Abstract
We explored the feasibility of studying loss of heterozygosity (LOH) by using exome sequencing and compared the differences in genetic LOH between primary breast tumors and metastatic lesions. Exome sequencing was conducted to investigate the genetic LOH in the peripheral blood, a primary tumor, and a metastatic lesion from the same patient. LOH was observed in 30 and 48 chromosomal loci of the primary tumor and metastatic lesion, respectively. The incidence of LOH was the highest on chromosome 19, followed by chromosomes 14, 3, and 11 in the metastatic lesion. Among these ‘hot' regions, LOH was observed for multiple genes of the CECAM, MMP and ZNF families. Therefore, the use of exome sequencing for studying LOH is feasible. More members of gene families appeared with LOH in ‘hot' regions, suggesting that these gene families had synergistic effects in tumorigenesis.
Breast cancer metastasis is an intricate process involving the interplay of multiple gene products. Tumor progression is the major cause of breast cancer mortality. Loss of heterozygosity (LOH) was first proposed to cause cancer by Cavenee1 in a hereditary retinoblastoma study. He deemed that the fertilized egg was Rb/rb in heterozygous individuals. Genetic damage to the somatic cell would result in Rb gene mutation or deletion, and the subsequent formation of rb/rb recessive homozygotes might lead to tumor formation. This scenario raised the concept of genetic LOH. The mechanisms of LOH include mitotic nondisjunction, chromosome loss, homologous mitotic recombination, and recombination (translocation) between two non-homologous chromosomes2,3. Chromosome loss is a major mechanism of LOH that results in an abnormal number of chromosomes; thus, aneuploidy develops. Genetic instability is associated with carcinogenesis and poor prognosis. LOH in a high-frequency region usually includes one or more tumor suppressor genes.
Currently, there are several methods of searching for genes that cause disease or for susceptibility genes, including genome-wide association study (GWAS), exome sequencing, and whole-genome sequencing4. Although using Gene Chips has proven to be effective, they are insensitive to rare variants (MAF < 5%) and other structural variations. An increasing number of studies have shown that many complex diseases originate from such rare mutations5,6. In theory, whole-genome sequencing can identify all human genome sequence variations7; however, the high cost is prohibitive for this technique to be widely used. Exome sequencing is the use of sequencing technology to selectively capture the exon coding regions of the whole genome and to enrich for high-throughput genomic sequencing analysis. To a certain extent, the technique overcomes the limitations of genotyping chips and traditional sequencing technology with substantially lower costs, typically approximately 1/50 of the cost for whole genome sequencing, a shorter processing time, and more accurate data. More importantly, this technique can produce a more detailed analysis of the genetic variation in the coding region; therefore, it has become an important tool in disease-related genetic research8,9.
Despite rapid research and development of the exome sequencing technique, the use of this technology for the study of LOH has rarely been reported. To explore the feasibility of using this technology to study LOH, differences in genetic LOH between primary and metastatic lesions were compared. We conducted a genome-scale study by investigating genetic LOH in the peripheral blood, a primary tumor, and a distant metastatic lesion from the same patient. Here, we present the preliminary results.
Results
Basic statistics
Exome sequencing of a total of three sets of samples – a peripheral blood sample, a primary tumor sample, and a metastatic lesion sample – was performed. After the removal of low-quality sequence reads, the alignment of the remaining high-quality sequence reads with the human reference genome was performed using Bowtie 2 software. The average coverage in the target region of the different samples was 56–62× with an average of 60×. The sequencing depth is displayed in Table 1.
Table 1. Sequencing reads of samples.
| Blood sample | Primary tumor sample | Metastatic lesion sample | |
|---|---|---|---|
| Bases | 3,076,304,580 | 2,585,696,220 | 2,800,547,460 |
| Bases aligned to hg19 | 3,035,541,354 | 2,562,943,093 | 2,554,099,283 |
| Bases in target region aligned to hg19 | 2,502,399,805 | 2,200,607,212 | 2,354,921,851 |
| Average depth | 62.88 | 56.86 | 60.87 |
| Target region coverage (%) | 90.45 | 87.95 | 87.92 |
| Depth ≥ 4 regions coverage (%) | 78.06 | 76.71 | 76.91 |
Single nucleotide variants identification
Our analytical pipeline identified 66,635 single nucleotide variants with a minimum coverage of 8× in each sample (Table S1). All further LOH analyses were based on this data file. The identification of somatic mutations in the tumor sample was performed subsequently, and three non-silent variants were identified, affecting three distinct genes: CUL7 and RLF (in both the primary tumor and the metastatic lesion) and KIAA2026 (in the metastatic lesion only; Table S2).
LOH distribution
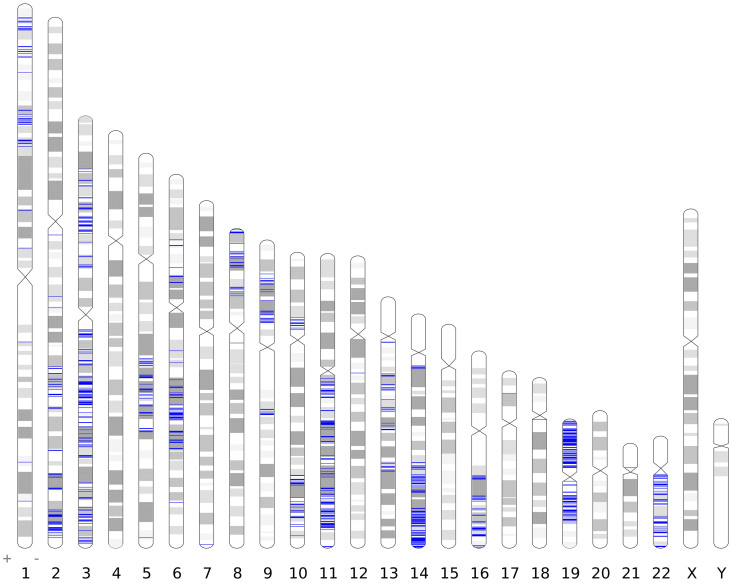
For LOH characterization, we calculated a heterozygosity score (HS) for each SNV locus in each sample using the following formula: HS = 1 − absolute(Ca/(Ca + Cb) − 0.5)/0.5, where Ca and Cb represent the number of reads of allele a and allele b, respectively. Then, a genome-wide scan of tumors based on the HS was performed. To ensure fidelity, an LOH region was identified only when the average HS of at least three consecutive loci was less than 0.1, and the probability of observing such a pattern by chance was smaller than 0.0001 (one-way-ANOVA). Based on genome-wide scanning, LOH in the primary tumor was found at 30 chromosomal loci involving 54 genes (Table S3), some of which may be associated with tumorigenesis (Table 2). In the metastatic lesion, LOH occurred at 48 chromosomal loci (Fig. 1), affecting 1157 genes distributed on 19 chromosomes (Table S4).
Table 2. Selected LOH regions in the primary tumor and genes involved in tumorigenesis.
| Gene | Full name | Position |
|---|---|---|
| ZNF148 | zinc finger protein 148 | 3q21.2 |
| EIF4G1 | eukaryotic translation initiation factor 4 gamma, 1 | 3q27.1 |
| RICTOR | RPTOR independent companion of MTOR, complex 2 | 5p13.1 |
| ELL2 | elongation factor, RNA polymerase II, 2 | 5q15 |
| ARHGEF10 | Rho guanine nucleotide exchange factor (GEF) 10 | 8p23.3 |
| RP1L1 | retinitis pigmentosa 1-like 1 | 8p23.1 |
| VPS13A | vacuolar protein sorting 13 homolog A | 9q21.2 |
| GAL | galanin/GMAP prepropeptide | 11q13.3 |
Figure 1. LOH across the genome in a metastatic lesion.
An LOH region was identified when the average heterozygosity score (HS) of at least three consecutive SNVs was less than 0.1. The blue lines show the LOH regions.
Metastasis
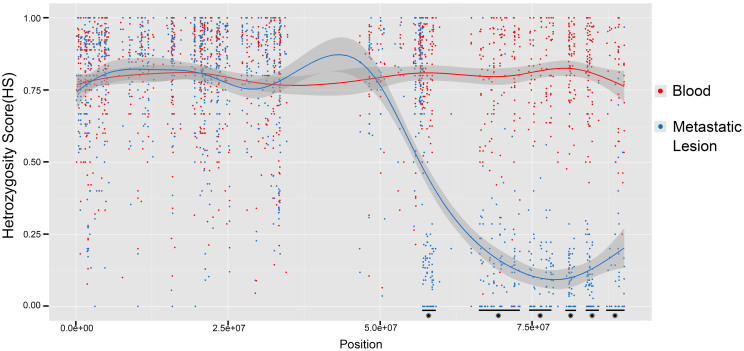
All LOH regions detected in the metastatic lesion are listed in Table 3, and a comparison shows that ~40% of these regions overlapped with those found in the primary tumor; the remaining ~60% were novel. Some regions observed in this study were consistent with previously reported regions: 3p21.3; 13q12.11; 13q13.2--13q13.3; and 16q22-2310,11,12. Our study found that the incidence of LOH in these regions were 69%, 20–40%, and 36–67%, respectively. Additionally, certain hot regions, such as 11q22.2 and 19q13.2, were observed. The heterozygosity score (HS) profile of chromosome 16 is displayed in Fig. 2, and the genes involved in the above-mentioned regions are shown in Table 4.
Table 3. Comparison of genome-wide LOH regions in two tumor sets.
| Loss of heterozygosity region in the metastatic lesion | Compared with the primary tumor |
|---|---|
| 1p36.31--1p36.22, 1p35.2, 1p33, 1p32.3, 1p31.3, 1p22.1, 1p13.2, 1q42.132q11.2, 2q14.3, 2q21.2--2q31.2, 2q33.1--2q37.33p24.2--3p21.1, 3q11.2--3q12.3,5q21.1--5q35.2,6q14.1--6q14.3, 6q16.1--6q22.31, 6q25.1,7q36.3,9p22.3--9p13.2,10p11.23--10p11.21,12q13.13,13q12.11, 13q13.2--13q13.3, 13q14.11--13q14.3, 13q21.1--13q31.1,17p13.1,18q11.2, | Novel |
| 1p36.13--1p36.11, 1q21.3, 1q32.23p14.3--3p14.1, 3q13.12--3q13.33, 3q21.1--3q295q156p22.1--6p12.38p23.3--8p129q21.13--9q21.210q24.31--10q26.211q12.1--11q2514q11.2--14q12, 14q24.1--14q32.3316q13--16q22.1, 16q22--16q2319p13.3--19q13.3221p11.122q11.1--22q13.31, 22q13.33 | Selected from the primary tumor |
Figure 2. The heterozygosity score (HS) profile of chromosome 16, comparing the metastatic lesion to the blood sample.
There are six “hot” LOH regions, which are marked by stars (P < 2.2e-16; one-way ANOVA test).
Table 4. Selected LOH regions in the metastatic lesion.
| Loss of heterozygosity region | Gene | Full name |
|---|---|---|
| 3p21.3 | RBM5 | RNA binding motif protein 5 |
| SEMA3F | semaphorin 3F | |
| PRSS45 | protease, serine, 45 | |
| PLXNB1 | plexin B1 | |
| FBXW12 | F-box and WD repeat domain containing 12 | |
| ALS2CL | ALS2 C-terminal like | |
| 13q12.11, 13q13.2--13q13.3 | NBEA | neurobeachin |
| ELF1 | E74-like factor 1 | |
| 16q22-23 | CNOT1 | CCR4-NOT transcription complex, subunit 1 |
| CDH3 | cadherin 3, type 1, P-cadherin | |
| PLEKHG4 | pleckstrin homology domain containing, family G (with RhoGef domain) member 4 | |
| EDC4 | enhancer of mRNA decapping 4 | |
| DPEP3 | dipeptidase 3 | |
| ACD | adrenocortical dysplasia homolog | |
| EXOC3L1 | exocyst complex component 3-like 1 | |
| PMFBP1 | polyamine modulated factor 1 binding protein 1 | |
| 19q13.2 | CEACAM family | Carcinoembryonic antigen-related cell adhesion molecule family |
| ZNF family | zinc finger protein family | |
| 11q22.2 | MMP family | matrix metalloproteinase family |
Discussion
Recently, SNP chip technology has generally been used for LOH-related studies of particular regions of interest, but this technology is prone to false positives and negatives and therefore has certain limitations. Today, the cost of exome sequencing is lower than in which time period, and the processing time is shorter. Moreover, exome sequencing can identify genetic variation at the base-pair level and is regarded as an important tool for disease-related genetic research8,9. We used exome sequencing technology in a genome-scale search to examine genetic LOH in the peripheral blood, a primary tumor, and a distant metastatic lesion in the same patient. The results of the study illustrate the credibility of exome sequencing.
The current study indicated that breast cancer metastasis is a highly selective process that involves a complex interaction between the tumor and the host13. Metastasis includes cancer cell separation from the primary tumor, tumor-driven matrix invasion, microvascular invasion and metastasis, and colonization and growth at another organ site. The accumulation of genetic alterations increases genetic instability, and this process involves a variety of genes, such as oncogenes, tumor or metastasis suppressor genes, and genes involved in DNA repair functions14. In this study, a higher incidence of LOH was observed in the metastatic tumor than in the primary tumor. The LOH involved cell differentiation, proliferation, adhesion, and motility genes, among which a considerable number were tumor or metastasis suppressor genes.
In the present study, we found that LOH was observed in certain gene families, suggesting that there may be synergistic effects of the family during metastasis, e.g., the carcinoembryonic antigen-related cell adhesion molecule (CEACAM) family, which is located at the 19q13.1-19q13.2 region. The CEACAM family belongs to the immunoglobulin superfamily, which is important for cell adhesion and signal transduction15. It has been reported that CEACAM1 expression levels are significantly lower in breast cancer tissue than in normal breast tissue16. CEACAM1 and other tumor suppressor genes might be jointly involved in apoptosis17. Additionally, it has been reported that CEACAM6 is associated with breast cancer metastasis and recurrence18; however, the relationship to breast cancer has not been observed in other family members. In our study, LOH was observed in CEACAM1, CEACAM3, CEACAM5, CEACAM6,CEACAM7, and CEACAM8, indicating that the structure and function of this family might be closely linked, synergistically regulating carcinogenesis and metastasis.
We also found that the LOH incidence in a zinc finger gene family located at 19q13.1-19q13.3 was the highest, with 40 members of this gene family showing LOH. The gene products of these family members bind to DNA, RNA, double-stranded DNA-RNA hybrid molecules and other zinc finger proteins, and they homodimerize19 to regulate gene expression at the transcriptional and translational level. Zinc finger family members play important roles in gene expression regulation, cell differentiation, and embryonic development and are associated with many diseases. For example, ZNF303 is associated with the occurrence and metastasis of hepatocellular carcinoma, and ZNF202 is associated with leukemia. Additionally, some studies have found that many zinc finger proteins inhibit the transcriptional activity of AP-1 and SRE genes and control the MAPK signaling pathway. Several zinc finger genes, such as ZNF383, ZNF480, ZNF411, ZNF466, and ZNF649, play important roles in tumorigenesis. The LOH of zinc finger genes might cause a loss of gene function, leading to the activation of AP-1 and MMPs. Such activation would, in turn, lead to the degradation of extracellular matrix proteins and, subsequently, to breast cancer recurrence and metastasis19,20,21.
Three members of the matrix metalloproteinase (MMP) family, located at 11q22.3, showed LOH: MMP1; MMP8; and MMP10. These gene products of this family can degrade extracellular matrix proteins, leading to tumor angiogenesis and metastasis. According to a study of breast cancer progression, MMP over-expression degrades the basement membrane and extracellular matrix, leading to angiogenesis and ultimately breast cancer metastasis22.
Similarly to previously reported findings,we found that metastasis-related LOH regions, such as the 3p21.3 locus containing three genes, RBM5, SEMA3F, and ALS2CL, showed a corresponding tumor inhibitory effect. The RBM5 gene not only is associated with tumor formation but also is important for tumor proliferation, differentiation, and apoptosis. RBM5 is a negative regulator of tumor cell proliferation. Its overexpression can induce apoptosis and G1-phase cell cycle arrest and enhance p53-mediated tumor suppression.
Genes located at the 13q13.2-13q14.3 region include NBEA and ELF1. The downregulation of the NBEA gene has been observed in certain multiple myeloma studies; therefore, the NBEA gene may be a novel tumor suppressor gene23. It was also found that the gene may be associated with breast cancer24. ELF1 expression was found to be associated with breast cancer stage, histological grade, and lymph node metastasis and is regarded as an independent factor for invasiveness and prognosis in breast cancer. Although the gene has been considered to be involved in the mechanism of tumor metastasis, more evidence has shown that breast cancer invasion and metastasis are mediated through MMP2 gene activation.
In our study, we discovered that the CDH3 gene, located at chromosome 16q22-23, might be associated with tumor metastasis. The CDH3 gene, also named P-cadherin, and E-cadherin belong to the calcium-dependent cell adhesion factors; however, there are differences in function and tissue expression layers. It has been found that P-cadherin is more commonly detected in the basal layer of stratified epithelia and on the myoepithelial cells of the adult mammary gland25. Some studies have found that P-cadherin expression is associated with tumor invasiveness and that the stimulation of tumor cell growth by paracrine growth factors is associated with MMP upregulation. P-cadherin is strongly associated with the invasiveness and metastasis of malignant tumors25. In addition, Madhavan et al.26 found that the expression of the P-cadherin gene is negatively associated with lymph node metastasis, suggesting that this gene is essential for maintaining the structural integrity and stability of the breast cancer tissue, as well as for preventing the shedding of tumor cells and metastasis.
Because this was an exploratory, preliminary study, our initial results only indicate the feasibility of exome sequencing for studying LOH. Additional samples should be analyzed in future studies to increase the reliability of these results.
Methods
Subject characteristics
In this study, the breast cancer patient had a pathological diagnosis of a luminal A type invasive ductal carcinoma that was ER and PR positive (50% and 20%, respectively), HER2 negative (8%), and Ki-67 positive at a low level (8%) after treatment with a modified radical mastectomy before any distant metastases were observed. Post-operative CAF chemotherapy was administered for a total of 6 cycles. One year after the adjuvant chemotherapy, skin and liver metastases were observed. A skin biopsy was performed to confirm the metastasis. The patient's blood, primary tumor sample, and skin metastatic lesion were obtained for analysis.
Sample collection, handling and storage
At the time of the first surgery, 10 mL of peripheral blood from the subject was collected in an EDTA blood tube and stored at −80°C. The surgical tumor samples, once resected, were immediately snap-frozen in liquid nitrogen and stored at −80°C. Tumor specimens were assessed and confirmed for the presence of over 75% of cancer cells in the sample by a pathologist. All samples were collected with the consent of the patient and her family, and the study was approved by the hospital's ethics committee.
DNA extraction procedure
DNA was extracted from peripheral blood samples and tumor specimens using the QIAGEN Genomic-Tip in accordance with the user manual.
DNA quality, purity and concentration measurements
The OD260/OD280 values of the extracted samples were measured using a UV spectrophotometer. A value of approximately 1.8 indicates DNA of high purity; a value greater than 1.9 suggests contaminating RNA; and a value less than 1.6 indicates the presence of proteins, phenol, and other contaminants. The qualifying DNA samples were sent to Shenzhen BGI for exome sequencing.
Exome sequencing procedures and data analysis
First, genomic DNA was randomly cleaved into a fragment library, which was purified and subsequently enriched by NimbleGen 2.1M-probe sequence capture array. The enriched library targeting the exome was sequenced on the HiSeq 2000 platform to acquire paired-end reads with a read length of 90 base pairs. After removing reads containing sequencing adapters and low-quality reads with more than five unknown bases, high-quality reads were aligned with the human genome reference sequence (hg19/GRCh37) using Bowtie2 software27 with default parameters. The PCR duplicates detected from Alignment files were subsequently removed with Picard (http://picard.sourceforge.net/) to improve alignment accuracy. The Genome Analysis Toolkit (GATK)28 was then employed for base quality recalibration, local realignment around the potential insertion/deletion (Indel) sites and variant calling. The raw single nucleotide variants were filtered for low mapping quality, low coverage, snp clusters, etc. All loci with more than three alleles were filtered. Then, the filtered variants were annotated using ANNOVAR29 for the following parameters: function (exonic or splicing); gene; exonic function (synonymous, nonsynonymous, stop gain, non-frameshift or frameshift indels); amino acid change; conservation; dbSNP (version 135) reference number; allele frequency in 1000 Genomes Project (2012 February version).
LOH characterization
The blood sample was used as a control, and SNVs with a homozygous genotype in the control were removed. All remaining SNVs with a minimum coverage of 8× in each sample were selected, including the common SNPs reported in dbSNP and 1000 Genomes Project. We calculated a heterozygosity score (HS) for each SNV locus in each sample using the following formula: HS = 1 − absolute(Ca/(Ca + Cb) − 0.5)/0.5, where Ca and Cb stand for the number of reads of allele a and allele b, respectively. Then, a genome-wide scan of tumors based on the HS was performed. To ensure fidelity, a LOH region was identified only when the average HS of at least three consecutive loci was less than 0.1, and the probability (P) of observing such a pattern by chance was smaller than 0.0001 (one-way-ANOVA). Finally, all LOH regions were double-checked manually and annotated using ANNOVAR. The LOH distribution ideogram was generated by Idiographica30.
Supplementary Material
Table S1
Table S2
Table S3
Table S4
Acknowledgments
We want to thank Dr. Xionglei He of School of Life Sciences, Sun Yat-Sen University for the advices on the study design, and Dr. Tao Luo for the advices on DNA testing. This work was supported by the Science and Technology Planning Project of Guangdong Province, China (Project No.2010B031600147 and No.2012B010900044), Guangzhou Municipal University Research Project (Project no. 10A191), and the Major Special Project of Guangzhou Medical University Cancer Institute and Hospital (Project No.2011-yz-11). DNA extraction, detection, and analysis were performed in the lab of Dr. He of the School of Life Sciences, Sun Yat-sen University. Exome sequencing was performed by BGI Shenzhen.
Footnotes
The authors declare no competing financial interests.
Author Contributions H.L. conceived the idea and designed the experiment. B.W., W.H. and J.Z. collected experimental specimen, B.Y. and B.W. performed the experiments, K.X. analyzed the data and prepared figures and tables. Z.C. and N.Y. prepared the manuscript. All authors reviewed the manuscript.
References
- Cavenee W. K. et al. Expression of recessive alleles by chromosomal mechanisms in retinoblastoma. Nature 305, 779–84 (1983). [DOI] [PubMed] [Google Scholar]
- Thiagalingam S. et al. Mechanisms underlying losses of heterozygosity in human colorectal cancers. Proc. Natl. Acad. Sci. U. S. A. 98, 2698–702 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop A. J. & Schiestl R. H. Homologous recombination as a mechanism of carcinogenesis. Biochim. Biophys. Acta 1471, M109–21 (2001). [DOI] [PubMed] [Google Scholar]
- Kahvejian A., Quackenbush J. & Thompson J. F. What would you do if you could sequence everything? Nat. Biotechnol. 26, 1125–33 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClellan J. M., Susser E. & King M. C. Schizophrenia: a common disease caused by multiple rare alleles. Br. J. Psychiatry 190, 194–9 (2007). [DOI] [PubMed] [Google Scholar]
- McClellan J. & King M. C. Genetic heterogeneity in human disease. Cell 141, 210–7 (2010). [DOI] [PubMed] [Google Scholar]
- Lalonde E. et al. Unexpected allelic heterogeneity and spectrum of mutations in Fowler syndrome revealed by next-generation exome sequencing. Hum. Mutat. 31, 918–23 (2010). [DOI] [PubMed] [Google Scholar]
- Zaghloul N. A. & Katsanis N. Functional modules, mutational load and human genetic disease. Trends Genet. 26, 168–76 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. C. et al. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science 305, 869–72 (2004). [DOI] [PubMed] [Google Scholar]
- Maitra A. et al. High-resolution chromosome 3p allelotyping of breast carcinomas and precursor lesions demonstrates frequent loss of heterozygosity and a discontinuous pattern of allele loss. Am. J. Pathol. 159, 119–30 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins N. et al. Consistent loss of the wild type allele in breast cancers from a family linked to the BRCA2 gene on chromosome 13q12–13. Oncogene 10, 1673–5 (1995). [PubMed] [Google Scholar]
- Cleton-Jansen A. M. et al. Loss of heterozygosity mapping at chromosome arm 16q in 712 breast tumors reveals factors that influence delineation of candidate regions. Cancer Res. 61, 1171–7 (2001). [PubMed] [Google Scholar]
- Fidler I. J. Cancer metastasis. Br. Med. Bull. 47, 157–77 (1991). [DOI] [PubMed] [Google Scholar]
- Hampl M. et al. Loss of heterozygosity accumulation in primary breast carcinomas and additionally in corresponding distant metastases is associated with poor outcome. Clin. Cancer Res. 5, 1417–25 (1999). [PubMed] [Google Scholar]
- Czepczynska-Krezel H. & Krop-Watorek A. [Human carcinoembryonic antigen family proteins, structure and function]. Postepy Hig Med Dosw (Online) 66, 521–33 (2012). [DOI] [PubMed] [Google Scholar]
- Wang J. L. et al. Clinicopathological significance of CEACAM1 gene expression in breast cancer. Chin. J. Physiol. 54, 332–8 (2011). [PubMed] [Google Scholar]
- Bamberger A. M. et al. Expression of the adhesion molecule CEACAM1 (CD66a, BGP, C-CAM) in breast cancer is associated with the expression of the tumor-suppressor genes Rb, Rb2, and p27. Virchows Arch. 440, 139–44 (2002). [DOI] [PubMed] [Google Scholar]
- Ihnen M. et al. Protein expression analysis of ALCAM and CEACAM6 in breast cancer metastases reveals significantly increased ALCAM expression in metastases of the skin. J. Clin. Pathol. 64, 146–52 (2011). [DOI] [PubMed] [Google Scholar]
- Laity J. H., Lee B. M. & Wright P. E. Zinc finger proteins: new insights into structural and functional diversity. Curr. Opin. Struct. Biol. 11, 39–46 (2001). [DOI] [PubMed] [Google Scholar]
- Guruvayoorappan C. & Kuttan G. Amentoflavone inhibits experimental tumor metastasis through a regulatory mechanism involving MMP-2, MMP-9, prolyl hydroxylase, lysyl oxidase, VEGF, ERK-1, ERK-2, STAT-1, NM23 and cytokines in lung tissues of C57BL/6 mice. Immunopharmacol. Immunotoxicol. 30, 711–27 (2008). [DOI] [PubMed] [Google Scholar]
- El-Dahr S. S., Dipp S. & Baricos W. H. Bradykinin stimulates the ERK --> Elk-1 --> Fos/AP-1 pathway in mesangial cells. Am. J. Physiol. 275, F343–52 (1998). [DOI] [PubMed] [Google Scholar]
- Vizoso F. J. et al. Study of matrix metalloproteinases and their inhibitors in breast cancer. Br. J. Cancer 96, 903–11 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neal J. et al. Neurobeachin (NBEA) is a target of recurrent interstitial deletions at 13q13 in patients with MGUS and multiple myeloma. Exp. Hematol. 37, 234–44 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T. C. et al. Up-regulation of the cAMP/PKA pathway inhibits proliferation, induces differentiation, and leads to apoptosis in malignant gliomas. Lab. Invest. 78, 165–74 (1998). [PubMed] [Google Scholar]
- Peralta Soler A. et al. P-cadherin expression in breast carcinoma indicates poor survival. Cancer 86, 1263–72 (1999). [DOI] [PubMed] [Google Scholar]
- Madhavan M. et al. Cadherins as predictive markers of nodal metastasis in breast cancer. Mod. Pathol. 14, 423–7 (2001). [DOI] [PubMed] [Google Scholar]
- Langmead B. & Salzberg S. L. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K. et al. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38, e164 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kin T. & Ono Y. Idiographica: a general-purpose web application to build idiograms on-demand for human, mouse and rat. Bioinformatics 23, 2945–2946 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1
Table S2
Table S3
Table S4