later. These superovulated female mice were mated with C57BL/6N(+/Y) or Ftx-deficient mice (−/Y) and blastocysts were collected from the uterus on 3.5 dpc from around 92–100 h post-hCG.
5′-RACE
Total RNA was prepared from male and female wild-type blastocysts using TRIzol (Invitrogen, Carlsbad, CA, USA). 5′-RACE was performed on RNA prepared from blastocysts with GSP1 and GSP2 (Supplementary Table S2) using SMARTer RACE cDNA Amplification kits (Clontech, Mountain View, CA, USA). To separate male and female blastocysts, we used a non-invasive sexing method by tagging X chromosomes with an enhanced green fluorescent protein (EGFP) transgene18,19,20. With this method, the female (XXEGFP) blastocysts can be separated from the non-fluorescent male (XY) blastocysts by mating XEGFPY male mice to wild-type female (XX) mice. The RACE products were subsequently cloned into pCR4-TOPO (Invitrogen) for sequencing. Sequencing was performed using BigDye Terminator v3 Cycle Sequencing kits (ABI, Life Technologies, Carlsbad, CA, USA).
Construction of the Ftx-targeting vector
The targeting vector was designed to replace exons 1–4 of Ftx with a pNT1.1 Neo cassette. A 5.3 kb NotI–XhoI fragment as a short chromosomal arm and a 5.0 kb AscI–PacI fragment as a long arm were obtained by PCR using genomic DNA of a bacterial artificial chromosome clone (RP23, #239G16) as a template. Primers used for constructing the Ftx targeting vector are listed in Supplementary Table S2.
Generation of Ftx-deficient mice
The targeting vector was linearized with NotI digestion and electroporated into ES cells (EGR-G101) derived from C57BL/6N mice (Crea, Inc.). ES cell clones that were candidates for possessing the targeted allele were isolated by positive selection with G418. Correct targeting of the Ftx allele in ES cell clones by homologous recombination was confirmed by PCR. Positive ES cell clones were subsequently used to generate chimeras by injection into 8-cell stage ICR strain embryos. Injected embryos were transferred to ICR strain pseudopregnant foster mothers, resulting in the birth of chimeric mice. Male chimeric mice were crossed with C57BL/6N female mice to obtain F1 heterozygous female offspring, and then crossed with CAG-FLPe transgenic mice (B6.Cg-Tg (CAG-FLPe)) to delete the Neo cassette. The Ftx-deficient mice used in the present study were from a C57BL/6N background. The primers used for confirming homologous recombination are listed in Supplementary Table S2.
q-PCR analysis
Total RNA of embryos was extracted using TRIzol (Invitrogen) and contaminating DNA was digested using RNase-free DNase (Nippon Gene, Tokyo, Japan). First-strand cDNA was synthesized using reverse transcriptase SuperScript III (Invitrogen). For miRNA expression analysis, total RNA was reverse transcribed using the TaqMan MicroRNA Reverse Transcription Kit (ABI). The TaqMan MicroRNA assays used (with identification numbers in parentheses), were as follows: mmu-miR-374-5p (001319), mmu-miR-421-3p (002700), and mmu-miR-295 (000189). Mature miRNAs were amplified with TaqMan Universal PCR Master Mix II (ABI) using a LightCycler 480 Instrument (Roche, Basel, Switzerland). The q-PCR reactions were performed using samples collected from three independent preparations.
FISH
Xist-specific probes were prepared by nick translation with Cy3-dCTP (GE Healthcare, Little Chalfont, UK) from pXist 1986–9498. Blastocysts were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 10 min on ice and incubated in 0.1% Triton X-100 in PBS for 10 min on ice. Hybridisation was carried out at 42 °C overnight. The nuclei of embryos were stained with DAPI. Xist signals were observed using a confocal scanning laser microscope (LSM 510; Carl Zeiss, Jena, Germany).
Polymorphic analysis
(J × B) or (J × KO) F1 embryos were obtained from mating 8- to 12-week-old wild-type JF1 (J) female mice with wild-type C57BL/6N (B) or Ftx-deficient male mice (KO). Polymorphisms of X-linked genes between JF1 and C57BL/6N were detected by RFLP analysis. Primers and restriction enzymes used for polymorphic analysis are summarized in Supplementary Table S2.
Supplementary Material
Supplementary infomation
Acknowledgments
We thank Dr Kuniya Abe at RIKEN BioResource Center for providing the pXist 1986–9498 plasmid, Y. Esaki, S. Nishioka, Y. Koreeda and K. Kawata for technical assistance in generating Ftx-deficient mouse lines, Y. Hosoi for assistance in FISH analysis, Dr Takashi Kohda, and Dr Daisuke Endo for critical reading and discussion. This work was supported by the Japan Science and Technology Agency, Precursory Research for Embryonic Science and Technology (PRESTO) and by a Grant- in-Aid for Scientific Research from The Ministry of Education, Culture, Sports, Science and Technology and the Sumitomo Foundation to S.K. The funders had no roles in the study design, data collection and analysis, decision to publish or preparation of the manuscript.
Footnotes
The authors declare no competing financial interests.
Author Contributions M.S. and S.K. performed most experiments, assisted by Y.F., M.O. and I.F.; S.K., Y.F. and M.O. generated mutant mice. M.S. and S.K. analysed the data. M.S. and S.K. wrote the manuscript and all authors discussed the results and commented on the manuscript.
References
- Huynh K. D. & Lee J. T. Inheritance of a pre-inactivated paternal X chromosome in early mouse embryos. Nature 426, 857–862, 10.1038/nature02222 (2003). [DOI] [PubMed] [Google Scholar]
- Mak W. et al. Reactivation of the paternal X chromosome in early mouse embryos. Science 303, 666–669, 10.1126/science.1092674 (2004). [DOI] [PubMed] [Google Scholar]
- Okamoto I., Otte A. P., Allis C. D., Reinberg D. & Heard E. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science 303, 644–649, 10.1126/science.1092727 (2004). [DOI] [PubMed] [Google Scholar]
- Rastan S. Non-random X-chromosome inactivation in mouse X-autosome translocation embryos--location of the inactivation centre. J. Embryol. Exp. Morphol. 78, 1–22 (1983). [PubMed] [Google Scholar]
- Rastan S. & Robertson E. J. X-chromosome deletions in embryo-derived (EK) cell lines associated with lack of X-chromosome inactivation. J. Embryol. Exp. Morphol. 90, 379–388 (1985). [PubMed] [Google Scholar]
- Marahrens Y., Panning B., Dausman J., Strauss W. & Jaenisch R. Xist- deficient mice are defective in dosage compensation but not spermatogenesis. Genes Devel. 11, 156–166 (1997). [DOI] [PubMed] [Google Scholar]
- Namekawa S. H., Payer B., Huynh K. D., Jaenisch R. & Lee J. T. Two-step imprinted X inactivation: repeat versus genic silencing in the mouse. Mol. Cell. Biol. 30, 3187–3205, 10.1128/MCB.00227-10 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S. et al. Identification of an imprinted gene cluster in the X-inactivation center. PloS One 8, e71222, 10.1371/journal.pone.0071222 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. T. Disruption of imprinted X inactivation by parent-of-origin effects at Tsix. Cell 103, 17–27 (2000). [DOI] [PubMed] [Google Scholar]
- Sado T., Wang Z., Sasaki H. & Li E. Regulation of imprinted X-chromosome inactivation in mice by Tsix. Development 128, 1275–1286 (2001). [DOI] [PubMed] [Google Scholar]
- Okamoto I. et al. Evidence for de novo imprinted X-chromosome inactivation independent of meiotic inactivation in mice. Nature 438, 369–373, 10.1038/nature04155 (2005). [DOI] [PubMed] [Google Scholar]
- Schulz E. G. & Heard E. Role and control of X chromosome dosage in mammalian development. Curr. Opin. Genet. Devel. 23, 109–115, 10.1016/j.gde.2013.01.008 (2013). [DOI] [PubMed] [Google Scholar]
- Chureau C. et al. Ftx is a non-coding RNA which affects Xist expression and chromatin structure within the X-inactivation center region. Hum. Mol. Genet. 20, 705–718, 10.1093/hmg/ddq516 (2011). [DOI] [PubMed] [Google Scholar]
- Kay G. F., Barton S. C., Surani M. A. & Rastan S. Imprinting and X chromosome counting mechanisms determine Xist expression in early mouse development. Cell 77, 639–650 (1994). [DOI] [PubMed] [Google Scholar]
- Zuccotti M. et al. Mouse Xist expression begins at zygotic genome activation and is timed by a zygotic clock. Mol. Reprod. Dev. 61, 14–20, 10.1002/mrd.1126 (2002). [DOI] [PubMed] [Google Scholar]
- Sado T. & Sakaguchi T. Species-specific differences in X chromosome inactivation in mammals. Reproduction 146, R131–139, 10.1530/REP-13-0173 (2013). [DOI] [PubMed] [Google Scholar]
- Tian D., Sun S. & Lee J. T. The long noncoding RNA, Jpx, is a molecular switch for X chromosome inactivation. Cell 143, 390–403, 10.1016/j.cell.2010.09.049 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi T. et al. FISH analysis of 142 EGFP transgene integration sites into the mouse genome. Genomics 80, 564–574 (2002). [DOI] [PubMed] [Google Scholar]
- Kobayashi S. et al. Comparison of gene expression in male and female mouse blastocysts revealed imprinting of the X-linked gene, Rhox5/Pem, at preimplantation stages. Curr. Biol. 16, 166–172, 10.1016/j.cub.2005.11.071 (2006). [DOI] [PubMed] [Google Scholar]
- Kobayashi S. et al. The X-linked imprinted gene family Fthl17 shows predominantly female expression following the two-cell stage in mouse embryos. Nucl. Acids Res. 38, 3672–3681, 10.1093/nar/gkq113 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary infomation