Abstract
The yellow dwarf disease associated with phytoplasmas is one of the most devastating diseases of mulberry and the pathogenesis involved in the disease is poorly understood. To analyze the molecular mechanisms mediating gene expression in mulberry-phytoplasma interaction, the comprehensive sRNA changes of mulberry leaf in response to phytoplasma-infection were examined. A total of 164 conserved miRNAs and 23 novel miRNAs were identified, and 62 conserved miRNAs and 13 novel miRNAs were found to be involved in the response to phytoplasma-infection. Meanwhile, target genes of the responsive miRNAs were identified by sequencing of the degradome library. In addition, the endogenous siRNAs were sequenced, and their expression profiles were characterized. Interestingly, we found that phytoplasma infection induced the accumulation of mul-miR393-5p which was resulted from the increased transcription of MulMIR393A, and mul-miR393-5p most likely initiate the biogenesis of siRNAs from TIR1 transcript. Based on the results, we can conclude that phytoplasma-responsive sRNAs modulate multiple hormone pathways and play crucial roles in the regulation of development and metabolism. These responsive sRNAs may work cooperatively in the response to phytoplasma-infection and be responsible for some symptoms in the infected plants.
Mulberry tree is a perennial woody plant that has long been planted, and is of considerable economic importance to sericulture1. It is susceptible to a number of diseases2, among which yellow dwarf disease associated with aster yellows phytoplasma is one of the most devastating diseases3. Phytoplasmas are obligate endocellular parasites lacking cell walls, and have devastating effects on more than several hundred plant species worldwide4. Infected plants show dramatic changes in morphology including yellowing, phyllody, stunting, proliferation, and witches' broom5. These abnormal developments in plants can cause devastating losses in agriculture and forestry6. Because phytoplasmas were unculturable in pure cultures in laboratory broth, these organisms remain one of the most poorly characterized plant pathogens, and the mechanisms underlying their pathogenicities are not yet understood7.
When faced with pathogen invasion, plants activate sophisticated response mechanisms to reprogram gene expression8. MicroRNAs (miRNAs) and endogenous small interfering RNAs (siRNAs) are two main categories of small regulatory RNAs which globally regulate plant immunity by inhibiting target gene expression at transcriptional or post-transcriptional level9. Increasing evidence indicated that miRNAs serve as one important mechanism for mediating gene expression in plant-microorganism interactions10,11,12,13,14. As for the phytoplasma-responsive miRNAs, only one study has explored in Mexican lime (Citrus aurantifolia L.) infected by ‘Candidatus Phytoplasma aurantifolia’15. Though some of the pathogen-responsive miRNA families are deeply conserved among various plant species, their function may be specie specific16. Moreover, every plant has its own specie-specific miRNAs which play important roles in the regulatory networks associated with the stress resistance, and individual miRNAs of the same family may be expressed differentially and have different functions in response to the same stress13,16. Therefore, the phytoplasma responsive miRNAs have not been fully explored, and how their gene expression reprogrammings achieved are largely unknown. As far as we know, there was no miRNA information reported in mulberry so far. Characterization of the phytoplasma responsive miRNAs in mulberry may provide a novel platform to better understand the plant-phytoplasma interactions.
SiRNAs are distinguished with miRNAs by the structure of their precursors and by their targets, and they arise from a long double-stranded RNAs and typically direct the cleavage of transcripts to which they are completely complementary17. Though thousands of endogenous siRNAs have been sequenced18,19,20, their biological roles are largely unknown except for the functions of some chromatin-associated siRNAs in DNA methylation and transcriptional gene silencing and the roles of transacting siRNAs (ta-siRNA) in plant development and hormone signaling21. Except for the nat-siRNAATGB2, which was specifically induced by the effector avrRpt2 of Pseudomonas syringae pv. tomato DC3000 (Pst DC3000)21, the AtlsiRNA1, which was strongly induced by Pst (avrRpt2), and some siRNAs were found differentially expressed in response to Heterodera schachtii infection22, limited siRNAs involved in plant biotic stress has been reported. Therefore, identification of entire set of siRNAs and their expressions in response to biotic stress is needed to unravel the complex siRNA-mediated regulatory networks controlling gene expression in plant-microorganism interactions.
In the present study, based on the transcriptome information attained, we employed the next generation high throughput sequencing technology to characterize the miRNAs and siRNAs in mulberry. By comparing the normalized expression levels of sRNAs from the healthy and infected leaf libraries, the phytoplasma-responsive miRNAs and siRNAs were identified. In addition, the potential target genes of the responsive miRNAs were identified by sequencing of the degradome library and their functions involved in the response of mulberry to phytoplasma infection were discussed.
Results
Illumina sequencing of small RNAs
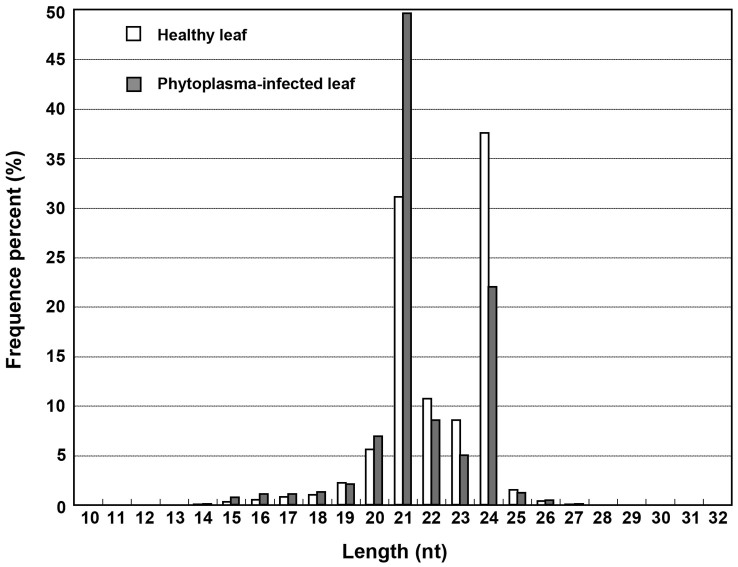
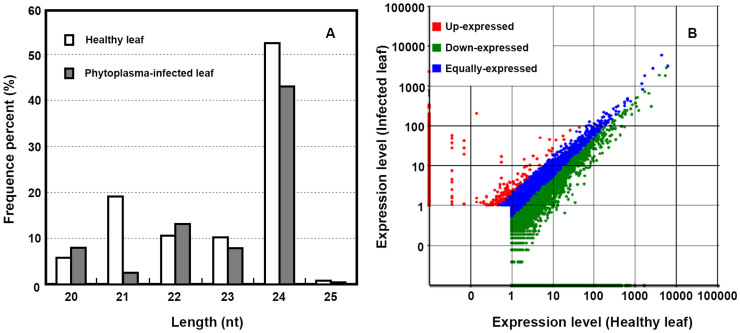
To examine the phytoplasma-responsive sRNAs, two sRNA libraries were constructed from phytoplasma-infected leaves and healthy leaves, respectively, and were then subjected to Solexa deep sequencing. The resulting raw sequence reads were cleaned and yielded 28 208 781 and 26 814 061 transcriptome-matching reads from the healthy and phytoplasma-infected leaf libraries, respectively. More than 60% of these mapped small RNAs were 20–24 nt in length with 24 and 21 nt as the major size groups (Fig. 1), consistent with the size of Dicer-like cleavage products. The matched sRNA sequences were clustered into several RNA categories such as known miRNAs, repeats, rRNAs, tRNAs, snRNAs, snoRNAs and unannotatated sRNAs. The known miRNAs account for 41.25% of all sequence reads in the healthy leaf library and 41.85% in the phytoplasma-infected leaf library. However, the proportion of sRNA unique sequences derived from known miRNAs represented only a very small fraction (0.64% and 0.79%, respectively) of the total unique sequences (Table 1). The highest proportion of unique sequences was unannotatated sRNA sequence, which probably include some novel miRNA candidates and lots of siRNAs as well as other type RNAs.
Figure 1. Length distribution of the small RNA in mulberry.
Table 1. Distribution of the sequence reads in the small RNA libraries of mulberry leaves.
| Healthy leaf | Phytoplasma-infected leaf | |||
|---|---|---|---|---|
| RNA class | Reads | Unique sequences | Reads | Unique sequences |
| Total | 28208781 (100%) | 7513906 (100%) | 26814061 | 4812269 |
| miRNAs | 11636122 (41.25%) | 48348 (0.64%) | 11222483 (41.85%) | 38056 (0.79%) |
| Repeats | 10 (0.00%) | 7 (0.00%) | 5 (0.00%) | 4 (0.00%) |
| rRNA | 3212490 (11.39%) | 104485 (1.39%) | 2370547 (8.84%) | 108431 (2.25%) |
| tRNA | 537157 (1.90%) | 13514 (0.18%) | 659539 (2.46%) | 16373 (0.34%) |
| snRNA | 24485 (0.09%) | 3861 (0.05%) | 18057 (0.07%) | 3820 (0.08%) |
| snoRNA | 4054 (0.01%) | 1396 (0.02%) | 2936 (0.01%) | 1019 (0.02%) |
| Unanna | 12794463 (45.36%) | 7342295 (97.72%) | 12540494 (46.77%) | 4644566 (96.53%) |
aContains all of the unclassified sequences that possibly include new miRNAs.
Identification and expression profiling of conserved miRNAs
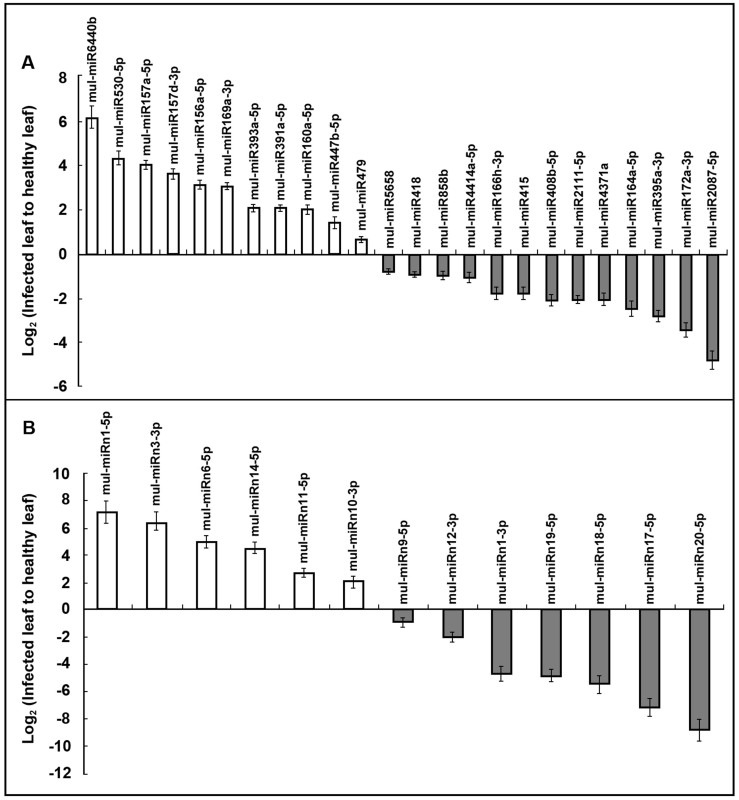
When the sRNA sequences obtained from our libraries were aligned to plant miRNAs in the miRBase database, 164 miRNA members showed perfect matches to the known miRNAs. These miRNAs identified have been shown conserved in many plant species. These conserved miRNAs varied significantly in expression abundances in both libraries (Table 2), and a total of 62 conserved miRNAs identified were found to be significantly responsive to phytoplasma infection, among which, 37 miRNAs were significantly decreased and 25 miRNAs were increased in the infected leaf (P < 0.05, fold 2.0). These differentially expressed miRNAs included some highly expressed miRNAs such as mul-miR156a-5p and mul-miR157a-5p, also some low abundance miRNAs such as mul-miR394a-5p and mul-miR4343a. Moreover, many differentially expressed miRNA-3p sequences, such as mul-miR1023b-3p, mul-miR1157-3p and mul-miR157d-3p, were identified, and these miRNAs (3p) may be the authentic miRNAs since the corresponding miRNA-5p sequences were not detected (Table 2). Real-time PCR analysis for 24 individual miRNAs covering different expression patterns were performed, and the results demonstrated that there was a very strong correlation between PCR data and read frequencies, which indicated that the sequencing profiles are quantitative and reliable (Fig. 2A).
Table 2. Expression profiling of conserved in mulberry miRNAs.
| Normalized value | ||||||
|---|---|---|---|---|---|---|
| MiRNA-name | Sequence (5′-3′) | Healthy leaf (HL) | Infected leaf (IL) | Fold-change (log2 IL/HL) | P-value | Significance lable |
| mul-miR1023b-3p | ACAGAACUGAAGAAGAGUGCAUA | 25.7721 | 13.3885 | −0.94482 | 1.55E-25 | |
| mul-miR1030a | UCUGCAUUUGCACCUGCACCU | 1.2053 | 8.9505 | 2.892576 | 6.38E-42 | ** |
| mul-miR1122 | UCAGAUACAUCCGGAUUUGCA | 0.9571 | 1.5663 | 0.710619 | 0.0444562 | |
| mul-miR1134 | GAAGAACAAAAGAAUGAAGAAGAU | 3.4386 | 2.4987 | −0.46064 | 0.0438324 | |
| mul-miR1144b | UGCGGAAGUGUGGCGGAACGGCAG | 1.6661 | 0.9323 | −0.83761 | 0.01748 | |
| mul-miR1157-3p | UUCAGGUAGUGGGAACCAGGC | 221.8813 | 103.6769 | −1.09769 | 1.10E-263 | ** |
| mul-miR1310 | AGGCAUCGGGGGCGCAACGC | 16.1297 | 15.477 | −0.05959 | 0.5436935 | |
| mul-miR1521a | CUAUUAUGGACAAUGUUGGA | 43.2489 | 28.8282 | −0.58518 | 4.43E-19 | |
| mul-miR156a-5p | UGACAGAAGAGAGUGAGCAC | 867.3186 | 6523.48 | 2.911008 | 0 | ** |
| mul-miR157a-5p | UUGACAGAAGAUAGAGAGCAC | 5519.133 | 322078.7 | 5.866828 | 0 | ** |
| mul-miR157d-3p | GCUCUCUAUGCUUCUGUCAUCC | 1.418 | 31.1404 | 4.456858 | 1.34E-203 | ** |
| mul-miR159a-3p.1 | UUUGGAUUGAAGGGAGCUCUG | 1764.699 | 1693.253 | −0.05962 | 1.89E-10 | |
| mul-miR160a-5p | UGCCUGGCUCCCUGUAUGCCA | 4.7148 | 10.5542 | 1.162549 | 2.03E-15 | ** |
| mul-miR160b-3p | GCGUAUGAGGAGCCAUGCAUA | 56.4363 | 58.6633 | 0.055835 | 0.2761912 | |
| mul-miR162-3p | UCGAUAAACCUCUGCAUCCA | 103.6911 | 97.7472 | −0.08516 | 0.0281548 | |
| mul-miR162a-3p | UCGAUAAACCUCUGCAUCCAG | 110.1785 | 103.0057 | −0.09712 | 0.0100218 | |
| mul-miR164a-5p | UGGAGAAGCAGGGCACGUGCA | 616.7583 | 140.7471 | −2.1316 | 0 | ** |
| mul-miR164c-3p | CAUGUGCCCGUCUUCGCCAUC | 5.1757 | 0.4848 | −3.41629 | 1.87E-28 | ** |
| mul-miR165a | UCGGACCAGGCUUCCCCCC | 132.5828 | 104.3109 | −0.346 | 5.59E-22 | |
| mul-miR165a-3p | UCGGACCAGGCUUCCCCC | 69.8364 | 53.8523 | −0.37497 | 4.68E-14 | |
| mul-miR166a-3p | UCGGACCAGGCUUCAUUCCCC | 22928.82 | 18439.32 | −0.31438 | 0 | |
| mul-miR166h-3p | UCUCGGACCAGGCUUCAUUCC | 7061.206 | 3301.962 | −1.09659 | 0 | ** |
| mul-miR167d-5p | UGAAGCUGCCAGCAUGAUCUG | 5533.773 | 3360.961 | −0.71939 | 0 | |
| mul-miR167h-3p | AGGUCAUCUUGCAGCUUCAAC | 19.2493 | 17.2671 | −0.15678 | 0.0857988 | |
| mul-miR168a-5p | UCGCUUGGUGCAGGUCGGGAA | 1734.637 | 1280.858 | −0.43752 | 0 | |
| mul-miR168a-3p | CCCGCCUUGCAUCAACUGAAU | 30.9124 | 24.0918 | −0.35964 | 1.41E-06 | |
| mul-miR169a-3p | UGGCAAGUUGUUCUUGGCUAC | 12.3366 | 41.247 | 1.741344 | 8.12E-101 | ** |
| mul-miR169q-5p | UGAGCCAGGAAUGACUUGCCG | 100.9969 | 104.7585 | 0.052756 | 0.168986 | |
| mul-miR170-5p | UAUUGGCCUGGUUCACUCAGA | 1.3825 | 1.3799 | −0.00272 | 0.9963935 | |
| mul-miR171b | UGAUUGAGCCGUGCCAAUAUC | 101.5287 | 60.9009 | −0.73735 | 3.22E-63 | |
| mul-miR171b-3p | CGAGCCGAAUCAAUAUCACUC | 8.2953 | 15.477 | 0.89976 | 6.79E-15 | |
| mul-miR172a-3p | AGAAUCUUGAUGAUGCUGCAU | 2493.763 | 365.3307 | −2.77105 | 0 | ** |
| mul-miR172e-3p | GAAUCUUGAUGAUGCUGCAU | 2461.326 | 359.662 | −2.77472 | 0 | ** |
| mul-miR1854-5p | UGUGAGUUUUGUAGAUUCGGA | 24.7086 | 11.0017 | −1.16729 | 5.43E-34 | ** |
| mul-miR1863a | CGCUCUGAUACCAUGUUAGUUUAC | 23.8933 | 13.8733 | −0.7843 | 9.08E-18 | |
| mul-miR1873 | ACUAACAUGGUAUCAGAGCGGGAG | 71.7507 | 39.2704 | −0.86955 | 1.95E-59 | |
| mul-miR1919a | AGAGAGUCUUCUGUGGACGGG | 6.7 | 3.5056 | −0.9345 | 1.39E-07 | |
| mul-miR2078 | GGUUCGCUGCCUGUGACGU | 65.157 | 40.0536 | −0.70199 | 5.94E-38 | |
| mul-miR2086-3p | UACACUGAAUGCAGAAAUGGACA | 7.7635 | 5.967 | −0.3797 | 0.0111255 | |
| mul-miR2087-5p | GAAGAAAGAACCGGCAGUCAU | 12.2302 | 0.3356 | −5.18756 | 2.86E-86 | ** |
| mul-miR2108a | UUAAUAGUGUUUGUAAGUCGG | 38.9949 | 28.1196 | −0.47171 | 3.21E-12 | |
| mul-miR2111a-5p | UAAUCUGCAUCCUGAGGUUUA | 4.2185 | 1.1188 | −1.91478 | 5.67E-13 | ** |
| mul-miR2119 | CGAAAGGGAGCUUGUAGGGAA | 4.7857 | 3.8413 | −0.31714 | 0.0928318 | |
| mul-miR2199 | UGAUAACUCGACGGAUCGC | 715.4864 | 607.7035 | −0.23556 | 1.88E-54 | |
| mul-miR2595 | UACAGUUUUCUUCUUUUUUCC | 2.0915 | 0.7459 | −1.48748 | 2.36E-05 | ** |
| mul-miR2610a | CGAUGUGAGACUGUACGGCUU | 11.9466 | 10.1812 | −0.23069 | 0.0494168 | |
| mul-miR2645 | UUUAUAGAUGAUGAGCAUUAU | 1.9143 | 3.2819 | 0.777714 | 0.0015907 | |
| mul-miR2661 | UGAGGUUUAAGAAAAUGGCAC | 2.5169 | 2.0139 | −0.32166 | 0.2182677 | |
| mul-miR2670f | AGGGUCUGUUUGGUUGGGGGA | 75.6857 | 38.8229 | −0.96311 | 5.57E-74 | |
| mul-miR2873c | CAAUAUGAGUUGUGUUUGGAA | 53.9903 | 46.4682 | −0.21646 | 8.34E-05 | |
| mul-miR2916 | UGGGGGCUCGAAGACGAUCAG | 243.6475 | 224.6582 | −0.11706 | 4.22E-06 | |
| mul-miR319g | UUGGACUGAAGGGAGCUCCUC | 26.8002 | 27.3364 | 0.02858 | 0.7018003 | |
| mul-miR3437-5p | AAAAAACACAGGAUCAACGGACA | 5.1402 | 2.9089 | −0.82135 | 3.59E-05 | |
| mul-miR3515 | AAUGUGAGCAAAGAACGGUAU | 1.1698 | 0.6713 | −0.80123 | 0.0562362 | |
| mul-miR3522b | UGAGACCUAAAUGAAGAAGAUGAC | 0.8862 | 1.2307 | 0.473775 | 0.2161161 | |
| mul-miR3626-5p | GGUAGUUCGACCGUGAAAUUUAA | 9105.604 | 10815.07 | 0.248217 | 0 | |
| mul-miR3627-5p | UGUCGCAGGAGAGAUGGCGAAU | 2.3397 | 11.7849 | 2.332544 | 2.85E-43 | ** |
| mul-miR3628-3p | CCAAGCAGAGCUCUUCGCAUC | 1.737 | 2.2003 | 0.341102 | 0.2213622 | |
| mul-miR3710 | UGCGGCACGUGACGGGCCUCC | 3.9704 | 2.797 | −0.5054 | 0.0183083 | |
| mul-miR390a-5p | AAGCUCAGGAGGGAUAGCGCC | 308.0956 | 229.3573 | −0.42578 | 4.23E-71 | |
| mul-miR390a-3p | CGCUAUCUAUCCUGAGUUUCA | 5.4593 | 4.3261 | −0.33565 | 0.058158 | |
| mul-miR391-5p | UGUCGCAGGAGAGAUGGCGAA | 3.4032 | 15.6634 | 2.202434 | 3.40E-53 | ** |
| mul-miR3933 | AGAAGACAAAAUGCACGACUCUA | 34.032 | 20.0268 | −0.76496 | 1.14E-23 | |
| mul-miR393-5p | UCCAAAGGGAUCGCAUUGAUCC | 1.4534 | 3.9531 | 1.443553 | 9.68E-09 | ** |
| mul-miR393a-3p | AUCAUGCUAUCUCUUUGGAUU | 5.8847 | 9.5472 | 0.698109 | 9.02E-07 | |
| mul-miR3946-5p | UUGUAGACAGAGAGAGAGAGAGAC | 24.7086 | 28.5298 | 0.207456 | 0.0059993 | |
| mul-miR394a-5p | UUGGCAUUCUGUCCACCUCC | 1.0989 | 8.5403 | 2.958227 | 5.74E-41 | ** |
| mul-miR3954 | CUGUACAGAGAAAUCACAGCA | 139.7437 | 103.043 | −0.43954 | 4.06E-35 | |
| mul-miR395a-3p | UGAAGUGUUUGGGGGAACUCC | 10.5995 | 0.8951 | −3.5658 | 7.59E-59 | ** |
| mul-miR396b-5p | UUCCACAGCUUUCUUGAACUU | 153.3211 | 132.5051 | −0.21051 | 1.08E-10 | |
| mul-miR396b-3p | GCUCAAGAAAGCUGUGGGAGA | 954.0292 | 107.6301 | −3.14795 | 0 | ** |
| mul-miR397a-5p | UCAUUGAGUGCAGCGUUGAUG | 29.5653 | 53.1065 | 0.844984 | 1.26E-42 | |
| mul-miR397b-3p | UCAGCGCUGCACUCAAUUAUG | 1.8788 | 1.1561 | −0.70055 | 0.0304262 | |
| mul-miR398a-5p | GGCGUGACCCCUGAGAACACAAG | 13.6482 | 1.8274 | −2.90085 | 2.22E-62 | ** |
| mul-miR399k-3p | UGCCAAGAAGAGUUGCUCUGU | 14.3927 | 7.3469 | −0.97013 | 1.52E-15 | |
| mul-miR403a-3p | UUAGAUUCACCCACAAACUCG | 15.7752 | 19.542 | 0.30892 | 0.0008772 | |
| mul-miR408b-5p | CAGGGAACGGACAGAGCAUGG | 1508.963 | 530.1323 | −1.50913 | 0 | ** |
| mul-miR415 | AAGGAGCAGAGCAGAGCAG | 17.1223 | 5.6687 | −1.59479 | 1.12E-37 | ** |
| mul-miR418 | UAAUCUGAUGAUAGAUGGACG | 62.0729 | 20.9219 | −1.56895 | 5.93E-129 | ** |
| mul-miR419 | UGAUGAAUGUAUGGAUGAUGGAU | 9.8551 | 7.2723 | −0.43846 | 0.0010722 | |
| mul-miR4343a | AAAAAACUUACGGACAAGACGACU | 2.3042 | 1.0442 | −1.14187 | 0.0002923 | ** |
| mul-miR4371a | AAGAGAGGACAGUGACAAGCAAGU | 8.1535 | 3.6921 | −1.14298 | 7.09E-12 | ** |
| mul-miR4376-5p | UACGCAGGAGAGAUGACGCUGU | 2564.095 | 1769.519 | −0.53509 | 0 | |
| mul-miR4376a-3p | AGCAUCAUUUUCCUGCAUAGU | 1.2053 | 1.0442 | −0.20699 | 0.579823 | |
| mul-miR4385 | AAUCGAUGGUAGAAAGUGAUGGGC | 1.1698 | 0.5967 | −0.97118 | 0.024715 | |
| mul-miR4401b | UCACAAGACCUUGCUGAAGAA | 9.6069 | 6.1535 | −0.64266 | 4.93E-06 | |
| mul-miR4414a-3p | AUCCAACGAUGCAGGAGCUAGCC | 92.7371 | 37.3312 | −1.31276 | 3.71E-147 | ** |
| mul-miR4414a-5p | AGCUGCUGACUCGUUGGUUCA | 162.8216 | 71.4923 | −1.18743 | 8.25E-220 | ** |
| mul-miR447b-5p | ACUCUCACUCAAGGGCUUCA | 4.0767 | 8.5403 | 1.066885 | 2.71E-11 | ** |
| mul-miR472b-3p | UUUUCCCAACACCACCCAUACC | 60.903 | 87.2676 | 0.518933 | 4.45E-30 | |
| mul-miR473a-5p | ACUCUCCCCCUUAAGGCUUCCA | 181.5392 | 697.6191 | 1.942158 | 0 | ** |
| mul-miR477a-5p | ACUCUCACUCAAGGGCUUCAG | 4.2894 | 9.3981 | 1.131593 | 2.29E-13 | ** |
| mul-miR479 | CGUGAUAUUGAUUCGGCUCAUA | 4.8921 | 14.7684 | 1.593988 | 5.47E-33 | ** |
| mul-miR482a-3p | UUCCCAAGGCCGCCCAUUCCGA | 201.8166 | 361.3403 | 0.840313 | 1.57E-276 | |
| mul-miR482a-5p | GGAAUGGGCUGUUUGGGAAGA | 10799.72 | 20081.4 | 0.894866 | 0 | |
| mul-miR5020a | CUGGAAGAAGCUGAGCGUGCA | 27.4383 | 11.6357 | −1.23763 | 4.89E-41 | ** |
| mul-miR5021 | UGAGGAAGAAGAAGAAAUGA | 52.1114 | 35.8394 | −0.54005 | 7.94E-20 | |
| mul-miR5029 | AUGAGAGAAAAACACUGCAUA | 4.4312 | 2.8716 | −0.62584 | 0.0024964 | |
| mul-miR5054 | GUGCCCCACGGUGGGCGCCA | 1.1344 | 3.5056 | 1.627732 | 3.68E-09 | ** |
| mul-miR5059 | CGUUCCUGGGCAGCAACACCA | 35.45 | 31.849 | −0.15454 | 0.0214399 | |
| mul-miR5065 | UAGGCAAUUCACUUAGAUCUG | 2.3397 | 4.1769 | 0.836109 | 0.0001483 | |
| mul-miR5072 | GUUCCCCAGUGGAGUCGCCA | 2.2333 | 5.333 | 1.25577 | 2.07E-09 | ** |
| mul-miR5077 | UUCACGUCGGGUUCACCA | 203.6954 | 291.4143 | 0.516658 | 1.49E-95 | |
| mul-miR5085 | AAGGACAUUUGGUUGUGGCUC | 83.3074 | 92.9736 | 0.158376 | 0.0001338 | |
| mul-miR5138 | AAACGAAUCGUUGGCCGCUA | 1.2762 | 1.7528 | 0.457807 | 0.1517183 | |
| mul-miR5139 | AACCUGGCUCUGAUACCA | 51.0479 | 34.5341 | −0.56383 | 6.68E-21 | |
| mul-miR5224a | UUGAUGGACAUGAAGACGUUAU | 119.5372 | 160.1772 | 0.422209 | 2.38E-37 | |
| mul-miR5225a | ACUGUCGCAGGAGAGAUGACGC | 81.5703 | 33.4153 | −1.28754 | 6.81E-126 | ** |
| mul-miR5234 | GUCUUGUUAUGGAUGGCAGAA | 6.0619 | 12.0086 | 0.986226 | 1.29E-13 | |
| mul-miR5236a | UGAAUUUUCGGGCAGAUCGGGUU | 4.1476 | 1.8647 | −1.15333 | 8.88E-07 | ** |
| mul-miR5247 | GCAGGAGCCAUCUCUGAUCGA | 1.2053 | 0.6713 | −0.84436 | 0.0426108 | |
| mul-miR5266 | CGGGGGACGGACUGGGGG | 530.8631 | 339.1504 | −0.64641 | 3.68E-256 | |
| mul-miR5280 | AAUUAUUAAACGGGCCGUGACGGG | 43.8161 | 20.9592 | −1.06388 | 5.08E-51 | ** |
| mul-miR5291a | GUUUGGUAGGAUGGAUGGAUGGAG | 2.7651 | 2.0139 | −0.45734 | 0.0729034 | |
| mul-miR529-3p | GCUGUACCCCCUCUCUUCUCC | 3.1905 | 6.2654 | 0.973624 | 1.23E-07 | |
| mul-miR529b | AGAAGAGAGAGAGUACAGCUU | 275.7297 | 874.2055 | 1.664718 | 0 | ** |
| mul-miR530-3p | AGGUGCAGAUGCAGAUGCAG | 15.208 | 44.2678 | 1.541427 | 5.81E-91 | ** |
| mul-miR530-5p | UGCGUUUGCACCUGCACCUUA | 4.3603 | 62.7656 | 3.847475 | 0 | ** |
| mul-miR535a | UGACAACGAGAGAGAGCACGC | 54.0257 | 592.2266 | 3.454432 | 0 | ** |
| mul-miR5368 | AGGGACAGUCUCAGGUAGACA | 7.8699 | 15.5515 | 0.982637 | 4.25E-17 | |
| mul-miR5369 | UGAGAACAGUAGGAUGUCAUC | 7.1254 | 4.3634 | −0.70752 | 1.87E-05 | |
| mul-miR5386 | CGUCAGCUGUCGGCGGACUG | 81.0386 | 161.6316 | 0.996028 | 1.41E-165 | |
| mul-miR5485 | UGACAGAGUGGUAUCAGAGCA | 17.4414 | 6.8994 | -1.33797 | 6.69E-30 | ** |
| mul-miR5500 | AUCACUGAUGAAACUUCGGGCGGC | 2.2688 | 0.9696 | −1.22647 | 0.000143 | ** |
| mul-miR5509 | AAGGCCUUUUGCUCUUGGCAU | 1.418 | 0.9323 | −0.60499 | 0.0997003 | |
| mul-miR5519 | UGGUAGACGCUACGGACUUAG | 26.6584 | 68.9937 | 1.371874 | 1.99E-118 | ** |
| mul-miR5521 | AAGUGGCUGGAUGUUGAUGAG | 2.2688 | 2.3868 | 0.073148 | 0.7729617 | |
| mul-miR5559-5p | UACUUGGUGAAGUGUUGGAUAA | 0.1772 | 7.3096 | 5.366342 | 6.12E-54 | ** |
| mul-miR5564c-5p | CAAUUCGUCGAACGCCGCACC | 1.5243 | 0.7459 | −1.03109 | 0.0068859 | ** |
| mul-miR5565g-3p | ACACAAGUGGAUUGAUGAUGAAUC | 1.5243 | 0.5221 | −1.54575 | 0.0002156 | ** |
| mul-miR5658 | AUGAUGCUGAUAAUGAUGACAA | 1.5598 | 0.4848 | −1.6859 | 6.78E-05 | ** |
| mul-miR5665 | AUUGGUGAACAAGAUCGUGAU | 5.7429 | 1.7901 | −1.68174 | 1.35E-14 | ** |
| mul-miR5741a | AACGAGGACUAAGUUGAUGGUUU | 3.8995 | 2.536 | −0.62073 | 0.0048851 | |
| mul-miR5751 | UUAAUUCUGAUUGAGAUGGUUUU | 21.4827 | 12.2324 | −0.81247 | 4.83E-17 | |
| mul-miR5813 | AGCAGGACGGUGGUCAUGGA | 314.5829 | 295.3674 | −0.09093 | 4.52E-05 | |
| mul-miR6021 | UUGGAAGAGGAUGCAUUGAAC | 10.0678 | 0.1119 | −6.49139 | 1.86E-77 | ** |
| mul-miR6145b | UAUGAGACGUAUGCACUAGCC | 0.5317 | 1.0442 | 0.973714 | 0.032467 | |
| mul-miR6171 | ACUGUGGAUCGCAGAAGGUUU | 21.4827 | 25.6209 | 0.254146 | 0.0015546 | |
| mul-miR6196 | AGGCGAGUGAGACGGAGAUGA | 7.0545 | 3.804 | −0.89103 | 2.07E-07 | |
| mul-miR6203 | AGAGAGAUUAAGAAGACCUGGUAA | 2.2333 | 0.7832 | −1.51172 | 9.71E-06 | ** |
| mul-miR6204 | AGGAGAAGAAUUAGAAGCUUUCGA | 5.991 | 2.9835 | −1.00579 | 1.21E-07 | ** |
| mul-miR6213 | AAGCAGAUUGAUACAGACUGGUU | 20.561 | 9.2862 | −1.14675 | 8.30E-28 | ** |
| mul-miR6223-5p | UUCUUGAGGAGGAGCGUACUG | 5.8492 | 3.8786 | −0.5927 | 0.0009253 | |
| mul-miR6300 | GUCGUUGUAGUAUAGUGGU | 112.0573 | 127.3213 | 0.184237 | 2.27E-07 | |
| mul-miR6429 | GAGUAGAAAAUGCAGUACUAG | 3.7931 | 2.4614 | −0.6239 | 0.0053003 | |
| mul-miR6440b | GGAGUUUGAUUGAGUUCGGUU | 0.6026 | 29.4249 | 5.609693 | 3.52E-217 | ** |
| mul-miR6444 | AGGAAAAUCAAGAGAUAAUGU | 7.1254 | 4.4007 | −0.69524 | 2.51E-05 | |
| mul-miR6449 | UCAUGAUUCGGAUCACGGUUU | 8.6498 | 8.0928 | −0.09603 | 0.4769051 | |
| mul-miR6462c-5p | AAGUGGACAGAAAAUGGAAUAAAA | 10.7768 | 5.706 | −0.91738 | 4.59E-11 | |
| mul-miR6463 | UGUGAUGAUCAUUGGACAACC | 0.1418 | 3.5056 | 4.627732 | 1.19E-24 | ** |
| mul-miR6464 | UAAUGCUUGUUGGGUAUUU | 8.9688 | 5.1838 | −0.7909 | 1.21E-07 | |
| mul-miR6466-5p | UUUGGAUGACAUUUGACGA | 2.5169 | 1.7901 | −0.49161 | 0.0676547 | |
| mul-miR6475 | UCUUGAGAAGUAGAGACGUCU | 1.8079 | 10.2931 | 2.509291 | 2.81E-41 | ** |
| mul-miR6478 | CCGACCUUAGCUCAGUUGGUA | 22.298 | 20.2506 | −0.13895 | 0.1001727 | |
| mul-miR6483 | UAUUGUAGAAAUCUUCGGGAU | 3.9704 | 3.6175 | −0.13429 | 0.5044449 | |
| mul-miR774b-5p | UGAGAUGAAGAUUAUAGUGAA | 38.4632 | 44.7153 | 0.21729 | 0.0003217 | |
| mul-miR780.2 | UUUUCGUGAAUAUAUGGUCGU | 56.5781 | 23.4205 | −1.27247 | 2.43E-86 | ** |
| mul-miR827-5p | UUUGUUGAUGGUCAUUUAACU | 6.5228 | 6.8994 | 0.08098 | 0.5892211 | |
| mul-miR829.1 | CCUCUGAUAACAAAUGAUGGACAU | 1.0989 | 0.1119 | −3.29578 | 8.66E-07 | ** |
| mul-miR845b-3p | CGCUCUGAUACCAACUGUGACG | 126.9817 | 80.7412 | −0.65324 | 6.83E-64 | |
| mul-miR858b | UUCGUUGUCUGUUCGACCUUG | 52.0051 | 25.2479 | −1.04249 | 2.55E-58 | ** |
| mul-miR893 | AUUGGGUACUUGUGGUUGGC | 3.4741 | 2.9835 | −0.21963 | 0.3141346 | |
| mul-miR894 | GUUUCACGUCGGGUUCACCA | 584.1443 | 788.9517 | 0.433612 | 1.40E-185 | |
| mul-miR952a | AACAAGCAUCAUCGUUGGUU | 12.372 | 11.673 | −0.0839 | 0.4559723 | |
Figure 2. MiRNA abundance analysis by real-time PCR.
(A) Conserved miRNA abundance analysis by real-time PCR. (B) Novel miRNA abundance analysis by real-time PCR. The relative miRNA abundance was evaluated using comparative Ct method taking U6 as the reference. The log2 values of the ratio of phytoplasma-infected samples to the healthy samples are plotted. Values are given as mean ± SD of three experiments in each group.
Identification and expression profiling of novel miRNAs
All the non-annotated clean tag sequences were mapped to our mulberry transcriptome database, and only 23 sequences perfectly matched transcriptome sequences able to fold into hairpin structures. These sequences comprise 20 putative new miRNA families (Table 3). The lengths of novel miRNAs identified vary from 20 to 23 nt with a peak at 21 nt, and most begin with a 5′ uridine which is a characteristic feature of miRNAs. These newly identified mulberry miRNA precursors have negative folding free energies (from −21.0 to −105.0 kcal mol−1 with an average of about −46.9 kcal mol−1) according to Mfold, which are similar to the free energy values of rice and Arabidopsis23 and much lower than the folding free energies of tRNA (−27.5 kcal mol−1) and rRNA (−33 kcal mol−1)24. Of these miRNAs, 3 candidates contained both miRNA-5p and miRNA-3p sequences. Moreover, other 5 miRNA-3p sequences were also detected (Table 3). The detection of miRNA-3p is a strong clue for the formation of precursor hairpin structures and represents further evidence supporting them as novel miRNAs. Compared to the conserved miRNAs, most of the novel miRNAs exhibited relatively low abundance as indicated by their frequencies (Table 2, 4). Among the 23 novel miRNAs, only 7 had at least 20 transcripts per million (TPM) in both libraries with the highest abundance 506 TPM for mul-miRn12-5p. Therefore, the expression levels of the majority of the novel miRNAs identified were low.
Table 3. Novel miRNAs in mulberry by Illumina sequencing.
| MiRNA-name | Sequence (5′-3′) | Precursor sequence (5′-3′) | Energy (kcal mol−1) |
|---|---|---|---|
| mul-miRn1-5p | UAUAACAAUGACUUUAUAAAG | GACAUUUAUUUUAUAUAACAAUGACUUUAUAAAGGUGUAACAAAAAAUUUAUCUAUUGUAAACUGAUGUUGCAUCUUUUUGAAGUCGUUGUUAUACAGAACAAAUGU | −39.70 |
| mul-miRn1-3p | UUUUGAAGUCGUUGUUAUACA | ||
| mul-miRn2-5p | UCUGGGACAAGUAACACUACAU | CCAAAGUCAUUCUGGGACAAGUAACACUACAUUAAAUUAGAUCUUUAAUAUUACAUUUUCAAUUUCAGGGCUAAAUCUUAGUUGAAUAAGCGAAUUUAGUAAUUCAAACAUACAUGCGAUUUUUAGCCAAAACAAAUGUAAGAGAUACAUUGCUUCAGUCUUUAA | −23.20 |
| mul-miRn3-3p | GUGGUGAUCAAUUGGACCUUU | UGGAAUCGUCUUAGGUCUAAUUCCUAUUACUUUGGCUGGAUUAUUCGUAACGGCAUAUUUACAAUAUAGACGUGGUGAUCAAUUGGACCUUUGAUUAAUUAA | −21.80 |
| mul-miRn4-5p | CGGGCCUGGGAGGUUUGGUA | UUUGAGCUUUAUGAAGUUGUCGGGCCUGGGAGGUUUGGUAGGAGUAAUAAGUAAUUACCAUUUAGUUUUUUGUUCACUUAAUUGAUAUUAUAAUUGUAUGUUUUAAUUUAGUUCUCCUUCCAAAUCCACCCAUGCCCACAAUUUCCUCAGGCUUCUCUC | −51.80 |
| mul-miRn4-3p | UUCCAAAUCCACCCAUGCCCAC | ||
| mul-miRn5-3p | UCCCUUUGGAUGUCGUCCUGC | CUGUGGCUUUUGCUGGGGACAGUGACAAAGGGAUGCUUUAUGUGUUGAGAGGGUGUAAGAAACUUCGCAAGCUUGAGAUCAGGGACUGCCCCUUUGGUGAUGUUGCUCUCUUGACGGACGUGGGAAAGUAUGAAACAAUGCGAUCCCUUUGGAUGUCGUCCUGCGAAGUUACCC | −58.30 |
| mul-miRn6-5p | UAAGGCCGCGUACAUAUGGGAUU | GUAAGGCCGCGUACAUAUGGGAUUAAGCUUUCUCCCGAGCCUGUCAUAGUACGGGAGCCUUUU | −21.00 |
| mul-miRn7-3p | UUGUAGAAGUCUUAAGAGAAG | AUGGUUUUUUCUUCUCUUAACACUUCUAUAAAUAUCUUUAUCCGUCAGAUGAUCAAGUAAAUUGCGGGGAGCGAAAUUUGUAGAAGUCUUAAGAGAAGAGAAAAACCA | −42.30 |
| mul-miRn8-5p | UCAUGAUUCGUGGUUCGCUA | UCAUGAUUCGUGGUUCGCUACAAUAUUUUCUCUUACAAAAAAUACACAAAUUAAAAAUAAUUCGAAUCACAUAGUUUGAAUCUACCAACUAAACGUCUUUUUUUUUUAGUUGAUCAUAAUUGUACUCUUUUUUUUUCUCUCUUGUUAAAUCAGAGAACUCUACUCCUUACGAAACACUCCAUUGGAGGAAGUGCCUUUAAGUUUGAAAGUGCAAUCCGUGGCUGGCUAGACGAAACUAUCAUGUGAUGCAUAUGGACCCCCCAUGCCUGAUGCCACACAAGUUAAACAGCAGGAUACGAAUCUUGU | −60.80 |
| mul-miRn9-5p | GGGGUUACCUGAGAAUACAUG | GAGUCUCACAGGGGUUACCUGAGAAUACAUGAAUAAAUUAUUUAUAUUUAUUUAUCUCACAAUGUGGUGGUCGUUUCAUGUGUUCUCAGGUCGCCCCUGUAUGGCUUU | −50.70 |
| mul-miRn10-3p | GGUGCAGAUGCAGAUGCAGG | GUCGCCUUUAUCUGCGUUUGCACCUGCACCUUAAUUGGCUUCUUAAUUUUCUUCAUCUCAUUUGCCGAUCACUAACUCUACUUGAGAUCAGCCAAACCCUCAAACUAAUCAACGGCGAUAAGACGAAGGACCAUAGCAAGAGGUGCAGAUGCAGAUGCAGGUGAAUGCCAU | −60.52 |
| mul-miRn11-5p | UGUUAGGAUCCUGGUUGAGUC | UAUGUCACUCUGUUAGGAUCCUGGUUGAGUCUCGAGUCCUGGGACUCAACUAGGAUCCUAACACACUCUUACU | −40.60 |
| mul-miRn12-5p | UGUGGGGGGAUCGGUAAGUGU | GUUUAGCAGGAAGUUAUAGGUGUGGGGGGAUCGGUAAGUGUUAAAUCUGUCGUGUUUUUAGCGGAUAAUUCUUGCCGAGACCUCCCAUACCGGUCAUUUCCUGCUGUUUCU | −55.80 |
| mul-miRn12-3p | UCUUGCCGAGACCUCCCAUACC | ||
| mul-miRn13-3p | UUCUGUGUUCUUCUACGGCGA | CCUUGGUGAGUAGUAGUAGUAUAGAACAAGUGAAACAAAACUGUGUUUGUUAGAAAUGGUUUUGAUUGAUGAGGAAUGAAUGAAAGAAAGAGAGAGAGUGAUUAGAGAACUGACGGUGCCAUCUCUCCUGCGACAGUUUUUUCGUUUCUGUGUUCUUCUACGGCGACGAUGACAAG | −42.70 |
| mul-miRn14-5p | UCUACCAGCCGAGUAGAAGUA | ACAUUGAAUCUCUACCAGCCGAGUAGAAGUAACAAUCGAGGGAAAUGAGAUACUUACUUCUACUCGGCUGGUCGAGAUCCAAUAU | −45.10 |
| mul-miRn15-5p | CAGAGUGCACAAAUACAUUAG | CAGAGCCAGAGUGCACAAAUACAUUAGAAGACAAUUGGACUUCUACUGCUUUUGUGCACUCUGGCUCUGA | −44.70 |
| mul-miRn16-3p | UAAUGUAUUUGCACACUCUGG | AUCAUCAGGUUCUCUUUUCUCCAACCGUAACAUGUCUAAAAGAGGGUGCGAAUCUGGCAGCUCAUAUACUUGGUGCUCUGUUCUUAGUCGGUUCACAUUCUUCAGCUUGGGCAUAGGAAUAGAAGCAGCUUCUGGUCCUAAAGCAACUAAGGCCUUCGACAUUUCAGCACCUUGAAGUUCCAUGUUUCUUUGCAUGUAGUUCUGUACAUUCUGAGUGAACUCUUCAAUGUUAAGCUUGAUUGUAGGAAUUUCAUUAGGAUCUUCAGAGAAAAAGUCCUCUAUGUCCCCUAAUGUAUUUGCACACUCUGGCUCUGGUGUU | −76.05 |
| mul-miRn17-5p | AUGACACAAGAUCAUCCUCCA | AUUUCACUUUAUGACACAAGAUCAUCCUCCAAAUAGAAAUAGAAACAAAUCAUUGGUUUACCUUGAUUAUUCUUAUUGGCUUACGCCUAGAAGUUAGAGGGAUAAUAUCAAGAUAACGGGGAAGUCUAGGGGUUUUGCGGUUUUGUGGGUGUUUAUAGAGUUACAACAUGGCGGUUGAUUGUUGAGUGUUGAUUUGUUUCUAUUUCUAUUUGAAGGAUGAUCUUGUGUCAUGUAGUGAAAUG | −100.5 |
| mul-miRn18-5p | UUCUGCAAUCAUAACUCGGAG | UCAUUAGUUAUUCUGCAAUCAUAACUCGGAGGAAAGAUUCUGGUGUUGGCCGGAUUGCAGAUUGCUUUACA | −22.7 |
| mul-miRn19-5p | AACUCGGUGCUGGAAUCGGUC | UGGAACUCGGUGCUGGAAUCGGUCAUGUCGGGCGUGCCAAUGCUAGCUUGGCCCAUGGGAGCCGACCAAUUCAUAAACGCGACAUUAUUAGACCAAUUGAAGUUGGCAAUUAGAGUGGGUGAAGGAGAUCGAAAUGUUUCUGAUUCGGUCGAUUUGACUCGAGUUUUG | −49.42 |
| mul-miRn20-5p | AGGAGGAGGAGGAGGAGUUU | GAUGACGAGGAGGAGGAGGAGGAGGAGUUUGAGAUGGACGAGUUGUCUUGUUUCAGAGGUCUUGUUCUCGAUACCUCCUACAGGCCAAU | −30.7 |
Table 4. Novel miRNA abundance analysis by Illumina sequencing.
| Normalized value | |||||
|---|---|---|---|---|---|
| miRNA-name | Healthy leaves (HL) | Infected leaves (IL) | Fold-change (log2 IL/HL) | P-value | Significance lable |
| mul-miRn1-3p | 1.0989 | 0.01 | −6.77992 | 0.132485 | ** |
| mul-miRn1-5p | 0.01 | 0.7086 | 6.14690 | 0.143462 | ** |
| mul-miRn2-5p | 21.6954 | 32.8932 | 0.6004 | 1.46E-15 | |
| mul-miRn3-3p | 0.01 | 29.1638 | 11.50996 | 7.30E-245 | ** |
| mul-miRn4-5p | 290.7251 | 219.9219 | −0.40266 | 9.14E-61 | |
| mul-miRn4-3p | 94.4398 | 78.91425 | −0.25911 | 1.42E-32 | |
| mul-miRn5-3p | 52.1114 | 48.4447 | −0.10526 | 0.055345 | |
| mul-miRn6-5p | 0.01 | 1.268 | 6.986411 | 2.37E-11 | ** |
| mul-miRn7-3p | 17.9731 | 16.4466 | −0.12805 | 0.173085 | |
| mul-miRn8-5p | 7.3381 | 5.8178 | −0.33493 | 0.028247 | |
| mul-miRn9-5p | 41.6891 | 19.0572 | −1.12933 | 2.22E-53 | ** |
| mul-miRn10-3p | 12.372 | 37.9652 | 1.617599 | 7.55E-84 | ** |
| mul-miRn11-5p | 0.01 | 1.0442 | 6.706254 | 1.77E-09 | ** |
| mul-miRn12-5p | 506.0126 | 267.5089 | −0.91959 | 0 | |
| mul-miRn12-3p | 156.38231 | 72.49928 | −1.1090 | 3.54E-42 | ** |
| mul-miRn13-5p | 16.7324 | 15.9618 | −0.06802 | 0.480784 | |
| mul-miRn14-5p | 0.01 | 3.804 | 8.571373 | 1.40E-32 | ** |
| mul-miRn15-5p | 1.5243 | 0.8578 | −0.82943 | 0.024299 | |
| mul-miRn16-3p | 58.1379 | 31.9235 | −0.86486 | 4.54E-48 | |
| mul-miRn17-5p | 6.0619 | 0.01 | −9.24363 | 2.48E-50 | ** |
| mul-miRn18-5p | 1.9852 | 0.01 | −7.63314 | 5.78E-17 | ** |
| mul-miRn19-5p | 1.0989 | 0.01 | −6.77992 | 1.04E-09 | ** |
| mul-miRn20-5p | 2.3397 | 0.01 | −7.87018 | 7.25E-20 | ** |
In order to identify the novel miRNAs involved in response to phytoplasma-infection, normalized expression profiles of the novel miRNAs in healthy and infected leaves were compared (Table 4). Among 13 differential novel miRNAs, 7 miRNAs were significantly decreased and 6 miRNAs were up-regulated more than 2.0 fold changes (P < 0.05) in the infected leaf. Real-time PCR analysis to determine the expression patterns of the 13 differential miRNAs (Fig. 2B) reveals a very strong correlation between qRT-PCR data and read frequencies, demonstrating that the profiles of these novel miRNAs detected by Illumina sequencing are reliable.
Targets of phytoplasma-responsive miRNAs
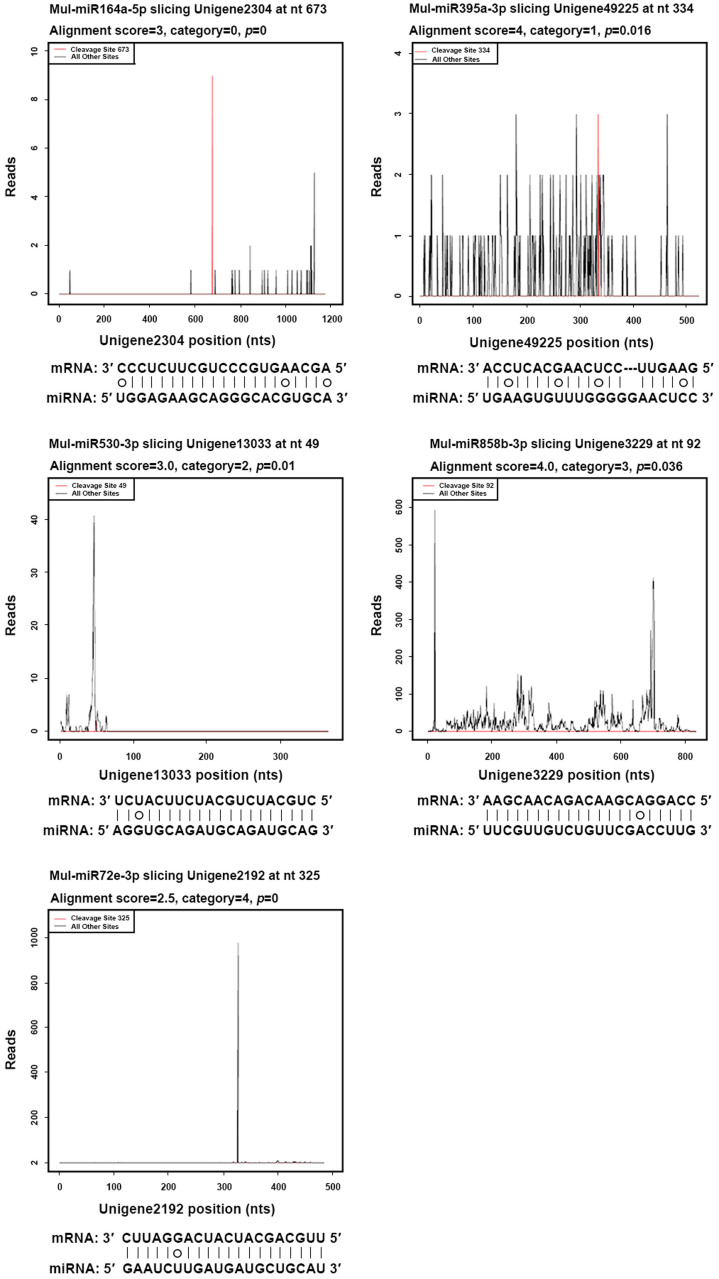
To understand the functions of phytoplasma-responsive miRNAs, target genes of the phytoplasma-responsive miRNAs including conserved and novel miRNAs were predicted. In total, 42 differentially expressed miRNAs were predicted to match 122 target genes (Table 5). The predicted miRNA targets were further experimentally verified by mRNA degradome sequencing. In total, we obtained 25 850 468 raw reads from the degradome libraries. After removing the reads without the adaptor, 25 721 355 clean reads were obtained, among which the percent of 20–21 nt reads was 99.39%. The distinct sequences were aligned to the mulberry transcriptome database, and 6913 912 sequences could be matched to the transcriptome without mismatch. The matched sequences were further analyzed by the CleaveLand pipeline, and 69 target genes of 28 phytoplasma-responsive miRNAs were qualified (Table 5), of which 8 genes were cleaved by two unconserved miRNAs and 61 genes were cleaved by 26 conserved miRNAs. Among these identified targets, 27 belonged to category 0, 5 were in category I, 13 were in category II, 15 were in category III and 9 were in category VI. These results indicated that most of these targets were efficiently cleaved by miRNA. The representative ‘target plots’ (tplots) of identified targets included all five categories were showed in Figure 3.
Table 5. Predicted targets for the differential miRNAs.
| MiRNA-name | Predicted target annotations in mulberry transcriptome data | Putative GO_process | Degradome sequencing | ||
|---|---|---|---|---|---|
| Read | Score | Class | |||
| mul-miR1030a | SWIB complex BAF60b domain-containing protein | Transcription regulation | 52 | 4 | 0 |
| mul-miR156a-5p | Homeobox protein BEL1-like protein | Transcription regulation | 57 | 4 | 0 |
| Squamosa promoter-binding-like protein 7 | Transcription regulation | 62 | 3 | I | |
| Promoter-binding protein SPL9 | Transcription regulation | 26 | 3 | 0 | |
| Promoter-binding protein SPL10 | Transcription regulation | 334 | 2.5 | III | |
| Homolog A-like transcriptional coactivator KELP | Transcription regulation | No | |||
| UDP-glycosyltransferase-like protein | Metabolic process | No | |||
| mul-miR157a-5p | SPL domain class transcription factor | Transcription regulation | 119 | 1 | |
| Squamosa promoter-binding-like protein 7 | Transcription regulation | 45 | 3 | 0 | |
| Promoter-binding protein | Transcription regulation | 54 | 2 | 0 | |
| Promoter-binding protein SPL10 | Transcription regulation | 1435 | 3 | III | |
| Dioxygenase-like protein transcriptional coactivator KELP | Transcription regulation | No | |||
| mul-miR160a-5p | Auxin response factor 10 (ARF10) | Auxin signaling; transcription regulation | 65 | 0.5 | 0 |
| ARF18-like | Auxin signaling; transcription regulation | 56 | 1 | 0 | |
| ARF16 | Auxin signaling; transcription regulation | 1282 | 3 | IV | |
| Beta-amylase | Metabolic process | 1611 | 2 | II | |
| mul-miR164a-5p | NAC domain protein | Transcription regulation | 139 | 3 | 0 |
| CBL-interacting serine/threonine-protein kinase 25-like | Signalling pathway; stress response | 4353 | 4 | III | |
| mul-miR164c-3p | C2 domain-containing protein | Calcium-mediated signalling | No | ||
| mul-miR166h-3p | Class III HD-Zip protein 6 | Transcription regulation; development | 102 | 2 | 0 |
| Homeobox leucine-zipper protein | Transcription regulation; transcription | No | |||
| mul-miR169a-3p | Phosphatidylinositol 4-kinase | Signalling pathway | No | ||
| Serine/threonine-protein kinase BRI1-like 1 | Signalling pathway | No | |||
| O-fucosyltransferase family protein | Hormone metabolism | No | |||
| Nuclear transcription factor Y subunit A-1 | Transcription regulation | 109 | 3 | 0 | |
| Nuclear transcription factor Y subunit A-9-like | Transcription regulation | 1091 | 3 | III | |
| Nuclear transcription factor Y subunit A-3-like | Transcription regulation | 3524 | 3 | IV | |
| mul-miR172a-3p | Ethylene-responsive transcription factor RAP2-7-like | Transcription regulation | 979 | 1.5 | IV |
| AP2 domain-containing transcription factor | Transcription regulation | 82 | 2.5 | 0 | |
| S-locus lectin protein kinase-like protein | Signalling pathway | 1979 | 1.5 | III | |
| mul-miR172e-3p | Transcription factor bHLH123 | Transcription regulation | No | ||
| S-locus lectin protein kinase-like protein | Signalling pathway | 234 | 3 | II | |
| NAC domain protein, IPR003441 | Transcription regulation | No | |||
| Trehalose 6-phosphate synthase | Carbohydrate metabolic process | No | |||
| Ethylene-responsive transcription factor RAP2-7-like | Transcription regulation | 979 | 2.5 | IV | |
| AP2 domain class transcription factor | Transcription regulation | 82 | 1.5 | 0 | |
| mul-miR2087-5p | PCI domain-containing protein-like | Transcription regulation; development | No | ||
| mul-miR2111a-5p | Myb-like transcription factor family protein | Transcription regulation | 123 | 3 | 0 |
| ACC oxidase 1 | Hormone metabolism | No | |||
| Disease resistance protein (TIR-NBS-LRR class) family | Stress response | 451 | 3 | 0 | |
| C2H2-like zinc finger protein | Transcription regulation | 2561 | 4 | III | |
| mul-miR2595 | RNA processing factor1 | RNA processing | No | ||
| MIF4G domain and MA3 domain-containing protein | RNA.processing | No | |||
| FAR1-related protein | Response to red or far red light | 478 | 2 | II | |
| Tir-nbs-lrr resistance protein | Stress response | 3271 | 2.5 | IV | |
| FKBP12 interacting protein 37 | Development | No | |||
| Pentatricopeptide repeat-containing protein | RNA processing; stress response | 373 | 3 | III | |
| Transcription factor MYB884 | Transcription regulation | 422 | 2 | III | |
| mul-miR393-5p | Transport inhibitor response protein | Signaling pathway | 678 | 4 | III |
| Auxin receptor 1 (TIR1) | Signaling pathway | 721 | 3.5 | II | |
| mul-miR394a-5p | OSIGBa0145C02.4 | Unknown | No | ||
| Aluminum-activated, malate transporter 12 | Transportion; stomatal movement | No | |||
| Methyltransferase PMT18-like | DNA processing | No | |||
| F-box family protein | Signaling pathway; development | 1522 | 2.5 | II | |
| mul-miR395a-3p | AP2 domain-containing transcription factor | Transcription regulation | 334 | 4 | I |
| Zinc finger (CCCH-type) family protein | Transcription regulation | 1724 | 3 | IV | |
| mul-miR415 | ARF domain class transcription factor (IAA2) | Transcription regulation | 77 | 4 | 0 |
| NBS-LRR resistance protein gene | Stress response | 142 | 3 | 0 | |
| Transcription factor BPE | Transcription regulation | 1150 | 4 | 0 | |
| Ubtilisin-like protease-like | Metabolic process | 2951 | 3 | IV | |
| ADP-ribosylation factor 1-like, transcript variant 2 | Carbohydrate metabolic process | 77 | 4 | 0 | |
| mul-miR4414a-3p | Uncharacterized protein | Unknown | No | ||
| mul-miR4414a-5p | Peptide deformylase | Protein biosynthesis | No | ||
| mul-miR447b-5p | Psb P-like protein | Photosystem II reaction | No | ||
| ATP binding microtubule motor family protein | Microtubule-based movement | No | |||
| Chitinase-like protein LA-b | Stress response | 38 | 4 | 0 | |
| mul-miR473a-5p | Aminopeptidase M1 | Hormone metabolism | 1251 | 2.5 | II |
| mul-miR477a-5p | Nucleic acid binding protein | Spliceosome assembly; stimulation of apoptosis | No | ||
| mul-miR5020a | RNA recognition motif-containing protein | Stress response | 631 | 3 | II |
| Galactosyltransferase family protein | Protein biosynthesis | No | |||
| mul-miR5072 | Heteroglycan glucosidase 1 | Carbohydrate metabolic process | No | ||
| mul-miR529b | Xyloglucan endotransglycosylase/hydrolase precursor | Carbohydrate metabolic process | 5556 | 4 | III |
| Chlorophyll synthase | Chlorophyll biosynthesis | 9002 | 4 | IV | |
| Cysteine-rich repeat secretory protein 6 | Stress response | No | No | ||
| SPL domain class transcription factor | Transcription regulation | 119 | 0.5 | 0 | |
| mul-miR530-3p | AWPM-19-like family protein | Unknown | No | ||
| Zinc finger family protein | Transcription regulation | 732 | 4 | III | |
| Esterase/lipase/thioesterase family protein | Metabolic process | 323 | 3 | II | |
| Beige-related and WD-40 repeat-containing protein | Unknown | No | |||
| Transcription regulator | Transcription regulation | 541 | 2.5 | II | |
| TIR-NBS-LRR type disease resistance protein | Stress response | 152 | 3 | II | |
| mul-miR530a | SWIB complex BAF60b domain-containing protein | Unknown | 1152 | 4 | 0 |
| Zinc knuckle (CCHC-type) family protein | Transcription regulation; growth | 101 | 3 | I | |
| mul-miR5565g-3p | Unknown | Unknown | No | ||
| mul-miR5658 | Anthranilate N-benzoyltransferase protein | Metabolic process | 63 | 4 | I |
| C2H2L domain class transcription factor | Transcription regulation | 53 | 3 | 0 | |
| Uncharacterized protein | Unknown | No | |||
| Eukaryotic translation initiation factor 2c | Transcription regulation; protein synthesis | No | |||
| Putative RNA recognition motif-containing protein | Stress response; ethylene biosynthetic process | No | |||
| Trihelix transcription factor GT-2-like | Transcription regulation | No | |||
| mul-miR5665 | RAB geranylgeranyl transferase alpha subunit 1 | Protein synthesis, response to cadmium ion | 65 | 2 | I |
| Shikimate kinase | Development | No | |||
| mul-miR6021 | Transcription factor | Transcription regulation | No | ||
| mul-miR6204 | SAUR-like auxin-responsive protein family | Hormone metabolism | No | ||
| E3 ubiquitin-protein ligase LOG2-like | Metabolic process | 2011 | 4 | III | |
| mul-miR6213 | ARM repeat-containing protein-like protein | Unknown | No | ||
| mul-miR6463 | Lactosylceramide 4-alpha-galactosyltransferase | Protein synthesis | No | ||
| mul-miR6475 | Tir-nbs-lrr resistance protein | Stress response | 79 | 3 | 0 |
| mul-miR858b | Transcription factor MYB32-like | Transcription regulation | 76 | 3.5 | 0 |
| MYB domain protein 51-2 | Transcription regulation | 23040 | 3.5 | III | |
| mul-miRn17-5p | Bystin-like | Ribosome biogenesis and assembly | No | ||
| ATP-dependent RNA helicase DHX8/PRP22 | RNA processing | No | |||
| mul-miRn19-5p | 40S ribosomal protein S20-2 | Protein biosynthesis | No | ||
| mul-miRn10-3p | Auxin response factor 19 | Auxin signaling; transcription regulation | 1271 | 3 | II |
| Squamosa promoter binding protein-like 14 | Transcription regulation | 656 | 2 | III | |
| Leucine-rich receptor-like protein kinase family protein | Signaling pathway | 155 | 2 | 0 | |
| S-formylglutathione hydrolase | Response to cadmium;methanal catabolism | No | |||
| Photosystem II reaction center PsbP family protein | PS.lightreaction.photosystem II | No | |||
| Protein kinase superfamily protein | Signaling pathway | No | |||
| mul-miRn11-5p | Nucleoside-triphosphatase | Catalysis of the hydrolysis of various bonds | No | ||
| mul-miRn18-5p | Haloacid dehalogenase-like hydrolase superfamily protein | Unknown | No | ||
| mul-miRn20-5p | Aminoacyl-tRNA synthetase-like | Protein synthesis | 231 | 3.5 | 0 |
| Chaperone protein dnaJ 13-like | Development | 1518 | 3.5 | III | |
| C2H2-type zinc finger protein | Transcription regulation | 42 | 3.5 | 0 | |
| Galactose oxidase | Carbohydrate metabolic process | No | |||
| Calmodulin binding protein | Transcription; stress response | No | |||
| F-box/kelch-repeat protein SKIP6 | Unknown | 322 | 2 | II | |
| Splicing factor 1 | RNA.processing | No | |||
| Dicer-like protein | RNA.processing | No | |||
| AP2 domain class transcription factor | Transcription regulation | No | |||
| Zinc finger-homeodomain protein 1 | Transcription regulation | 4541 | 2 | IV | |
| Pentatricopeptide repeat-containing protein | Stress response | No | |||
Figure 3. Target plots (t-plots) of miRNA targets in different categories confirmed by degradome sequencing.
(A) T-plot (top) and mRNA: miRNA alignments (bottom) for category 0 target, Unigene 2304 (NAC domain protein) transcripts. (B) As in (A) for Unigene 49225 (AP2 domain-containing transcription factor), a category I target for mul-miR395a-3p. (C) As in (A) for Unigene 13033 (TIR-NBS-LRR type disease resistance protein), a category II target for mul-miR530-3p. (D) As in (A) for Unigene 3229 (MYB domain protein 51-2), a category III target for mul-miR858b. (E) As in (A) for Unigene 2192 (Ethylene-responsive transcription factor RAP2-7-like), a category IV target for mul-miR72e-3p. The solid lines and dot in mRNA: miRNA alignments indicate matched and mismatch RNA base pairs, respectively.
In most cases, the identified miRNAs were predicted to cleave two or more different targets. For example, three members of SPL transcription factor family genes were predicted to be cleaved by mul-miR157a-5p, and mul-miR160a-5p was predicted to slice three genes belonging to the auxin response factor. Inconstantly, some miRNAs were only predicted to cleave one target such as mul-miR1030a, mul-miR164c-3p, mul-miR6463, and mul-miR6475. Unfortunately, we could not detect the cleavage signature for most of miRNAs in this degradome library. The low identification frequency might be because that their target mRNAs have not been annotated in our mulberry transcriptome data.
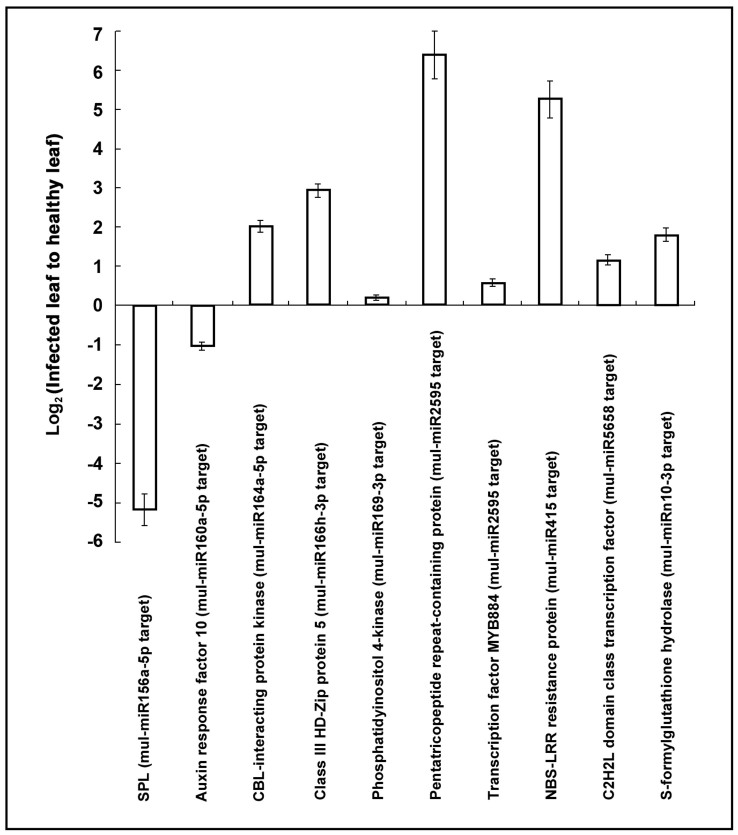
The expressions of the targets of phytoplasma-responsive miRNAs were investigated by real-time PCR in response to phytoplasma-infection (Fig. 4). The results indicated that most of the target genes showed a negative correlation with the corresponding miRNA expression, and this is consistent with miRNA function in guiding the cleavage of target mRNAs. But it should be noted that a similar expression pattern between the target gene and its corresponding miRNAs was also observed. For instance, mul-miR169a-3p was positively correlated with its target gene encoding phosphatidylinositol 4-kinase. Of course, the putative target genes were bioinformatically predicted, and are subjected to experimental verification.
Figure 4. Predicted target abundance analysis by real-time PCR.
The relative gene expression was evaluated using comparative Ct method taking actin (Accession No. DQ785808) as the reference gene. The log2 values of the ratio of phytoplasma-infected leaves to the healthy leaves are plotted. Values are given as mean ± SD of three experiments in each group.
GO function analysis of targets
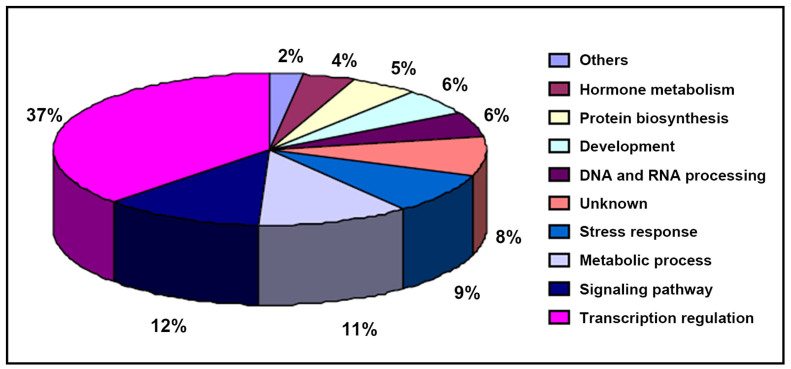
To better understand miRNA functions, we subjected the identified target genes to Gene Ontology (GO) analysis. The result of GO analysis demonstrated that the target genes of the phytoplasma-responsive miRNAs were classified into 10 categories according to their ontologies in Arabidopsis based on KEGG functional annotations (Fig. 5). The first category of the predicted target genes encoding an array of transcription factors involved in regulation of gene expression. In the second category, target genes associated with signaling pathway, indicating that there were many signaling pathways involved in phytoplasma infection. Target genes involved in metabolic process belong to the third category, suggesting that phytoplasma infection may change diverse metabolic processes in infected plant. The other categories included stress response, development, DNA and RNA processing, and so on. In most cases, targets of the same miRNA were with similar functions (Table 5), however, some miRNAs, such as mul-miR394a-5p, target many genes with different functions. So these miRNAs might be play potential roles in the expression control of genes related to diverse biologic processes, and the regulation networks of miRNAs involved in the response of mulberry to phytoplasma-infection were intricate.
Figure 5. Percentage distribution of the predicted target genes for the differentially expressed miRNAs in various categories.
Identification and expression profiling of siRNAs
According to the standard that siRNAs were a pair of perfectly complementary sRNAs with 2 nt overhangs at the 3′-end, 315 910 and 271 367 unique siRNA sequences were identified in the healthy and infected leaf small RNA libraries, respectively. The siRNAs identified varied in length from 20 to 24 nt, and the 24 nt siRNAs, which were considered as long siRNAs, were the major size groups (Fig. 6). In order to determine the siRNAs responsive to phytoplasma infection, we looked for differential siRNAs by comparing the normalized expression levels of the siRNAs between the healthy and infected leaf libraries. A total of 14 598 siRNAs were identified to be significantly responsive to phytoplasma infection. Among these differential siRNAs, 10 745 siRNAs were significantly decreased and 3853 siRNAs were up-regulated in the infected leaf (P < 0.05, fold 2.0) (Fig. 6; Supplementary Text).
Figure 6. Length distribution of the siRNAs in mulberry(A) and scatter plots display the levels of siRNAs in healthy and infected leaf libraries (B).
MulMIR393A is induced by phytoplasma infection
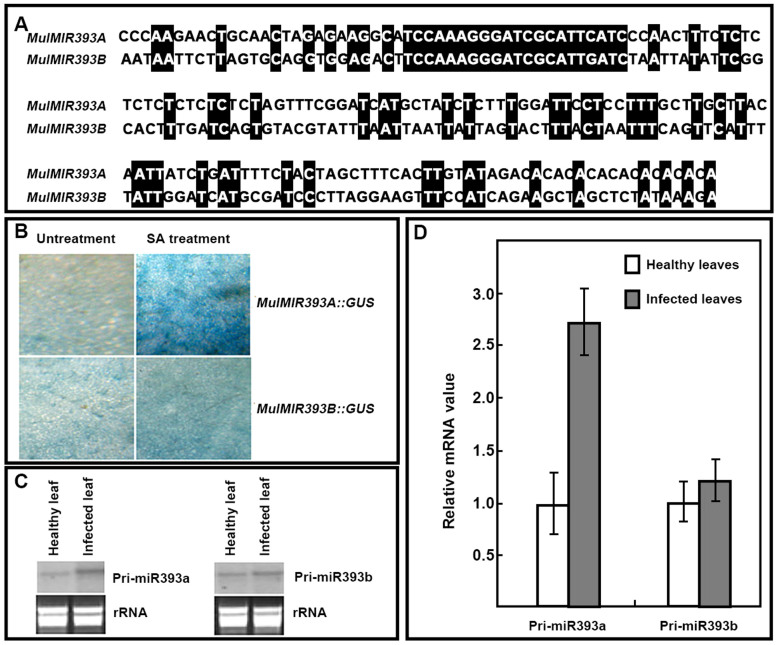
It was reported that Arabidopsis miR393 plays a role in plant PAMP-triggered immunity in Arabidopsis10. Though phytoplasma lacks cell wall and flagellum, and has no PAMPs like flg224,6, our data showed that mul-miR393 was also induced by phytoplasma infection (Table 2). Therefore, mul-miR393 was induced by phytoplasma through some pathway other than flg22. MiR393 is potentially encoded by two genes, AtMIR393A and AtMIR393B, in Arabidopsis18. To study mul-miR393 function in the response to phytoplasma infection, we cloned the two genes encoding mul-miR393, MulMIR393A and MulMIR393B, in mulberry. Sequence analysis showed that limited homology exists between the MulMIR393A and MulMIR393B, but the mature mul-miRNA393-5p sequences produced are identical (Fig. 7A). Meanwhile, the promoters of MulMIR393A and MulMIR393B were cloned, and cis-acting regulatory elements analysis revealed that the two promoters were different, such as the promoter of MulMIR393A has a cis-acting element involved in salicylic acid responsiveness, but this was not found in the promoter of MulMIR393B. GUS activity analysis indicated that MulMIR393A::GUS showed induced GUS activity, but in contrast MulMIR393B::GUS showed no induced GUS activity after application of salicylic acid (SA) (Fig. 7B), confirming the cis-acting regulatory elements analysis results above. Therefore, MulMIR393A and MulMIR393B have different regulators. Previous studies showed that miR393 is a developmentally regulated miRNA which arises predominantly from AtMIR393B in aerial organs of Arabidopsis25. But when the plants were treated with flg22, the expressed level of miR393 was up-regulated mainly from activated transcription of AtMIR393A10. Northern blot and real-time PCR analysis results showed that the transcript level of MulMIR393A was increased in the infected leaves, but the transcript level of MulMIR393B was unaltered (Fig. 7C, D). Therefore, the mul-miR393 accumulation resulted from the increased transcripts of MulMIR393A in responding to phytoplasma infection.
Figure 7. Comparison of nucleotide sequences of MulMIR393A and MulMIR393B, MulMIR393A::GUS and MulMIR393B::GUS activity, and pri-miR393a pri-miR393b abundance analysis.
(A) Comparison of nucleotide sequences of MulMIR393A and MulMIR393B. Identical nucleotides were black shaded. (B) MulMIR393A::GUS and MulMIR393B::GUS activity analysis by histochemical staining. Histochemical staining for GUS activity was performed 24 h later after application of SA with X-gluc as a substrate. (C) Pri-miR393a and pri-miR393b abundance analysis by northern blot. (D) Pri-miR393a and pri-miR393b abundance analysis by real-time PCR. The relative gene expression was evaluated using comparative Ct method taking actin (Accession No. DQ785808) as the reference gene. Values are given as mean ± SD of three experiments in each group.
Discussion
So far, miRNAs and their regulatory functions have been extensively characterized in model plant species, but to our knowledge, it was the first report about the sRNAs of mulberry. In present study, solexa high-throughput sequencing of small RNAs revealed a diverse and complex small RNA population. Among the 62 conserved phytoplasma-responsive miRNAs identified in mulberry, only 6 miRNAs, including miR157a, miR172a-3p, miR4414, miR473, miR477a, and miR479, were also differentially expressed in Candidatus Phytoplasma aurantifolia infected Mexican lime (Citrus aurantifolia L.)15. Therefore, the effect of phytoplasma infection on the expression of miRNAs depends on the pathogen and host types. Furthermore, it was found that the miRNAs of the same family were differentially regulated in the response to phytoplasma-infection. For instance, the expression level of mul-miR160a-5p was up-regulated by phytoplasma-infection, but the expression level of mul-miR160b-3p showed no obvious change in the infected leaves (Table 2). So the gene expression programmings of the miRNAs, which share a high degree of sequence similarity and belong to the same family, were controlled by different regulator16.
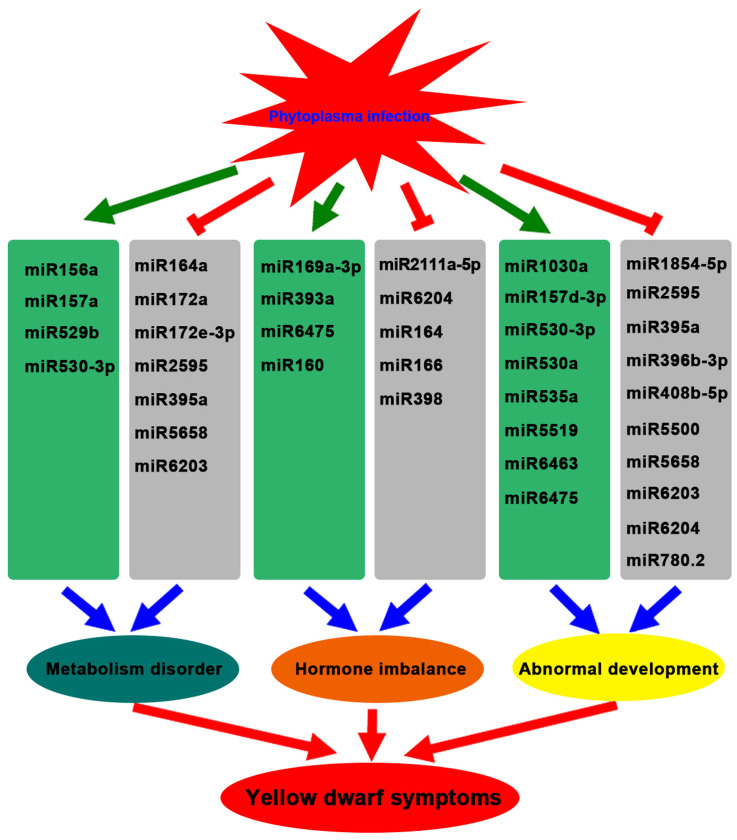
Phytoplasma-associated disorders are believed to be the auxinic diseases of plants26. It was reported that many auxin response factors were down-regulated by the effector (tengu) of Onion yellows phytoplasma, and suggested that the symptoms caused by phytoplasma may be involved in an auxin-related pathway5. In plants, a group of miRNAs regulate and fine-tune multiple hormone signaling pathways coordinately27. Our results showed some miRNAs, which target the genes associated with auxin signaling and metabolism, were differentially expressed between the healthy and infected leaves. MiR393 was the first miRNA identified which targets auxin receptor mRNAs and subsequently represses auxin signaling in Arabidopsis10, and we found that mul-miR393 was up-regulated in the infected leaves (Table 2). In addition to miR393, target analyses showed that mul-miRn10-3p was also involved in auxin signaling pathway by regulating auxin response factor (Table 5). Moreover, miR160, miR164 and miR166, which were associated with auxin signaling pathway by regulating different transcription factors13, which were found differentially expressed in healthy and infected leaves (Table 2). Furthermore, the differential mul-miR169a-3p and mul-miR6204 were predicted to target the genes coding for O-fucosyltransferase family protein and SAUR-like auxin-responsive protein family, respectively, which were associated with auxin metabolism (Table 5). Therefore, these differentially expressed miRNAs may play a role in altering auxin signaling and metabolism in infected plants.
Beside the auxin signaling pathway, miRNAs were also involved in regulating other hormone signaling pathways. For instance, miR164, miR166 and miR398 were reported involved in abscisic acid signaling pathway28,29,30. MiR166 and miR172 were found to be down-regulated by gibberellin31 and cytokinin32, respectively. The expression levels of these miRNAs were differential between healthy and infected leaves (Table 2). In addition, mul-miR2111a-5p was predicted to target the gene coding ACC oxidase 1 associated with ethylene metabolism, and mul-miR2595 and mul-miRn20-5p were predicted to target the pentatricopeptide repeat-containing protein gene involved in the response to SA stimulus (Table 5). Therefore, it is more likely that the phytoplasma infection-induced alteration in hormonal signaling triggers reprogramming of the plant growth and developmental pattern, leading to some symptoms in infected plants. This is in accord with early reports that phytoplasma infection disturbs hormonal balance in the host plants4,33,34,35,36.
Symptoms induced in infected plants suggest that phytoplasma infection may modulate developmental processes within the plant host37. This was confirmed by the reports that the expression of effector protein (phyllogen) of phytoplasmas in Arabidopsis can alter floral development, resulting in the production of leaf-like flowers38,39. In Arabidopsis, many Squamosa Promoter-Binding Protein-Like (SPL) genes are post-transcriptionally regulated by miR156. Meanwhile, some SPL genes, such as AtSPL9, in turn positively regulate the expression of miR172. This forms the miR156-AtSPL9-miR172 regulatory pathway40. Our results showed that mul-miR156 was up-regulated and its target gene SPL9 was down-regulated, meanwhile the mul-miR172 was down-regulated (Table 2; Fig. 2A). Therefore, the miR156-SPL9-miR172 regulatory pathway may be also conserved in mulberry and responsive to phytoplasma infection. SPL family of transcription factors play important roles in flower and fruit development, plant architecture and phase transition40. AP2 domain-containing transcription factor, which is the target of mul-miR172, takes part in regulating flowering time and floral organ identity41. Therefore, the expression changes of miR156 and miR172 might disturb the plant development process and be responsible for the mulberry yellow dwarf symptoms such as development of green leaf-like structures instead of flowers and sterility of flowers.
In addition, our results showed some other miRNAs, which target the genes associated with plant growth and development, were also differentially expressed. For example, the expression level of miR166h-3p, which targets the class III HD-Zip protein 6 (Table 2, 5) involved in the regulation of shoot meristem initiation, vascular development42, was down-regulated by phytoplasma infection, and this probably resulted in an abnormal plant development such as meristem defects and was partially responsible for the symptoms such as dwarf and witches' broom. Meanwhile, miR160, miR162, miR164, miR165, miR168, and miR319 were reported to modulate plant development through different pathway41,43,44,45,46,47,48. These up or down-regulation of these miRNAs (Table 2) may disorder the expressions of many genes involved in diverse development processes and change the architecture of infected plants causing some symptoms in the infected plants.
Our data showed that some differentially expressed miRNAs target the genes associated with CHO metabolism, protein metabolism, lipid metabolism, and so on. For example, the up-regulated mul-miR160a-5p may down-regulate its target, beta-amylase gene (Table 2, 5), and responsible for the accumulation of starch in the infected leaves. Similarly, the up-regulated mul-miR529b may down-regulate its target, chlorophyll synthase gene (Table 2, 5), resulting in the blockage of the biosynthesis of chlorophyll and the decoloration symptoms. Thus, the differential expressions of these miRNAs might disturb the normal metabolism process and partly responsible for some of the mulberry yellow dwarf symptoms. Based on the information obtained, the complex regulatory networks of miRNAs involved in the response of mulberry to phytoplasma-infection is outlined in Figure 8.
Figure 8. Complex regulatory networks of phytoplasma-responsive microRNAs.
Phytoplasma-responsive miRNAs target different genes involved in diverse biological processes, contribute to fine-tuning in defense responses and trigger metabolism, hormone and development perturbation and promote disease development.
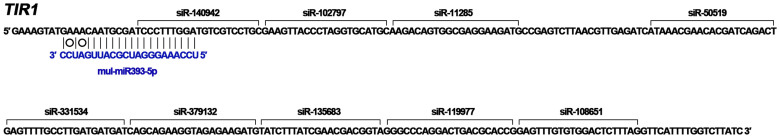
Unlike miRNAs, the lengths of siRNAs are more variable, ranging from 21 to 30 nt. The biogenesis of the siRNAs depends on specific RNA-dependent RNA polymerases and Dicer-like (DCL) proteins and RNA polymerase IV49. The siRNAs produced by different DCLs may have different lengths and functions50. In addition to the relatively limited number of miRNAs, thousands of differentially expressed siRNAs were involved in the response of mulberry to phytoplasma-infection. The lengths of these siRNAs are more variable (Fig. 6), and the targeted genes of these differentially expressed siRNAs were associated with a wide range of functions in hormone networks, development and metabolism. It was reported that the 22-nt forms of miRNAs are important for triggering secondary siRNA production in plants51,52. Previous research showed that miR393 is necessary to initiate the production of the secondary siRNAs from the transport inhibitor response 1/auxin signaling F-box 2 (TIR1/AFB2) clade of auxin receptors (TAARs) transcripts in Arabidopsis25. Sequencing results showed that mul-miR393 is 22-nt long, so we wondered whether it can trigger the production of the secondary siRNAs. Our small RNA library sequencing data showed there were some siRNAs matching to the TIR1 mRNA downstream of the mul-miR393-5p binding sites (Figure 9). Thus, mul-miR393 might also trigger the production of the secondary siRNAs. However, we can't predict which one is necessary to trigger the production of these siRNAs, mul-miR393a or mul-miR393b. Future studies will be carried out to explore the functions of these phytoplasma-responsive siRNAs, and identify the pathways connected with the response against phytoplasma-infection. In conclusion, the information provided here would be particularly useful for understanding the function of sRNAs in mulberry and be aid to reveal the mechanisms underlying of phytoplasma pathogenicity and the symptoms they cause in host plants.
Figure 9. The production of secondary siRNAs triggered by mul-miR393-mediated cleavage of TIR1 mRNA.
Methods
Plant material
One-year-old cutting seedlings derived from the same mother tree, Husang 32 (Morus multicaulis Perr.), were used as rootstocks which were grafted with the scions collected from healthy and phytoplasma-infected mulberry trees (Husang 32), respectively. All the grafts were incubated in a growth chamber at 26°C, humidity 90% and under 12 h of light. Six weeks after infection, the plant showing typical symptoms as yellowing of the leaves, stunting, and witches'-broom were confirmed by PCR assay with an amplified fragment of the 16S rRNA gene (GenBank Accession No. EF532410) and TEM detection as described before1. The plants that were phytoplasma-positive by PCR assay and TEM detection were used as treatments while the plants grafted with the scions collected from healthy trees were used as controls.
Small RNA library construction and high-throughput sequencing
Leaves were collected from the infected plants and the controls, and RNAs were isolated from the leaves of healthy and infected plants separately using TRIzol kit (Invitrogen) following the manufacturer's instructions. Total RNAs were separated on 15% denaturing PAGE and small RNAs between 14 and 30 nt were enriched and ligated with 5′ and 3′ adaptors. The ligation products were purified and amplified after reverse transcription. The amplification products were purified and used for sequencing by Solexa machine.
Conserved miRNAs in mulberry
The raw sequences tags from HiSeq sequencing were cleaned by getting rid of adaptors, low-quality tags, as well as contaminants to generate clean reads. Length distribution of clean tags is then summarized, and the trimmed sequences longer than 18 nt were used for further analyses. The remaining clean reads were annotatated into different categories through standard bioinformatics analysis. Subsequently, the sequences were compared with a miRBase database v19.0 (http://www.mirbase.org/). The sequences with identical or related sequences from other plants were identified as conserved miRNAs, and the other sequences were analyzed for novel miRNA candidates.
Novel miRNA candidates
To discover novel miRNAs, all non-annotated clean tag sequences were mapped to our mulberry transcriptome database. The characteristic hairpin structure of miRNA precursor was used to predict novel miRNA using the Mireap algorithm (http://sourceforge.net/projects/mireap/) by exploring the secondary structure, the dicer cleavage site, the minimum free energy, and other criteria provided by before53.
Identification of siRNAs
To identify siRNAs, tags from clean reads were aligned with each other. A pair of perfectly complementary sRNAs with 2 nt overhangs at the 3′-end were considered as siRNAs.
Differential expression analyses of miRNAs and siRNAs
The expression of miRNAs and siRNAs in healthy and infected leaf samples were normalized to obtain the expression of transcripts per million, and only the sRNA with an expression value of ≥1, it was considered in future differential expression analysis. Based on the normalized expression analysis, the fold-change and P-value were calculated according to the following equation. Normalized expression = (Actual sRNA sequencing reads count/Total clean reads count) ×1,000,000; Fold change = Log2 (x/y); P-value formula:
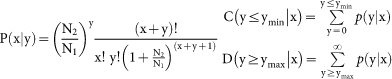 |
The N1 and N2 represent total count of clean reads of a given sRNA in the sRNA library of phytoplasma infected and healthy leaves, respectively. The x and y represent normalized expression level of a given sRNA in the sRNA library of phytoplasma infected and healthy leaves, respectively.
Prediction and validation of targets of differential miRNAs
The sequences from the mulberry transcriptome data were chosen to predict the miRNA targets, and the plant small RNA target analysis server, psRNATarget (http://plantgrn.noble.org/psRNATarget/), was used to predict the target genes following the rules suggested by Allen et al.54. All the predicted miRNA targets were experimentally verified by mRNA degradome sequencing. A degradome sequencing library was constructed using mRNAs isolated from the healthy and infected leaves according to the protocols described previously55,56 and then were submitted for parallel analysis of RNA end (PARE) sequencing on a Solexa Analyzer. All the adaptor sequences and low quality sequencing reads were removed from the raw sequencing reads to obtain the clean reads with the length of 20 and 21 nt, which were used to identify potentially cleaved targets with the software package, CleaveLand 3.0, as previously described55,56. The clean reads were mapped to the mulberry transcriptome unigene sequences obtained, and only the perfect matching reads were kept and extend to 35–36 nt by adding 15 nt of upstream of the sequence. All resulting reads were reverse-complemented and aligned to the miRNA identified and scored by previously described method54. All targets identified were classified into five categories based on the abundance of the resulting mRNA tag relative to the overall profile of degradome reads that matched the target57. When a target mRNA was predicted or identified, a BlastN search against a reference Arabidopsis thaliana database downloaded from TAIR (http://www.arabidopsis.org/; version TAIR10) was used to provide the gene ontologies, and then GO analysis was performed for the matched Arabidopsis accession entries based on their TAIR GO categories.
Quantitative real-time PCR analysis for miRNAs and mRNAs
RNA was extracted using the TRIzol® reagent following the manufacturer's recommendations (Invitrogen) and digested with DNase I. Real-time PCR was performed using the PrimeScript™ miRNA qPCR Starter Kit Ver.2.0 for miRNAs and the SYBR Premix Ex TaqTM kit for mRNA according to the manufacturer's protocol on the Rotor-Gene 3000A system. The U6 and actin genes were amplified as reference genes for miRNA and mRNA normalization, respectively. The primers (Supplementary table 1, 2) used to amplify the genes were designed based on our available mulberry transcriptome data. Comparative cycle threshold (Ct) method58 was used to evaluate the relative gene expression level. All samples were assayed in triplicate.
RNA gel blotting
Twenty micrograms of total RNA of each sample isolated as above were separated on 1.2% formaldehyde denatured agarose gel, and then were blotted onto nylon Hybond N membrane (Hybond N, Amersham). The blots were hybridized with digoxigenin-labeled RNA probes prepared using the PCR DIG Probe Synthesis Kit (Roche, Germany). Prehybridization, hybridization, membrane washing, and detection were carried out as described previously59.
Promoter analysis of MulMIR393A and MulMIR393B genes
The promoter sequences of MulMIR393A and MulMIR393B genes were cloned using Tail-PCR method and analyzed using the program online (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) to search for cis-acting motifs. Then they were cloned into the vector pBI121, which harbors the promoter 35S and the GUS gene, to replace 35S, respectively. The pBI121 vector containing MulMIR393A::GUS or MulMIR393B::GUS was introduced into Agrobacteriun tumefaciens strain GV3101. Tobacco (Nicotiana benthamiana) plants were used for transformation by infection of leaf sections with A. tumefaciens strain harboring the constructs described above. Selected transgenic shoots were treated with SA by dipping the leaves into a solution containing 1 mM SA, and the leaves treated with water were used as control. Histochemical staining for GUS activity was performed 24 h later using the method described by Jefferson60 with X-gluc as a substrate.
Supplementary Material
Supplementary Information
Acknowledgments
This work was funded by the national natural science foundation of China (No. 30972366, 31070573, 31100478) and science foundation for the excellent youth scholars of Shandong province of China (No. BS2009NY024, BS2010NY015).
Footnotes
The authors declare no competing financial interests.
Author Contributions Y.P. and X.L. were responsible for design, sample preparations, and contributed to write the manuscript. Y.Q. and F.Y. were responsible for high-throughput deep sequencing analysis. C.Z. and Y.Y. were responsible for PCR analysis. H.L. and H. were responsible for mul-miR393 analysis. All authors edited the manuscript and reviewed the manuscript.
References
- Ji X. L., Gai Y. P., Zheng C. C. & Mu Z. M. Comparative proteomic analysis provides new insights into mulberry dwarf responses in mulberry (Morus alba L.). Proteomics. 9, 5328–5339 (2009). [DOI] [PubMed] [Google Scholar]
- Kumar V. & Gupta V. P. Scanning electron microscopy on the perithecial development of Phyllactinia corylea on mulberry-II. Sexual stage. J. Phytopathol. 152, 169–173 (2004). [Google Scholar]
- Jiang H. et al. Distribution patterns of mulberry dwarf phytoplasma in reproductive organs, winter buds, and roots of mulberry trees. J. Gen. Plant Pathol. 70, 168–173 (2004). [Google Scholar]
- Lee I. M., Davis R. E. & Gundersen-Rindal D. E. Phytoplasma: Phytopathogenic mollicutes. Annu. Rev. Microbiol. 54, 221–255 (2000). [DOI] [PubMed] [Google Scholar]
- Hoshi A. et al. A unique virulence factor for proliferation and dwarfism in plants identified from a phytopathogenic bacterium. Proc. Natl. Acad. Sci. U. S. A. 106, 6416–6421 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertaccini A. & Duduk B. Phytoplasma and phytoplasma diseases: a review of recent research. Phytopathol. Mediterr. 48, 355–378 (2009). [Google Scholar]
- Christensen N. M., Axelsen K. B., Nicolaisen M. & Schulz A. Phytoplasmas and their interactions with hosts. Trends Plant Sci. 10, 526–535 (2005). [DOI] [PubMed] [Google Scholar]
- Hill M. K., Lyon K. J. & Lyon B. R. Identification of disease response genes expressed in Gossypium hirsutum upon infection with the wilt pathogen Verticillium dahliae. Plant Mol. Biol. 40, 289–296 (1999). [DOI] [PubMed] [Google Scholar]
- Chen D. et al. Plant siRNAs from introns mediate DNA methylation of host genes. RNA 17, 1012–1024 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro L. et al. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312, 436–439 (2006). [DOI] [PubMed] [Google Scholar]
- Agorio A. & Vera P. ARGONAUTE4 is required for resistance to Pseudomonas syringae in Arabidopsis. Plant Cell 19, 3778–3790 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S., Sun Y. H., Amerson H. & Chiang V. L. MicroRNAs in loblolly pine (Pinus taeda L.) and their association with fusiform rust gall development. Plant J. 51, 1077–1098 (2007). [DOI] [PubMed] [Google Scholar]
- Xin M. et al. Diverse set of microRNAs are responsive to powdery mildew infection and heat stress in wheat (Triticum aestivum L.). BMC Plant Biol. 10, 123 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J. P. et al. Phylogenetic analysis and molecular evolution patterns in the MIR482-MIR1448 polycistron of Populus L. PLoS One 7, e47811 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehya F. et al. Phytoplasma-responsive microRNAs modulate hormonal, nutritional, and stress signalling pathways in Mexican lime trees. PLoS One 8, e66372 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y. D., Gan Q. H., Chi X. Y. & Qin S. Roles of microRNA in plant defense and virus offense interaction. Plant Cell Rep. 27, 1571–1579 (2008). [DOI] [PubMed] [Google Scholar]
- Dunoyer P. et al. An endogenous, systemic RNAi pathway in plants. EMBO J. 29, 1699–1712 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Sunkar R. & Zhu J. K. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16, 2001–2019 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z. et al. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2, e104 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C. et al. Elucidation of the small RNA component of the transcriptome. Science 309, 1567–1569 (2005). [DOI] [PubMed] [Google Scholar]
- Katiyar-Agarwal S. et al. A pathogen-inducible endogenous siRNA in plant immunity. Proc. Natl. Acad. Sci. U. S. A. 103, 18002–18007 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katiyar-Agarwal S., Gao S., Vivian-Smith A. & Jin H. A novel class of bacteria-induced small RNAs in Arabidopsis. Genes Dev. 21, 3123–3134 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B. H., Pan X. P., Cox S. B., Cobb G. P. & Anderson T. A. Evidence that miRNAs are different from other RNAs. Cell Mol. Life Sci. 63, 246–254 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet E., Wuyts J., Rouzé P. & Van de Peer Y. Evidence that microRNA precursors, unlike other non-coding RNAs, have lower folding free energies than random sequences. Bioinformatics 20, 2911–2917 (2004). [DOI] [PubMed] [Google Scholar]
- Si-Ammour A. et al. MiR393 and secondary siRNAs regulate expression of the TIR1/AFB2 auxin receptor clade and auxin-related development of Arabidopsis leaves. Plant Physiol. 157, 683–691 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musetti R. Biochemical changes in plants infected by phytoplasmas. Phytoplasmas: genomes, plant hosts and vectors (eds Weintraub, P. G. & Jones, P. CABI press, Wallingford, UK.), 132–146 (2010). [Google Scholar]
- Zhang W. et al. Bacteria-responsive microRNAs regulate plant innate immunity by modulating plant hormone networks. Plant Mol. Biol. 75, 93–105 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R., Chinnusamy V., Zhu J. & Zhu J. K. Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci. 12, 301–309 (2007). [DOI] [PubMed] [Google Scholar]
- Jagadeeswaran G., Saini A. & Sunkar R. Biotic and abiotic stress down-regulate miR398 expression in Arabidopsis. Planta 229, 1009–1014 (2009). [DOI] [PubMed] [Google Scholar]
- Khraiwesh B., Zhu J. K. & Zhu J. Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim. Biophys. Acta 1819, 137–148 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q. et al. Expression analysis of phytohormone-regulated microRNAs in rice, implying their regulation roles in plant hormone signaling. FEBS Lett. 583, 723–728 (2009). [DOI] [PubMed] [Google Scholar]
- Jung J. H., Yun J., Seo Y. H. & Park C. M. Characterization of an Arabidopsis gene that mediates cytokinin signaling in shoot apical meristem development. Mol. Cells 19, 342–349 (2005). [PubMed] [Google Scholar]
- Davey J. E., Van Staden J. & De Leeuw G. T. N. Endogenous cytokinin levels and development of flower virescence in Catharanthus roseus infected with mycoplasmas. Physiol. Plant Pathol. 19, 193–200 (1981). [Google Scholar]
- Pertot I., Musetti R., Pressacco L. & Osler R. Changes in indole-3-acetic acid level in micropropagated tissues of Catharanthus roseus L. infected by the agent of the clover phyllody and effect of exogenous auxins on phytoplasma morphology. Cytobios 95, 13–23 (1998). [Google Scholar]
- Tan P. Y. & Whitlow T. Physiological response of Catharanthus roseus (periwinkle) to ash yellows phytoplasmal infection. New Phytol. 150, 757–769 (2001). [Google Scholar]
- Gai Y. P. et al. Metabolomic analysis reveals the potential metabolites and pathogenesis involved in mulberry yellow dwarf disease. Plant Cell Environ. 37, 1474-1490 (2014). [DOI] [PubMed] [Google Scholar]
- Wei W., Davis R. E., Nuss D. L. & Zhao Y. Phytoplasmal infection derails genetically preprogrammed meristem fate and alters plant architecture. Proc. Natl. Acad. Sci. U. S. A. 110, 19149–19154 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean A. M. et al. Phytoplasma effector SAP54 induces indeterminate leaf-like flower development in Arabidopsis plants. Plant Physiol. 157, 831–841 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maejima K., Oshima K. & Namba S. Exploring the phytoplasmas, plant pathogenic bacteria. J. Gen. Plant Pathol. 80, 210–221 (2014). [Google Scholar]
- Chen X. et al. SQUAMOSA promoter-binding protein-like transcription factors: star players for plant growth and development. J. Integr. Plant Biol. 52, 946–951 (2010). [DOI] [PubMed] [Google Scholar]
- Chen X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303, 2022–2025 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawker N. P. & Bowman J. L. Roles for class III HD-Zip and KANADI genes in Arabidopsis root development. Plant Physiol. 135, 2261–2270 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palatnik J. F. et al. Control of leaf morphogenesis by microRNAs. Nature 425, 257–263 (2003). [DOI] [PubMed] [Google Scholar]
- Xie Z., Kasschau K. D. & Carrington J. C. Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA guided mRNA. Curr. Biol. 13, 784–789 (2003). [DOI] [PubMed] [Google Scholar]
- Juarez M. T., Kui J. S., Thomas J., Heller B. A. & Timmermans M. C. MicroRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature 428, 84–88 (2004). [DOI] [PubMed] [Google Scholar]
- Laufs P., Peaucelle A., Morin H. & Traas J. MicroRNA regulation of the CUC genes is required for boundary size control in Arabidopsis meristems. Development 131, 4311–4322 (2004). [DOI] [PubMed] [Google Scholar]
- Vaucheret H., Vazquez F., Crété P. & Bartel D. P. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 18, 1187–1197 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T. W., Xue L. G. & An L. Z. Functional diversity of miRNA in plants. Plant Sci. 172, 423–432 (2007). [Google Scholar]
- Ghildiyal M. & Zamore P. D. Small silencing RNAs: an expanding universe. Nat. Rev. Genet. 10, 94–108 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y., Denli A. M. & Hannon G. J. Biochemical specialization within Arabidopsis RNA silencing pathways. Mol. Cell 19, 421–428 (2005). [DOI] [PubMed] [Google Scholar]
- Chen H. M. et al. 22-Nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc. Natl. Acad. Sci. U. S. A. 107, 15269–15274 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuperus J. T. et al. Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis. Nat. Struct. Mol. Biol. 17, 997–1003 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers B. C. et al. Criteria for annotation of plant microRNAs. Plant Cell 20, 3186–3190 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen E., Xie Z., Gustafson A. M. & Carrington J. C. MicroRNA directed phasing during trans-acting siRNA biogenesis in plants. Cell 121, 207–221 (2005). [DOI] [PubMed] [Google Scholar]
- Addo-Quaye C., Eshoo T. W., Bartel D. P. & Axtell M. J. Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr. Biol. 18, 758–762 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- German M. A. et al. Global identification of microRNA–target RNA pairs by parallel analysis of RNA ends. Nat. Biotechnol. 26, 941–946 (2008). [DOI] [PubMed] [Google Scholar]
- Yang J. et al. Identification of miRNAs and their targets using high-throughput sequencing and degradome analysis in cytoplasmic male-sterile and its maintainer fertile lines of Brassica juncea. BMC Genomics 14, 9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J. & Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative RT-PCR and the 2−ΔΔCT method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- Matsuda N., Tsuchiya T., Kishitani S., Tanaka Y. & Toriyama K. Partial male sterility in transgenic tobacco carrying antisense and sense PAL cDNA under the control of a tapetum-specific promoter. Plant Cell Physiol. 37, 215–222 (1996). [Google Scholar]
- Jefferson R. A. Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol. Biol. Rep. 5, 387–405 (1987). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information