Abstract
RNA recombination is a major driving force for the evolution of RNA viruses and is significantly implicated in the adaptation of viruses to new hosts, changes of virulence, as well as in the emergence of new viruses including drug-resistant and escape mutants. However, the molecular details of recombination in animal RNA viruses are only poorly understood. In order to determine whether viral RNA recombination depends on translation of viral proteins, a nonreplicative recombination system was established which is based on cotransfection of cells with synthetic bovine viral diarrhea virus (family Flaviviridae) RNA genome fragments either lacking the internal ribosome entry site required for cap-independent translation or lacking almost the complete polyprotein coding region. The emergence of a number of recombinant viruses demonstrated that IRES-mediated translation of viral proteins is dispensable for efficient recombination and suggests that RNA recombination can occur in the absence of viral proteins. Analyses of 58 independently emerged viruses led to the detection of recombinant genomes with duplications, deletions and insertions in the 5′ terminal region of the open reading frame, leading to enlarged core fusion proteins detectable by Western blot analysis. This demonstrates a remarkable flexibility of the pestivirus core protein. Further experiments with capped and uncapped genome fragments containing a luciferase gene for monitoring the level of protein translation revealed that even a ∼1,000-fold enhancement of translation of viral proteins did not increase the frequency of RNA recombination. Taken together, this study highlights that nonreplicative RNA recombination does not require translation of viral proteins.
Keywords: virus evolution, nonreplicative RNA recombination, nonhomologous, Flaviviridae, Bovine Viral Diarrhea Virus
Introduction
RNA recombination plays a central role in the evolution of animal, plant, and bacterial RNA viruses as it facilitates the elimination of debilitating or lethal mutations. The importance of RNA recombination is further illustrated by the resulting alterations of the biological properties of RNA viruses including changes in virulence and host range, and even by the emergence of new pathogens and diseases (Worobey and Holmes 1999; Becher and Tautz 2011). To explain the mechanism of RNA recombination, two major models have been suggested. For several positive-strand RNA viruses, it has been demonstrated in cell-free systems that the viral RNA-dependent RNA polymerases (RdRp) are able to perform a template switch during viral RNA synthesis (Biebricher and Luce 1992; Arnold et al. 1999; Kim and Kao 2001; Cheng and Nagy 2003). In contrast, only few studies have described nonreplicative RNA recombination. Experiments in tissue culture cells with synthetic RNAs derived from bovine viral diarrhea virus (BVDV) and poliovirus clearly proved the existence of a nonreplicative RNA recombination mechanism in the absence of a functioning viral RdRp (Gmyl et al. 2003; Gallei et al. 2004). Similar results have been recently reported for analogous experiments with synthetic genome fragments derived from Hepatitis C virus (HCV) (Scheel et al. 2013). Furthermore, for poliovirus and BVDV efficient nonreplicative joining of RNA fragments is significantly influenced by the 5′ and 3′ terminal groups of the recombining RNA molecules (Gmyl et al. 2003; Austermann-Busch and Becher 2012). This supports the hypothesis that nonreplicative RNA recombination is based on endoribonucleolytic cleavage or cryptic ribozyme activity of RNA molecules, followed by a ligation reaction of the RNA fragments. Hence, it is conceivable that viral proteins might play an important role in nonreplicative RNA recombination, in particular when considering the fact that some of the pestiviral proteins directly interact with RNA (Zhong et al. 1998; Iqbal et al. 2004; Ivanyi-Nagy et al. 2008; Murray et al. 2008; Gladue et al. 2011).
Pestiviruses like BVDV-1, BVDV-2, classical swine fever virus (CSFV), and border disease virus (BDV) are economically important livestock pathogens worldwide. The genus Pestivirus belongs to the family Flaviviridae together with the genera Hepacivirus, Flavivirus, and Pegivirus (Simmonds et al. 2012). The positive-sense single-stranded RNA genome of pestiviruses has a length of about 12.3 kb and consists of one single open reading frame (ORF) coding for one large polyprotein, which is co and posttranslationally processed by viral and cellular proteases into the structural proteins C (core), Erns, E1, and E2, and the nonstructural (NS) proteins Npro, p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B (Becher and Thiel 2011; Simmonds et al. 2012). The ORF is flanked by 5′ and 3′ nontranslated regions (NTRs) (Deng and Brock 1993). Translation of pestiviral proteins occurs cap-independent and is mediated by an essential internal ribosomal entry site (IRES) located in the 5′NTR (Poole et al. 1995; Pestova et al. 1998). The first protein encoded by the ORF is the unique pestivirus-specific N-terminal protease (Npro), which generates its own C-terminus plus the N-terminus of C protein by autoproteolytic cleavage (Wiskerchen et al. 1991; Stark et al. 1993). Npro is dispensable for viral replication (Tratschin et al. 1998). The C-terminus of C protein is produced by an intramembrane signal peptide peptidase cleavage (Heimann et al. 2006). The C protein is a highly basic and intrinsically disordered protein that binds RNA with a low affinity and specificity and possesses RNA chaperone activity (Ivanyi-Nagy et al. 2008; Murray et al. 2008). The glycoprotein Erns has endoribonuclease activity and represents the second unique protein exclusively encoded by members of the genus Pestivirus (Schneider et al. 1993). Erns could contribute to the generation of substrates for RNA recombination by endonucleolytic cleavage of RNA molecules and thus might be significantly implicated in viral RNA recombination. NS3 possesses helicase and NTPase activities (Tamura et al. 1993; Warrener and Collett 1995). For NS4B, an NTPase motif has been described (Gladue et al. 2011), whereas NS5B is the viral RNA-dependent RNA polymerase (Zhong et al. 1998; Kao et al. 1999; Steffens et al. 1999).
According to their effects on tissue culture cells, a cytopathogenic (cp) and a noncytopathogenic (ncp) biotype of pestiviruses can be distinguished (Lee and Gillespie 1957; Gillespie et al. 1960). The emergence of cp BVDV strains by nonhomologous RNA recombination in cattle persistently infected with ncp BVDV is directly linked to the onset of the fatal mucosal disease (Meyers et al. 1997; Becher and Tautz 2011). The existence of cp and ncp biotypes together with the availability of reverse genetics (Meyers et al. 1997; Pankraz et al. 2005) and a cell culture based RNA recombination system (Gallei et al. 2004; Austermann-Busch and Becher 2012) makes BVDV a particularly suited model to study fundamental aspects of RNA recombination. In the BVDV system, RNA recombination is monitored by the detection of newly emerged replicating viral RNA genomes that are amplified by the viral RdRp. Accordingly, the experimental design applied in the present study excludes the detection of artificial recombination events resulting from template-switching during RT-PCR driven amplification of RNA molecules. Although it has been demonstrated that viral RNA recombination can occur in the absence of a functional RdRp (Gallei et al. 2004), the role of other viral proteins for RNA recombination has not been investigated so far. The present study proves that efficient translation of pestiviral proteins is not required for frequent nonreplicative RNA recombination in cell culture. Moreover, characterization of selected recombinant viruses demonstrates a remarkable flexibility with regard to the structure of C protein.
Materials and Methods
Cells
Madin-Darby bovine kidney (MDBK) cells were obtained from the American Tissue Culture Collection (Manassas, VA). MDBK cells were grown in Dulbecco’s modified Eagle’s (EDulb) medium supplemented with 5% horse serum. Baby hamster kidney (BHK-21) cells were obtained from the DSMZ (Braunschweig, Germany) and maintained in EDulb medium supplemented with 5% fetal calf serum.
Construction of BVDV cDNA Clones
All recombination partners are based on the parental construct plasmid pCP7-388, which displays a cDNA copy of the complete genome of the BVDV-1 strain CP7 under the control of an SP6 RNA polymerase promoter (Pankraz et al. 2005). The nucleotide numbering included in this study refers to the published full-length genomic sequence of CP7-388 encompassing 12293 nucleotides (Pankraz et al. 2005). The plasmids encoding 5′ recombination partners CP7/1-686 (pCP7/1-686) and CP7/1-997 (pCP7/1-997) comprise the 5′ terminal 686 and 997 nucleotides of the BVDV CP7 genome downstream of an SP6 promoter, respectively. The plasmid encoding the 3′ recombination partner Ubi-CP7/887-12293 (pUbi-CP7/887-12293) contains a T7 promoter directly upstream of nucleotides 1-513 of p + ubi-SGT (Gallei et al. 2004), coding for five C-terminal amino acids (aa) of the NS2 gene of BVDV strain CP14 and a C-terminal fragment, as well as two complete ubiquitin monomers, followed by nucleotides 887-12293 of pCP7-388. This genomic region encodes the entire C protein, the three envelope proteins, and (with the exception of Npro) all nonstructural proteins and also comprises the complete 3′NTR. Plasmids pCP7/1-686, pCP7/1-997, and pUbi-CP7/887-12293 were cloned by standard cloning procedures. For construction of the 3′ recombination partner NLuc-Ubi-CP7/887-12293, a plasmid containing a NheI-site that is followed by a T7 promoter sequence, a noncoding sequence of 21 nucleotides (GCATCCCTGAGACAAGCCACC), the Nano-luciferase (NanoLuc) coding sequence, the porcine teschovirus-1 2A peptide encoding sequence (allowing efficient processing at the C-terminus of the 2A peptide (Kim et al. 2011)), and two complete ubiquitin monomers, followed by nucleotides 887-1916 of pCP7-388 was synthesized (Genscript), and further digested with NheI and HindIII in order to replace the corresponding region of plasmid pUbi-CP7/887-12293.
To analyze the processing of the C-ubi-C fusion proteins of recombinants R-2 and R-3, plasmids encoding the N-terminal part of Npro to the C-terminal part of C of R-2 and R-3 were constructed (pCITE_P-2 and pCITE_P-3) by standard cloning procedures (fig. 3C). The Npro and C coding sequence of R-1 served as control (pCITE_P-1). Plasmids pCITE_P-2 and pCITE_P-3 served as template for site-directed mutagenesis using phosphorylated primers in order to exchange the first amino acid of the full length C protein from a serine into a proline residue (aa 250 for R-2, aa 257 for R-3). The PCR products were excised from a 1% agarose-gel, purified, and subsequently self-ligated with T4 DNA ligase (NEB) resulting in plasmids pCITE_P-2P and pCITE_P-3P.
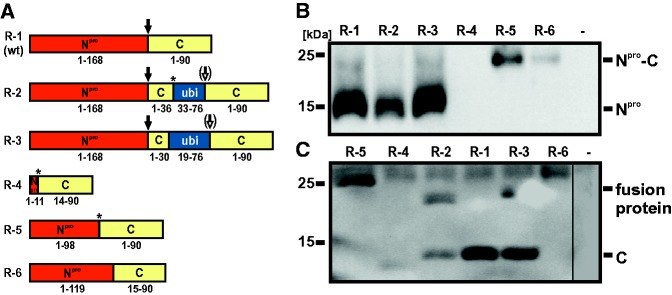
Fig. 3.—
Schematic representation and expression of the N-terminal part of the viral polyprotein encoded by selected emerged recombinants. (A) Illustration of the Npro and C protein region of recombinant viruses R-1 (corresponding to wt virus), R-2, R-3, R-4, R-5, and R-6. R-4 and R-5 emerged after cotransfection with CP7/1-686 and Ubi-CP7/887-12293, the four remaining recombinants derived from experiments with CP7/1-997 and Ubi-CP7/887-12293. Numbers below boxes indicate the presence of complete and partial proteins encoded by the recombinant viruses and refer to the authentic proteins encoded by wt virus (Npro: 168 aa, C protein: 90 aa) or a cellular ubiquitin monomer (consisting of 76 aa). The asterisks mark the positions of encoded amino acids differences of the recombinant genomes compared with the recombination partners. For R-2 four nucleotides at the junction site are identical to both recombination partners, and therefore, the recombination site could not be determined precisely. The black arrows indicate cleavage by the autoprotease Npro (R-1, R-2, and R-3). The putative cleavage site (cleavage mediated by cellular ubiquitin C-terminal hydrolase) within the C-ubi-C fusion proteins encoded by recombinants R-2 and R-3 is indicated by white arrows. (B and C) Detection of Npro and C protein in cell lysates of infected MDBK cells by Western blot analysis. Cells were infected at an MOI of 0.01 with the indicated recombinant viruses. Noninfected cells (−) served as negative control. Npro and Npro fusion proteins as well as C and C fusion proteins were detected 48 h p.i. with the Npro-specific monoclonal antibody 13B6 (B) and the C-specific monoclonal antibody 1F7 (C), respectively. An unspecific cellular protein with an apparent molecular weight of about 25 kDa, which was also detected in noninfected cells, was detected by the antibody 1F7.
All plasmids described in this chapter were tested by restriction enzyme mapping and the sequences of these plasmids were confirmed by nucleotide sequence analysis. Further details of the cloning strategies can be obtained from the authors upon request.
Synthesis of RNA Fragments
In vitro transcription of recombination partners and electroporation of RNA was performed as described previously (Gallei et al. 2004; Austermann-Busch and Becher 2012). Briefly, cDNA was transcribed into RNA using SP6- or T7-Megascript kit (Ambion) depending on the promoter of the respective plasmid. After TurboDNase (Ambion) digestion for 15 min at 37 °C, the RNA was extracted with Megaclear kit (Ambion) according to the manufacturer’s instructions. Concentrations of RNA were photometrically determined with NanoDrop (Thermo Scientific) and the quality of RNA was controlled by ethidium bromide staining of samples after agarose gel electrophoresis.
Electroporation, Detection of Recombinant Viruses, and Virus Rescue
The recombination partners were electroporated (180 V, 950 µF) with a GenePulser Xcell (Bio-Rad) (Becher et al. 2000) in a molar ratio of 5:1, namely 2 pmol of the 3′ recombination partner and 10 pmol of one of the 5′ recombination partners, into MDBK cells. Ten percent of the electroporated cells were subjected to a 10-fold dilution in a suspension of naïve cells. The undiluted electroporated cells as well as the diluted cells were distributed to 48 wells of two 24-well plates after electroporation. From day 3 to day 5 postelectroporation (p.e.) the cells were checked daily for cytopathic effect (CPE) by light microscopic inspection. The supernatants of each well were collected and after treatment with trypsin, the cells of each well were transferred separately to a well of a six-well plate and checked for CPE after an additional incubation time of 20–48 h. The cells were either harvested for RNA extraction or heat fixated for the detection of BVDV antigen by indirect immunofluorescence (IF) analysis with anti-NS3 monoclonal antibody BVD/C16 (diluted 1:25 in PBS with 0.02% Tween 20 [PBS-Tween]) (Peters et al. 1986) and Cy3-conjugated goat antimouse IgG (diluted 1:800 in PBS-Tween; Dianova). The 10-fold diluted electroporated cells were directly heat fixated 5 days p.e. and analyzed by IF. For first virus passage, 5 × 105 MDBK cells were infected with 500 µl of the collected supernatants of the recombinant viruses. Three to four days later, 1 ml tissue culture supernatant was used to infect 3 × 106 MDBK cells for further virus passages. This procedure was repeated and the virus titers were determined as 50% tissue culture infectious doses (TCID50) per ml as described previously (Pankraz et al. 2005). Depending of the reached titers, the second or third passage of the recombinant viruses was used for the experiments described in this study.
Analysis of Recombinant Genomes
Total cellular RNA was prepared using the RNeasy Mini kit (Qiagen). The cDNA synthesis and subsequent PCR (applying 2 μl cDNA) were performed as described previously (Postel et al. 2012). Cycling conditions were as follows: 98 °C, 30 s; 35 × (98 °C, 15 s; 58 °C, 30 s; 72 °C, 35 s); 72 °C, 5 min; 4 °C. For PCR amplification of the 5′-terminal part of the recombinant viral genomes BVDV specific oligonucleotides Ol-1400R (nucleotides 1427–1445; antisense primer) and Ol-100 (nucleotides 105–125; sense primer) were used (Becher et al. 1997, 1998). For amplification of the NS3 gene, 2 μl cDNA each were subjected to two PCRs with primer pairs Ol_CP7_5133F (nucleotides 5133 to 5153; sense primer) and Ol_CP7_6137R (nucleotides 6137–6156; antisense primer) and Ol_CP7_6025F (nucleotides 6025–6045, sense primer) and Ol_CP7_7328R (nucleotides 7328–7348; antisense primer), respectively, using the Maxima Hot Start Green PCR Master Mix (Thermo Scientific) according to the manufacturer’s instructions.
Nucleotide sequencing was carried out by commercial sequencing services (Qiagen or LGC). The obtained sequences were analyzed by using the GCG software package included in HUSAR (DKFZ, Heidelberg, Germany) (Devereux et al. 1984). In addition, the Npro and C coding sequences of the recombinant viruses were confirmed by sequence analysis at the end of the experiments used for determination of viral growth kinetics and viral RNA synthesis.
Transient Protein Expression
For transient expression of recombinant proteins, the MVA-T7 system was applied as previously described (Meyer et al. 2012). Briefly, BHK-21 cells (7.5 × 105) were infected with recombinant MVA-T7 virus and 2 h later transfected with the respective plasmid (pCITE_P-1, pCITE_P-2, pCITE_P-2P, pCITE_P-3, pCITE_P-3P) using Lipofectamine 2000 (Thermo Fisher Scientific). Recombinant MVA-T7 virus was kindly provided by Gerd Sutter (Ludwig-Maximilians-Universität München, Munich, Germany). Twenty hours posttransfection the cells were lysed using NP40 lysis buffer (50 mM Tris/HCl [pH 7.5], 150 mM NaCl, 0.5% [w/v] sodium deoxycholate, 1% [v/v] NP-40, complete protease inhibitor [Roche]). Protein concentration was determined by using the BCA protein assay kit (Pierce Biotechnologies).
SDS-Page and Western Blotting
Cells were lysed using NP40 lysis buffer and proteins were separated in 12% or 15% polyacrylamide gels. After sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) proteins were transferred to nitrocellulose membrane or PVDF membrane. The membrane was blocked with 2% ECL Blocking Buffer (GE Healthcare Bio-Sciences AB). For detection of Npro and C the mouse monoclonal antibodies (MAb) 13B6 and 1F7 were used, both diluted 1:5 in PBS containing 0.02% Tween 20 (PBS-Tween), respectively. Both antibodies were generated according to standard procedures previously described (Lamp et al. 2011). Antibody binding was detected by using a secondary rabbit anti-mouse peroxidase-coupled antibody diluted 1:1000 in PBS-Tween (Dako) and ECL Advanced Western Blotting Detection kit or ECL Select Western Blotting Detection kit (GE Healthcare Bio-Sciences AB).
Determination of Growth Kinetics
MDBK cells (1 × 106) were infected in a six-well dish with infectious supernatants of the recombinant viruses R-1, R-2, R-3, R-4, and R-5 at a multiplicity of infection (MOI) of 0.01. After adsorption for 1 h at 37 °C, the cells were washed six times with PBS, overlaid with medium containing 5% horse serum, and incubated at 37 °C and 5% CO2. At 0, 6, 12, 24, 36, 48, 72, and 96 h postinfection (p.i.) aliquots (230 µl) of the supernatant were removed and replaced by fresh medium. The supernatants of three independent experiments were used individually for titration on MDBK cells. After 72 h incubation at 37 °C, virus detection was carried out by NS3-specific IF analysis as described above. Titers were determined as 50% tissue culture infectious doses (TCID50) per ml as described previously (Pankraz et al. 2005)
Quantification of Viral RNAs by Real-Time RT-PCR
Real-time RT-PCR was performed in a qPCR system Mx3005P apparatus (Agilent). For comparative analysis of the quantities of accumulated viral RNA genomes, MDBK cells (1 × 106) were infected with supernatants of the recombinant viruses R-1, R-2, R-3, R-4, and R-5 at an MOI of 0.01. Total cellular RNA was prepared 10 h p.i., marking the end of one replication cycle (Gong et al. 1996). Total cellular RNA from three independent experiments was prepared using the RNeasy Mini kit (Qiagen). The RNA was diluted in a ratio of 1:50 in RNA-safe buffer (50 ng/μl poly[A] carrier RNA, 0.05% Tween 20, and 0.05% sodium azide in nuclease-free water). For comparative quantification of viral RNA, 5 µl of the diluted RNAs as well as five serial 10-fold dilutions of a BVDV RNA standard and a glyceraldehyde-3-phosphate dehydrogenase (GAPDH) standard were subjected to a one-step duplex real-time RT-PCR (qRT-PCR) using the QuantiTect Probe PCR kit (Qiagen). BVDV genome load and GAPDH mRNA abundance were simultaneously detected in the same reaction tube in triplicates. For amplification of BVDV genomes the primer pair Pesti.100F (5′-CTAACCATGCCCTTAGTAG-3′) and Pesti.206R (5′-CGTCGAACCACTGACGACT-3′), and the probe BVDV.139p (5′-FAM-TAGCAACAGTGGCGAGTTCGTTGGATGGCT-BHQ1-3′), which were adapted from a previous publication (Baxi et al. 2006) and target the 5′NTR, were used. For amplification of the GAPDH mRNA, the primers GAPDH.465F (5′-GGCGTGAACCACGAGAAGTATAA-3′) and GAPDH.583R (5′-CCCTCCACGATGCCAAAGT-3′), and the probe GAPDH.490p (5′-HEX-ACACCCTCAAGATTGTCAGCAATGCCTCCT-BHQ1-3′), which were adapted from a previous publication (Leutenegger et al. 2000), were used. Cycling conditions were as follows: 50 °C, 30 min; 95 °C, 15 min; 40 × (95 °C, 30 s; 60 °C, 30 s; 72 °C, 30 s). The quantity of viral RNA was normalized to the abundance of bovine GAPDH mRNA. For comparing the viral RNA quantities of the recombinant viruses R-1, R-2, R-3, R-4, and R-5 in cells 10 h p.i., the viral RNA amount of R-1 was set 100%.
Determination of Translational Activity of the 3′ Recombination Partner
For the luciferase reporter assay, a portion of the electroporated cells used for the RNA recombination experiments and detection of recombinant viruses was seeded in quadruplicates into 96-well plates (4 × 104 cells per well). Four hours p.e. the cells were washed with PBS, overlaid with 50 µl PBS and expression of luciferase was measured with the Nano-Glo Luciferase Assay System (Promega) according to the manufacturer’s instructions in a Genios Pro chemiluminometer (Tecan).
Statistical Methods
Statistical analysis was performed using the t-test. Significance was considered with P values <0.05. Statistical tests were run using the freeware program R.
Results
Experimental Design and Structure of Recombination Partners
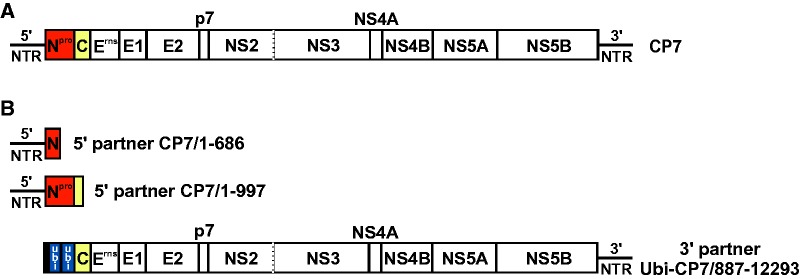
In order to determine if translation of viral proteins is essential for RNA recombination, a recombination system based on cotransfection of cells with two synthetic replication-incompetent RNA genome fragments (5′ and 3′ recombination partner) derived from cp BVDV strain CP7 was established (fig. 1A and B). The two constructed 5′ recombination partners (fig. 1B) comprise the complete 5′NTR including the IRES element essential for pestiviral translation plus a short part of the Npro coding region (5′ recombination partner CP7/1-686) or the genomic region encoding the complete Npro and part of C protein (5′ recombination partner CP7/1-997). Accordingly, both 5′ recombination partners lack major parts of the entire polyprotein coding region and the 3′NTR and thus are replication incompetent. The 3′ recombination partner Ubi-CP7/887-12293 represents a 5′ truncated BVDV genome, lacking the 5′NTR with the IRES element and the Npro coding region (fig. 1B). This partner encompasses the entire genetic information for all other viral structural and nonstructural proteins and also includes the complete 3′NTR, which is essential for viral replication (Pankraz et al. 2005). At the 5′ end of this construct, an additional sequence derived from BVDV strain CP14 (Gallei et al. 2004), which encodes five C-terminal amino acids of NS2, a C-terminal ubiquitin fragment (14 aa), as well as two complete ubiquitin monomers (76 aa each), was included. The ubiquitin coding sequence was introduced into the 3′ partner in order to facilitate appropriate processing of the N-terminus of the downstream located C protein in putatively generated recombinants. As the 3′ recombination partner lacks the IRES element, IRES-mediated translation of this viral RNA fragment cannot occur. Several independently performed experiments demonstrated that transfection of MDBK cells with the individual recombination partners did not result in translation of detectable amounts of viral protein NS3 and did not produce replicating viruses (data not shown).
Fig. 1.—
Structure of BVDV recombination partners. (A) Genome organization of BVDV strain CP7, which represents the genetic basis of the recombination partners used in this study (details are described in the introduction). (B) Schematic representation of the 5′ and 3′ recombination partners applied for RNA recombination experiments. The two depicted 5′ recombination partners comprise a complete 5′ NTR (nt 1-382) with the IRES element and either nt 383-686 encoding an N-terminal part of Npro (CP7/1-686) or nt 383-997 encoding the complete Npro and a short N-terminal part of C (CP7/1-997). The 3′ recombination partner Ubi-CP7/887-12293 contains a fragment derived from BVDV strain CP14, which encodes the five C-terminal aa of NS2, the C-terminal 14 aa of ubiquitin as well as two complete ubiquitin monomers (76 aa each), fused to a long sequence fragment corresponding to nt 887-12293 of BVDV CP7 wt virus. Nucleotide numbering refers to the genomic sequence of CP7-388 (Pankraz et al. 2005).
Emergence of Recombinant Viruses in the Absence of IRES-Mediated Translation of Viral Proteins
After cotransfection of cells with 2 pmol of the 3′ recombination partner and 10 pmol of either 5′ recombination partner CP7/1-686 or partner CP7/1-997, recombinant viruses emerged and were detectable by their cytopathic effect on cell culture. In three cotransfection experiments with each 5′ partner a total of at least 70 independently emerged recombinant viruses were recovered and subsequently analyzed by RT-PCR and nucleotide sequencing. For 58 recombinant genomes (29 from each 5′ recombination partner), the sequences corresponding to nucleotide positions 126–1426 of the BVDV CP7 genome were analyzed for determination of the recombination sites. For the remaining 12 virus-positive RNAs, limited amounts of the obtained PCR products did not allow determination of nucleotide sequences. The emergence of recombinant viruses after cotransfection of MDBK cells with two BVDV RNA fragments either lacking almost the complete genomic region encoding the viral proteins (5′ partner) or the IRES element (3′ partner) demonstrates that RNA recombination can occur in the absence of IRES-mediated translation of viral proteins.
Genome Structure of the Recombinant Viruses
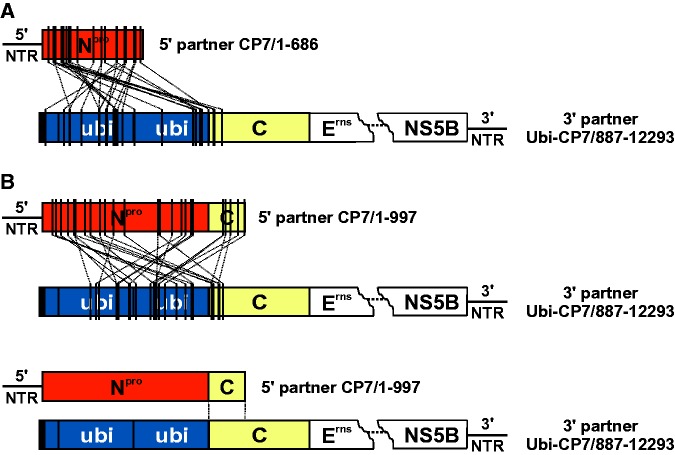
Analyses of the cross-over sites of 58 recombinant viruses revealed, that with the exception of one homologous recombinant (R-1) all other viruses emerged by nonhomologous RNA recombination, which occurred exclusively at internal positions of the recombining RNA molecules (fig. 2A and B). Further analysis of the recombination sites did not provide evidence for a sequence bias. The distribution of nucleotides and dinucleotides situated at the recombination sites resembles the distribution of dinucleotides in the particular genome stretches. The extent of nucleotide identity between the recombining RNA molecules at the cross-over site was not significantly higher than the expected random level. The recombination sites in both 5′ partners were exclusively located within the coding regions, which was in line with the requirement of a complete 5′NTR for preserving a functional IRES element. Although the recombination sites in the 3′ recombination partner were predominantly located in the ubiquitin coding region (47 of 58 recombinants), for eleven recombinants the 3′ recombination sites were located within the C coding region. Furthermore, 20 analyzed recombinant genomes did not encode for a complete ubiquitin monomer. As it was previously shown that an incomplete ubiquitin monomer leads to abrogation of processing by cellular proteases (Tautz et al. 1993), it was assumed that the respective recombinant viruses (e.g., R-2 and R-3) express C fusion proteins of up to 178 aa instead of the authentic C protein consisting of 90 aa (fig. 3A). These proteins would comprise C-terminal parts of either Npro or C protein fused to an N-terminally truncated ubiquitin followed by a complete C protein (Npro-ubi-C and C-ubi-C fusion proteins [figs. 3 and 4]). Accordingly, duplications of C protein-coding sequences of up to 109 nucleotides (R-3) were found in recombinant genomes. Furthermore, other recombinant virus genomes encode for Npro-C fusion proteins (e.g., R-4, R-5, and R-6). Their size varied from 89 aa (R-4) to 195 aa (R-6), which was the largest putative fusion protein predicted for the analyzed recombinant viruses (fig. 3A).
Fig. 2.—
Recombination sites within the 5′ recombination partners and the 3′ recombination partner. The cross-over sites for 29 independently emerged recombinants, derived from cotransfection experiments with the 3′ partner and either with the 5′ recombination partner CP7/1-686 (A) or CP7/1-997 (B) are schematically displayed by lines between the bars. Npro coding sequences are highlighted in red, ubiquitin coding sequences in blue and C protein coding sequences in yellow. For the 3′ recombination partner, the genomic region encoding proteins Erns to NS5B is indicated by dots. Bottom: One homologous recombinant (R-1). The two vertical lines between the recombination partners indicate the genomic region in which the cross-over site is located.
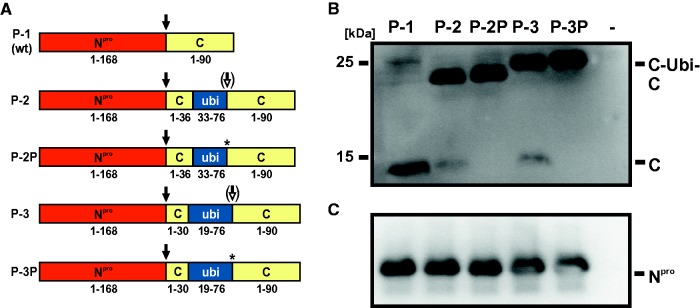
Fig. 4.—
Processing of fusion proteins of recombinants R-1, R-2, and R-3. (A) Schematic representation of authentic (P-1, P-2, P-3) and mutated (P-2P, P-3P) fusion proteins transiently expressed in the MVA-T7 system. P-1 encompasses the wt proteins Npro and C, P-2 and P-3 comprise Npro and the C fusion proteins encoded by the recombinant viruses R-2 and R-3, respectively. In P-2P and P-3P, the asterisks mark the serine to proline substitutions directly downstream of the ubiquitin fragments. Black arrows indicate autoproteolytic cleavage mediated by Npro. White arrows (P-2 and P-3) indicate partial processing directly downstream of the ubiquitin fragments. (B and C) Western blot analysis. BHK-21 cells were infected with MVA-T7 and transfected with pCITE-Npro-C constructs encoding P-1, P-2, P-2P, P-3, and P-3P, respectively. Nontransfected cells (−) served as negative control. Cells were lysed 20 h posttransfection and analyzed by Western blot using MAb 1F7 (panel B) and MAb 13B6 (panel C) to detect C (C-ubi-C) and Npro, respectively.
Six representative recombinant viruses (R-1 to R-6) were selected (fig. 3A) and subjected to further analyses with regard to protein expression, growth kinetics, and viral RNA synthesis. Virus propagation up to passage 12 and subsequent RT-PCR analyses of RNA obtained after passage 2, 5, 9, and 12 revealed an unaltered size of amplified PCR products (5′ NTR, Npro, and C coding region) for each of the individual recombinants (R-1 to R-6) over time (data not shown).
Expression of Npro and C
In order to determine if the detected genome alterations of the selected recombinants R-1 to R-6 correlated with expression of the predicted fusion proteins, Western blot analysis was performed. In terms of Npro expression, a protein with the expected apparent molecular weight of 20 kDa was detected for the homologous recombinant R-1 (corresponding to BVDV CP7 wild-type [wt] virus) as well as for the nonhomologous recombinants R-2 and R-3 (fig. 3B). For recombinant virus R-4, an Npro-specific protein was not detectable as it probably did not display the respective epitope due to its shortened sequence. For recombinants R-5 and R-6 Western blot analysis revealed significantly enlarged proteins with a molecular weight of ∼25 kDa corresponding to the length of the predicted Npro-C fusion proteins.
Regarding the expression of C protein, a specific protein with the size of approximately 14 kDa was detected by Western blot analysis for recombinant R-1 (fig. 3C). For R-4 a protein with a slightly lower molecular weight was detected and thus confirmed the predicted shorter fusion protein of 89 aa. The fusion proteins’ sizes of recombinants R-2, R-3, and R-5 corresponded to the predicted lengths of about 25 kDa. Also for recombinant R-6 an enlarged fusion protein of about 25 kDa was detected, though merely a weak band was visible. The size of the detected fusion protein corresponds to that detected in the Npro blot. Surprisingly, for R-2 and R-3 an additional protein comigrating with mature C protein was detected. This finding can be explained by a hypothetical processing event downstream of the incomplete ubiquitin monomer mediated by cellular ubiquitin carboxy-terminal hydrolases (UCH), although this processing appeared to be inefficient.
To confirm this hypothesis, the C-ubi-C fusion proteins of R-2 and R-3 and derivates of these fusion proteins with a serine to proline mutation directly downstream of the N-terminally truncated ubiquitin were transiently expressed using the MVA-T7 system (fig. 4A). It has been described that the presence of a proline residue at this position inhibits UCH cleavage (Butt et al. 1988). Comparative analysis of the recombinant C-ubi-C fusion proteins of R-2 and R-3 with or without proline substitution revealed that the fusion proteins containing the serine residue were partially cleaved, whereas no cleavage occurred if the serine residue was replaced by a proline residue (fig. 4B). Processed C and C-ubi-C fusion proteins were detected at the same molecular weights as previously observed after infection of permissive cells with the recombinant viruses. However, the relative amount of processed C was lower after transient expression (fig. 4B). This might be due to differences in protein expression after infection of bovine MDBK cells with recombinant virus and transient expression using the MVA-T7 system in BHK-21 cells or due to the different time points of the analysis (48 vs. 20 h, respectively). The detected amounts of Npro protein confirmed similar transient expression levels for each construct (fig. 4C).
Growth Characteristics and RNA Synthesis of Recombinant Viruses
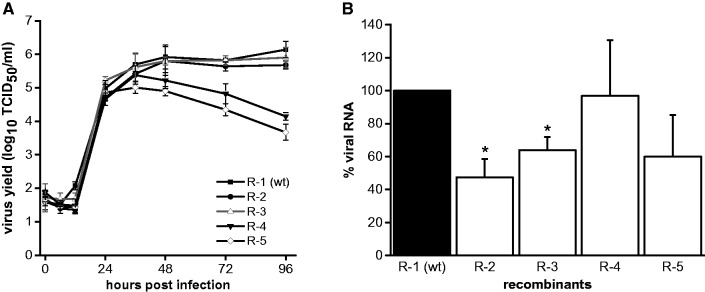
Growth characteristics and RNA synthesis of the selected recombinant viruses were compared by infection of MDBK cells at an MOI of 0.01 (fig. 5A and B). R-6 had a very low replication rate resulting in maximum titers of about 7 × 103 TCID50/ml. Therefore, it was not possible to obtain sufficient amounts of virus to include R-6 in the analysis of viral growth characteristics and RNA synthesis. Virus titers were analyzed at different time points (0 to 96 h p.i.). For recombinants R-2 and R-3, the obtained growth curves and maximum virus yields were comparable to recombinant R-1 (fig. 5A). In contrast, for R-4 and R-5 a significant decrease in viral titers was observed 36 h p.i. Regarding the maximum titers reached by recombinants R-4 (2.8 × 105 TCID50/ml) and R-5 (1.1 × 105 TCID50/ml), a reduction of about 5-fold (R-4) and about 14-fold (R-5) compared with R-1 (1.5 × 106 TCID50/ml) was detected. For analysis of viral RNA production, total cellular RNA was prepared at 10 h p.i. to exclude the effects of potential differences in viral spread, as this time point correlates with the end of the first viral replication cycle (Gong et al. 1996). The relative amounts of accumulated viral RNA were determined by quantitative real-time RT-PCR and normalized to the abundance of GAPDH RNA (fig. 5B). Compared with the amount of RNA accumulated in cells infected with R-1, the RNA amount detected for recombinants R-2 and R-3 was slightly reduced (2-fold and 1.5-fold, respectively). Although the mean value of R-5 was reduced by 1.7-fold, no significant difference in terms of viral RNA level was detected for R-4 and R-5 compared with R-1.
Fig. 5.—
Infectious virus production and RNA synthesis of selected recombinant viruses. MDBK cells were infected with the recombinant viruses R-1, R-2, R-3, R-4, and R-5 at an MOI of 0.01. (A) Growth kinetics of the recombinant viruses. Viral titers were determined over a 4-day period. Depicted are mean values and standard deviations of three independent experiments. (B) Relative amounts of accumulated viral genomic RNA. Total cellular RNA was prepared 10 h p.i. and subjected to quantitative real-time RT-PCR. The viral RNA amounts were normalized to the abundance of GAPDH RNA. The data were plotted relatively to R-1, which corresponds to the wt virus, and the amount of viral RNA of R-1 was set as 100%. Data show the mean values and standard deviations of three independent experiments, each analyzed in triplicate. The asterisks mark a significant reduction in viral RNA level compared with R-1.
It was previously shown that efficient replication of CSFVs, with massively enlarged or almost completely deleted C proteins, depends on single amino acid substitutions within the NS3 gene (Riedel et al. 2010, 2012). Therefore, the NS3 genes of the six analyzed recombinant viruses were amplified by RT-PCR and subsequently sequenced. Except from R-3, which revealed a single amino acid exchange from valine to alanine at position 2278 (position within CP7 strain), no adaptive mutations were identified. Considering that the replication of R-4, R-5, and R-6 is reduced, it appears likely that these recombinant viruses undergo further evolution to increase their replication efficiencies if passaged sufficiently. Additional passages of these recombinant viruses were not performed so far as a further detailed analysis of the genetic stability and evolution of these viruses was not within the scope of this study.
Efficient Translation of Viral Proteins Does Not Enhance Nonreplicative RNA Recombination
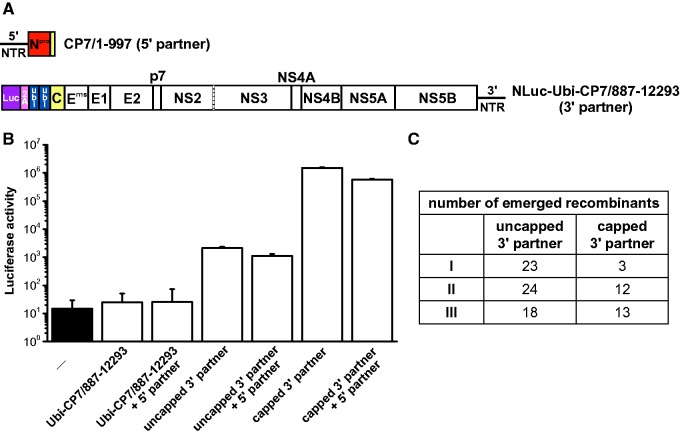
The results of the RNA recombination experiments described above demonstrated that IRES-mediated translation of viral proteins is not required for RNA recombination (figs. 1 and 2). However, it cannot be excluded that low level of IRES-independent translation of viral proteins occurred and contributed to RNA recombination. Therefore an additional set of RNA recombination experiments including quantification of translation efficiency was performed. To allow quantification of translation of the 3′ recombination partner, a luciferase reporter gene was introduced into the 3′ recombination partner at its 5′ terminus (termed NLuc-Ubi-CP7/887-12293) (fig. 6A). For efficient translation of viral proteins, the 3′ recombination partner NLuc-Ubi-CP7/887-12293 was capped prior electroporation. After cotransfection of MDBK cells with the 5′ recombination partner CP7/1-997 (10 pmol) and either a capped or uncapped 3′ recombination partner NLuc-Ubi-CP7/887-12293 (2 pmol each), the number of emerged recombinant viruses was determined for each set-up in three independent experiments (fig. 6A). In parallel, a portion of the (co)transfected cells was subjected to a luciferase reporter assay 4 h p.e. for quantitative detection of translation (fig. 6B). Luciferase reporter assay analysis demonstrated that capping of the 3′ recombination partner resulted in a 200- to 600-fold increase of translation compared with the uncapped 3′ recombination partner (fig. 6B).
Fig. 6.—
Efficient translation of viral proteins does not enhance nonreplicative RNA recombination. (A) Schematic representation of the applied 5′ and 3′ recombination partners. The 5′ recombination partner CP7/1-997 (5′ partner) has been described in figure 1. The 3′ recombination partner NLuc-Ubi-CP7/887-12293 (3′ partner) is a modified version of the 3′ recombination partner Ubi-CP7/887-12293 (see fig. 1) with a replacement of the 5′-terminal sequence coding the C-terminal 14 aa of ubiquitin by a luciferase reporter gene and the coding sequence for the porcine teschovirus-1 2A peptide. Nucleotide numbering refers to the positions in the genome of the wt virus. (B) Luciferase activity. Absolute values of luciferase activity measured 4 h p.e. of the 3′ partner (capped and uncapped) alone or in combination with the 5′ partner. Reaction set-ups with the 3′ recombination partner Ubi-CP7/887-12293 (without luciferase reporter gene) and nontransfected cells (−) served as control. Data show the mean values and standard deviations of one of three independent experiments. (C) Number of recombinant viruses emerged after cotransfection of cells with the 5′ recombination partner CP7/1-997 and the capped or uncapped 3′ recombination partner NLuc-Ubi-CP7/887-12293. Data of three independent experiments are shown.
Three independent cotransfection experiments revealed that capping and subsequent enhanced translation of viral proteins encoded by the 3′ recombination partner did not result in an increased RNA recombination frequency (fig. 6C). In more detail, 28 recombinants emerged after cotransfection with the capped template compared with 65 recombinants produced in the three control experiments with the uncapped template. The reason for the observed slight decrease in the yield of recombinants produced with the capped template remains elusive. One possible explanation is that the capping reaction might have a negative effect on the integrity of the RNA. Alternatively, it can be speculated that ribosomal binding of the capped RNA template might limit its accessibility for nonreplicative joining of RNA molecules Nine independently emerged recombinant viruses were analyzed regarding their cross-over sites, which were comparable to those of the recombinant viruses shown in figure 2 (data not shown). Taken together, the results of the present study demonstrate that nonreplicative RNA recombination can occur in the absence of IRES mediated translation of pestiviral proteins and is not enhanced by efficient translation of viral proteins (fig. 6).
Discussion
Nonreplicative RNA recombination has been proven for several plus-strand RNA viruses including poliovirus, BVDV, and HCV (Gmyl et al. 2003; Gallei et al. 2004; Scheel et al. 2013). Although it has previously been shown that RNA recombination in pestiviruses can occur in the absence of a functional RNA-dependent RNA polymerase (RdRp), the role of other viral proteins remained unclear (Gallei et al. 2004). Several pestiviral proteins (C, Erns, NS3, NS4B, and NS5B) were shown to interact with the viral RNA (Zhong et al. 1998; Iqbal et al. 2004; Ivanyi-Nagy et al. 2008; Murray et al. 2008; Gladue et al. 2011). It was therefore interesting to investigate whether viral proteins other than the RdRp are involved in RNA recombination. By the detection of over 70 independently emerged recombinant viruses the results of the present study demonstrate that IRES-mediated translation of the pestiviral proteins is not required for nonreplicative RNA recombination. Moreover, our conclusion that RNA recombination can occur independent from translation of viral proteins is further supported by the observation that increasing the efficiency of translation of viral proteins by capping of the 3′ recombination partner (carrying an almost complete polyprotein encoding sequence) did not enhance RNA recombination (fig. 6).
With regard to the recombination system applied in this study, one might argue that the possibility of noncanonical initiation of translation might result in the expression of small amounts of viral proteins and a small background RNA-dependent RNA polymerase (RdRp) activity in the very early stages of transfection might lead to replication-driven recombination. A strong argument against such a scenario of a possible early RdRp activity in the experiments described in the present study is provided by the results of previously reported experiments. Two independent studies in the poliovirus and BVDV system have demonstrated that transfection of cells with two synthetic overlapping subgenomic transcripts each lacking a functional RdRp gene resulted in the generation of recombinant RNA genomes and thereby proved the existence of nonreplicative recombination in the absence of a viral RdRp (Gallei et al. 2004; Gmyl et al. 2003). The recombination system applied in the present study is very similar to the one used for the proof of nonreplicative RNA recombination (Gallei et al. 2004) as both are based on cotransfection of cells with overlapping nonreplicative BVDV subgenomic transcripts resulting in the emergence of recombinant infectious viral RNA genomes. The similar number of recombinant viruses generated in the experiments of these two studies strongly suggests that RNA recombination in both systems results from nonreplicative RdRp-independent reactions.
Nonreplicative RNA recombination in the absence of translation of viral proteins was also suggested in a study with poliovirus genomic RNA fragments, in which the 5′ fragments carried the IRES element and a spacer sequence and each of the three 3′ fragments encompassed the complete authentic poliovirus coding sequence and 3′NTR as well as a modified 5′NTR with different lethal deletions and mutations in the essential cis-acting elements, resulting in abrogation of translation and replication activity (Gmyl et al. 1999). Similar to the results of the present study in the BVDV system, cotransfection of the poliovirus fragments led to the emergence of infectious virus progeny. In contrast to the results with BVDV and poliovirus RNA fragments, experiments with subgenomic transcripts derived from HCV, which did not allow translation of viral proteins prior to an RNA recombination event, did not result in the emergence of infectious virus progeny (Scheel et al. 2013). This difference might be attributed to the fact that the first protein in the polyprotein of HCV, the C protein, is less tolerant for genomic changes than pestivirus Npro, which is dispensable for viral replication (Tratschin et al. 1998). The present study demonstrates that significantly enhanced translation of viral proteins did not increase the frequency of RNA recombination in BVDV, whereas the effect of enhanced translation of viral proteins on RNA recombination was not analyzed in the studies with poliovirus and HCV (Gmyl et al. 1999; Scheel et al. 2013). In conclusion, the results of the present study in the BVDV system together with the previously reported results obtained with poliovirus RNA fragments show that nonreplicative RNA recombination does not require efficient translation of viral proteins.
Alternatively, it can be hypothesized that mechanisms involving cellular factors are implicated in nonreplicative RNA recombination leading to the emergence of recombinant viruses. It was previously proposed that the recombining RNA substrates might be provided by physical forces, cryptic ribozyme activity, or endoribonucleolytic cleavage (Chetverin et al. 1997; Gmyl et al. 2003; Gallei et al. 2004). Furthermore, it was shown that modification of the 5′ triphosphate and 3′ hydroxyl ends of the recombining RNA molecules to 5′ hydroxyl and 3′ monophosphoryl ends resulted in a significantly increased RNA recombination frequency (Gmyl et al. 1999; Austermann-Busch and Becher 2012). RNA fragments with such RNA termini are typically produced by cellular endoribonucleases (Li et al. 2010). The hypothesis of a ribonucleolytic involvement is underlined by the observed preferential occurrence of RNA recombination in single-stranded regions of the RNA substrates and by the fact that many endoribonucleases specifically cleave single-stranded RNA substrates (Li et al. 2010; Austermann-Busch and Becher 2012). Indeed, cellular exo- and endoribonucleases were shown to influence the frequency of viral RNA recombination in plant and yeast cells (Serviene et al. 2005; Cheng et al. 2006; Jaag and Nagy 2009; Jaag et al. 2011). Therefore, ribonucleases might play an important role for BVDV RNA recombination. Although pestiviruses encode the unique viral endoribonuclease Erns (Schneider et al. 1993), the present study revealed that such a potential ribonucleolytic involvement must have derived from the host cell. Cellular components that are enriched with ribonucleases, like processing bodies (P-bodies) and stress granules (SG), were shown to be involved in viral life cycles of several RNA viruses (Beckham and Parker 2008). Thus, host ribonucleases may also be important for the intracellular generation of viral RNA templates that are similar to the ones described in the present study and may finally contribute to the emergence of viral recombinants during pestivirus infection. Further studies will address the role of cellular ribonucleases for providing the recombining RNA substrates. Another major goal of future studies on nonreplicative RNA recombination is the identification of the factors involved in the ligation reaction responsible for joining of the RNA fragments.
In the experiments described in the present study, high amounts of RNA were transfected which resulted in a low yield of recombinant viruses. Accordingly, it can be concluded that the translation-independent, nonreplicative process of RNA recombination is a rare event. For poliovirus, it has been reported that the frequency of replicative recombination is about 25-fold higher than the frequency of nonreplicative recombination (Lowry et al. 2014). In the BVDV system, the frequencies of replicative and nonreplicative RNA recombination have not been compared so far. Under natural conditions, viral RNA genomes present in an infected cell cannot serve as substrates for nonreplicative recombination and subgenomic RNA molecules contributing to nonreplicative joining of RNA molecules have first to be generated, e.g., by ribonucleolytic cleavage as mentioned above. This may represent a significant limitation of the efficiency of nonreplicative RNA recombination. Nevertheless, if such an inefficient process results in the generation of infectious recombinant viruses with novel biological features, the biological consequences of nonreplicative RNA recombination might be dramatic. The emergence of cytopathogenic BVDV by RNA recombination in persistently infected cattle finally leads to lethal mucosal disease and thus represents an illustrative example of the biological significance of RNA recombination (Becher and Tautz 2011).
The analyzed recombinant viruses demonstrated a remarkable flexibility regarding the structure of the pestiviral C protein (fig. 3A). Several emerged recombinant viruses express C-fusion proteins ranging from 89 to 195 aa instead of wt C protein consisting of 90 aa (fig. 2A and B). The C protein is a structural viral protein, putatively involved in condensation of viral RNA and formation of the nucleocapsid (Ivanyi-Nagy et al. 2008; Murray et al. 2008). Evidence for the flexibility of C protein regarding increase and decrease of size has been described in the context of infectious cDNA clones of the related pestivirus CSFV (Riedel et al. 2010, 2012). In line with the results reported for CSFV C protein, the variety of differently sized BVDV C-fusion proteins described in this study stands in stark contrast to a rigid structure like an icosahedral nucleocapsid, as the described alterations presumably result in a highly different nucleocapsid shape. However, in contrast to the studies with CSFV (Riedel et al. 2010, 2012), no adaptive mutations within the NS3 gene were identified except one mutation in R-3 at aa position 2278. Regarding virus yield and RNA production, the results show that replacement of the C protein by enlarged Npro-C fusion proteins (R-4 and R-5) resulted in a substantial decrease in viral titers without significantly affecting viral RNA production (fig. 5A and B). Also R-6, encoding an Npro-C fusion protein of more than twice the size of the parental C protein, reached only very low titers. It can be assumed that either the incorporation of large-scale fusion proteins into the virions or the lack of C protein in virions results in an altered structure of the virus particle, which is expected to influence virus assembly and entry, representing essential steps of the viral life cycle. Furthermore, also the uncoating of the viral RNA during the entry process might be impaired, resulting in reduced amounts of infectious virus progeny. In contrast, the genomic alterations of R-2 and R-3 only slightly affect viral RNA synthesis, which are negligible, as they have no relevant effect on efficient production of infectious viruses (fig. 5A and B). It can be speculated that those recombinants have incorporated the unaltered C protein which was shown to be cleaved from C-ubi-C fusion proteins (figs. 3B and 4B), leading to the release of virions that presumably have the same shape as wt virus particles.
Several analyzed recombinant genomes encode truncated ubiquitin monomers, leading to the expression of large C-ubi-C fusion proteins with a length of up to 178 aa. According to the literature, one complete ubiquitin monomer is a prerequisite for N-terminal and C-terminal processing of the fusion proteins by cellular ubiquitin C-terminal hydrolases (Agell et al. 1988; Tobias and Varshavsky 1991; Baker et al. 1992; Tautz et al. 1993). Furthermore, it has been reported that the four N-terminal aa of a truncated ubiquitin can be functionally replaced by a host-derived oligopeptide for maintaining processing at the C-terminus of this ubiquitin fusion protein, whereas replacement by a randomly selected virus encoded peptide did not result in processing of the respective ubiquitin fusion protein (Becher et al. 1998). It is therefore a remarkable result of the expression studies presented here (fig. 4) that the lack of up to 32 N-terminal aa of ubiquitin can be functionally replaced by viral C protein derived sequences, yet still allow for processing of the fusion proteins by cellular proteases, though in an inefficient manner.
Taken together, the results of the present study demonstrate that nonreplicative viral RNA recombination does not require the efficient translation of viral proteins and provide evidence for a remarkable flexibility regarding the structure of the pestivirus C protein. Future studies will focus on the identification of cellular factors putatively involved in nonreplicative RNA recombination.
Acknowledgments
Part of this work was performed by M.K.B. in partial fulfillment of the requirements for the doctoral degree from University of Veterinary Medicine, Hannover, Germany. This study was supported by a grant from the Deutsche Forschungsgemeinschaft (BE 2333/2-2). P.B. was supported by a Heisenberg professorship (BE 2333/1-2) from the Deutsche Forschungsgemeinschaft. We thank Norbert Tautz for helpful discussions and critical reading of the manuscript.
Literature Cited
- Agell N, Bond U, Schlesinger MJ. 1988. In vitro proteolytic processing of a diubiquitin and a truncated diubiquitin formed from in vitro-generated mRNAs. Proc Natl Acad Sci U S A. 85:3693–3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold JJ, Ghosh SK, Cameron CE. 1999. Poliovirus RNA-dependent RNA polymerase (3D(pol)). Divalent cation modulation of primer, template, and nucleotide selection. J Biol Chem. 274:37060–37069. [DOI] [PubMed] [Google Scholar]
- Austermann-Busch S, Becher P. 2012. RNA structural elements determine frequency and sites of nonhomologous recombination in an animal plus-strand RNA virus. J Virol. 86:7393–7402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker RT, Tobias JW, Varshavsky A. 1992. Ubiquitin-specific proteases of Saccharomyces cerevisiae. Cloning of UBP2 and UBP3, and functional analysis of the UBP gene family. J Biol Chem. 267:23364–23375. [PubMed] [Google Scholar]
- Baxi M, et al. 2006. A one-step multiplex real-time RT-PCR for detection and typing of bovine viral diarrhea viruses. Vet Microbiol. 116:37–44. [DOI] [PubMed] [Google Scholar]
- Becher P, et al. 1997. Phylogenetic analysis of pestiviruses from domestic and wild ruminants. J Gen Virol. 78 (Pt 6):1357–1366. [DOI] [PubMed] [Google Scholar]
- Becher P, Orlich M, Thiel H-J. 1998. Complete genomic sequence of border disease virus, a pestivirus from sheep. J Virol. 72:5165–5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becher P, Orlich M, Thiel HJ. 2000. Mutations in the 5′ nontranslated region of bovine viral diarrhea virus result in altered growth characteristics. J Virol. 74:7884–7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becher P, Orlich M, Thiel HJ. 1998. Ribosomal S27a coding sequences upstream of ubiquitin coding sequences in the genome of a pestivirus. J Virol. 72:8697–8704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becher P, Tautz N. 2011. RNA recombination in pestiviruses: cellular RNA sequences in viral genomes highlight the role of host factors for viral persistence and lethal disease. RNA Biol. 8:216–224. [DOI] [PubMed] [Google Scholar]
- Becher P, Thiel H-J. 2011. Genus pestivirus (Flaviviridae) In: Tidona CA, Darai G, editors. Springer index of viruses, 2nd ed. Heidelberg, Germany: Springer Verlag; p. 483–488. [Google Scholar]
- Beckham CJ, Parker R. 2008. P bodies, stress granules, and viral life cycles. Cell Host Microbe 3:206–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biebricher CK, Luce R. 1992. In vitro recombination and terminal elongation of RNA by Q beta replicase. EMBO J. 11:5129–5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt TR, Khan MI, Marsh J, Ecker DJ, Crooke ST. 1988. Ubiquitin-metallothionein fusion protein expression in yeast. A genetic approach for analysis of ubiquitin functions. J Biol Chem. 263:16364–16371. [PubMed] [Google Scholar]
- Cheng C-P, Nagy PD. 2003. Mechanism of RNA recombination in carmo- and tombusviruses: evidence for template switching by the RNA-dependent RNA polymerase in vitro. J Virol. 77:12033–12047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C-P, Serviene E, Nagy PD. 2006. Suppression of viral RNA recombination by a host exoribonuclease. J Virol. 80:2631–2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chetverin AB, Chetverina HV, Demidenko AA, Ugarov VI. 1997. Nonhomologous RNA recombination in a cell-free system: evidence for a transesterification mechanism guided by secondary structure. Cell 88:503–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng R, Brock KV. 1993. 5′ and 3′ untranslated regions of pestivirus genome: primary and secondary structure analyses. Nucleic Acids Res. 21:1949–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J, Haeberli P, Smithies O. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallei A, Pankraz A, Thiel H-J, Becher P. 2004. RNA recombination in vivo in the absence of viral replication. J Virol. 78:6271–6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie JH, Baker JA, McEntee K. 1960. A cytopathogenic strain of virus diarrhea virus. Cornell Vet. 50:73–79. [PubMed] [Google Scholar]
- Gladue DP, et al. 2011. Identification of an NTPase motif in classical swine fever virus NS4B protein. Virology 411:41–49. [DOI] [PubMed] [Google Scholar]
- Gmyl AP, et al. 1999. Nonreplicative RNA recombination in poliovirus. J Virol. 73:8958–8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gmyl AP, Korshenko SA, Belousov EV, Khitrina EV, Agol VI. 2003. Nonreplicative homologous RNA recombination: promiscuous joining of RNA pieces? RNA 9:1221–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y, et al. 1996. Characterization of RNA synthesis during a one-step growth curve and of the replication mechanism of bovine viral diarrhoea virus. J Gen Virol. 77:2729–2736. [DOI] [PubMed] [Google Scholar]
- Heimann M, Roman-Sosa G, Martoglio B, Thiel H-J, Rümenapf T. 2006. Core protein of pestiviruses is processed at the C terminus by signal peptide peptidase. J Virol. 80:1915–1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal M, Poole E, Goodbourn S, McCauley JW. 2004. Role for bovine viral diarrhea virus Erns glycoprotein in the control of activation of beta interferon by double-stranded RNA. J Virol. 78:136–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanyi-Nagy R, Lavergne J-P, Gabus C, Ficheux D, Darlix J-L. 2008. RNA chaperoning and intrinsic disorder in the core proteins of Flaviviridae. Nucleic Acids Res. 36:712–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaag HM, Lu Q, Schmitt ME, Nagy PD. 2011. Role of RNase MRP in viral RNA degradation and RNA recombination. J Virol. 85:243–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaag HM, Nagy PD. 2009. Silencing of Nicotiana benthamiana Xrn4p exoribonuclease promotes tombusvirus RNA accumulation and recombination. Virology 386:344–352. [DOI] [PubMed] [Google Scholar]
- Kao CC, Del Vecchio AM, Zhong W. 1999. De NovoInitiation of RNA synthesis by a recombinant flaviridae RNA-dependent RNA polymerase. Virology 253:1–7. [DOI] [PubMed] [Google Scholar]
- Kim JH, et al. 2011. High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS One 6:e18556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MJ, Kao C. 2001. Factors regulating template switch in vitro by viral RNA-dependent RNA polymerases: implications for RNA-RNA recombination. Proc Natl Acad Sci U S A. 98:4972–4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamp B, et al. 2011. Biosynthesis of classical swine fever virus nonstructural proteins. J Virol. 85:3607–3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KM, Gillespie JH. 1957. Propagation of virus diarrhea virus of cattle in tissue culture. Am J Vet Res. 18:952–953. [PubMed] [Google Scholar]
- Leutenegger CM, Alluwaimi AM, Smith WL, Perani L, Cullor JS. 2000. Quantitation of bovine cytokine mRNA in milk cells of healthy cattle by real-time TaqMan® polymerase chain reaction. Vet Immunol Immunopathol. 77:275–287. [DOI] [PubMed] [Google Scholar]
- Li WM, Barnes T, Lee CH. 2010. Endoribonucleases—enzymes gaining spotlight in mRNA metabolism. FEBS J. 277:627–641. [DOI] [PubMed] [Google Scholar]
- Lowry K, Woodman A, Cook J, Evans DJ. 2014. Recombination in enteroviruses is a biphasic replicative process involving the generation of greater than genome length ‘imprecise’ intermediates. PLoS Pathog. 10(6):e1004191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer D, et al. 2012. New insights into the antigenic structure of the glycoprotein Erns of classical swine fever virus by epitope mapping. Virology 433:45–54. [DOI] [PubMed] [Google Scholar]
- Meyers G, Tautz N, Becher P, Thiel HJ, Kümmerer BM. 1997. Recovery of cytopathogenic and noncytopathogenic bovine viral diarrhea viruses from cDNA constructs. J Virol. 71:1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray CL, Marcotrigiano J, Rice CM. 2008. Bovine viral diarrhea virus core is an intrinsically disordered protein that binds RNA. J Virol. 82:1294–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankraz A, Thiel H-J, Becher P. 2005. Essential and nonessential elements in the 3′ nontranslated region of Bovine viral diarrhea virus. J Virol. 79:9119–9127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, Hellen CUT. 1998. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 12:67–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters W, Greiser-Wilke I, Moennig V, Liess B. 1986. Preliminary serological characterization of bovine viral diarrhoea virus strains using monoclonal antibodies. Vet Microbiol. 12:195–200. [DOI] [PubMed] [Google Scholar]
- Poole TL, et al. 1995. Pestivirus translation initiation occurs by internal ribosome entry. Virology 206:750–754. [DOI] [PubMed] [Google Scholar]
- Postel A, et al. 2012. Improved strategy for phylogenetic analysis of classical swine fever virus based on full-length E2 encoding sequences. Vet. Res. 43:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel C, et al. 2012. The core protein of classical Swine Fever virus is dispensable for virus propagation in vitro. PLoS Pathog. 8:e1002598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel C, Lamp B, Heimann M, Rümenapf T. 2010. Characterization of essential domains and plasticity of the classical Swine Fever virus Core protein. J Virol. 84:11523–11531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheel TKH, et al. 2013. Productive homologous and non-homologous recombination of hepatitis C virus in cell culture. PLoS Pathog. 9:e1003228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider R, Unger G, Stark R, Schneider-Scherzer E, Thiel HJ. 1993. Identification of a structural glycoprotein of an RNA virus as a ribonuclease. Science 261:1169–1171. [DOI] [PubMed] [Google Scholar]
- Serviene E, et al. 2005. Genome-wide screen identifies host genes affecting viral RNA recombination. Proc Natl Acad Sci U S A. 102:10545–10550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmonds P, et al. 2012. Family flaviviridae In: King A, Lefkowitz E, Adams MJ, Carstens EB, editors. Virus taxonomy. Ninth report of the International Committee on Taxonomy of Viruses. San Diego, CA: Academic Press; p 1003–1020. [Google Scholar]
- Stark R, Meyers G, Rümenapf T, Thiel HJ. 1993. Processing of pestivirus polyprotein: cleavage site between autoprotease and nucleocapsid protein of classical swine fever virus. J Virol. 67:7088–7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffens S, Thiel H-J, Behrens S-E. 1999. The RNA-dependent RNA polymerases of different members of the family Flaviviridae exhibit similar properties in vitro. J Gen Virol. 80:2583–2590. [DOI] [PubMed] [Google Scholar]
- Tamura JK, Warrener P, Collett MS. 1993. RNA-stimulated NTPase activity associated with the p80 protein of the pestivirus bovine viral diarrhea virus. Virology 193:1–10. [DOI] [PubMed] [Google Scholar]
- Tautz N, Meyers G, Thiel HJ. 1993. Processing of poly-ubiquitin in the polyprotein of an RNA virus. Virology 197:74–85. [DOI] [PubMed] [Google Scholar]
- Tobias JW, Varshavsky A. 1991. Cloning and functional analysis of the ubiquitin-specific protease gene UBP1 of Saccharomyces cerevisiae. J Biol Chem. 266:12021–12028. [PubMed] [Google Scholar]
- Tratschin JD, Moser C, Ruggli N, Hofmann MA. 1998. Classical swine fever virus leader proteinase Npro is not required for viral replication in cell culture. J Virol. 72:7681–7684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrener P, Collett MS. 1995. Pestivirus NS3 (p80) protein possesses RNA helicase activity. J Virol. 69:1720–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiskerchen M, Belzer SK, Collett MS. 1991. Pestivirus gene expression: the first protein product of the bovine viral diarrhea virus large open reading frame, p20, possesses proteolytic activity. J Virol. 65:4508–4514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worobey M, Holmes EC. 1999. Evolutionary aspects of recombination in RNA viruses. J Gen Virol. 80:2535–2543. [DOI] [PubMed] [Google Scholar]
- Zhong W, Gutshall LL, Vecchio AMD. 1998. Identification and characterization of an RNA-dependent RNA polymerase activity within the nonstructural protein 5B region of bovine viral diarrhea virus. J Virol. 72:9365–9369. [DOI] [PMC free article] [PubMed] [Google Scholar]