Abstract
The adaptive significance of human brain evolution has been frequently studied through comparisons with other primates. However, the evolution of increased brain size is not restricted to the human lineage but is a general characteristic of primate evolution. Whether or not these independent episodes of increased brain size share a common genetic basis is unclear. We sequenced and de novo assembled the transcriptome from the neocortical tissue of the most highly encephalized nonhuman primate, the tufted capuchin monkey (Cebus apella). Using this novel data set, we conducted a genome-wide analysis of orthologous brain-expressed protein coding genes to identify evidence of conserved gene–phenotype associations and species-specific adaptations during three independent episodes of brain size increase. We identify a greater number of genes associated with either total brain mass or relative brain size across these six species than show species-specific accelerated rates of evolution in individual large-brained lineages. We test the robustness of these associations in an expanded data set of 13 species, through permutation tests and by analyzing how genome-wide patterns of substitution co-vary with brain size. Many of the genes targeted by selection during brain expansion have glutamatergic functions or roles in cell cycle dynamics. We also identify accelerated evolution in a number of individual capuchin genes whose human orthologs are associated with human neuropsychiatric disorders. These findings demonstrate the value of phenotypically informed genome analyses, and suggest at least some aspects of human brain evolution have occurred through conserved gene–phenotype associations. Understanding these commonalities is essential for distinguishing human-specific selection events from general trends in brain evolution.
Keywords: adaptive evolution, brain size, Cebus, comparative genomics, molecular evolution, primate evolution
Introduction
Relative to body size the mass of the human brain surpasses all other species (Jerison 1973; Martin 1981). The energetic expense of a large brain (Aiello and Wheeler 1995) imposes significant costs that must be outweighed by fitness benefits. The rapid, directional expansion of the hominin brain is therefore commonly interpreted as a response to strong selective pressure favoring behavioral adaptations and enhanced cognitive performance (Jerison 1973).
Identifying the molecular changes that enabled the evolutionary increase in human brain mass has the potential to shed light on the selection pressures that shaped our distant past, and highlight species-specific adaptations that contribute to our uniquely derived condition. Molecular evidence of positive selection acting on brain-expressed genes has been sought by comparing our genome with that of closely related species. Many of these studies identified human-specific accelerations at the level of protein coding genes (Doan et al. 2004; Dorus et al. 2004; Goldberg et al. 2003; Grossman et al. 2004; Nielsen et al. 2005; Schmidt et al. 2005; Shi et al. 2006; Wang et al. 2006), regulatory, or noncoding elements (Pollard et al. 2006; McLean et al. 2011). Other studies have sought to uncover human-specific divergence in gene expression (Caceres et al. 2003; Enard et al. 2002, 2009,Khaitovich et al. 2004; Uddin et al. 2004) and protein expression (Bauernfeind et al. 2015), or to identify gene duplications or losses specific to the human lineage (Fortna et al. 2004; McLean et al. 2011; Dennis et al. 2012). While these analyses have successfully identified potentially important adaptations, some of which have been explored functionally (Enard et al. 2002; Pulvers et al. 2010; McLean et al. 2011), they typically involve only a small number of species and largely focus on the terminal human lineage (i.e., human descent after divergence from the lineage leading to chimpanzees). These studies therefore suffer from the limitation of not being able to test whether all of the important evolutionary changes in human brain evolution are human-specific. This limitation can only be addressed by incorporating the evolution of phenotypic diversity among living primates.
In primates, brain expansion was not limited to the human lineage, but occurred throughout primate evolution, across independent lineages (Montgomery et al. 2010; Boddy et al. 2012). Additionally, the expansion of different regions of the brain, organization, and shape (Aristide et al. 2016), and scaling of the neurons within each brain structure likely varies independently across lineages (Barton and Harvey 2000). These independent elaborations underpin the convergent evolution of increased cognitive performance (McLean et al. 2011; Reader et al. 2011) in association with the development of complex social ecology (Symington 1990) and/or tool use (Chiang 1967; Lawick-Goodall 1968; Phillips 1998). For example, the mass of capuchin monkey (Cebus sp.) brains are approximately four times larger than expected for a mammal of its body mass, ranking capuchins among the most highly encephalized nonhuman mammals (Jerison 1973; Montgomery et al. 2010; Boddy et al. 2012). Capuchins demonstrate skilled tool use (Ottoni and Izar 2008; Phillips 1998), proficient social learning (Truppa et al. 2009; Addessi et al. 2010) and, like humans, display high rates of early postnatal brain growth (Courchesne et al. 2000; Phillips and Sherwood 2008).
Despite evidence that primate brain expansion has occurred in parallel in multiple independent lineages, large-scale genomic comparisons among primates have mostly ignored these independent episodes of brain expansion in analyses seeking to understand the unique aspects of human evolution. To identify human-specific adaptations it is necessary to contextualize these adaptations in relation to changes shared among other lineages. The failure to incorporate phenotypic diversity in brain size among primates may result in the incorrect identification of human-specific molecular changes. Previous genome-wide analyses have demonstrated that parallelism in rates of molecular evolution may be relatively common among protein-coding genes (Scally et al. 2012), and may underpin the convergent evolution of large brain size in mammals (Goodman et al. 2009; McGowen et al. 2012). These results suggest patterns of evolution on the terminal human lineage may not be atypical compared with other anthropoid primates, or other relatively large brained mammals.
In this study, we present analyses designed to exploit the parallel expansion of brain size across anthropoids. By combining publically available data with newly sequenced transcriptome data from Cebus apella neocortical tissue, we contrast patterns of selection during independent episodes of brain expansion in an ape (Homo sapiens), an Old World monkey (OWM) (Papio anubis), and a New World monkey (NWM) (Cebus apella) with related smaller-brained species, chosen to reflect similar differences in brain mass and divergence date. Using these three species-pairs, we performed a series of analyses to test for different genomic signatures that: (i) examine the overlap between genes with accelerated rates of evolution on lineages leading to the three large-brained species (fig. 1); and (ii) identify genes with rates of evolution across all six species that suggest a persistent coevolutionary relationship with interspecific variation in brain size (fig. 2). We then test the robustness of our results by reanalyzing patterns of molecular evolution among the highlighted genes after adding additional taxa.
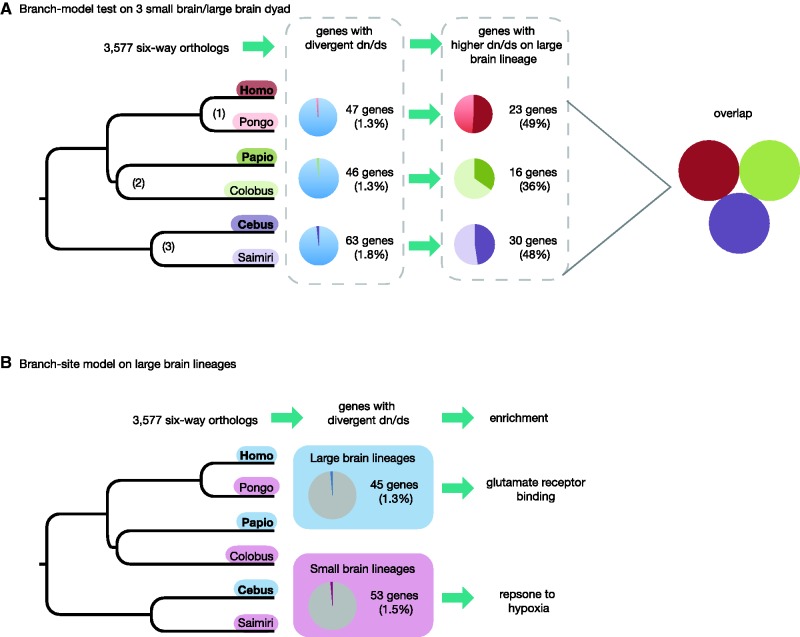
Fig. 1.—
Testing for parallel evolution in three large-brain lineages. We examined the overlap among genes (n = 3,577) with accelerated rates of evolution on three large-brain lineages using a branch-model and a branch-site model test. The branch-model compared each large-brain/small brain dyad independently (A), including Ape (1), Old World monkeys (OWM) (2), and New World monkeys (NWM) (3), while the branch-site model compared all three large-brain lineages (blue) against the three small-brain lineages (pink) (B). There was no overlap in genes with higher dN/dS on large-brain lineages in the branch-model test (A). When examining divergent dN/dS in all three large-brain lineages together we find enrichment for glutamate receptor binding, while the genes with divergent dN/dS in the small-brain subset had enrichment for response to hypoxia.
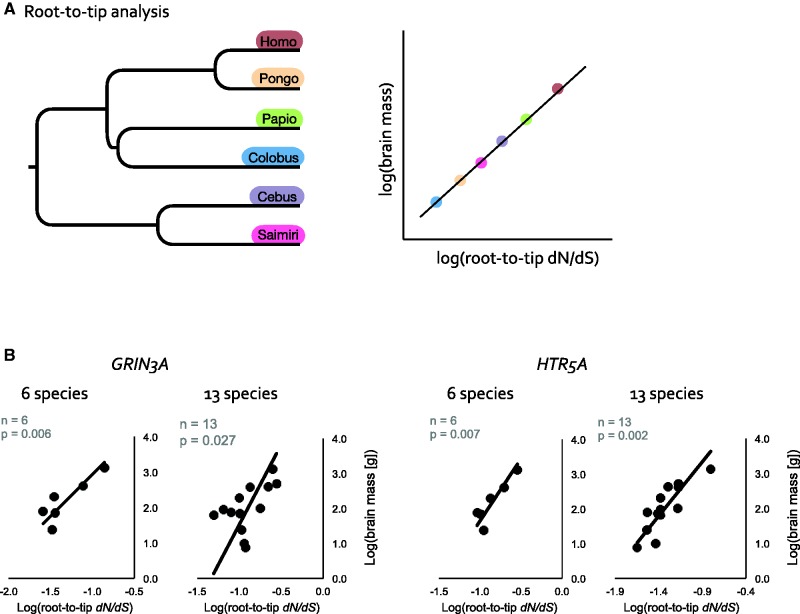
Fig. 2.—
Tests for coevolution of dN/dS and brain size across primates. (A) An illustrated example of our test for coevolution of gene–phenotype. We calculated root-to-tip dN/dS ratios for each species, for each gene (indicated by the individual colors on the tip of the tree). Using a phylogenetically controlled method, we tested for a relationship between the gene (i.e., root-to-tip dN/dS) and the phenotype (e.g., brain mass or EQ). A linear relationship between gene and phenotype provides support for coevolution, as hypothetically illustrated in (A). This test for gene–phenotype coevolution would show small patterns of change across genes. (B) A linear relationship was found between GRIN3A and brain mass using the original six-species data set and the expanded 13-species data set (left), and between HTR5A and brain mass using the original six-species data set and the expanded 13-species data set (right). The raw data is displayed with the phylogenetically controlled regression overlaid.
Materials and Methods
Sequencing and De Novo Assembly of Cebus apella Neocortical Transcriptome
Brain tissue samples were extracted from the frontal pole (homologous to human area 10) of the left hemisphere from fresh frozen whole brain of one male infant capuchin monkey, Cebus apella (genus also known as Sapajus apella; Alfaro et al. 2012). Total RNA was isolated and the sample had RNA integrity number (RIN) of 9. We performed paired-end RNA sequencing of the neocortical tissue (FASTQ files available at Sequence Read Archive, accession number SRP058420). Sequencing was performed at Wayne State University’s Applied Genomics Technology Center using Illumina’s paried-end RNA-seq protocol with an insert size of 200 bp and a read length of 76 bp. Raw sequencing reads were assembled de novo using the RNA-seq assembler, Trinity (r2012-10-05), with a kmer length of k = 25 (Grabherr et al. 2011). Likely coding sequences of the assembled contigs were extracted by identifying the longest open reading frame (ORF) within the transcript. Reads were mapped back to assembled contigs to remove poor quality sequences. Transcripts were annotated using BLASTn to the human transcriptome (see supplementary methods, Supplementary Material online for full sequencing and assembly methods).
Genomic Data, Orthology Detection, and Alignment
We inferred 4,770 six-way one-to-one orthologs from Cebus apella, published data for H. sapiens, P. abelii, P. anubis, C. angolensis, and S. boliviensis (supplementary methods, Supplementary Material online) (6 way alignment data available from the Dryad Digital Repository, doi:10.5061/dryad.qt834). These were aligned using PRANK (Löytynoja and Goldman 2008), with an input guide tree obtained from the Primate 10K Trees Project (Arnold et al. 2010). Multiple sequence alignments were filtered using SWAMP (Harrison et al. 2014) to remove short alignments, and to mask potential alignment and sequencing errors that can inflate dN/dS estimates (see supplementary methods, Supplementary Material online for full parameters). Alignments with dS > 2 were also removed. After filtering, we obtained a final set of 3,577 six-way 1:1 orthologs; supplementary table S4, Supplementary Material online). Patterns of molecular evolution were analysed using the CODEML package in PAML (Yang 2007). PAML infers selection pressures acting on coding regions of genes by estimating the ratio of rates of nonsynonymous to synonymous fixed base changes (measured as dN/dS ratios or the ω parameter in PAML).
Testing Alternative Gene–Phenotype Association Tests: Branch-Specific Shifts and Phylogenetic Regressions
We performed two types of analyses to test for genes targeted by selection acting on brain size. In our first approach, we identified genes with accelerated rates of evolution on lineages leading to the large-brained species (fig. 1) using a branch-model analysis (see below) that tests for significant differences in the rate of molecular evolution on the small and large brained lineages. In our second approach, we calculated the average rate of evolution during the lineage leading to each species, referred to as the root-to-tip dN/dS, and use this to test for an association with continuous variation in brain mass across all six species.
These two approaches make different assumptions. The branch-model tests treat phenotypic variation as a binary trait and identify significant shifts on an individual branch associated with changes in that binary state. This approach is widely used in human–chimpanzee comparisons, and between other taxon-pairs (Gilbert et al. 2005). However, the test is limited to identifying genes with substantial rate heterogeneity across only two branches, therefore ignoring a substantial amount of information. Essentially, the test is for “episodic” or branch-specific, rate shifts. However, by comparing results across the three species pairs, we can estimate the rate of convergent shifts in the rate of evolution of individual genes on branches leading to large-brained species. In the second approach, we test for an association between the molecular evolution of a gene and variation in the phenotype of interest across a phylogenetic tree (fig. 2). This treats both molecular and phenotypic data as continuous traits and can detect more subtle variation between species. By treating data in this way, the test detects associations between gene–phenotype evolution that are conserved across the phylogeny. The different approaches also reflect the possibility that selection on brain size acts either through “episodic” positive selection on different genes on specific branches, or continuous selection on the same gene(s) across large phylogenetic distances. As such, the comparison between tests allows us to assess the assumption that selection on the human lineage has acted on a unique set of genes, and identify genes that may play a recurrent role in primate brain evolution.
Evolutionary Analyses: Testing for Evidence of Discrete Shifts in Selection Regime on Large Lineages Leading to Large Brained Species
To test for discrete shifts in selection regime on lineages leading to large brained species we performed two analyses: (1) a branch-model test (Model = 0; NSsites = 2), and (2) a branch-site model test (Model = 2; NSsites = 2). Branch models allow dN/dS to vary across branches in the phylogeny but not across sites. For each small brain/large brain dyad, we performed a test to identify divergent rates of evolution between the two species. Specifically, we compared the likelihood of a model in which a pair of branches was estimated to have a single ω to another model where ω on individual branches was independently estimated. In each case, the remaining four species were included in a separate ω category. (2) Branch-site models also test for episodic positive selection but instead assume positive selection is restricted to a subset of sites on a subset of branches determined a priori, in this case each large-brained lineage (Yang and Nielsen 2002; Yang 2005; Zhang et al. 2005; Anisimova and Yang 2007) (see supplementary methods, Supplementary Material online for full methods).
Evolutionary Analyses: Testing for Evidence of Gene–Phenotype Coevolution across Anthropoids
Our second approach sought to identify genes where the strength of selection acting on them is associated with interspecific variation across anthropoids. Tests for coevolution between dN/dS and brain size were performed using PGLS regressions in BayesTraits (Pagel 1999). We calculated root-to-tip dN/dS ratios for each species for each gene using the branch models. These were used in phylogenetically controlled genotype–phenotype association tests (Montgomery et al. 2011) with log10-transformed EQ and brain mass. We chose to analyze both EQ and brain mass as it is unclear to what extent brain and body coevolution is caused by a common genetic basis. Trajectories of brain and body mass evolution suggest both traits can respond to separate selection pressures (Montgomery et al. 2010) and data from both intra and interspecific analyses suggest at least a partially independent genetic basis (Montgomery et al. 2016). In addition, genes associated with either variable may reflect different aspects of brain evolution.
To test whether the associations with brain mass did not reflect an association with body mass, we also repeated the analyses with body mass data. Phenotypic associations that were specific to brain mass were identified when there was no significant association with body mass in this analysis, or there was a difference in Akaike Information Criterion (AIC) score of >2 between the likelihood PGLS regression with brain mass and body mass. All phenotypic data were taken from published data sets (Stephan et al. 1981). Functional information on specific genes discussed in the results were taken from online catalogues of protein function (OMIM: http://www.omim.org/; UNIPROT: http://www.uniprot.org; GeneCards: http://www.genecards.org/). For sets of significant genes under each analysis we tested for an enrichment of particular functions/gene ontology (GO) terms using GOrilla (Eden et al. 2009) with the full gene list as the background selection.
Additional Statistical Analyses
Small data set comparisons may be prone to false positives (Anisimova and Yang 2007; Zhai et al. 2012). Given the large number of genes and low statistical power with six species we report nominal P-values unless otherwise stated. Accordingly, we sought to assess how robust our results and conclusions are, both generally and for individual genes, through a series of additional analyses.
Genome-Wide Rates of Evolution
Interspecific variation in effective population size could alter the rate of neutral substitution and the efficacy of selection to remove or fix nonsynonymous substitutions. This effect could bias our phylogenetic regression analyses if it co-varies with brain size. To address this effect, we calculated an estimate of genome-wide root-to-tip dN/dS for each species by concatenating the alignments from all 3,577 genes and re-running the branch models to estimate a “transciptome-wide” dN/dS for each lineage. We then used these values in a PGLS regression to test for an association with brain mass, body mass or EQ.
Reanalysis Including Seven Additional Anthropoid Primates Species
We focused our analyses on three small-large brain pairs to facilitate (i) comparisons between the branch-model tests from different species pairs, and (ii) comparisons between the results of branch-model tests and phylogenetic regressions. However, we were able to test the reliability, repeatability and robustness of our key results by re-analyzing the observed patterns by including a wider range of anthropoid primates. Using publically available genome (www.ensembl.org) or exome (George et al. 2011) data we obtained orthologs for an additional three apes (Pan troglodytes, Gorilla gorilla, and Nomascus leucogenys), two Old World monkeys (Chlorocebus aethiops, Macaca mulatta), and two New World monkeys (Callithrix jacchus, Saguinus midas) for as many genes with significant nominal P-values under the tests described above as possible (13-way alignment data available from the Dryad Digital Repository, doi:10.5061/dryad.qt834). We then used these data to validate the six-taxon analyses. To confirm any signal was not driven exclusively by the original six species we also re-ran the analyses on the additional seven species.
Permutation Tests
We performed two permutation tests to assess whether the results of our phylogenetic regressions are substantially different from random expectations. First we used the species-specific root-to-tip dN/dS values from all 3,577 genes in the n = 6 data set to create a library of values for each species. We randomly selected a value from these libraries for each species independently and then repeated our test for an association with the three phenotypes. This was repeated 1,000 times to get a proportion of results that are significant at P = 0.05, here referred to as the false positive rate of our “global” permutation test. Second, for genes associated with brain mass or EQ in our n = 6 and n = 13 data sets that showed a significant gene–phenotype associations, we added a further “gene-specific” permutation test for each individual gene in the analyses in which the values of brain mass or EQ were shuffled between species before re-running the test for a phenotypic association. This test was again repeated 1,000 times. We calculated the permutation-P-value as the proportion of the 1,000 tests that have a lower P-value than the unpermutated data.
Results
Transcriptome Sequencing
Capuchin monkeys are the most encephalized nonhuman primate (Montgomery et al. 2010; Boddy et al. 2012). Although capuchins are an ideal candidate to study brain size evolution among primates, few comparative genomics studies have included capuchin monkeys, likely due to lack of genomic data. Here, we performed RNA-sequencing of neocortical tissue from an infant male capuchin monkey using Illumina’s paired-end RNA-seq protocol (insert size = 200 bp). We assembled the transcripts de novo using the RNA-seq assembler, Trinity (Grabherr et al. 2011). This resulted in 145,708 contigs with a N50 of 1,809 bp, with the longest contigs of 17,354 bp and a total assembly size of ∼114 Mb (supplementary fig. S1, Supplementary Material online). We extracted the likely protein coding regions (i.e., open reading frames) from the brain transcriptome. This procedure resulted in 50,716 protein-coding regions with an N50 of 1,365 bp and a maximum length of 16,299 bp (supplementary table S2, Supplementary Material online). These transcripts were annotated via interrogation of the human genome using BLASTn.
Molecular Evolution of Brain-Expressed Genes during Independent Episodes of Brain Expansion
Increases in primate brain size have occurred independently multiple times (i.e., they are phylogenetically dispersed). In order to capture this phylogenetic diversity, we selected three independent episodes of brain expansion in the following clades (1) hominid (Homo sapiens), (2) an OWM (Papio anubis), and (3) a NWM (Cebus apella) (fig. 1 and supplementary fig. S2, Supplementary Material online). We then selected a related smaller-brained species, chosen to reflect similar differences in brain size, divergence date, and the availability of genome sequence data (1) Pongo abelii (hominid), (2) Colobus angolensis (OWM), and (3) Saimiri boliviensis (NWM), respectively. These three small-brain-large-brain primate dyads were then used to compare the level of genomic convergence among the dyads. Using 3,577 6-way 1:1 orthologous, brain-expressed genes, we applied two approaches to assess the level of parallelism during these three independent episodes of brain expansion.
Accelerated Rates of Evolution on Terminal Branches: Limited Evidence of Convergent Shifts in Selection
We first used codon-based branch models to identify genes with significant accelerations in dN/dS on each of the large brained lineages (Homo, Papio, and Cebus) in comparison to their smaller brained sister-genera (Pongo, Colobus, and Saimiri, respectively) (fig. 1). We found a similar number of genes (c. 50–60) with evidence of divergent dN/dS (P > 0.05) within each large-brain: small-brain pair (supplementary table S5, Supplementary Material online). The percentage of these genes with higher rates of evolution in the larger-brained species is also comparable (Apes: 49%, OWM: 36%, NWM: 48%). We found no overlap between genes with accelerated rates of evolution along the terminal Homo, Papio, and Cebus lineages. A similar conclusion is drawn from two branch-site tests where either the three larger-brained species, or three smaller-brained species, were set as the foreground branches. Forty-five genes (1.3%) show evidence of episodic (branch-specific) positive selection on the large-brained terminal branches, while a similar number (55 genes, 1.5%) is obtained for the small-brained terminal branch-site test (supplementary table S6, Supplementary Material online, without correction for multiple testing). These results therefore provide little evidence of large, discrete, or convergent shifts in dN/dS along the terminal branches leading to large-brained species (fig. 1).
Coevolution of dN/dS and Brain Size across the Primate Phylogeny: Evidence for Consistent Gene–Phenotype Associations
An alternative approach to identifying selection processes relevant to a particular phenotype is to test for signatures of coevolution between a gene and a phenotype across a phylogeny. The presence of such a pattern of coevolution suggests selection at a locus varies in association with phenotypic variation across the phylogeny, potentially indicating a functional role in brain evolution. Using phylogenetically controlled regressions we tested for an association between root-to-tip dN/dS (Montgomery et al. 2011) and three continuous phenotypic traits; brain mass, body mass, and encephalization quotient (EQ), a measure of relative brain size (Jerison 1973). Results from the root-to-tip association studies identified a much greater proportion of genes with potentially consistent or conserved roles in brain evolution than was found using analyses focusing on terminal branches (fig. 2A). From our 1:1 6-way orthologs data set of brain expressed genes (n = 1,616 after removing genes with dN/dS of “999” or “0.0001” see supplementary information, Supplementary Material online), we found 179 genes (11.1%) that showed evidence of an association between brain mass and dN/dS (supplementary table S7, Supplementary Material online). The majority of these 179 genes do not show any evidence of an association with body mass (107 genes), and/or have a difference in AIC > 2 suggesting a closer phylogenetic association with variation in brain mass than body mass (120 genes). About 273 genes (16.9%) showed evidence of a coevolutionary relationship with EQ (supplementary table S8, Supplementary Material online). Of these 107 (39.2%) also show evidence for an association with brain mass and provide a list of candidate genes which may be responding to selection for increased brain size relative to body size by altering the development of brain mass.
We estimated genome-wide root-to-tip dN/dS to test whether variation in global rates of evolution caused by differences in effective population size could bias our phylogenetic tests. The resulting values are similar across species (Cebus: 0.114, Colobus: 0.111, Homo: 0.118, Papio: 0.107, Pongo: 0.119, Saimiri: 0.115). We found no significant association between them and either brain mass (t4 = 0.561, P = 0.302), body mass (t4 = 0.244, P = 0.409), or EQ (t4 = 0.809, P = 0.232) in individual PGLS regressions. We also performed multiple regressions for each gene including the average root-to-tip values as a covariate. Despite the reduced degrees of freedom the majority of genes showing an association with brain size remained significant. For brain mass, 106/179 (59%) genes were significant after including genome-wide root-to-tip dN/dS as a covariate. For EQ, 199/273 (73%) remained significant. The t values for the single regression with gene-specific root-to-tip dN/dS and the multiple regressions including genome-wide root-to-tip dN/dS are also strongly correlated (Spearman correlation: brain mass, r = 0.938; EQ, r = 0.921).
Functional Insights from Explicit Gene–Phenotype Associations
Among the 45 genes with evidence for episodic positive selection limited to lineages leading to the larger brained species in each pair (fig. 1B, supplementary table S6, Supplementary Material online), there is enrichment for “glutamate receptor binding” (CACNG2, DRD2; nominal-P < 0.001). The branch-site test for episodic positive selection limited to the smaller brained species are not enriched for this function, but instead show evidence for enrichment for genes related to the “response to hypoxia” (CDK4, CAT, PDGFRB; nominal-P < 0.001).
Genes with evidence of a coevolutionary relationship with brain mass (fig. 2, N = 179, supplementary table S7, Supplementary Material online) are enriched for “negative regulation of cell projection organization” (OMG, CIB1, PTPRG, LRP4 genes; nominal-P < 0.001). This enrichment is not found among genes with a dN/dS associated with body mass, which show no enrichment for GO terms (P > 0.05). Genes with the strongest evidence for an association with brain mass (P < 0.001) include genes functionally related to neurodevelopment (e.g., CHD6, MCM7), neurite outgrowth and cell migration (e.g., TNN, CRADD), and collagen binding or angiogenesis (e.g., RNF213, THOP1, and LUM). Additionally, we find support for several loci with suspected roles in neuropsychiatric disorders affecting typical developmental trajectories, including Autism Spectrum Disorders (ASD) and schizophrenia (e.g., HTR5A and GRIN3A). We found no enrichment for GO terms when testing the coevolutionary relationship between dN/dS and EQ (supplementary table S8, Supplementary Material online). However, we again identify a number of genes linked to brain development, including ARHGEF1, a Rho guanine nucleotide exchange factor, which shows a strong coevolutionary association with EQ that is robust to correction for multiple testing (P < 0.001; corrected-P = 0.029). Other genes with high significance before multiple test correction (P <0.001) include SPICE1, NEDD9, and GPR37.
Reanalysis with Additional Taxa Supports Gene–Phenotype Associations for Key Loci
Our results suggest a relatively large percentage of orthologous genes show signatures of molecular evolution that are consistent with being continuously targeted by selection to bring about increases in brain mass, or adaptations associated with that increase, during anthropoid evolution. In these cases, the linear relationship between dN/dS and brain mass (fig. 2B) may suggest gene–phenotype associations are maintained in parallel across independent anthropoid lineages. A common problem in large scale comparative analyses is a lack of power due to the large number of genes and small number of species (Enard 2014). Indeed, our estimates of the “global” false-positive rate in our data set are high, but variable, across the three phenotypes; 14.4%, 2.4%, and 27.6% for brain mass, body mass, and EQ, respectively. We therefore cannot rule out the possibility that a number of our associations with the six species data set are false positives. However, the “gene-specific” permutation tests suggest a large proportion of the phenotypic associations we detect with the n = 6 data set are robust. About 146/179 (81.6%) of genes associated with brain mass in the n = 6 data set remained significant in our permutation test. For EQ, this figure was 115/273 (42.1%). All of the individual genes mentioned in our summary of potential functional affects (see above) were significant in the permutation test.
We further tested the reproducibility and robustness of our key results by reanalyzing our data after the inclusion of an increased number of anthropoid primates (total n = 13; supplementary information, Supplementary Material online). We obtained 13-way 1:1 orthologs for 55 out of 107 genes that showed evidence of a coevolutionary relationship with brain mass, but not body mass. We reproduced the phenotypic association for 24% of these genes (n = 13) before multiple test correction (table 1b;supplementary table S9, Supplementary Material online). We obtained 13-way 1:1 orthologs for 115 out of 273 genes with evidence of a coevolutionary relationship with EQ. An association with EQ was reproduced for 23% of these genes (n = 27) across this expanded data set, before multiple test correction (table 1a;supplementary table S10, Supplementary Material online). Two loci remained significant after strict multiple test-correction (HTR5A and ZBTB16).
Table 1.
Top Genes with Evidence of a Coevolutionary Relationship with (a) EQ and (b) Brain Mass in the Expanded 13-Species Data Set
| Gene | Gene Name | t11 | R2 | Nominal-P | Permutation-P |
|---|---|---|---|---|---|
| (a) EQ | |||||
| HTR5A | 5-Hydroxytryptamine (Serotonin) Receptor 5A | 6.267 | 0.751 | <0.001 | 0.007 |
| ZBTB16 | Zinc Finger And BTB Domain Containing 16 | 4.784 | 0.638 | <0.001 | 0.006 |
| PPIL2 | Peptidylprolyl Isomerase (Cyclophilin)-Like 2 | 3.570 | 0.495 | 0.002 | 0.067 |
| DHDH | Dihydrodiol Dehydrogenase | 3.453 | 0.478 | 0.003 | 0.045 |
| TMEM86A | Transmembrane Protein 86A | 3.441 | 0.477 | 0.003 | 0.033 |
| ACOT8 | Acyl-CoA Thioesterase 8 | 3.367 | 0.466 | 0.003 | 0.037 |
| DRP2 | Dystrophin Related Protein 2 | 3.147 | 0.432 | 0.005 | 0.047 |
| NUFIP2 | Nuclear Fragile X Mental Retardation Protein Interacting Protein 2 | 3.119 | 0.428 | 0.005 | 0.054 |
| RNASEH2A | Ribonuclease H2, Subunit A | 2.983 | 0.406 | 0.006 | 0.018 |
| DARS2 | Aspartyl-TRNA Synthetase 2, Mitochondrial | 2.805 | 0.377 | 0.009 | 0.111 |
| ST3GAL1 | ST3 Beta-Galactoside Alpha-2,3-Sialyltransferase 1 | 2.746 | 0.367 | 0.010 | 0.100 |
| (b) Brain mass | |||||
| HTR5A | 5-Hydroxytryptamine (Serotonin) Receptor 5A | 3.533 | 0.490 | 0.002 | 0.006 |
| SPRY4 | Sprouty Homolog 4 | 3.353 | 0.464 | 0.003 | 0.003 |
| ZBTB11 | Zinc Finger And BTB Domain Containing 11 | 2.808 | 0.377 | 0.009 | 0.013 |
| LMBR1L | Limb Development Membrane Protein 1-Like | 2.343 | 0.297 | 0.019 | 0.036 |
| FBN1 | Fibrillin 1 | 2.254 | 0.281 | 0.023 | 0.010 |
| LUM | Lumican | 2.195 | 0.270 | 0.025 | 0.018 |
| ECI2 | Enoyl-CoA Delta Isomerase 2 | 2.178 | 0.267 | 0.026 | 0.013 |
| GRIN3A | Glutamate Receptor, Ionotropic, N-Methyl-d-Aspartate 3A | 2.149 | 0.262 | 0.027 | 0.034 |
| FKBP10 | FK506 Binding Protein 10 | 2.063 | 0.247 | 0.032 | 0.028 |
| TRAFD1 | TRAF-Type Zinc Finger Domain Containing 1 | 2.037 | 0.242 | 0.033 | 0.122 |
| ZCCHC6 | Zinc Finger, CCHC Domain Containing 6 | 2.033 | 0.241 | 0.033 | 0.027 |
| BDH1 | 3-Hydroxybutyrate Dehydrogenase, Type 1 | 1.863 | 0.211 | 0.045 | 0.031 |
| SERPIND1 | Serpin Peptidase Inhibitor, Clade D (Heparin Cofactor), Member 1 | 1.834 | 0.205 | 0.047 | 0.017 |
The top two genes associated with EQ (HTR5A and ZBTB16) have been implicated in increasing susceptibility to neurodevelopmental disorders in humans, including ASD, schizophrenia and bipolar disorder (Yosifova et al. 2009; Sun et al. 2010; Winchester et al. 2014). Among those genes with the strongest evidence for a coevolutionary relationship with EQ (nominal P < 0.01) several more (e.g., DRP2, RNASEH2A) have been putatively linked to neurological disorders (Hong et al. 2005; Crow et al. 2006). The serotonin receptor HTR5A shows a particularly noteworthy pattern of evolution as it shows evidence of an association with brain mass (nominal P-value = 0.002; fig. 2B) but not body mass (nominal P-value = 0.130) and has the strongest association with brain mass, ahead of SPRY4 (nominal P-value = 0.005). SPRY4 has also been putatively linked to schizophrenia (Zaharieva et al. 2008). GRIN3A, a glutamate receptor, also demonstrates evidence for a positive association with brain mass before multiple-test correction (nominal P-value = 0.027; fig. 2B) and is again linked to susceptibility to schizophrenia (Takata et al. 2013), reinforcing the potential importance of genes associated with glutamatergic function and developmental disorders.
For each gene that showed a gene–phenotype association across both the n = 6 and n = 13 data set we performed a permutation test, shuffling values of dN/dS among the 13 species for each gene separately. For the n = 13 data set, all of the genes that show a significant association with brain mass are significant under the permutation test. For genes that showed an association with EQ, 34.6% are significant under the permutation test. These include the top seven genes and all genes referred to individually in in the text above and the discussion (e.g., HTR5A, ZBTB16, DRP2, and RNASEH2A; table 1). We also repeated the phylogenetic regressions using only the additional seven species (supplementary table S11, Supplementary Material online). Among the additional seven species, 23.6% of genes that showed a significant association with brain mass in the n = 6 data set have the same association at a nominal P-value of 0.05, with 45.45% at P < 0.1. About 14.5% genes that showed a significant association with EQ in the n = 6 data set also do so across the additional seven species, with 25.6% at P < 0.1. The signal in the n = 13 data set is therefore not solely due to the signal in the n = 6 data set.
Finally, we performed additional analyses to test for a specific association with pre- or post-natal brain growth in a multiple regression (supplementary information, table S9, Supplementary Material online). Pre- and post-natal brain growths are evolutionarily dissociable suggesting they are under distinct genetic control (Barton and Capellini 2011). This reflects the differing developmental processes active before and after birth (Uddin et al. 2008), with neocortical neurogenesis being restricted to prenatal brain growth (Bhardwaj et al. 2006), whilst postnatal brain growth is largely due to axonogenesis, gliagenesis, and myelination. This analysis highlights HTR5A as showing a strong association with postnatal brain growth (P <0.009), along with LMBR1L (P = 0.032) and FKBP10 (P = 0.037). In contrast, GRIN3A is strongly associated with pre-natal brain growth (P = 0.008).
Discussion
By adopting a comparative approach to analyzing patterns of molecular evolution that embraces the phenotypic diversity of our close primate relatives, genomic and transcriptomic data were used to dissect the shared and species-specific aspects of human evolution. In the present study, we provide a newly sequenced neocortical transcriptome from a primate with a large brain size relative to body size, the tufted capuchin monkey. Additionally, we took advantage of the convergent increase in brain size during the evolution of three distinct primate lineages to examine patterns of selection in the lineage leading to humans in the wider context of anthropoid primate evolution.
We performed tests for two different patterns of genomic change. First we tested for high rates of accelerated evolution on large-brain lineages. Focusing on terminal branches, only a small proportion of genes show accelerated rates of evolution in lineages leading to large brained species. In addition, we found no evidence for an overlap in genes with discrete shifts in evolutionary rate in the terminal Homo, Papio and Cebus lineages. Our second test, which adopted a more continuous, phylogenetic approach, identified many more genes with a more quantitative, coevolutionary relationship with interspecific variation in brain size among primates. Although the small number of species in the analysis limits statistical power and may inflate the false positive rate, if we take only the genes that survive our filtering from the n = 6 to the n = 13 data sets, and the permutation tests in the n = 13 data set, we still find a higher number of genes with a phylogenetic association with brain mass (15) and EQ (9) than identified using the branch-model or branch-site tests to identify genes with convergent accelerations in dN/dS in the three large brained lineages. Hence, we conclude that (i) there is evidence for at least a partially conserved molecular response to selection on brain size, and (ii) phylogenetic approaches to testing gene–phenotype associations offer promising avenues of research.
While comparative functional data are necessary to demonstrate that these associations reflect causative relationships among loci and brain evolution, our results suggest pairwise analyses between humans and a nonhuman relative may not detect all genes that are important for phenotypic evolution in this lineage. Additionally, our analyses indicate the proportion of genes with species-specific roles in brain evolution may be smaller than the proportion that has a conserved role in regulating changes in brain size across primates. An alternative explanation for the discrepancy in results from our two tests is that the contribution of different brain structures to the expansion of overall size varies across the three lineages studied. Local variability in neuronal number in rodents and primates has been reported, suggesting there is diversity across species and within species-specific brain tissue (Charvet et al. 2015). However, our study focused on the phenotype of total brain mass, and we provide new insights into functions targeted by selection during convergent episodes of brain expansion and support several notable hypotheses concerning the production, advantages, and costs of large brains.
Neural Proliferation, the Centriole, and Brain Size Evolution
The centriole plays a key role in the proliferation of neural progenitor cells by moderating key cell-fate switches (Rakic 2009; Fietz and Huttner 2011). A number of well-studied genes, such as microcephaly associated loci and ninein, have such a role (Thornton and Woods 2009), and are implicated as persistent targets of selection during changes in primate brain size (Montgomery et al. 2011; Montgomery and Mundy 2012a,b). Our analysis highlights a number of genes that have known roles in spindle or cytoskeletal function and show a pattern of coevolution with EQ or brain size. These include SPICE1 which is known to interact with several proteins linked to microcephaly (Comartin et al. 2013), NEDD9 which has an essential role in neuronal cell fate (Vogel et al. 2010), and MCM7, which interacts with a key regulator of neural cell proliferation, CDK4 (Lange et al. 2009). A role for internal reorganization of neural structures during changes in brain size (Barton and Harvey 2000) is suggested by an enrichment of genes involved in cell projection organization among those genes that coevolve with brain mass, including CIB1 and PTPRG (Bouyain and Watkins 2010; Sobczak et al. 2011). Early molecular switches that control cell proliferation may have profound effects on brain size. For example, a recently identified human-specific Rho-GTPase activating protein (ARHGAP11B) promotes basal progenitor generation and self-renewal when expressed in the embryonic mouse neocortex (Florio et al. 2015). We provide support for another gene in the Rho-GTPase family, ARHGEF1, in multiple episodes of primate brain size increase. Our results suggest these loci merit further investigation as candidates in regulating neural proliferation, and the evolution of brain size.
Genes Associated with Human Neurological Disorders Coevolve with Brain Size
Several genes that have been linked to an increased risk of human neurological disorders in humans (HTR5A, GRIN3A, DRP2, and RNASEH2A) show evidence of coevolutionary relationship with brain size in anthropoids (fig. 2). This finding supports previous hypotheses that link neurodevelopmental disorders and brain expansion (Burns 2006; Crespi et al. 2007; Khaitovich et al. 2008). HTR5A is a serotonin receptor implicated in a wide range of psychiatric conditions (Coutinho et al. 2007; Yosifova et al. 2009), but more generally the wider serotonergic system is consistently implicated with susceptibility to Autistic Spectrum Disorders (Zafeiriou et al. 2009). GRIN3A is a glutamate NMDA receptor involved in synapse formation (Takata et al. 2013), DRP2 is expressed principally in the brain and spinal cord (Roberts et al. 1996), while mutations in RNASEH2A cause Aicardi–Goutieres syndrome (AGS), an autosomal recessive neurological disorder characterized by progressive microcephaly (Coffin et al. 2011). Comparisons between modern human and Neanderthal genomes suggest that genes involved in cognitive disorders may have been targeted by selection during the recent evolution of modern humans (Green et al. 2006). Our analysis extends the potential evolutionary role of genes or neural pathways associated with neurological disorders across anthropoid primates.
Autistic Spectrum Disorder (ASD) and schizophrenia are associated with a suite of deficits in social behaviors and neuroanatomical differences (Burns 2006; Baron-Cohen 2009). ASD is defined as “persistent deficits in social communication and social interaction across multiple contexts” and “restricted, repetitive patterns of behavior, interests, or activities” (American Psychiatric Association 2013). ASD Symptoms must appear in early development and, although heterogeneous in their clinical presentation, ASD and schizophrenia have some common features of particular interest. ASD is often associated by an increase in total brain volume caused by increased postnatal growth (Redcay and Courchesne 2005) affecting white and gray matter, and cortical connectivity (Hazlett et al. 2005; Palmen et al. 2005). In contrast, schizophrenia has diametrically opposed effects on brain development (Crespi and Badcock 2008; Crespi et al. 2010) and is often associated with a decreased brain volume (Haijma et al. 2013). Whether the gene networks disrupted in these disorders were preferentially targeted during primate brain expansion is an open question. Given the hypothesized link between selection for social cognition and brain expansion in primates (Dunbar 2009), and the social deficits characteristic of ASD and schizophrenia (Penn et al. 1997; Baron-Cohen 2009), it remains a tantalizing hypothesis that the remodeling of the primate brain has targeted suites of genes that influence this neurological continuum. It is also notable that we find evidence that some genes associated with neurodevelopmental disorders show contrasting associations with pre and postnatal brain growth. For example, HTR5A shows a strong association with postnatal brain growth, consistent with its assocation with bipolar disorder and ASD which are thought to develop postnatally (Coutinho et al. 2007; Yosifova et al. 2009). A similar association with postnatal brain growth has also been reported for variation in DUF1220 genome content, which is also associated with the severity of ASD (Zimmer and Montgomery 2015). In contrast, GRIN3A, shows a specific association with pre-natal brain growth. GRIN3A is linked to schizophrenia (Takata et al. 2013), a disorder which may instead have its route in prenatal neurodevelopment, and in some cases associated with a reduced head circumference at birth (Davis et al. 1995; Kunugi et al. 1996; Lewis and Levitt 2002; Rapoport et al. 2012). These results provide support for previous evidence that there are different selection pressures acting on the pre- vs. post-natal environment (Uddin et al. 2008; Barton and Capellini 2011).
A further link is provided by evidence that selection targeted glutamate receptor binding genes during episodes of primate brain expansion. Dysfunction in glutamate transmission has been linked to ASD, putatively through a disruption in excitatory/inhibitory balance in glutamate signaling leading to abnormalities in synapse maturation and microcircuit development (Canitano and Scandurra 2014). This could provide one explanation for the abnormal development of neocortical minicolumns (Buxhoeveden et al. 2006) and overall connectivity (Belmonte et al. 2004) in individuals with ASD. Glutamatergic genes are consistently highlighted by comparative analyses of both protein-coding genes and gene expression in primates (Burki and Kaessmann 2004; Fu et al. 2011; Somel et al. 2013; Muntané et al. 2014). Understanding the phenotypic relevance of this consistant signal may reveal what functional aspects of brain connectivity were modified by selection during episodes of brain expansion.
Comparative Transcriptomics in a Phenotypic Framework
In this study, we have assessed the degree of conservation, convergence, and divergence underpinning parallel episodes of primate brain expansion. By adopting a conservative alignment-filtering regime and by grounding our analyses in a clear phenotypic framework, we demonstrate the potential for this approach to provide novel insights and highlight new candidate genes for further study. Although our power to detect coevolutionary gene–phenotype associations is limited due to the small number of species, the results of our reanalysis using a larger data set suggest a large fraction of our initial results reflect consistent and robust gene–phenotype associations across anthropoids. This provides encouragement that, as the number of sequenced genomes increases, comparative genomics will provide a powerful tool for investigating phenotypic variation in a phylogenetic context.
We adopted a conservative filtering approach to limit our analyses to high quality alignments between well-supported 1:1 orthologs (n = 3,577). As a result our data reflects a sub-sample of genes expressed in the postnatal developing brain, and may miss genes whose expression is limited to early embryogenesis. However, our approach has the advantage of analyzing molecular evolution in a clear phenotypic framework. In drawing conclusions from our data, we assume analyses of these genes reflect global selection regimes acting across the genome, but a major contribution of this work is to show how phenotypic variation can inform studies of molecular evolution. Protein-coding evolution is one of multiple avenues through which selection can bring about phenotypic change. Evolutionary changes in promoter regions, and other regulatory changes affecting gene expression, may play a significant role in phenotypic evolution (King and Wilson 1975; Carroll 2005). The general conservation of mammalian brain development is reflected in interspecific analyses of gene-expression (Khaitovich et al. 2005; Brawand et al. 2011). Despite the generally high level of conservation, a number of human–chimpanzee comparisons have revealed potentially non-neutral patterns of divergence among brain-expressed genes (Khaitovich et al. 2004, 2005; Haygood et al. 2010; Sholtis and Noonan 2010; McLean et al. 2011). This suggests adopting a phenotypically informed, comparative approach to both sequence and expression evolution may yield further insights into the molecular basis of brain evolution. The need for age, sex, and environmentally matched tissue samples in gene expression studies (Harrison et al. 2012) may limit the potential for large-scale comparative analyses across primates at developmentally relevant stages. The continued genome and transcriptome sequencing of a wider range of species and the phylogenetic analysis of promoter, or conserved noncoding regions may offer a feasible alternative. Recent advances in models analyzing selection on promoter regions (Hoffman and Birney 2010) and tests of gene–phenotype associations in noncoding sequences (O’Connor and Mundy 2013) provide useful tools to pursue these aims.
Conclusion
Whether adaptations governing increases in brain size are shared amongst primates or have a species-specific origin is unknown. To address this, we analyzed patterns of molecular evolution across three independent episodes of brain expansion. We have demonstrated the utility of incorporating phenotypic diversity into genomic comparisons. We find no evidence that the number and type of genes targeted by selection along the terminal human lineage are atypical (fig. 1). Instead we find a number of genes with a linear relationship between dN/dS and brain size, suggesting a conserved or parallel response to phenotypic change. Given the small number of genes with species-specific shifts relative to the larger number of genes with a conserved phenotypic association, there is little a priori reason to justify the assumption that all changes on the human lineage represent human-specific adaptations. Our results suggest genes with potential functions in cell proliferation, migration, and neurological disorders have conserved roles in the evolution of anthropoid brain size. Incorporating phenotypic variation into the design of comparative genomics can facilitate the inference of macro-evolutionary gene–phenotype associations. Further, the addition of distinct brain regions could provide insights into the genetic control of species specific events. To fully understand human evolution, we must embrace the phenotypic diversity of our close relatives, and utilize this diversity to design more informative genome analyses.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by the National Science Foundation, grant award numbers BCS-0751508, BCS-0827546, and BCS-1061370 for AMB and DEW. Professor Chet Sherwood (The George Washington University) provided useful guidance in the initial stages of this project. S.H.M. is grateful for support from a BBSRC doctoral training grant and a Research Fellowship from the Royal Commission for the Exhibition of 1851. N.I.M. is grateful for support from the Leverhulme Trust and Murray Edwards College, Cambridge.
Literature Cited
- Addessi E, Mancini A, Crescimbene L, Ariely D, Visalberghi E. 2010. How to spend a token? Trade-offs between food variety and food preference in tufted capuchin monkeys (Cebus apella). Behav Processes 83:267–275. [DOI] [PubMed] [Google Scholar]
- Aiello LC, Wheeler P. 1995. The expensive-tissue hypothesis: the brain and the digestive system in human and primate evolution. Curr Anthropol. 36:199–221. [Google Scholar]
- Alfaro JWL, Silva JDSE, Rylands AB. 2012. How different are robust and gracile capuchin monkeys? An argument for the use of Sapajus and Cebus. Am J Primatol. 74:273–286. [DOI] [PubMed] [Google Scholar]
- American Psychiatric Association. 2013. Diagnostic and Statistical Manual of Mental Disorders 5 2013. Arlington, VA: American Psychiatric Association 299.00 (F84.0).
- Anisimova M, Yang Z. 2007. Multiple hypothesis testing to detect lineages under positive selection that affects only a few sites. Mol Biol Evol. 24:1219–1228. [DOI] [PubMed] [Google Scholar]
- Aristide L, et al. 2016. Brain shape convergence in the adaptive radiation of New World monkeys. Proc Natl Acad Sci U S A. 113(8):2158–2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold C, Matthews LJ, Nunn CL. 2010. The 10kTrees website: a new online resource for primate phylogeny. Evol Anthropol Issues News Rev. 19:114–118. [Google Scholar]
- Baron-Cohen S. 2009. Autism: the empathizing–systemizing (E‐S) theory. Ann N Y Acad Sci. 1156:68–80. [DOI] [PubMed] [Google Scholar]
- Barton RA, Capellini I. 2011. Maternal investment, life histories, and the costs of brain growth in mammals. Proc Natl Acad Sci U S A. 108:6169–6174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton RA, Harvey PH. 2000. Mosaic evolution of brain structure in mammals. Nature 405:1055–1058. [DOI] [PubMed] [Google Scholar]
- Bauernfeind AL, et al. 2015. Evolutionary divergence of gene and protein expression in the brains of humans and chimpanzees. Genome Biol Evol. 7:2276–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmonte MK, et al. 2004. Autism and abnormal development of brain connectivity. J Neurosci. 24:9228–9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj RD, et al. 2006. Neocortical neurogenesis in humans is restricted to development. Proc Natl Acad Sci U S A. 103:12564–12568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy AM, et al. 2012. Comparative analysis of encephalization in mammals reveals relaxed constraints on anthropoid primate and cetacean brain scaling. J Evol Biol. 25:981–994. [DOI] [PubMed] [Google Scholar]
- Bouyain S, Watkins DJ. 2010. The protein tyrosine phosphatases PTPRZ and PTPRG bind to distinct members of the contactin family of neural recognition molecules. Proc Natl Acad Sci U S A. 107:2443–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brawand D, et al. 2011. The evolution of gene expression levels in mammalian organs. Nature 478:343–348. [DOI] [PubMed] [Google Scholar]
- Burki F, Kaessmann H. 2004. Birth and adaptive evolution of a hominoid gene that supports high neurotransmitter flux. Nat Genet. 36:1061–1063. [DOI] [PubMed] [Google Scholar]
- Burns J. 2006. The social brain hypothesis of schizophrenia. World Psychiatry 5:77.. [PMC free article] [PubMed] [Google Scholar]
- Buxhoeveden D, et al. 2006. Reduced minicolumns in the frontal cortex of patients with autism. Neuropathol. Appl. Neurobiol. 32:483–491. [DOI] [PubMed] [Google Scholar]
- Caceres M, et al. 2003. Elevated gene expression levels distinguish human from non-human primate brains. Proc Natl Acad Sci U S A. 100:13030–13035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canitano R, Scandurra V. 2014. Glutamatergic agents in Autism Spectrum disorders: current trends. Res Autism Spectrum Disord. 8:255–265. [Google Scholar]
- Carroll SB. 2005. Evolution at two levels: on genes and form. PLoS Biol. 3:e245.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charvet CJ, Cahalane DJ, Finlay BL. 2015. Systematic, cross-cortex variation in neuron numbers in rodents and primates. Cereb Cortex 25:147–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang M. 1967. Use of tools by wild macaque monkeys in Singapore. Nature 214:1258–1259. [DOI] [PubMed] [Google Scholar]
- Coffin SR, Hollis T, Perrino FW. 2011. Functional consequences of the RNase H2A subunit mutations that cause Aicardi-Goutieres syndrome. J Biol Chem. 286:16984–16991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comartin D, et al. 2013. CEP120 and SPICE1 cooperate with CPAP in centriole elongation. Curr Biol. 23:1360–1366. [DOI] [PubMed] [Google Scholar]
- Courchesne E, et al. 2000. Normal brain development and aging: quantitative analysis at in vivo MR imaging in healthy volunteers. Radiology 216:672–682. [DOI] [PubMed] [Google Scholar]
- Coutinho AM, et al. 2007. Evidence for epistasis between SLC6A4 and ITGB3 in autism etiology and in the determination of platelet serotonin levels. Hum Genet. 121:243–256. [DOI] [PubMed] [Google Scholar]
- Crespi B, Badcock C. 2008. Psychosis and autism as diametrical disorders of the social brain. Behav Brain Sci. 31:241–261. [DOI] [PubMed] [Google Scholar]
- Crespi B, Stead P, Elliot M. 2010. Comparative genomics of autism and schizophrenia. Proc Natl Acad Sci U S A. 107:1736–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespi B, Summers K, Dorus S. 2007. Adaptive evolution of genes underlying schizophrenia. Proc R Soc B Biol Sci. 274:2801–2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow YJ, et al. 2006. Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutieres syndrome and mimic congenital viral brain infection. Nat Genet. 38:910–916. [DOI] [PubMed] [Google Scholar]
- Davis JO, Phelps JA, Bracha HS. 1995. Prenatal development of monozygotic twins and concordance for schizophrenia. Schizophr Bull. 21:357–366. [DOI] [PubMed] [Google Scholar]
- Dennis MY, et al. 2012. Evolution of human-specific neural SRGAP2 genes by incomplete segmental duplication. Cell 149:912–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan JW, et al. 2004. Coadaptive evolution in cytochrome c oxidase: 9 of 13 subunits show accelerated rates of nonsynonymous substitution in anthropoid primates. Mol Phylogenet Evol. 33:944–950. [DOI] [PubMed] [Google Scholar]
- Dorus S, et al. 2004. Accelerated evolution of nervous system genes in the origin of Homo sapiens. Cell 119:1027–1040. [DOI] [PubMed] [Google Scholar]
- Dunbar RI. 2009. The social brain hypothesis and its implications for social evolution. Ann Hum Biol. 36:562–572. [DOI] [PubMed] [Google Scholar]
- Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. 2009. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10:48.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enard W. 2014. Comparative genomics of brain size evolution. Front Hum Neurosci. 8:345.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enard W, et al. 2009. A humanized version of Foxp2 affects cortico-basal ganglia circuits in mice. Cell 137:961–971. [DOI] [PubMed] [Google Scholar]
- Enard W, et al. 2002. Intra- and interspecific variation in primate gene expression patterns. Science 296:340–343. [DOI] [PubMed] [Google Scholar]
- Fietz SA, Huttner WB. 2011. Cortical progenitor expansion, self-renewal and neurogenesis—a polarized perspective. Curr Opin Neurobiol. 21:23–35. [DOI] [PubMed] [Google Scholar]
- Florio M, et al. 2015. Human-specific gene ARHGAP11B promotes basal progenitor amplification and neocortex expansion. Science 347:1465–1470. [DOI] [PubMed] [Google Scholar]
- Fortna A, et al. 2004. Lineage-specific gene duplication and loss in human and great ape evolution. PLoS Biol. 2:e207.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X, et al. 2011. Rapid metabolic evolution in human prefrontal cortex. Proc Natl Acad Sci U S A. 108:6181–6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George RD, et al. 2011. Trans genomic capture and sequencing of primate exomes reveals new targets of positive selection. Genome Res. 21:1686–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert SL, Dobyns WB, Lahn BT. 2005. Genetic links between brain development and brain evolution. Nat Rev Genet. 6:581–590. [DOI] [PubMed] [Google Scholar]
- Goldberg A, et al. 2003. Adaptive evolution of cytochrome c oxidase subunit VIII in anthropoid primates. Proc Natl Acad Sci U S A. 100:5873–5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman M, et al. 2009. Phylogenomic analyses reveal convergent patterns of adaptive evolution in elephant and human ancestries. Proc Natl Acad Sci U S A. 106:20824–20829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 29:644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RE, et al. 2006. Analysis of one million base pairs of Neanderthal DNA. Nature 444:330–336. [DOI] [PubMed] [Google Scholar]
- Grossman LI, Wildman DE, Schmidt TR, Goodman M. 2004. Accelerated evolution of the electron transport chain in anthropoid primates. Trends Genet. 20:578–585. [DOI] [PubMed] [Google Scholar]
- Haijma SV, et al. 2013. Brain volumes in schizophrenia: a meta-analysis in over 18 000 subjects. Schizophr Bull. 39:1129–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison PW, Jordan GE, Montgomery SH. 2014. SWAMP: sliding window alignment masker for PAML. Evol Bioinform Online 10:197.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison PW, Wright AE, Mank JE. 2012. The evolution of gene expression and the transcriptome-phenotype relationship. Semin Cell Dev Biol. 23:222–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haygood R, Babbitt CC, Fedrigo O, Wray GA. 2010. Contrasts between adaptive coding and noncoding changes during human evolution. Proc Natl Acad Sci U S A. 107:7853–7857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazlett HC, et al. 2005. Magnetic resonance imaging and head circumference study of brain size in autism: birth through age 2 years. Arch Gen Psychiatry 62:1366–1376. [DOI] [PubMed] [Google Scholar]
- Hoffman MM, Birney E. 2010. An effective model for natural selection in promoters. Genome Res. 20:685–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong LE, et al. 2005. Dihydropyrimidinase‐related protein 2 (DRP‐2) gene and association to deficit and nondeficit schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 136:8–11. [DOI] [PubMed] [Google Scholar]
- Jerison HJ. 1973. Evolution of the brain and intelligence. New York: Academic Press. [Google Scholar]
- Khaitovich P, et al. 2005. Parallel patterns of evolution in the genomes and transcriptomes of humans and chimpanzees. Science 309:1850–1854. [DOI] [PubMed] [Google Scholar]
- Khaitovich P, et al. 2008. Metabolic changes in schizophrenia and human brain evolution. Genome Biol. 9:R124.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaitovich P, et al. 2004. Regional patterns of gene expression in human and chimpanzee brains. Genome Res. 14:1462–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King M-C, Wilson AC. 1975. Evolution at two levels in humans and chimpanzees. Science 188:107–116. [DOI] [PubMed] [Google Scholar]
- Kunugi H, Takei N, Murray RM, Saito K, Nanko S. 1996. Small head circumference at birth in schizophrenia. Schizophr Res. 20:165–170. [DOI] [PubMed] [Google Scholar]
- Lange C, Huttner WB, Calegari F. 2009. Cdk4/cyclinD1 overexpression in neural stem cells shortens G1, delays neurogenesis, and promotes the generation and expansion of basal progenitors. Cell Stem Cell 5:320–331. [DOI] [PubMed] [Google Scholar]
- Lawick-Goodall V. 1968. The behaviour of free-living chimpanzees in the Gombe Stream Reserve. Anim Behav Monogr. 1:161-IN112. [Google Scholar]
- Lewis DA, Levitt P. 2002. Schizophrenia as a disorder of neurodevelopment. Annu Rev Neurosci. 25:409–432. [DOI] [PubMed] [Google Scholar]
- Löytynoja A, Goldman N. 2008. Phylogeny-aware gap placement prevents errors in sequence alignment and evolutionary analysis. Science 320:1632–1635. [DOI] [PubMed] [Google Scholar]
- Martin RD. 1981. Relative brain size and basal metabolic rate in terrestrial vertebrates. Nature 293:57–60. [DOI] [PubMed] [Google Scholar]
- McGowen MR, Grossman LI, Wildman DE. 2012. Dolphin genome provides evidence for adaptive evolution of nervous system genes and a molecular rate slowdown. Proc R Soc B Biol Sci. 279:3643–3651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, et al. 2011. Human-specific loss of regulatory DNA and the evolution of human-specific traits. Nature 471:216–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery S, Mundy N. 2012a. Positive selection on NIN, a gene involved in neurogenesis, and primate brain evolution. Genes Brain Behav. 11:903–910. [DOI] [PubMed] [Google Scholar]
- Montgomery SH, Capellini I, Barton RA, Mundy NI. 2010. Reconstructing the ups and downs of primate brain evolution: implications for adaptive hypotheses and Homo floresiensis. BMC Biol. 8:9.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery SH, Capellini I, Venditti C, Barton RA, Mundy NI. 2011. Adaptive evolution of four microcephaly genes and the evolution of brain size in anthropoid primates. Mol Biol Evol. 28:625–638. [DOI] [PubMed] [Google Scholar]
- Montgomery SH, Mundy NI. 2012b. Evolution of ASPM is associated with both increases and decreases in brain size in primates. Evolution. 66:927–932. [DOI] [PubMed] [Google Scholar]
- Montgomery SH, Mundy NI, Barton RA. 2016. Brain evolution and development: adaptation, allometry and constraint. Proc Roy Soc B. 283(1838):20160433.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muntané G, et al. 2014. Analysis of synaptic gene expression in the neocortex of primates reveals evolutionary changes in glutamatergic neurotransmission. Cereb Cortex 25:1596–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R, et al. 2005. A scan for positively selected genes in the genomes of humans and chimpanzees. PLoS Biol. 3:e170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor TD, Mundy NI. 2013. Evolutionary modeling of genotype-phenotype associations, and application to primate coding and non-coding mtDnA rate variation. Evol Bioinform Online 9:301.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottoni EB, Izar P. 2008. Capuchin monkey tool use: overview and implications. Evol Anthropol Issues News Rev. 17:171–178. [Google Scholar]
- Pagel M. 1999. Inferring the historical patterns of biological evolution. Nature 401:877–884. [DOI] [PubMed] [Google Scholar]
- Palmen SJ, et al. 2005. Increased gray-matter volume in medication-naive high-functioning children with autism spectrum disorder. Psychol Med. 35:561–570. [DOI] [PubMed] [Google Scholar]
- Penn DL, Corrigan PW, Bentall RP, Racenstein J, Newman L. 1997. Social cognition in schizophrenia. Psychol Bull. 121:114.. [DOI] [PubMed] [Google Scholar]
- Phillips KA. 1998. Tool use in wild capuchin monkeys (Cebus albifrons trinitatis). Am J Primatol. 46:259–261. [DOI] [PubMed] [Google Scholar]
- Phillips KA, Sherwood CC. 2008. Cortical development in brown capuchin monkeys: a structural MRI study. Neuroimage 43:657–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, et al. 2006. An RNA gene expressed during cortical development evolved rapidly in humans. Nature 443:167–172. [DOI] [PubMed] [Google Scholar]
- Pulvers JN, et al. 2010. Mutations in mouse Aspm (abnormal spindle-like microcephaly associated) cause not only microcephaly but also major defects in the germline. Proc Natl Acad Sci U S A. 107:16595–16600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P. 2009. Evolution of the neocortex: a perspective from developmental biology. Nat Rev Neurosci. 10:724–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapoport J, Giedd J, Gogtay N. 2012. Neurodevelopmental model of schizophrenia: update 2012. Mol Psychiatry 17:1228–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reader SM, Hager Y, Laland KN. 2011. The evolution of primate general and cultural intelligence. Phil Trans R Soc B Biol Sci. 366:1017–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redcay E, Courchesne E. 2005. When is the brain enlarged in autism? A meta-analysis of all brain size reports. Biol Psychiatry 58:1–9. [DOI] [PubMed] [Google Scholar]
- Roberts RG, et al. 1996. Characterization of DRP2, a novel human dystrophin homologue. Nat Genet. 13:223–226. [DOI] [PubMed] [Google Scholar]
- Scally A, et al. 2012. Insights into hominid evolution from the gorilla genome sequence. Nature 483:169–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt TR, et al. 2005. Rapid electrostatic evolution at the binding site for cytochrome c on cytochrome c oxidase in anthropoid primates. Proc Natl Acad Sci U S A. 102:6379–6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi P, Bakewell MA, Zhang J. 2006. Did brain-specific genes evolve faster in humans than in chimpanzees? Trends Genet. 22:608–613. [DOI] [PubMed] [Google Scholar]
- Sholtis SJ, Noonan JP. 2010. Gene regulation and the origins of human biological uniqueness. Trends Genet. 26:110–118. [DOI] [PubMed] [Google Scholar]
- Sobczak A, et al. 2011. Calmyrin1 binds to SCG10 protein (stathmin2) to modulate neurite outgrowth. Biochim Biophys Acta (BBA) Mol Cell Res. 1813:1025–1037. [DOI] [PubMed] [Google Scholar]
- Somel M, Liu X, Khaitovich P. 2013. Human brain evolution: transcripts, metabolites and their regulators. Nat Rev Neurosci. 14:112–127. [DOI] [PubMed] [Google Scholar]
- Stephan H, Frahm H, Baron G. 1981. New and revised data on volumes of brain structures in insectivores and primates. Folia Primatol. 35:1–29. [DOI] [PubMed] [Google Scholar]
- Sun J, et al. 2010. Schizophrenia gene networks and pathways and their applications for novel candidate gene selection. PLoS One 5:e11351.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington MM. 1990. Fission-fusion social organization inAteles andPan. Int J Primatol. 11:47–61. [Google Scholar]
- Takata A, et al. 2013. A population-specific uncommon variant in GRIN3A associated with schizophrenia. Biol Psychiatry 73:532–539. [DOI] [PubMed] [Google Scholar]
- Thornton GK, Woods CG. 2009. Primary microcephaly: do all roads lead to Rome? Trends Genet. 25:501–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truppa V, Spinozzi G, Stegagno T, Fagot J. 2009. Picture processing in tufted capuchin monkeys (Cebus apella). Behav Processes 82:140–152. [DOI] [PubMed] [Google Scholar]
- Uddin M, et al. 2004. Sister grouping of chimpanzees and humans as revealed by genome-wide phylogenetic analysis of brain gene expression profiles. Proc Natl Acad Sci U S A. 101(9):2957–2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uddin M, et al. 2008. Distinct genomic signatures of adaptation in pre- and postnatal environments during human evolution. Proc Natl Acad Sci U S A. 105:3215–3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel T, Ahrens S, Büttner N, Krieglstein K. 2010. Transforming growth factor β promotes neuronal cell fate of mouse cortical and hippocampal progenitors in vitro and in vivo: identification of Nedd9 as an essential signaling component. Cereb Cortex 20:661–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H-Y, et al. 2006. Rate of evolution in brain-expressed genes in humans and other primates. PLoS Biol. 5:e13.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winchester CL, Pratt JA, Morris BJ. 2014. Risk genes for schizophrenia: translational opportunities for drug discovery. Pharmacol Ther. 143:34–50. [DOI] [PubMed] [Google Scholar]
- Yang Z. 2005. Bayesian inference in molecular phylogenetics. Math Evol Phylogeny 63–90. [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24:1586–1591. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R. 2002. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol Biol Evol. 19:908–917. [DOI] [PubMed] [Google Scholar]
- Yosifova A, et al. 2009. Case-control association study of 65 candidate genes revealed a possible association of a SNP of HTR5A to be a factor susceptible to bipolar disease in Bulgarian population. J Affect Disord. 117:87–97. [DOI] [PubMed] [Google Scholar]
- Zafeiriou D, Ververi A, Vargiami E. 2009. The serotonergic system: its role in pathogenesis and early developmental treatment of autism. Curr Neuropharmacol. 7:150–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaharieva I, et al. 2008. Association study in the 5q31-32 linkage region for schizophrenia using pooled DNA genotyping. BMC Psychiatry 8:11.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai W, Nielsen R, Goldman N, Yang Z. 2012. Looking for Darwin in genomic sequences—validity and success of statistical methods. Mol Biol Evol. 29:2889–2893. [DOI] [PubMed] [Google Scholar]
- Zhang J, Nielsen R, Yang Z. 2005. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 22:2472–2479. [DOI] [PubMed] [Google Scholar]
- Zimmer F, Montgomery SH. 2015. Phylogenetic analysis supports a link between DUF1220 domain number and primate brain expansion. Genome Biol Evol. 7:2083–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.