Abstract
Chlamydia suis is an endemic pig pathogen, belonging to a fascinating genus of obligate intracellular pathogens. Of particular interest, this is the only chlamydial species to have naturally acquired genes encoding for tetracycline resistance. To date, the distribution and mobility of the Tet-island are not well understood. Our study focused on whole genome sequencing of 29 C. suis isolates from a recent porcine cohort within Switzerland, combined with data from USA tetracycline-resistant isolates. Our findings show that the genome of C. suis is very plastic, with unprecedented diversity, highly affected by recombination and plasmid exchange. A large diversity of isolates circulates within Europe, even within individual Swiss farms, suggesting that C. suis originated around Europe. New World isolates have more restricted diversity and appear to derive from European isolates, indicating that historical strain transfers to the United States have occurred. The architecture of the Tet-island is variable, but the tetA(C) gene is always intact, and recombination has been a major factor in its transmission within C. suis. Selective pressure from tetracycline use within pigs leads to a higher number of Tet-island carrying isolates, which appear to be lost in the absence of such pressure, whereas the loss or gain of the Tet-island from individual strains is not observed. The Tet-island appears to be a recent import into the genome of C. suis, with a possible American origin.
Keywords: phylogenetics, farming, whole genome sequencing, tetracycline resistance, recombination
Introduction
Bacteria within the genus Chlamydia are biologically unique, obligate intracellular pathogens with a wide range of eukaryotic hosts. The species Chlamydia suis is an endemic pig pathogen that is associated with conjunctivitis, rhinitis, pneumonia, enteritis, and reproductive disorders (Schautteet and Vanrompay 2011). This pathogen is primarily found in the porcine gastrointestinal tract as an inapparent infection (Schautteet and Vanrompay 2011), with one recent longitudinal investigation of Swiss fattening pig farms revealing evidence of endemic shedding rates of >90% (Hoffmann et al. 2015). Whereas the animal health impact of these high rates of infection is still unclear, the same study found a positive correlation between the intestinal presence of C. suis and diarrhea (Hoffmann et al. 2015).
A very interesting aspect of this pathogen, in addition to its pathogenic potential, is that this chlamydial species is the only one known to have naturally acquired antibiotic resistance. The Austrian type strain C. suis S45 is sensitive to tetracycline (Kaltenboeck et al. 1992; Perez-Martinez and Storz 1985). Resistance was first reported within US strains of C. suis in 1998 (Lenart et al. 2001), enabled by the tetracycline efflux pump-encoding gene tetA(C) present on a Tet-island (Dugan et al. 2004). The presence of tetA(C) has since also been confirmed in isolates from Italy (Di Francesco et al. 2008), Switzerland (Borel et al. 2012), Belgium, Cyprus, and Israel (Schautteet et al. 2013), indicating that this phenomenon is globally distributed. Very recently, it was shown that tet operon expression in C. suis occurs even in the absence of tetracycline, which is unusual within Gram-negative bacteria (Donati et al. 2016).
Transfer of tetracycline resistance between C. suis, Chlamydiatrachomatis and Chlamydiamuridarum, has been demonstrated in vitro (Jeffrey et al. 2013; Marti et al. 2017; Suchland et al. 2009), yet genetic manipulation of Chlamydia species is still largely in its infancy (Wang et al. 2011, 2012). As such, genomic studies represent a critical method of investigation and have provided valuable insights into the lifestyle and evolution of human restricted C.trachomatis and avian-associated Chlamydia psittaci (Harris et al. 2012; Joseph et al. 2012; Read et al. 2013). The first draft genome of an Italian isolate of C. suis (Donati et al. 2014) has recently been complemented by a genomic study of eleven isolates, including the reference strain S45 and historic tetracycline resistant strains from the United States (Joseph et al. 2016). A greater understanding of the diversity of C. suis circulating within livestock, and the relation of the genomic backbone to the Tet-island, is now required.
Building on evidence for the presence of both tetracycline sensitive and resistant C. suis strains in Europe (Borel et al. 2012; Di Francesco et al. 2008; Schautteet et al. 2013), we aimed to provide a first genomic description of currently circulating European strains, investigating the diversity within the natural livestock reservoir and the presence and architecture of the Tet-island. A survey of 29 fattening pig farms in Switzerland between 2013 and 2014 was conducted to evaluate the prevalence of C. suis in pigs and the impact of prophylactic antibiotic oral group treatment on the presence of tetracycline resistance in these strains (Hoffmann et al. 2015). The farms were divided into three treatment groups: farms without antibiotic treatment; farms with prophylactic oral antibiotic treatment of the whole herd consisting of trimethoprime, sulfadimidin and sulfathiazole (TSS); and farms giving herd treatment with chlortetracycline with or without tylosin and sulfadimidin (CTS). Pigs were sampled at the beginning (age ∼12 weeks, before antibiotic treatment) and at the end of the fattening treatment (age approx. six months, after oral group treatment). In previous work, swab samples and cultured isolates were tested for the presence of tetA(C) by conventional PCR, and isolates further investigated for tetracycline susceptibility in vitro (Wanninger et al. 2016). Clear evidence was found for the effect of tetracycline selective pressure, as treatment resulted in a greater number of tetracycline resistant isolates. We performed whole genome sequencing on a cohort of 29 of these strains in order to investigate the dynamics of the C. suis genome. In particular, we focus on the association of the Tet-island with the genomic backbone of these strains, illustrating the flexibility of the genome and the effect of selective antibiotic pressure on carriage.
Materials and Methods
Strain Collection, Culture, Tetracycline Resistance Determination and DNA Isolation
Samples used in this study are from rectal swabs (FLOQSwabs® Copan Italia, Brescia, Italy) taken in nine fattening pig farms. Sampling was performed as previously described (Wanninger et al. 2016); briefly, the fattening period occurs from 12 weeks to six months of age, with antibiotic treatment administered orally for 5–12 days at the beginning of this period. Samples, farms and antibiotic regimes used on the farms (none; TSS; CTS; unknown) are shown in table 1. All culturing, antibiotic susceptibility testing and DNA extraction was performed as described (Wanninger et al. 2016). No ethical approval was required, as the samples originated from a previous study that was approved by the Veterinary Office of Canton Luzern (authorization no. LU03/14) and all efforts were made to minimize the discomfort of the animals during sampling (Hoffmann et al. 2015).
Table 1.
Strains Sequenced in This Study: Sources, Sequencing Data and tetA(C) Presence
| Isolatea | Farm # | Treatment | Mean Cov | Plasmid Cov | Pl:Chr | Suspected Mixed | # Contigs | Draft Length (bp) | TetRb | tetA(C) PCR | Tet-island |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 9-1 a | 1 | None | 20 | 66 | 4 | Only plasmid | 2 | 1082098 | S | N | N |
| 9-1 b | 1 | None | 134 | 462 | 4 | N | 1 | 1082175 | S | N | N |
| 1-25 a | 25 | None | 31 | 183 | 7 | Some read heterogeneity | 2 | 1088920 | S | N | N |
| 1-25 b | 25 | None | 75 | 536 | 8 | N | 2 | 1088751 | S | N | N |
| 3-25 a | 25 | None | 73 | 388 | 6 | Y – one dominant | 3 | 1092018 | S | N | N |
| 3-25 b | 25 | None | 28 | 146 | 6 | N | 1 | 1075010 | S | N | N |
| 9-25 a | 25 | None | 154 | 755 | 5 | N | 5 | 1093713 | S | N | N |
| 2-26 b | 26 | TSS | 27 | 134 | 6 | Y – one dominant | 1 | 1083861 | S | Y | N |
| 10-26 b | 26 | TSS | 29 | 109 | 4 | N | 1 | 1080931 | S | N | N |
| 14-23 b | 23 | TSS | 12 | 39 | 4 | Low coverage | 23 | 1081871 | S | N | N |
| 5-27 b | 27 | CTS | 45 | 326 | 8 | N | 1 | 1101914 | R | Y | Y |
| 1-28 a | 28 | Unknown | 119 | 633 | 6 | N | 2 | 1084542 | S | N | N |
| 1-28 b | 28 | Unknown | 129 | 872 | 7 | N | 2 | 1096577 | I | Y | Y, TetRfusion |
| 8-29 b | 29 | CTS | 93 | 427 | 5 | N | 1 | 1098010 | R | Y | Y |
| 6-17 a | 17 | CTS | 32.3 | 171 | 5 | Y – one dominant | 3 | 1072079 | S | N | N |
| 8-17 a | 17 | CTS | 16.5 | 135 | 8 | N | 2 | 1079515 | S | N | N |
| 11-17 a | 17 | CTS | 19.1 | 129 | 7 | N | 3 | 1106365 | R | Y | Y |
| 11-17 b | 17 | CTS | 24 | 123 | 5 | N | 3 | 1107636 | R | Y | Y |
| 5-22 b | 22 | CTS | 45 | 188 | 4 | Only plasmid | 1 | 1094277 | R | Y | Y |
| 22-22 b | 22 | CTS | 14.4 | 91 | 6 | Y – one dominant | 8 | 1092471 | R | Y | Y |
| 30-22 b | 22 | CTS | 37.6 | 213 | 6 | Y – one dominant | 2 | 1095150 | R | Y | Y |
| 15-27 b | 27 | CTS | 29.5 | 145 | 5 | N | 3 | 1103721 | R | Y | Y |
| 17-23 a | 23 | TSS | 12.3 | 66 | 6 | N | 5 | 1102774 | R | Y | Y |
| 17-23 b | 23 | TSS | 13.8 | 83 | 6 | N | 9 | 1072633 | S | Y | N |
| 19-23 a | 23 | TSS | 13.3 | 76 | 6 | N | 8 | 1099448 | R | Y | Y |
| 19-23 b | 23 | TSS | 6.1 | 21 | 3 | N | 344 | 1045494 | S | N | N |
| 3-29 b | 29 | CTS | 20.3 | 103 | 5 | N | 2 | 1096059 | R | Y | Y |
| 4-29 b | 29 | CTS | 15.7 | 67 | 4 | N | 1 | 1094002 | R | Y | Y |
| 9-29 b | 29 | CTS | 65.9 | 266 | 4 | Only plasmid | 5 | 1092041 | R | Y | Y |
| mean 5 |
Isolate refers to the pig ID followed by either: a, indicating first sample at beginning of fattening period, before antibiotic treatment; or b, indicating second sample at end of fattening period, after antibiotic treatment. Paired samples from the same pig at these timepoints are shaded together. Cov = coverage. bR, resistant; S, sensitive; I, intermediate. Anomalous tetA(C) PCR results are shown with a grey background.
Sequencing, Mapping, Assembly and Annotation
Sequencing of samples was performed on the Illumina Miseq platform with 250bp PE reads at the FGCZ Zurich, following NEBNext library creation. Initial mapping, to determine coverage statistics, was performed against the previously sequenced strain, C. suis MD56, using BWA (Li and Durbin 2009). Assembly was performed using SPAdes in multi-cell mode (Bankevich et al. 2012), followed by ordering of the contigs against MD56 using ABACAS (Assefa et al. 2009). Manual improvement of all assemblies was performed using Artemis and ACT (Carver et al. 2005; Rutherford et al. 2000) to resolve as many contig joins as possible. Several assemblies were found to be in single contigs and were used as references for subsequent mapping where necessary. Automated annotation was performed using AnnotateBacteria (https://github.com/sanger-pathogens/Bio-AutomatedAnnotation/; last accessed December 15, 2016) with further manual curation in Artemis. All read data, with associated assembly and annotation where relevant, has been submitted to ENA under project PRJEB17986.
Alignments, Phylogenies and Recombination Analysis
Parsnp within the Harvest suite (Treangen et al. 2014) was used to generate alignments and phylogenies from the optimized assemblies. Alignments used for subsequent analysis comprised at least 89% of the genome. Gubbins (Croucher et al. 2015) was used to determine putative recombinations, based on a midpoint root. Additional alignments and phylogenies were created in Seaview (Gouy et al. 2010) using Muscle and PhyML. Comparisons of phylogenies were performed using Compare2Trees (Nye et al. 2006). Single nucleotide polymorphism (SNP) density was calculated using 1 kb windows.
Results and Discussion
Sequencing of C.suis Livestock Isolates
Full genome sequence data were obtained from 29 isolates of C. suis, from pig farms across Switzerland (table 1). Following assembly of each genome with manual improvement, eight of the genome drafts were resolved into single contigs and a further 19 each contain fewer than ten contigs. These represent C. suis genomes at a standard of Improved High-Quality Draft (Chain et al. 2009). One sample (19-23b) provided a mean genome coverage of only 6×, yet it was possible to reconstitute the vast majority of the genome in 344 contigs, allowing comparison with other isolates. As these were isolates from recent swab samples, only subject to three passages in tissue culture for minimal laboratory manipulation, some samples were found to contain mixed strains (heterogeneous sequence reads), but in most cases a dominant strain is represented in the assembly. Sample 14-23b is an exception to this, containing mixed reads and also with only 12× genome coverage.
Automated annotation with subsequent manual curation was performed on the genome of isolate 8-29b, comprising the largest single contig genome. Within this genome, 910 coding sequences (CDSs), two rRNA operons, and 37 tRNA synthases were identified. This is lower than the number of CDSs (n = 933) found in the partial genome of reference strain MD56, largely due to the merging of partial CDSs annotated on MD56 contig ends. The genome is syntenic with that of C.trachomatis, with the most variable regions identified as: the plasticity zone (PZ; locus tag CS829B_1590-1780, containing multiple phospholipase D pld genes, toxin gene(s) and an intact trp operon); Inc proteins (CS829B_2340-2410); a locus carrying a highly repetitive helicase and putative restriction enzyme (CS829B_3450-3500); and around the rRNA operons (CS829B_8000-8160) (fig. 1).
Fig. 1.—
Comparison of the genome of C. suis 8-29b (top) with that of C. trachomatis SW2 (bottom; EMBL accession FN652779). This comparison, with tblastx hits between the genomes shown as grey lines, illustrates the synteny of the genomes. CDSs are shown on each genome as arrows, with several regions of difference as described in the text. Image produced using Easyfig (Sullivan et al. 2011).
Diversity within Isolate Genomes
Comparisons of assembled C. suis genomes from Switzerland show variability in key regions around the PZ, helicase region and one rRNA operon, which can be observed on a genome scale (see supplementary fig. S1, Supplementary Material online). Within the PZ, different isolates exhibit variable numbers of pld genes, and several of the strains carry a second, full-length (3224 aa) cytotoxin gene (see supplementary fig. S1, Supplementary Material online). A common site of assembly contig breaks is at the highly repetitive putative helicases or restriction enzymes (CS829B_3480 and 3500) (see supplementary fig. S1, Supplementary Material online).
The genome of C. suis carries an extra element, the cryptic plasmid, which was found in all our sequenced isolates. Several samples appear to possess mixed populations of plasmids (heterogeneous reads), which could either be indicative of mixed strains in the sample, or possibly mixed plasmids within a single isolate. The chromosomes show less evidence of heterogeneity, offering the presence of multiple, variable plasmids within an isolate as an intriguing possibility. From comparison of coverage levels, the mean copy number of the plasmid is five copies per chromosome, with a range from three-eight per sample (table 1), similar to that observed in C. trachomatis (Last et al. 2013; Pickett et al. 2005; Seth-Smith et al. 2009).
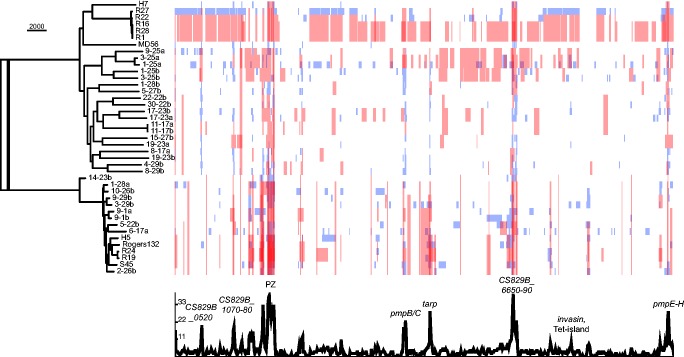
To assess the diversity of strains circulating in Switzerland, within the context of other available genomes from Italy, Austria and the United States, we determined a phylogeny by aligning all available genome drafts (fig. 2). Two major clades can be identified, in agreement with previous results (Joseph et al. 2016), with clusters of strains from the United States and from Europe present in both. The vast majority of the observed diversity derives from the genomes of the Swiss farm isolates, despite these having been collected over a shorter timeframe and within a smaller area. Indeed, in several cases, isolates located in both clades derive from a single farm, indicating no geographical clustering of strains within Switzerland. The previously sequenced strain MD56, with an Italian origin, shares a common ancestor with one Swiss clade. The Austrian reference strain S45 shares a common ancestor with one of the USA clades, indicating a possible European origin of this clade.
Fig. 2.—
Phylogeny of C. suis isolates. The genome drafts from US strains (Joseph et al. 2016), reference strain S45 and strain MD56 (Donati et al. 2014) are included. Indicated alongside Swiss samples are the farms from which the isolates originate, showing the diversity within each farm, and the antibiotic regimen used. Presence of tetC in the genome is indicated. Seven paired isolates taken from the same pig before and after the fattening period are also indicated in matched colors. The location of the assembly of sample 14-23b in the phylogeny suggests that it contains mixed isolates from both clades, in addition to being of low coverage (mean 12×). Bootstraps on all branches are in excess of 97% except where shown. Scale bar indicates number of substitutions per site.
The variable sites used to construct the phylogeny cover 51,096 nucleotides (5.25% of the 971,482 bp alignment), a similarly high level of diversity to that described in a study covering all nine serotypes of C. psittaci (Read et al. 2013). Approximately 19,000 SNPs separate the two major clades (18,962 variable sites between isolates 9-1a and 4-29b): four times more than the 4,860 SNPs separating the LGV and trachoma clades in C. trachomatis (Harris et al. 2012). This indicates that C. suis may be the most genomically diverse species of Chlamydia yet studied. That such a degree of diversity comes between samples from two relatively limited studies of Swiss and USA isolates indicates either that much more diversity will be found in further studies, or that strains have mixed globally and that this phylogeny is representative of the whole species.
Recombination and Genome Flexibility within C.suis Isolates
Genomes of other species of Chlamydia have been found to undergo recombination (Harris et al. 2012; Joseph et al. 2012; Read et al. 2013). In light of the high degree of genetic diversity amongst the C. suis strains analyzed in this study, and recent findings in C. suis (Joseph et al. 2016), we performed an analysis to determine whether recombination contributes to this diversity. Substantial levels of recombination were identified (fig. 3), although this was not found to affect the tree topology greatly, with an overall topology score of 82.8% between the two phylogenies (see supplementary fig. S2, Supplementary Material online). This strain collection exhibits a mean r/m value of 0.37 (indicating the relative impact of recombination to mutation on causing genome diversity) and mean ρ/θ of 0.007 (measuring the frequency of occurrence of recombination relative to mutation). These values were derived using different software compared with that used previously (Joseph et al. 2016), hence these cannot be truly compared; the r/m value for C. suis is slightly greater than that seen in the recombinogenic C. trachomatis using the same method (0.31, Hadfield et al. submitted).
Fig. 3.—
Recombinations detected within C. suis. The recombination-adjusted phylogeny is shown on the left, with genome tracks to the right showing blocks of recombination that have been identified in a single strain in blue, and in a clade ancestor in red. The plot below shows recombination density along the genome. This figure is based on alignment data from all assemblies, covering 89% of the genome. A further analysis of all but two strains is shown in supplementary fig. S3, Supplementary Material online, and highlights a further region of high recombination around ompA (CS829B_7350), which did not align in this dataset. Bootstraps are not provided post-removal of recombinations. Scale bar indicates the number of nucleotide substitutions.
Within the dataset, 759 recombination blocks were identified, comprising up to 380 kb (in sample 1-25a) of a single genome. Many recombinations affecting a single strain were identified (fig. 3, blue bars) as well as ancestral recombinations (fig. 3, red bars), indicating that this is an ongoing process within this species. Peaks of recombination frequency were identified across known variable regions including PZ, Pmp-encoding genes, tarp and ompA (see supplementary fig. S3, Supplementary Material online); similar regions to those identified in C. trachomatis (Harris et al. 2012). Other loci highly affected by recombination were identified at hypothetical proteins CS829B_0510-0530, CS829B_1070-1080, and putative type three secretion system (T3SS) effectors CS829B_6630-6660 (fig. 3). Analysis of SNP density along the genome also indicates that regions of high recombination fall at regions of high variability (data not shown), indicating possible pressure from the host immune system to generate variation, or import of variable regions from more diverse strains. Regions around the rRNA locus adjacent to an invasin-encoding gene have also been affected by recombination (see below).
The phylogenetic tree produced after the removal of recombinations shows much shorter branches to individual strains (fig. 3 compared with fig. 2). For example, several of the USA strains (R1, R16, R22, R27, and R28) now fall in a tighter clade, with R27 predicted to have undergone recombination over large parts of its genome. Interestingly, in the phylogeny with recombinations removed (fig. 3), Italian strain MD56 is located at the root of the USA isolates, suggesting that this branch too has a European origin. The long branches still seen between many of the Swiss strains also indicate a history of diversification by mutation. This implies that a large diversity of strains is maintained within the population, with no particularly successful clones expanding through selective advantage.
The plasmid appears to be exchangeable between isolates, as the phylogeny of the plasmid (defined using the dominant version in cases where reads are mixed) does not match that of the chromosome, with an overall phylogenetic comparison topology score of only 52.1% (see supplementary fig. S4, Supplementary Material online). Despite this predicted mobility of the plasmid, no plasmid swapping between the two major clades is seen, and no plasmid recombinations were detected, in contrast to the results of a recent study (Joseph et al. 2016).
The C.suis Tet-island
A genomic island carrying the tetA(C) gene is variably present among the samples, having been identified in 14 of the 29 Swiss genomes sequenced, as well as being found in all the USA isolates sequenced (Joseph et al. 2016). The Tet-island possesses a G + C content of 56.2%, compared with 42.2% for the rest of the genome (data from strain 8-29b). When the island is present, it is always found in the same genomic location, within an invasin-encoding gene (inv, 928b_7990) adjacent to an rRNA operon. The invasin protein is predicted to contain an N-terminal β-barrel domain (pfam11924), a central bacterial Ig-like domain (pfam02369) and three C-terminal Ig domains (pfam05345), altogether producing a protein putatively involved in adhesion. We found that the inv gene is inactivated in many of the strains analyzed, even when not disrupted by the Tet-island: central portions of inv are deleted in strains 1-25b, 3-25a, 3-25b, 9-25a, 8-17a, 17-23b, and 19-23b, meaning that full-length intact versions are present in only eight of the sequenced Swiss strains. This implies that there may be pressure to lose functionality in this region, either by deletion of key domains or acquisition of the Tet-island.
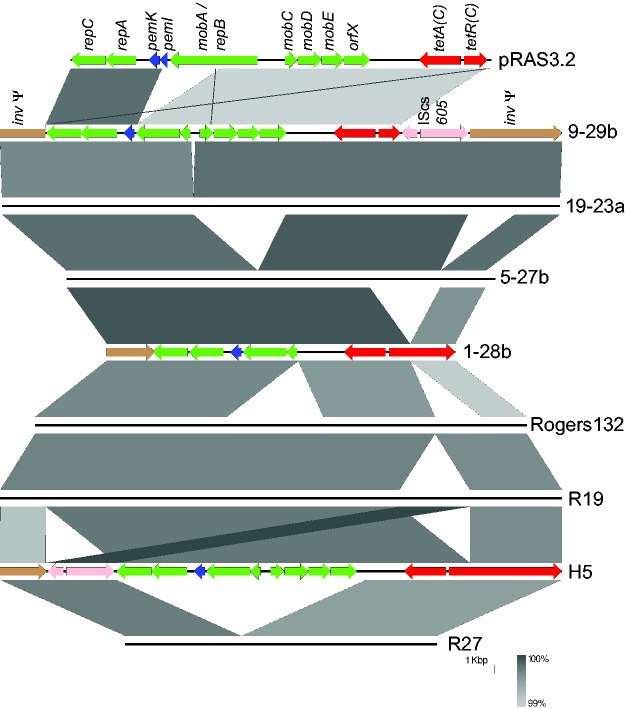
Annotation and comparison of the Tet-island between the international strains clearly shows that the architecture of the island can vary, but that the tetA(C) gene is always found intact and presumably functional (fig. 4). The origins of the Tet-island are, unusually, easily characterizable (Dugan et al. 2004). The Aeromonas salmonicida mobilizable plasmid pRAS3.2 has a high level of nucleotide identity (99.9%) and synteny with the Tet-island, including the tet operon with tetA(C) and its repressor-encoding tetR(C), followed by two transposases of the insertion sequence IScs605 from another source (Joseph et al. 2016) (figs. 4 and 5). It is possible that pigs came into contact with this fish pathogen through ingestion of fish meal (Sandoz and Rockey 2010).
Fig. 4.—
Variations of organization of TetC-carrying islands, compared against the putative ancestor pRAS3.2. Islands/plasmids are shown as lines, with the grey bars indicating nucleotide identity between island pairs. The top line represents pRAS3.2, followed by the island from isolate 9-29b (both of these are annotated with their encoded genes), which reflects the gene organization seen in strains 8-29b, 11-17a, 11-17b, 5-22b, 30-22b, 15-27b, 17-23a, 3-29b, 4-29b, and 22-22b (the latter from partial sequence information only) as well as the US strains R19 and R24. The island is inserted within an invasin gene (inv), splitting it into two pieces and rendering it a pseudogene (brown, left and right of the figure). The CDSs encoding plasmid-related Rep and Mob proteins (green), PemK (blue), and an insertion sequence (IScs605 with two transposase tnp genes; pink) are found adjacent to the tet operon (red). Strain 19-23a has a small deletion relative to this within the mob operon. Strain 5-27b has two deletions: comprising much of the mob operon and part of inv. Strain 1-28b (annotated) has these deletions and a further one which creates a fusion protein between the tetR (C) regulator and the C terminal portion of inv, indicated by the extended red CDS on the right. The organization of the US strain Tet-islands is shown in the lower part of the figure, with islands from Rogers132, R19 (full length island, also representing R24), H5 (annotated, showing translocation of IScs605 and tetR(C)-inv fusion), and R27 (reduced island, representing the gene organization in R1, R16, R22, R28, and H7) (Joseph et al. 2016). All nucleotide identities are in excess of 99% (see scale). Image produced using Easyfig (Sullivan et al. 2011).
Fig. 5.—
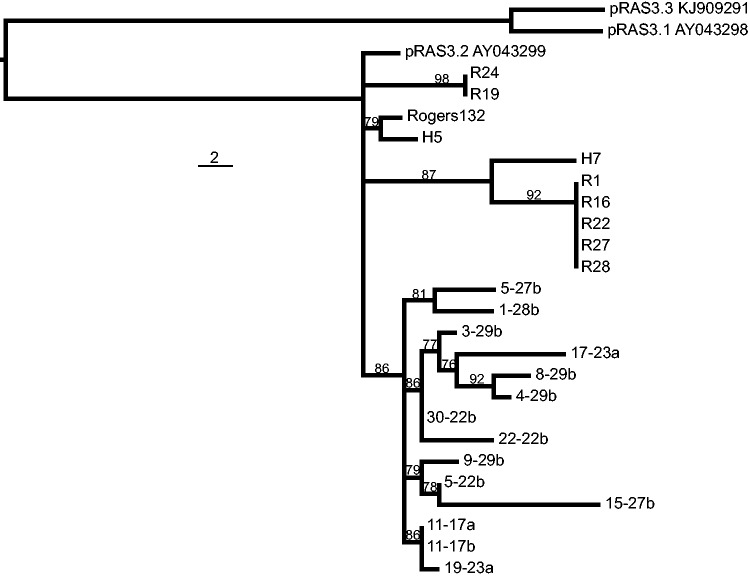
Phylogeny of Tet-island. Putative ancestral plasmids pRAS3 are used as outgroups. US strain Tet-islands were taken from (Joseph et al. 2016). Bootstraps (%) are shown. The topology of this phylogeny does not match that of the chromosome (see supplementary fig. S5, Supplementary Material online). Scale bar indicates number of nucleotide substitutions.
An intriguing comparison is between the Tet-island sequences from Swiss strains and those from USA strains (Dugan et al. 2004; Joseph et al. 2016). The island found within American strains R19 and R24 has the same architecture as the full-length versions of the island found within our Swiss strains, indicating that this genomic element has travelled globally, perhaps through livestock trade. The island from strains R27, R1, R16, R22, R28, and H7, however, represents a rearrangement not seen within our strain collection, with the translocation of IScs605 (also seen in the longer H5) and the production of a TetR-Inv fusion protein. One of our sequenced isolates (1-28b) demonstrates a deletion including IScs605, creating another fusion between tetR(C) and inv, with a larger internal deletion of the inv gene (fig. 4). Careful analysis of the MD56 genome draft (Donati et al. 2014) also shows that the assembly covers a small portion of this region, enough to indicate that the island is present and that a further alternative fusion between tetR(C) and inv has occurred.
The SNP-phylogeny of the Tet-island shows an intriguing polytomic topology (fig. 5), both globally and within the Swiss cohort, which indicates simultaneous diversification from a common ancestor. Three clades featuring USA isolates all share a most recent common ancestor (MRCA) with a single clade containing all the European isolates, possibly implying an American origin of the Tet-island in C. suis. This appears to have been followed by rapid divergence of the Tet-island within USA isolates, whereas all European isolates share two unique SNPs, after which further diversification occurred. The phylogeny does not support further movement of the Tet-island between European and USA strains. It must, however, be borne in mind that the phylogeny was built using very few SNPs, and that Tet-islands from Swiss strains show little diversification from the MRCA (separated by a single SNP from 30-22b, and 11 from 15-27b).
The Tet-island phylogeny contrasts with that of the chromosome (see supplementary fig. S5, Supplementary Material online); the presence of the Tet-island in strains not phylogenetically linked (fig. 2) implies that it has been gained or lost several times within the history of the species. The occurrence of the island always in the same genomic location, despite the presence of many matches within the genome to the IScs105 target location TTCAA (n = 3595) (Dugan et al. 2007) could imply that it moves horizontally by recombination rather than self-mobility of the island. Indeed, in vitro experiments involving recombination of the Tet-island show that it moves as part of larger recombinations (Marti et al. 2017; Suchland et al. 2009). Many of the Tet-island genes required for mobility remain intact in the majority of isolates, raising questions about the recentness of its insertion or its ongoing functionality. However, we were unable to demonstrate excision or circularization of the island by PCR (data not shown), suggesting that the island is not self-mobile through excision and integration.
The most parsimonious explanation of the Tet-island distribution involves its transfer by recombination, and indeed our recombination analysis goes some way to elucidating this. Regions of predicted recombination occur around the invasin gene in the MRCA of 13 strains from 1-28b to 8-29b, as well as that of seven strains from 5-22b to S45 (see supplementary fig. S3, Supplementary Material online). Two separate recombinations at inv in 3-29b and 9-29b are also identified. This explanation of the incoming transfer of the island by recombination invokes the subsequent loss of the Tet-island from strains 8-17a, 17-23b, 6-17a, and S45. A further insertion into the MRCA of H7-R16 would then complete the explanation of the presence of the Tet-island within the population studied.
In this scenario, the insertion of the island would appear to be a relatively ancient event (see supplementary fig. S3, Supplementary Material online). However, there are substantial discrepancies in the subsequent mutation rates of the chromosomes and the Tet-islands. The genomes within the clade containing 8-29b have accumulated ∼3.0 SNPs per kb by mutation since diversification (using branch lengths, SNPs between major clades and r/m values to calculate). The diversity within the 12 kb Tet-island, however, does not match this, with only 1–13 SNPs found within the conserved areas of these islands (0.1–1.1 SNPs per kb). This would then imply that the Tet-island is a more recent acquisition within C. suis, and its distribution within the phylogeny is due to recombination and local selection pressures. As previously noted, the provisional molecular dating of the C. suis species MRCA at 1400AD (Joseph et al. 2016) does not fit with the relatively recent industrial use of tetracycline, and its widespread use since 1950s (Castanon 2007; Chopra and Roberts 2001).
Selective Pressure on C.suis on Livestock Farms
The presence of the Tet-island in Swiss isolates can be related to selective host pressures in the different farms (fig. 2). Farms with no antibiotic treatment have no samples containing the Tet-island (7/7). Farms using TSS treatment have samples carrying the Tet-island at the first timepoint (2/2: possibly from breeder antibiotic use) but this is lost without selective pressure over the fattening period, and all isolates at the end did not carry the island (0/5). Farms which did use tetracycline treatment (CTS) saw their pigs retain or gain strains with the Tet-island from an initial point where 1/3 isolates were Tet-island carrying, to an endpoint where all were (9/9).
Following the strains present in pigs over time can also be done in best resolution using genomic data. We bear in mind that our samples may be subject to culture bias, with only those that grew well being sequenced, although we believe that due to the low number of passages, the sequences we obtained genuinely reflect the in vivo strains. In our dataset, we have seven paired samples from the same pig at the beginning and the end of the fattening period. In the farms with no tetracycline treatment (no antibiotics and TSS), highly diverse isolates were seen at the beginning and end timepoints (five paired samples). In two pigs (9-1 and 17–23), the strains carried at the beginning and end timepoints are distantly related, indicating a source with a common ancestor (fig. 2); both in pig 9-1 are tetracycline sensitive, whereas the first strain in pig 17–23 is tetracycline resistant, replaced by a tetracycline sensitive related strain. Given the genetic difference between these latter strains, even after recombinations have been removed (fig. 3), it is not hypothesized that this demonstrates in-host evolution, but infection with a related strain. Strains from pigs 1–25 and 3–25 also share a distant common ancestor. In farms with CTS treatment (one paired sample, pig 11–17), the pig retained the same Tet-island carrying strain. There is no evidence that individual isolates can lose or gain the Tet-island under selective pressure; rather strains are replaced. The loss of Tet-island carrying strains in the absence of selective pressure may indicate a metabolic burden associated with carrying the island.
The diversity of strains within pigs followed over time is high, and may reflect the effect of laboratory isolation of individual strains from the mixed porcine gut microbiota, rather than replacement of single strains. Our findings indicate that there is no obvious restriction to the movement of strains within Switzerland. The use of CTS does not seem to limit the high strain diversity, as the Tet island is carried by many distinct strains. The full scale of C. suis diversity on Swiss farms, the comparison of this to international diversity, and determination of the sources of the strains, can only be addressed in follow up studies. Our current snap-shot indicates that highly diverse strains are circulating in Switzerland, and even within individual farms.
Phenotypic assessment of tetracycline resistance in these samples indicates that the phenotype correlates with the presence or absence of tetA(C) in all cases (table 1). Strain 1-28b has an intermediate resistant phenotype and carries an island with a fusion of the tetA(R) gene to inv, which brings into question the function of the TetR within this context. All US Tet-island carrying strains were found to be resistant to tetracycline, regardless of the genomic context of tetR(C) (Dugan et al. 2004). Some discrepancies between the detection of tetA(C) by PCR and genomics were noted (table 1), corroborating previous findings (Donati et al. 2016; Wanninger et al. 2016). Contrasting results might be attributable to possible mixed strains within samples, or lack of PCR specificity. Caution should be taken when using this as a diagnostic PCR for C. suis, especially as this gene is found in other bacteria and cloning vectors and its presence is not firmly related to in vitro susceptibility characteristics (Donati et al. 2016; Wanninger et al. 2016).
Conclusion
C. suis has a very plastic genome, highly affected by recombination and plasmid exchange. Our data confirm and extend many of the findings of a recent, smaller study on C. suis genomes (Joseph et al. 2016). A large diversity of C. suis is found circulating in Swiss farms, where the infection might have occurred during breeding, transport or proximity of pigs on the fattening farms. New World isolates have more restricted diversity and appear to derive from European isolates, suggesting that C. suis originated around Europe and has been transferred to the United States on at least two occasions.
Selective antibiotic pressure can clearly be seen to affect the presence of the Tet-island in strains, as the larger study found 28/59 isolated strains to be phenotypically Tet resistant or intermediate, with obvious influence on this profile from farm treatment regimes (Wanninger et al. 2016). However, it is strain replacement, rather than loss or gain of the Tet-island, which occurs under selection, implying that the island is stably maintained within strains. In the absence of selection, isolates lacking the Tet-island appear to dominate (Wanninger et al. 2016), suggesting a fitness cost of carrying this cassette.
Our data give preliminary indications that the Tet-island within C. suis may have originated from the Americas. Yet there are still many questions to be answered regarding the timing of its introduction and its distribution within the species. Several pieces of evidence indicate that it moves by recombination, which relies on the occurrence of a co-infection with different strains: a highly likely scenario under common pig farming procedures. Moreover, the transfer of the Tet-cassette from C. suis to C. trachomatis and C. muridarum by recombination has been demonstrated in vitro (Jeffrey et al. 2013; Suchland et al. 2009). C. suis must be considered as a potentially zoonotic pathogen because C. suis has been detected on human ocular and nasal swabs (De Puysseleyr et al. 2014, 2015; Dean et al. 2013). This raises the real possibility of transfer of tetracycline resistance to other chlamydial species, through livestock or farmers. Complications associated with tetracycline-resistant C. trachomatis would be considerable, yet ongoing use of tetracycline in farms contributes to this scenario.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by the Swiss Federal Food Safety and Veterinary Office under grant number 1.13.03. We thank the Functional Genomics Centre Zurich, in particular Dr Lucy Poveda, for technical assistance.
Literature Cited
- Assefa S, Keane T, Otto T, Newbold C, Berriman M. 2009. ABACAS: algorithm-based automatic contiguation of assembled sequences. Bioinformatics 25:1968–1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borel N, et al. 2012. Selection for tetracycline-resistant Chlamydia suis in treated pigs. Vet Microbiol. 156:143–146. [DOI] [PubMed] [Google Scholar]
- Carver TJ, et al. 2005. ACT: the Artemis comparison tool. Bioinformatics 21:3422–3423. [DOI] [PubMed] [Google Scholar]
- Castanon J. 2007. History of the use of antibiotic as growth promoters in European poultry feeds. Poultry Sci. 86:2466–2471. [DOI] [PubMed] [Google Scholar]
- Chain P, et al. 2009. Genome project standards in a new era of sequencing. Science 326:236–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra I, Roberts M. 2001. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol Mol Biol Rev 65:232–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croucher NJ, et al. 2015. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 43:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Puysseleyr K, et al. 2014. Evaluation of the presence and zoonotic transmission of Chlamydia suis in a pig slaughterhouse. BMC Infect Dis 14:560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Puysseleyr L, et al. 2015. Assessment of Chlamydia suis infection in pig farmers. Transbound Emerg Dis. doi: 10.1111/tbed.12446 [DOI] [PubMed] [Google Scholar]
- Dean D, Rothschild J, Ruettger A, Kandel R, Sachse K. 2013. Zoonotic Chlamydiaceae species associated with trachoma, Nepal. Emerg Infect Dis 19:1948–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Francesco A, et al. 2008. Tetracycline-resistant Chlamydia suis isolates in Italy. Vet Rec 163:251–252. [DOI] [PubMed] [Google Scholar]
- Donati M, et al. 2016. Tetracycline susceptibility in Chlamydia suis pig isolates. PLOS ONE 11:e0149914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donati M, et al. 2014. Genome sequence of Chlamydia suis MD56, isolated from the conjunctiva of a weaned piglet. Genome Announc 2:00414–e00425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugan J, Andersen AA, Rockey DD. 2007. Functional characterization of IScs605, an insertion element carried by tetracycline-resistant Chlamydia suis. Microbiology 153:71–79. [DOI] [PubMed] [Google Scholar]
- Dugan J, Rockey DD, Jones L, Andersen AA. 2004. Tetracycline resistance in Chlamydia suis mediated by genomic islands inserted into the chlamydial inv-like gene. Antimicrob Agents Chemother 48:3989–3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M, Guindon S, Gascuel O. 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 27:221–224. [DOI] [PubMed] [Google Scholar]
- Hadfield J, et al. submitted. Comprehensive global genome dynamics of Chlamydia trachomatis show ancient diversification followed by contemporary mixing and recent lineage expansion. Gen Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris SR, et al. 2012. Whole-genome analysis of diverse Chlamydia trachomatis strains identifies phylogenetic relationships masked by current clinical typing. Nat Genet. 44:413–419. S411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann K, et al. 2015. Prevalence of Chlamydial infections in fattening pigs and their influencing factors. PLOS ONE 10:e0143576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey B, Suchland R, Eriksen S, Sandoz K, Rockey DD. 2013. Genomic and phenotypic characterization of in vitro-generated Chlamydia trachomatis recombinants. BMC Microbiol. 13:142.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph SJ, et al. 2012. Population genomics of Chlamydia trachomatis: insights on drift, selection, recombination, and population structure. Mol Biol Evol. 29:3933–3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph SJ, Marti H, Didelot X, Read TD, Dean D. 2016. Tetracycline selective pressure and homologous recombination shape the evolution of Chlamydia suis: a recently identified zoonotic pathogen. Genome Biol Evol. 8:2613–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltenboeck B, Kousoulas K, Storz J. 1992. Two-step polymerase chain reactions and restriction endonuclease analyses detect and differentiate ompA DNA of Chlamydia spp. J Clin Microbiol. 30:1098–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Last A, et al. 2013. Plasmid copy number and disease severity in naturally occurring ocular Chlamydia trachomatis infection. J Clin Microbiol. 52:324–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenart J, Andersen A, Rockey DD. 2001. Growth and development of tetracycline-resistant Chlamydia suis. Antimicrob Agents Chemother 45:2198–2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti H, et al. 2017. Tet(C) gene transfer between Chlamydia suis strains occurs by homologous recombination after co-infection: Implications for spread of tetracycline-resistance among Chlamydia. Front Microbiol. 8:156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nye T, Lio P, Gilks W. 2006. A novel algorithm and web-based tool for comparing two alternative phylogenetic trees. Bioinformatics 22:117–119. [DOI] [PubMed] [Google Scholar]
- Perez-Martinez J, Storz J. 1985. Antigenic diversity of Chlamydia psittaci of mammalian origin determined by microimmunofluorescence. Infect Immun 50:905–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett M, Everson J, Pead P, Clarke IN. 2005. The plasmids of Chlamydia trachomatis and Chlamydophila pneumoniae (N16): accurate determination of copy number and the paradoxical effect of plasmid-curing agents. Microbiology 151:893–903. [DOI] [PubMed] [Google Scholar]
- Read TD, et al. 2013. Comparative analysis of Chlamydia psittaci genomes reveals the recent emergence of a pathogenic lineage with a broad host range. MBio 4:e00604-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford KM, et al. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944–945. [DOI] [PubMed] [Google Scholar]
- Sandoz K, Rockey D. 2010. Antibiotic resistance in Chlamydiae. Future Microbiol. 5:1427–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schautteet K, et al. 2013. Tetracycline-resistant Chlamydia suis in cases of reproductive failure on Belgian, Cypriote and Israeli pig production farms. J Med Microbiol. 62:331–334. [DOI] [PubMed] [Google Scholar]
- Schautteet K, Vanrompay D. 2011. Chlamydiaceae infections in pig. Vet Res. 42:29.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth-Smith HM, et al. 2009. Co-evolution of genomes and plasmids within Chlamydia trachomatis and the emergence in Sweden of a new variant strain. BMC Genomics 10:239.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchland R, Sandoz K, Jeffrey B, Stamm W, Rockey D. 2009. Horizontal transfer of tetracycline resistance among Chlamydia spp. in vitro. Antimicrob Agents Chemother 53:4604–4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan M, Petty N, Beatson S. 2011. Easyfig: a genome comparison visualiser. Bioinformatics 27:1009–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treangen T, Ondov B, Koren S, Phillippy AM. 2014. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 15:524.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, et al. 2011. Development of a transformation system for Chlamydia trachomatis: restoration of glycogen biosynthesis by acquisition of a plasmid shuttle vector. PLoS Pathogens 7:e1002258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, et al. 2012. Genetic transformation of a clinical (genital tract), plasmid-free isolate of Chlamydia trachomatis: engineering the plasmid as a cloning vector. PLoS ONE 8:e59195.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanninger S, et al. 2016. Selective pressure promotes tetracycline resistance of Chlamydia suis in fattening pigs. PLOS ONE 11:e0166917. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.