Abstract
Geraniaceae are known for their unusual plastid genomes (plastomes), with the genus Pelargonium being most conspicuous with regard to plastome size and gene organization as judged by the sequenced plastomes of P. x hortorum and P. alternans. However, the hybrid origin of P. x hortorum and the uncertain phylogenetic position of P. alternans obscure the events that led to these extraordinary plastomes. Here, we examine all plastid reconfiguration hotspots for 60 Pelargonium species across all subgenera using a PCR and sequencing approach. Our reconstruction of the rearrangement history revealed four distinct plastome types. The ancestral plastome configuration in the two subgenera Magnipetala and Pelargonium is consistent with that of the P. alternans plastome, whereas that of the subgenus Parvulipetala deviates from this organization by one synapomorphic inversion in the trnNGUU–ndhF region. The plastome of P. x hortorum resembles those of one group of the subgenus Paucisignata, but differs from a second group by another inversion in the psaI–psaJ region. The number of microstructural changes and amount of repetitive DNA are generally elevated in all inverted regions. Nucleotide substitution rates correlate positively with the number of indels in all regions across the different subgenera. We also observed lineage- and species-specific changes in the gene content, including gene duplications and fragmentations. For example, the plastid rbcL–psaI region of Pelargonium contains a highly variable accD-like region. Our results suggest alternative evolutionary paths under possibly changing modes of plastid transmission and indicate the non-functionalization of the plastid accD gene in Pelargonium.
Keywords: plastid genome, Pelargonium, Geraniaceae, molecular evolution
Introduction
The chloroplast genome (plastome) of photosynthetic flowering plants is generally conserved with regard to genome size, gene order, and gene content (e.g., Wicke et al. 2011; Jansen and Ruhlman 2012). Coding for ca. 110–130 unique plastid genes that mostly function in photosynthesis-associated molecular processes and a few photosynthesis-unrelated pathways, plastomes are structured in a large and a small single copy region (LSC and SSC, respectively), which are separated by two large inverted repeat regions (IRs; hereafter used to refer exclusively to these plastome compartments). Notably divergent plastomes are found, however, in a handful of unrelated, fully photosynthetic angiosperm lineages (Jansen and Palmer 1987; Cosner et al. 2004; Kim et al. 2005; Cai et al. 2008; Greiner et al. 2008; Haberle et al. 2008; Magee et al. 2010; Guisinger et al. 2011; Knox 2014). Among those, Geraniaceae are clearly an exceptional example for variability in plastome size and structural rearrangements. Plastome sizes range from 117 kb in Erodium carvifolium to 218 kb in Pelargonium x hortorum in Geraniaceae, mainly because of the loss of the IR in Erodium and an unrelated expansion of the IR in Pelargonium (Chumley et al. 2006; Blazier et al. 2011). In the two other genera of Geraniaceae, Geranium and Monsonia, the IRs are reduced compared with other angiosperms (Guisinger et al. 2011). Plastid genome rearrangements in Geraniaceae include the disruption of two conserved operons (rpl23–rpoA and rps2–atpA), many full or partial gene duplications (Guisinger et al. 2011), and several major inversions (Chumley et al. 2006). The causes and mechanisms of these unique molecular evolutionary paths of plastome evolution in Geraniaceae are unknown to date. It is not known either, if plastid genomes are evolutionary stable within the different genera and subgenera, or if they undergo more idiosyncratic reconfigurations and gene losses.
Among Geraniaceae, the genus Pelargonium is a particularly well-suited model system to analyze the mechanisms and lineage-specific progression of plastid genome reconfiguration in photosynthetic flowering plants. With ca. 280 species, Pelargonium is the second largest genus in Geraniaceae. The infrageneric relationships are mostly well established, with molecular, morphological, and chemical evidence allowing to distinguish four subgenera: Pelargonium, Parvulipetala, Magnipetala, and Paucisignata (Röschenbleck et al. 2014). Apart from their importance as ornamental and medicinal plants, species of Pelargonium have been one of the earliest model plants for understanding the transmission of plastids and their genetic material. Among green plants, the genus is well known for its unusual evolutionary paths of its plastid genome.
The P. x hortorum plastome is characterized by an expanded IR (76 kb), making it three times larger than that of most angiosperms, and at least eight major inversions and 30 gene clusters in which the gene order resembles that of other rosids (Chumley et al. 2006). In contrast to P. x hortorum, the plastid genome of P. alternans has a smaller overall size (173 kb) and expansion of the IR (38 kb), and is less rearranged, lacking a 50-kb inversion (Weng et al. 2014). The closest relatives of P. alternans within the subgenus Pelargonium are still unclear (Jones et al. 2009; Albers and Becker 2010; Röschenbleck et al. 2014).
In this study, we examined the structural plastome variation on the basis of four selected rearrangement hotspots of P. x hortorum for 60 Pelargonium species, covering all subgenera. We inferred possible reconfigurations and their putative counterparts and possible transitions by analyzing the inversion breakpoint regions located in the LSC and within the expanded IR of P. x hortorum. The dense sampling allowed us to reconstruct the history of rearrangements events. We analyzed the association between nucleotide substitution rates and genomic traits such as length mutations and repetitive DNA across the different lineages and regions, and discuss in how far the extreme reconfigurations of the two so far fully sequenced Pelargonium plastomes are representative of the genus.
Materials and Methods
Taxon Sampling, PCR Screening, and Sequencing Strategies
Following Röschenbleck et al. (2014), we sampled every subgenus (i.e., Paucisignata, Magnipetala, Parvulipetala, and Pelargonium) with two to seven species per section, plus several additional species, whose phylogenetic position in the respective subgenera were ambiguous. The final set included 24 species of subgenus Pelargonium, 12 species of subgenus Parvulipetala, 14 of subgenus Paucisignata, and 10 of subgenus Magnipetala (supplementary table S1, Supplementary Material online). To better distinguish divergent clades within subgenus Paucisignata, we here introduce two informal clade names: “group C” will refer to the Paucisignata clade that includes section Ciconium, whereas “group S” addresses the clade that includes section Subsucculentia (fig. 1 graphically summarizes the different clade names).
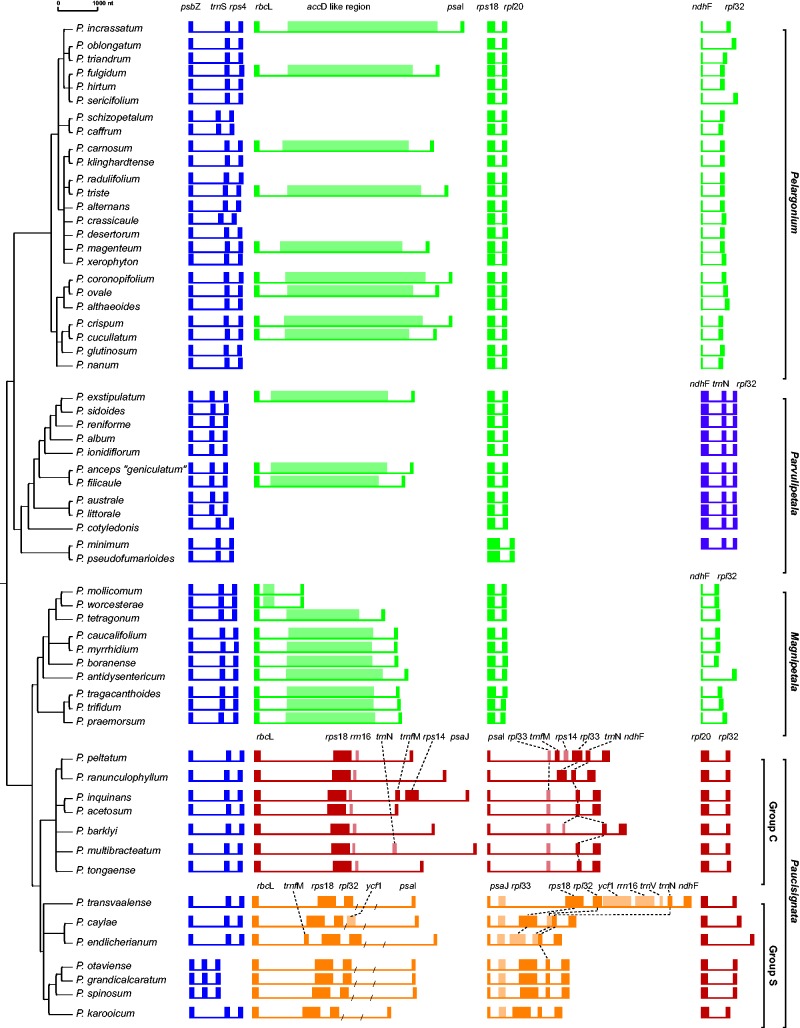
Fig. 1.—
Plastome rearrangements in Pelargonium. Differences in gene order between clades are color-coded and the gene content per region is given on top. Lighter colors demarcate pseudogenes. Lengths of inversion breakpoints and of coding and noncoding regions are to scale. The tree topology was inferred with a maximum likelihood analysis of the psbZ–trnSGAA–rps4 region. Names of subgenera and subgroups are shown to the right.
We used a PCR screen to diagnose the presence and absence of five rearrangement hotspots in the P. x hortorum genome, which are the result of several inversion and re-inversion events (Chumley et al. 2006). We investigated the trnEUUC–rps4 fragment that involves an inversion of trnGGCC–psbD, a re-inversion of psbD–psbZ, and an inversion of trnfM–ycf3 relative to other rosids by screening the inversion start (trnEUUC–trnGGCC–ycf3) and end point (psbZ–rps4). The second region was rbcL–rps18–psaJ at the LSC–IRB junction, which leads to a re-inversion of rps18, and we inspected the IRA–LSC (rbcL–psbA). The third and fourth regions involved the large 50-kb inversion of the P. x hortorum plastome from psaI to the start of the inverted trnNGUU–ndhF and the end of the 50-kb inversion of the IR (rpl20–rpl32), respectively. Fifth, we examined the inverted repeat boundaries to the LSC regions for subgenus Pelargonium (ycf2–rpoB and ycf2–petB). We used genomes of other rosids (e.g., Citrus sinensis, Vitis vinifera) to determine the standard (i.e., not reorganized) equivalents of these regions, namely rps4–ycf3, rbcL–psaI, rps18–rpl20, and ndhF–rpl32. The 27 primer combinations used in this study are summarized in supplementary table S2, Supplementary Material online. We also included standardly the IR boundaries as positive or negative controls in our PCR screens (supplementary table S2, Supplementary Material online).
DNA Extraction, Amplification, and Sequencing
Genomic DNA was extracted using the DNeasy Plant Mini Kit (Qiagen) according to the manufacturer’s protocol. PCR amplifications were performed as 50 µl reactions containing 60–120 ng/µl DNA template, 3 µl of a 2.5-mM dNTP mix, 10 mmol of each primer, and 1.5 U GoTaq polymerase (Promega). Thermocycling was carried out in a primus 96 advanced (PEQLAB), with an initial denaturation of 3 min at 94°C, 38–45 cycles of 20 s at 94°C, 30–90 s at 54–60°C, and 60–270 s at 72°C (depending upon the size of the target region; supplementary table S3, Supplementary Material online), and a final elongation of 5 min at 72°C. PCR products were purified using the NucleoSpin Extract II (Machery & Nagel, Düren, Germany), precipitated with 1/10th V 3 M Na-acetate and 2.5 V ethanol, and resuspended in 15 µl ultrapure water. Sanger sequencing was outsourced to Eurofins (Germany). We omitted sequencing of the highly variable trnEUUC–trnGGCC–ycf3 intergenic spacer. We restricted sequence validation to the psbZ–rps4 end point of the trnEUUC–rps4 region, and sequence-validated the rbcL–psaI spacer only for representatives of larger clades of Pelargonium and Parvulipetala. Due to the loss of one species (P. pseudofumarioides) from the Münster collection, we omitted sequence verification of the ndhF–rpl32 marker for this species, as it was inconspicuous in terms of amplicon size compared with other taxa.
Alignment and Gene Annotation
All sequence datasets were complemented with the reference sequences of P. alternans (Weng et al. 2014) and P. x hortorum (Chumley et al. 2006), and aligned using PRANK v.140603 (Löytynoja and Goldman 2005); all alignments are available on datadryad.org (doi: 10.5061/dryad.8g928). Except for the psbZ–rps4 dataset, which contained all 60 Pelargonium taxa, we build separate alignments for subsets of the four subgenera and according to the presence or absence of a region in the respective clades. As we were unable to assess homology in the rps18–psaI spacer in five of the seven species of group S of subgenus Paucisignata, we excluded this region (alignment positions 586–1,572) from all subsequent analyses. General sequence statistics such as the GC content across all datasets were computed with Seqstate v.1.4.1 (Müller 2005).
Sequences were annotated with DOGMA (Wyman et al. 2004) with an identity cutoff of 30% for coding genes, 80% for tRNAs, and an e-value of 1e− 5. We used BLASTN to detect short sequences of genes starts or ends and accD-like gene fragments. In addition, we searched for open reading frames (ORFs) of at least 30 codons in the rbcL–psaI spacer for potential protein coding segments using ORF finder tool of SMS v.2 (Stothard 2000). ORFs were characterized in detail by BLASTN and BLASTX searches against the nonredundant NCBI database.
Rearrangements and Structural Analyses
For all analyses we used the tree topology resulting from a maximum likelihood (ML) analysis for the universally conserved psbZ–rps4 region, because it was congruent with the phylogenetic relationships identified by earlier studies that dealt with organismal relationships in the genus (Röschenbleck et al. 2014, 2015). ML searches were performed using the GTR + G model in RaxML v.7.03 (Stamatakis 2006), rapid bootstrap analysis with 250 replicates, followed by 10 ML searches. Partial sequences (psbZ–trnSUGA spacer, trnSGGA–rps4) of Geranium palmatum, E . carvifolium, and Monsonia speciosa (Blazier et al. 2011; Guisinger et al. 2011) were used as outgroup.
The plastome rearrangement history was analyzed with MGR (Bourque and Pevzner 2002) under a linear and undirected model over the fixed ML topology. We first restricted this reconstruction to our PCR-screened regions, that is, psbZ, trnSGGA, rps4, rbcL, psaI, rps18, rpl20, ndhF, and rpl32. Second, we simulated the gene order of 111 Pelargonium plastid genes (supplementary table S4, Supplementary Material online), assuming that the gene order revealed in our PCR screen is indicative of either the plastid gene organizations of P. alternans or that of P. x hortorum. The gene order of Melianthus villosus (Weng et al. 2014) served as a reference for a standard genome and was used as outgroup. Two genes, trnTGGU, accD, and the truncated ycf1 of Melianthus were excluded because they were absent from the Pelargonium reference genomes. We excluded pseudogenes from our matrix, except for the ndhD, F, H, and K of Melianthus, and treated ORF574 of P. x. hortorum as rpoA.
Length mutations (here synonymous with short insertions or mutations—indels) were analyzed using simple indel coding (Simmons and Ochoterena 2000). We inferred the number of microstructural changes along branches by parsimony scoring via custom scripts in combination with the R-packages “ape” (Paradis et al. 2004) and “phangorn” (Schliep 2011) as described in Jansen et al. (2007) and modifications regarding the subsequent statistical test as detailed in Wicke et al. (2016). Di-, tri- and tetramer single sequence repeats (SSRs) with at least three times repetitions of the core sequence motif were analyzed and counted per species and regions occurring employing SSRIT (Temnykh et al. 2001). The number of forward (tandem) and palindromic (inverted) repeats of at least 12 bp were identified using the Tandem Repeats Finder (Benson 1999) and the Palindromic Sequences Finder (Bikandi et al. 2004), and further sorted by length; all tools were run at default settings.
Correlations of Nucleotide Substitution Rates and Genetic Traits
For all regions, substitution rate changes were analyzed in HyPhy v.2.22 (Kosakovsky Pond et al. 2005) based on alignments of each clade and the corresponding tree topology. We used an unconstrained likelihood function, with a full general time reversible model and local parameters to estimate relative substitution rates per branch. Correlations between substitution rates and microstructural changes were evaluated per dataset using Mantel tests, for which the ML tree and subsets thereof were scaled according to the nucleotide substitution rates, as well as to the different genetic traits of the various regions by tracing their histories over the tree using parsimony in R. The rescaled trees were transformed into patristic distance matrices, which were used for Mantel tests with phylogenetic permutation to evaluate a correlation between indels and substitution rates per gene region. Mantel tests were computed in R with 5,000 permutations each. The advantage of Mantel tests over other phylogenetic-comparative methods here was that it allowed us to compare indel- and substitution rate-scaled trees (and thus over all edges of the tree) as pairwise distances among taxa rather than treating substitution rates as ordinary traits (phenotypic or genotypic) of the terminal nodes. Besides the region- and clade-wise substitution rate/indel analyses, we evaluated global correlations between mean GC-content, substitution rates, indels, and repeat frequencies (number of repeats in relation to the length of the sequence) over all datasets (i.e., across all examined markers) via nonparametric Spearman tests to assess general trends between regions that show reorganization compared to those that exhibit a standard gene order; multiple testing was accounted for by sequential alpha error correction (Bonferroni–Holm correction).
Results
Structural Rearrangements in Pelargonium
We obtained 254 consensus sequences, totaling ca. 353 kb. The gene order analysis revealed one synapomorphic reconfiguration in the trnEUUC–rps4 region in the genus Pelargonium, and two rearrangements autapomorphic on the infrageneric level (fig. 1). Gene order in the rbcL–psaI, rps18–rpl20, and ndhF–rpl32 regions of the two Pelargonium subgenera Pelargonium and Magnipetala each resembles that of rosids with structurally regular plastomes. Subgenus Parvulipetala differed from these two subgenera by an inversion of ndhF and trnNGUU (figs.1 and 2A ). The IR/LSC boundaries in subgenus Pelargonium all were found to be located in the coding region of petD (as in P. alternans: Weng et al. 2014), which is located between rpoA and ycf2 at the IR-LSC border or petB and rpoA at the LSC-IR border, respectively.
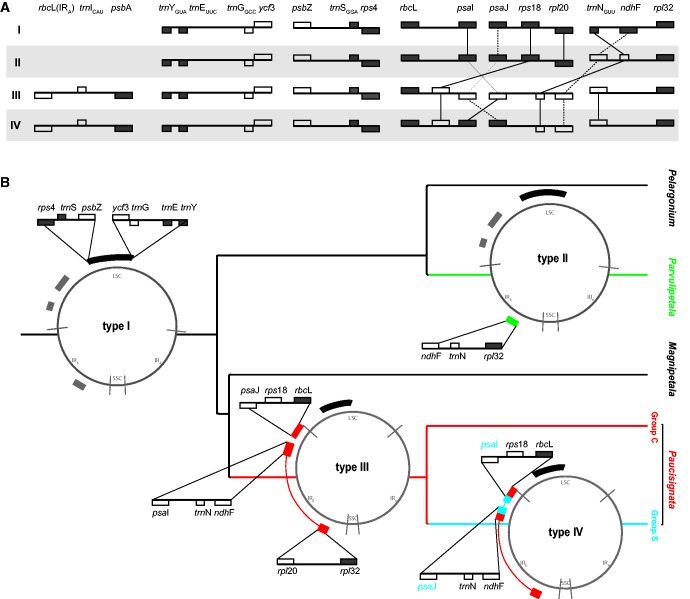
Fig. 2.—
Reconstruction of ancestral plastid genomes. (A ) Genes and their positions for breakpoints a–f are depicted in detail for plastome types I–IV in relation to the standard plastome of M. villosus (Geraniales). Square colors demarcate rearranged genes (white) and genes with a conserved position (gray). ( B ) Plastid genome types (i–iv) are shown as plastid circles along branches of the major Pelargonium lineages. The inferred mutational hotspots are highlighted by small insets, showing the series of structural changes leading to the distinct plastomes types. Branches are colored according to the dominating plastome type, showing the type I in black, II in green, III in blue, and IV in red. Regions with additional changes as detailed in (A) are marked in II and IV.
All examined species of subgenus Paucisignata shared the rearrangements at both the LSC/IRB junction (rbcL–rps18) and the 50-kb IR inversion. The two subclades C and S differed in the position of psaI and psaJ. These subclades were monophyletic in the psbZ–rps4 topology (fig. 1), but with low support (<50%) for the monophyly of group S. All species of Paucisignata, but none of other subgenera contained the rbcL–psbA and rbcL–trnICAU spacer (fig. 2A ).
We identified several duplications of genes and gene fragments in four of the five screened plastome regions (fig. 1). In subgenera Pelargonium, Parvulipetala, and Magnipetala, we discovered a highly variable accD-like region in the rbcL–psaI region, ranging between 234 and 3,743 bp in length. Subgenus Paucisignata showed numerous and uniquely fragmented duplications of genes and tRNAs (fig. 1). In group C, duplicated fragments of rrn16 and rpl33 are located between rps18–psaJ and psaI–ndhF. All species of group S have apparently intact copies of rpl32 in the rps18–psaI spacer, and of rps18 in the psaI–ndhF spacer. The three species P. caylae, P. endlicherianum, and P. transvaalense of group S have an additional rpl32 gene downstream of rps18.
Plastome Types in Pelargonium
The reconstruction of the evolution of plastome structure based on a nine-gene dataset revealed rearrangements ancestral to the subgenera Parvulipetala, Paucisignata, and group Sof Paucisignata (fig. 2B ). We identified additional rearrangements between trnE–rps4 in subgenus Pelargonium and group C of Paucisignata using an expanded 111 gene dataset (fig. 2A and B; supplementary table S4, Supplementary Material online). Based on this “simulated” whole plastome gene matrix that considered the results of our PCR screen, we inferred inversions within the SSC and at the IR junctions, in ycf1–ndhH of subgenus Paucisignata, as well as in both ycf1–trnLUAG and trnLCAA–ycf2 of the subgenera Pelargonium, Parvulipetala, and Magnipetala. These results suggest four distinct types of plastid genomes (types I–IV) in the genus Pelargonium. The ancestor of subgenera Pelargonium and Magnipetala (i.e., the root node of the genus Pelargonium) likely had a Melianthus-like gene order (type I) with 23% of its genes rearranged compared with the standard plastome of Melianthus (fig. 2B ). Subgenus Parvulipetala has a type II-plastome structure with 25% of plastid genes rearranged, and the subgenus Paucisignata and its group C both have a type-III gene order, in which 58% of plastid genes differ regarding their position relative to the Melianthus plastome. Plastomes of type IV, where 46% of genes are rearranged, are typical for the group S of Paucisignata.
Length Variation and Repetitive Plastid DNA
Differences in mean sequence lengths of the various examined markers between the different subgenera ranged from 52 to 1,651 bp (supplementary table S5, Supplementary Material online) in relation to the mean per region. Length mutations (indels) are found in all marker regions, particularly in the inverted regions and in the rbcL–psaI spacer (supplementary fig. S1, Supplementary Material online). Indels of 2–15 bp in length were the most abundant, but we also found indels of more than 100 bp in all regions. Subgenera Pelargonium, Parvulipetala, and Magnipetala differ notably in their indel frequencies in the ndhF–rpl32 region. The conserved ndhF–rpl32 spacer in subgenera Pelargonium and Magnipetala varied between 377 and 461 bp, respectively, in part due to indels of over 100 bp in both. The inverted ndhF–trnN–rpl32 region of subgenus Parvulipetala, however, differed only by 12 bp and indels of 1–7 bp. Both groups of the subgenus Paucisignata showed an increased number of indels larger than 100 bp, which are particularly frequent between rbcL and rps18, as well as between rpl20 and rpl32 in group S, and between rps18 and ndhF in group C (supplementary fig. S1 and table S5, Supplementary Material online).
The proportion of repetitive plastid DNA was generally high in the analyzed non-coding regions and in the rbcL–psaI region in particular (supplementary fig. S1, Supplementary Material online). In Paucisignata, SSRs with a tri- and tetramer motif were restricted to the rbcL–rps18 region and to the unique spacer regions rps18–psaJ/psaI and psaI/psaJ–ndhF of Paucisignata groups C and S, albeit with a different distribution of larger repeats (>12 bp). Direct repeats were more frequent in group C than in group S, which had a higher proportion of short inverted repeats.
We found three copies of a 64-bp motif in the coding region of the rps18 gene in P. minimum and P. pseudofumarioides, both belonging to subgenus Parvulipetala. One of the rpl32 copies between rps18 and psaJ in P. endlicherianum (group S of Paucisignata) shows a 21-bp motif that is tandemly repeated four times. Two or three segments of the accD-like region of the subgenera Pelargonium and Parvulipetala, respectively, are extremely rich in repeats, which vary species-specifically in both their size and copy number (supplementary fig. S2, Supplementary Material online).
Molecular Evolutionary Rates
Nucleotide substitution rates and GC content are, on average, elevated in all rearranged regions compared to regions with a conserved gene order (supplementary table S6, Supplementary Material online). The rbcL–psaI region, whose GC content is notably higher (37.9–39.8%) than that of all other regions with an unaltered gene order (26.1–36.5%; supplementary fig. S1, Supplementary Material online), has an average relative nucleotide substitution rate twice as high as other loci. Among clades, the substitution rates were conspicuously higher in subgenus Magnipetala and in group S of subgenus Paucisignata (supplementary table S6, Supplementary Material online) than elsewhere. In addition, Paucisignata group C revealed an increase of substitution rates and of the GC content in its unique rps18–psaJ spacer.
Correlations between Genetic Traits
All datasets, when analyzed separately with respect to both the different plastid regions and subgenera and groups, showed a significant positive correlation between indel and nucleotide substitution rates (P <0.001–0.003; table 1), with the exception of the rbcL–accD–psaI region in subgenus Parvulipetala (P = 0.326) and the rpl20–rpl32 region in Paucisignata group C (P = 0.256) (table 1). Indel-substitution rate evolution appears to differ slightly in regions that are commonly present in subgenus Paucisignata when single groups were tested separately. For example, group C shows no associations between microstructural and substitution rate changes in the rpl20– rpl32 region (P = 0.256), whereas these two traits are highly correlated in group S (P = 0.005). Analyses across the entire subgenus Paucisignata revealed a significant positive correlation of indels and substitution rates (P <0.001–0.006). Globally testing all plastid regions based on separate clade alignments (n = 22) indicated that nucleotide substitution rates are significantly associated with the number of both indels (P = 0.008) and inverted repeats larger than 12 bp (P = 0.023), but not with the amount of forward repeats and SSRs (table 2). Variation in the GC content is not correlated with substitution rates (P = 0.201). However, the GC content is associated positively with the number of SSRs (P = 0.033). In contrast to substitution rates, the elevated indel number coincides with an increase in GC content (P = 0.029).
Table 1.
Results of Mantel Tests Evaluating the Correlation of Indels and Substitution Rates Along Branches
| Region, Subgenus, Group | N | Z | P value |
|---|---|---|---|
| rps4–psbZ, genus-wide | 62 | 552.88 | <0.001 |
| psbZ–rps4, Pelargonium | 25 | 84.95 | <0.001 |
| psbZ–rps4, Parvulipetala | 12 | 13.79 | 0.004 |
| psbZ–rps4, Magnipetala | 10 | 21.34 | <0.001 |
| psbZ–rps4, Paucisignata | 15 | 41.67 | <0.001 |
| psbZ–rps4, Paucisignata, Group C | 8 | 10.62 | 0.032 |
| psbZ–rps4, Paucisignata, Group S | 7 | 9.76 | 0.005 |
| rbcL–psaI, Pelargonium | 10 | 23.37 | <0.001 |
| rbcL–psaI, Parvulipetala | 3 | 1.82 | 0.326 |
| rbcL–psaI, Magnipetala | 10 | 18.65 | <0.001 |
| rbcL–rps18, Paucisignata | 15 | 48.89 | <0.001 |
| rbcL–rps18, Paucisignata, Group C | 8 | 10.86 | 0.004 |
| rbcL–rps18, Paucisignata, Group S | 7 | 10.57 | 0.014 |
| 18s spacer–psaJ, Paucisignata, Group C | 8 | 13.25 | <0.001 |
| 18s spacer–psaI Paucisignata, Group S | 7 | 9.81 | 0.011 |
| psaI-ndhF, Paucisignata, Group C | 8 | 8.48 | <0.001 |
| psaJ-ndhF Paucisignata, Group S | 7 | 11.26 | 0.023 |
| rps18–rpl20, Pelargonium | 25 | 81.09 | <0.001 |
| rps18–rpl20, Parvulipetala | 12 | 13.67 | 0.001 |
| rps18–rpl20, Magnipetala | 10 | 14.24 | <0.001 |
| ndhF–rpl32, Pelargonium | 25 | 58.64 | <0.001 |
| ndhF–rpl32, Parvulipetala | 11 | 7.76 | 0.031 |
| ndhF–rpl32, Magnipetala | 10 | 15.35 | <0.001 |
| rpl20–rpl32, Paucisignata | 15 | 26.33 | <0.001 |
| rpl20–rpl32, Paucisignata, Group C | 8 | 9.00 | 0.256 |
| rpl20–rpl32, Paucisignata, Group S | 7 | 10.44 | 0.005 |
Table 2.
Results of Spearman Correlation Testing (Alpha Error-Corrected) for Global Correlation among Genetic Traits
| Trait 1 | Trait2 | ρ | P value |
|---|---|---|---|
| µ | #indels | 0.634 | 0.008 |
| µ | Forward Repeats | 0.433 | 0.133 |
| µ | Inverted Repeats | 0.568 | 0.023 |
| µ | %GC | 0.359 | 0.201 |
| µ | sum SSRs | 0.299 | 0.202 |
| indels | %GC | 0.465 | 0.118 |
| indels | sum SSRs | 0.452 | 0.118 |
| indels | Forward Repeats | 0.299 | 0.215 |
| indels | Inverted Repeats | 0.353 | 0.215 |
| %GC | sum SSRs | 0.531 | 0.033 |
| %GC | Forward Repeats | 0.294 | 0.366 |
| %GC | Inverted Repeats | 0.295 | 0.366 |
Pseudogenization of accD
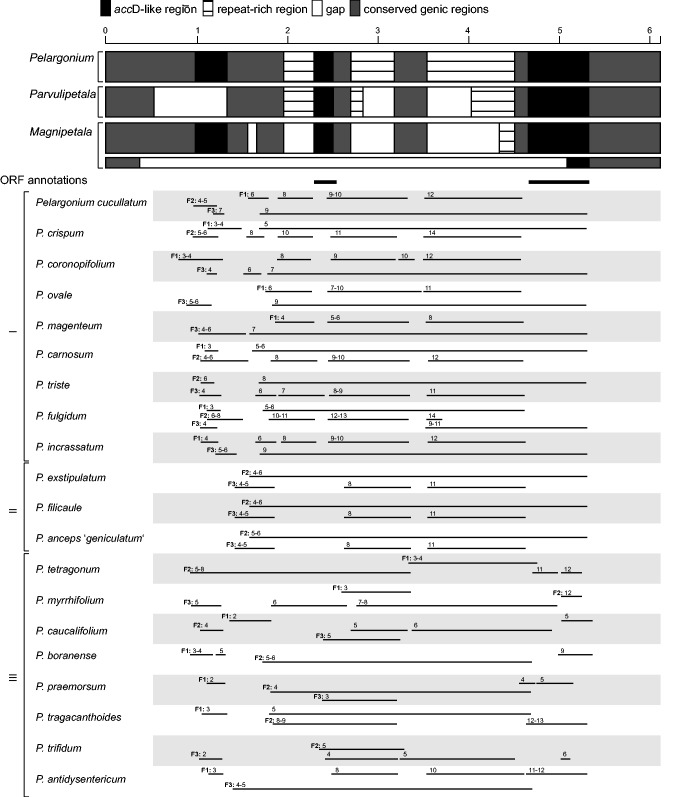
In the subgenera Pelargonium, Parvulipetala, and Magnipetala, the accD-like regions span ca. 70% of the rbcL–psaI spacer (figs. 1 and 3). The average length ranged between 2,304 and 3,285 bp (supplementary table S7, Supplementary Material online), except for one subclade in Magnipetala, which shows a substantial reduction of its lengths to only 234 bp. Sequence comparisons with other rosids, including Francoa and Melianthus of Geraniales, revealed three accD-like fragments (accD1–3) interspersed with one or two repeat-rich regions (fig. 3). AccD-1 has a length of ca. 300 bp in subgenera Pelargonium and Magnipetala, but is lacking in subgenus Parvulipetala (fig. 3). The 5′end of accD-1 resembles the authentic gene start of accD in Francoa, Melianthus and other rosids, whereas in subgenus Pelargonium we found only alternative start codons (GTG, TTG) nearby. The fragments accD-2 and accD-3, with a size of ca. 200 and 600 bp, respectively, are present in all three subgenera. All accD-like regions differ in their similarity to intact accD genes of other species (accD1: 69%, e-value = 1.E−04; accD2: 67%, e-value = 0.8; accD3: 78%, e-value = 3.E−119; supplementary table S8, Supplementary Material online). Annotations carried out in DOGMA matched only two of the three accD fragments (accD2 and accD3). ORF searches that considered all possible frames recovered all three regions as putative protein-coding regions (fig. 3), with larger ORFs (611–3,116 bp) starting between accD-1 and the first repeat-rich segment and ending with the 3′end of accD-3. In the subgenera Pelargonium and Magnipetala, we found short ORFs (173–443 bp) within accD-1, which match codons 50–88 of accD. In P. tetragonum, an ORF of 821 bp spanned from accD-1 to the region between accD-2 and accD-3. Large ORFs were also found on different frames for different species of the same subgenera. Because of these uncertainties and in the presence of lacking gene expression data, we prefer to refer to accD1-3 as accD-like region hereafter.
Fig. 3.—
AccD-like regions. Based on 22 Pelargonium species from three subgenera, the relative position of accD-like sequences, repeat-rich regions, and gaps are color-coded as detailed in the inset, genic regions are shown in gray blocks. The second (smaller) bar for subgenus Magnipetala represents the sequences of Pelargonium mollicomum and P. worcesterae. Bold lines indicate accD annotations obtained with DOGMA. Variation of selected ORFs within the accD-like region for subgenus (I) Pelargonium, (II) Parvulipetala, and (III) Magnipetala. F denotes frames 1–3, followed by the ORF number. Ranges on one line denote discontinuous ORFs, that is, ORFS interrupted by internal stop codons.
Discussion
The Evolution of Four Distinct Plastome Types in Pelargonium
Based on a genus-wide analysis of the mutational hotspots associated with massive plastome reconfiguration, we identified four distinct types (I, II, III, and IV) of plastid genomes in Pelargonium, and inferred a series of likely changes that lead to these extraordinary plastomes (fig. 2). Type I and II plastomes exist in 46 species of the 60 studied species, implying that, considering current species counts (e.g., Röschenbleck et al. 2014), 90% of all Pelargonium have a rather conservative plastome structure. Plastome types III and IV, which are characterized by several rearrangements, are confined to the subgenus Paucisignata, which comprises only 10% of extant Pelargonium species.
Similar to the gene arrangement of the completely sequenced P. alternans plastome (Weng et al. 2014), the gene order of the ancestral plastid genome (type I) of the genus Pelargonium is nearly unaltered relative to Melianthus, except for one autapomorphic inversion between trnEUUC and rps4 (fig. 2B ). We inferred this inversion for all subgenera based on both our gene scoring approaches. This inversion also exists in the P. x hortorum plastome, where it was explained earlier by a series of inversion events (Chumley et al. 2006). Although this region appears to be a general inversion hotspot in Geraniaceae showing differences both between and within genera (Weng et al. 2014), the co-localization of trnEUUC–trnGGCC–ycf3 and psbZ–trnSGAA–rps4 at both endpoints is unique for the genus Pelargonium (fig. 2B ) (Blazier et al. 2011; Guisinger et al. 2011; but see Weng et al. 2014). Contrary to P. x hortorum, gene duplication or pseudogenes were not reported in P. alternans outside the IR junction (Weng et al. 2014). Marginal differences concerning the IR boundaries are typical in plastomes with an otherwise unaltered genome structure (e.g., Camellia—Huang et al. 2014; Eucalyptus—Bayly et al. 2013). Therefore, we would expect only similar and less pronounced modifications of the IR boundaries in the type I-plastome of subgenera Pelargonium and Magnipetala. The conformity of gene clusters with Melianthus leads us to expect that possible variations in the gene order of those type I plastomes are restricted to the SSC region.
Plastomes of type II differ from type I by an inversion of the trnNGUU–ndhF region, which we found to be synapomorphic for subgenus Parvulipetala (fig. 2A and B ). Unlike in the subgenera Pelargonium and Magnipetala, the inverted ndhF–trnNGUU–rpl32 region shows little length variation and low nucleotide substitution rates in Parvulipetala (supplementary fig. S1, Supplementary Material online). The lower within-subgenus divergence in Parvulipetala perhaps could relate to its younger crown group age compared with other subgenera (subgenus Pelargonium: end of Miocene/beginning of Pliocene; Parvulipetala: end of Pliocene, Sytsma et al. 2014). A similar uniformity in genetic traits is also present in the rearranged rpl20–rpl32 spacer of group C of subgenus Paucisignata. The crown group age of group C was estimated to be 10 million years younger than that of group S (Bakker et al. 2005), which shows greater variation in both the length and the substitution rates (fig. 3; supplementary figs. S1 and S2, Supplementary Material online). However, formal statistical testing of the association between age and divergence remains to be performed in a subsequent study with denser sampling and thorough molecular dating analyses.
Type III plastomes apparently are collinear with the plastome of P. x hortorum (Chumley et al. 2006), provided no further rearrangements occur in regions that were not included in our PCR screening. A change of both the order and orientation of psaJ and psaI in two regions is the likely result of a re-inversion within the IR. This re-inversion distinguishes type IV from type III plastomes, and, thus, group S of subgenus Paucisignata from the rest (fig. 2A and B ). Based on our validation of the co-localization of rbcL with trnICAU and psbA at the IR boundaries (fig. 2A ), we may conclude that both type III and IV plastomes are similar to P. x hortorum in terms of size. Our inferred rearrangement history shows that the type III arrangement is ancestral in Paucisignata. In consequence, the re-inversion in group S must have happened later during the evolution of the subgenus. Relationships within group S are not fully resolved, but plastome structural features corroborate the so far weakly supported monophyly of the group (Jones et al. 2009; Weng et al. 2012; Röschenbleck et al. 2014).
It is important to note that our conclusion regarding the various plastome types are based on the reconstruction of ancestral gene orders using both a nine-gene dataset that reflects observed changes in gene organization seen in our PCR screen and a “simulated” matrix of 111 genes, which included our observations as well as those published earlier on the basis of complete plastome sequencing (Chumley et al. 2006; Blazier et al. 2011; Guisinger et al. 2011; Weng et al. 2014). The full gene matrix makes the (silent) assumption that no other reconfigurations occurred within clades, which might in fact underestimate the true diversity of plastome structures. The presented work thus must be seen as a starting point for future studies on the diversity of plastome genotypes in the genus Pelargonium.
Gene Duplications in the Extensively Rearranged Plastome Types III and IV
Direct or dispersed repeats found in the type III and type IV plastomes of Pelargonium (fig. 1) are typical features of highly rearranged plastid genomes (Chumley et al. 2006; Cai et al. 2008; Haberle et al. 2008; Knox 2014), potentially indicating a coevolution with nuclear-encoded genes for DNA replication, recombination, and repair (Zhang et al. 2016). In Geraniaceae, duplications of the same coding sequences are reported to occur independently in different genera (Chumley et al. 2006; Blazier et al. 2011; Guisinger et al. 2011), where they can be explained by repeated expansions and contractions of the IRs (Chumley et al. 2006; Guisinger et al. 2011). Insertions in the LSC of P. x hortorum likely originated by duplications of genes prior to these inversion events (Chumley et al. 2006), or by duplicative transposition (as in Oleaceae—Lee et al. 2007). In subgenus Paucisignata, the synapomorphic duplications of rps18 and rpl32 within the IR might also be the results of a combination of inversion events and changes within the dimensions of the IR.
Our dense taxon sampling revealed an even greater variation of gene duplications around the same inversion endpoints across several species of Paucisignata, irrespective of their plastome types (fig. 1). Those substantial differences within a single intergenic region are rare and often coincide with plastid gene loss as in Jasminum (Oleaceae—Lee et al. 2007) or parasitic Phelipanche species (Orobanchaceae—Wicke et al. 2013). The mechanisms of these duplications are as yet unknown, but it probably relates to errors during recombination-dependent replication and repair (Guisinger et al. 2008; Gray et al. 2009; Maréchal and Brisson 2010). The detected high variation of duplicated coding elements in species of Paucisignata indicates that DNA repair near breakpoints is mediated randomly. Such repeated and uncorrelated duplications of often the same genes are frequently observed in Geraniaceae (e.g., 3× trnfMCAU, 4× rpl33, and 2× rps14 in P. x hortorum, 8× rrn16 in Geranium, 3× trnfMCAU in Erodium).
Correlated Evolution of Molecular Evolutionary Rates and Genetic Traits
Rearranged plastome regions in the genus Pelargonium have higher rates of nucleotide substitutions and accumulate more indels and the repeats than conserved regions (supplementary fig. S1, Supplementary Material online). Genome reconfigurations are particularly strongly associated with higher rates of nucleotide substitutions and larger repeats in Geraniaceae (Weng et al. 2014). We also found correlations between substitution rates and occurrence of repeats, implying a relationship between primary DNA sequence evolution and genome structure. These correlations are in line with the general trend of plastid genome evolution across angiosperms (Jansen et al. 2007). One of the investigated spacers (rbcL–psaI), however, deviates from this general pattern, in that only the expanded accD-like region shows an elevated indel and repeat rate. This noncanonical behavior may be due to a change of selectional regimes and the pseudogenization of accD (see below).
With a GC content of 39 and 41% over the entire plastome or coding regions, respectively, Geraniaceae have a slightly higher GC content than other rosids with standard plastid genomes (35 and 38%— Weng et al. 2014). Although the GC content in rearranged regions of the taxa studied here resembles that of P. x hortorum, we detected a lower proportion of GC in the unaltered rps18–rpl20 and ndhF–rpl32 regions (supplementary fig. S1, Supplementary Material online), which both are also rare in indels. This is in contrast to observations in other plant lineages, and in particular differs from the evolution of plastomes in heterotrophic plants (Wicke et al. 2013, 2014), in which indels accumulate predominantly in GC poor regions. These findings point towards a potential mechanistic difference between DNA-substitution-rate/indel-rate associations in plastomes of Geraniaceae and plastomes that evolve under relaxed selective constraints and experience extensive physical and functional reductions.
We would like to stress that the conclusion base on results from correlation analyses that were based on the alignments of fast-evolving and, in part, repeat-rich noncoding DNA. Although we have inspected all data sets visually following automated alignment, we cannot exclude erroneous alignments and we did not conduct the correlation analyses on alternative alignment hypotheses. Therefore, the results on the molecular evolutionary trends within the genus Pelargonium should be viewed with some caution.
Evolution of Plastid accD in Pelargonium
Our screening reveals that the plastid accD gene is functionally lost in all taxa of the subgenus Paucisignata (figs. 1 and 2). Across all infrageneric lineages, the molecular evolution of the plastid accD region indicates neutral evolutionary processes, likely as a consequence of relaxed selective constraints on the plastid copy. The loss of accD from the plastomes of Geraniaceae was also reported for a few Erodium species, in M . speciosa, Hypseocharis bilobata, and P. x hortorum (Chumley et al. 2006; Blazier et al. 2011; Guisinger et al. 2011; Weng et al. 2014), and highly divergent accD genes were found in P. alternans and the genus Viviana of Geraniales (Weng et al. 2014). The accD gene encodes the β-subunit of the Acetyl–CoA carboxylase that catalyzes the irreversible conversion of acetyl-CoA to malonyl-CoA during fatty acid synthesis. This enzyme is essential in photosynthetic plants (Kode et al. 2005). The gene, therefore, is present in the plastome in most land plants (Wicke et al. 2011; but see Jansen et al. 2007), including in the majority of nonphotosynthetic species. These multiple independent losses of the plastid accD gene within Geraniaceae imply that the pseudogenization of plastid accD already started in a common ancestor of the family. It remains to be clarified whether this gene loss truly is a functional loss or if it is the result of an ancient functional transfer to the nuclear genome as seen in some other flowering plants (Jansen and Ruhlman 2012; Rousseau-Gueutin et al. 2013). The differing degrees of plastomic accD fragmentation in the single Pelargonium clades (fig. 3) resemble observations in lineages of Poales (Harris et al. 2013), Oleaceae (Lee et al. 2007), Campanulaceae sensu lato (Knox 2014), Jasmineae (Lee et al. 2007), Oenothera (Greiner et al. 2008), and some Fabaceae (Guo et al. 2007; Magee et al. 2010; Gurdon and Maliga 2014; Sveinsson and Cronk 2014). A functional transfer of accD to the nuclear genome is experimentally validated within two of these lineages (Trifolium repens, Fabaceae—Magee et al. 2010; Trachelium caeruleum, Campanulaceae—Rousseau-Gueutin et al. 2013). Length variations of accD in Fabaceae coincide with accelerated substitution rates (Guo et al. 2007; Magee et al. 2010), resembling the molecular evolutionary patterns of the rbcL–psaI region in Pelargonium.
Conclusions
By combining data from complete plastid genomes and a deep screening of mutational hotspots for 60 representatives, we here show the existence of at least four distinct plastome types in the genus Pelargonium and a series of plastome reconfigurations leading to those. Our data illustrate the complexity of rearrangement events and species-specific variation across the various plastome types coexisting within a single genus. This study thus contributes to our understanding of the evolution of plastid genomes in flowering plants in general and Geraniaceae in particular. Our study builds an essential starting point for future studies investigating the underlying mechanisms and trajectories of the remarkable plastome evolution in Pelargonium.
In angiosperms, the phylogenetic distribution of 206 genera that show evidence of biparental plastid inheritance is restricted to derived lineages (Hu et al. 2008), several of which exhibit some structural rearrangements in their plastomes (Jansen and Ruhlman 2012). Although it remains unclear how exactly the mode of transmission and the plastid genome itself contributes to destabilizing the standard plastome organization, a relationship between both was speculated about in multiple cases. For example, the genus Oenothera possesses structurally dynamic plastid genomes (Hupfer et al. 2000; Greiner et al. 2008). Distinguishable transmission patterns between the distinct plastome genotypes indicate cytoplasmic incompatibility on the one hand but also occasional recombination between genetically and structurally distinct plastome copies (Chiu and Sears 1985, 1993; Chiu et al. 1988), rather unseen in an uniparental inheritance. Biparental inheritance also has been experimentally shown several times within the genus Pelargonium (Metzlaff et al. 1981; Weihe et al. 2009). However, all of the species or hybrids used in these studies belong to group C of subgenus Paucisignata. Interestingly, the strongly reconfigured plastomes of subgenus Paucisignata shown here all derive from a more conserved plastome type, ancestral to the genus. The genus Pelargonium with it well-defined plastome genotypes, thus, could represent an important model for investigating the association of the mode of plastid transmission with the occurrence of plastid genomic reconfigurations and non-canonical plastome evolution in general.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
The authors would like to thank O. Lepping and H. Schwitte for excellent technical support. Many thanks are also due to the curators and staff of the Botanical Gardens of the Universities of Muenster and Dresden. This research was financed intramurally through endowment funds of the University of Muenster (to K.F.M, J. K.).
Literature Cited
- Albers F, Becker M. 2010. Phylogeny and speciation in succulent Geraniaceae (Geraniales). Schumannia 6:59–67. [Google Scholar]
- Bakker FT, Culham A, de Marais AB, Gibby M. 2005. Nested radiation in Cape Pelargonium In: Bakker FT, Chatrou LW, editors. Plant species—level systematics: new perspectives on pattern and process. Oberreifenberg: Regnum Vegetabile Koeltz Scientific Books; p. 75–100. [Google Scholar]
- Bayly MJ, et al. 2013. Chloroplast genome analysis of Australian eucalypts-Eucalyptus, Corymbia, Angophora, Allosyncarpia and Stockwellia (Myrtaceae). Mol Phylogenet Evol. 69:704–716. [DOI] [PubMed] [Google Scholar]
- Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27:573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bikandi J, San Millán R, Rementeria A, Garaizar J. 2004. In silico analysis of complete bacterial genomes: PCR, AFLP-PCR and endonuclease restriction. Bioinforma Oxf Engl. 20:798–799. [DOI] [PubMed] [Google Scholar]
- Blazier JC, Guisinger-Bellian MM, Jansen RK. 2011. Recent loss of plastid-encoded ndh genes within Erodium (Geraniaceae). Plant Mol Biol. 76:1–10. [DOI] [PubMed] [Google Scholar]
- Bourque G, Pevzner PA. 2002. Genome-scale evolution: reconstructing gene orders in the ancestral species. Genome Res. 12:26–36. [PMC free article] [PubMed] [Google Scholar]
- Cai Z, et al. 2008. Extensive reorganization of the plastid genome of Trifolium subterraneum (Fabaceae) is associated with numerous repeated sequences and novel DNA insertions. J Mol Evol. 67:696–704. [DOI] [PubMed] [Google Scholar]
- Chiu WL, Sears BB. 1985. Recombination between chloroplast DNAs does not occur in sexual crosses of Oenothera . Mol Gen Genet MGG. 198:525–528. [DOI] [PubMed] [Google Scholar]
- Chiu WL, Sears BB. 1993. Plastome-genome interactions affect plastid transmission in Oenothera . Genetics 133:989–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu WL, Stubbe W, Sears BB. 1988. Plastid inheritance in Oenothera: organelle genome modifies the extent of biparental plastid transmission. Curr Genet. 13:181–189. [Google Scholar]
- Chumley TW, et al. 2006. The complete chloroplast genome sequence of Pelargonium x hortorum: organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Mol Biol Evol. 23:2175–2190. [DOI] [PubMed] [Google Scholar]
- Cosner ME, Raubeson LA, Jansen RK. 2004. Chloroplast DNA rearrangements in Campanulaceae: phylogenetic utility of highly rearranged genomes. BMC Evol Biol. 4:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray B, Ahner B, Hanson M. 2009. Extensive homologous recombination between introduced and native regulatory plastid DNA elements in transplastomic plants. Transgenic Res. 18:559–572. [DOI] [PubMed] [Google Scholar]
- Greiner S, et al. 2008. The complete nucleotide sequences of the five genetically distinct plastid genomes of Oenothera, subsection Oenothera: I. Sequence evaluation and plastome evolution. Nucleic Acids Res. 36:2366–2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guisinger MM, Kuehl JV, Boore JL, Jansen RK. 2008. Genome-wide analyses of Geraniaceae plastid DNA reveal unprecedented patterns of increased nucleotide substitutions. Proc Natl Acad Sci U S A. 105:18424–18429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guisinger MM, Kuehl JV, Boore JL, Jansen RK. 2011. Extreme reconfiguration of plastid genomes in the angiosperm family Geraniaceae: rearrangements, repeats, and codon usage. Mol Biol Evol. 28:583–600. [DOI] [PubMed] [Google Scholar]
- Guo X, et al. 2007. Rapid evolutionary change of common bean (Phaseolus vulgaris L.) plastome, and the genomic diversification of legume chloroplasts. BMC Genomics 8:228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurdon C, Maliga P. 2014. Two distinct plastid genome configurations and unprecedented intraspecies length variation in the accD coding region in Medicago truncatula . DNA Res Int J Rapid Publ Rep Genes Genomes 21:417–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberle RC, Fourcade HM, Boore JL, Jansen RK. 2008. Extensive rearrangements in the chloroplast genome of Trachelium caeruleum are associated with repeats and tRNA genes. J Mol Evol. 66:350–361. [DOI] [PubMed] [Google Scholar]
- Harris ME, Meyer G, Vandergon T, Vandergon VO. 2013. Loss of the acetyl-CoA carboxylase (accD) gene in Poales. Plant Mol Biol Rep. 31:21–31. [Google Scholar]
- Huang H, Shi C, Liu Y, Mao S-Y, Gao L-Z. 2014. Thirteen Camellia chloroplast genome sequences determined by high-throughput sequencing: genome structure and phylogenetic relationships. BMC Evol Biol. 14:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hupfer H, et al. 2000. Complete nucleotide sequence of the Oenothera elata plastid chromosome, representing plastome I of the five distinguishable euoenothera plastomes. Mol Gen Genet. 263:581–585. [DOI] [PubMed] [Google Scholar]
- Hu Y, Zhang Q, Rao G., Sodmergen 2008. Occurrence of plastids in the sperm cells of Caprifoliaceae: biparental plastid inheritance in angiosperms is unilaterally derived from maternal inheritance. Plant Cell Physiol. 49:958–968. [DOI] [PubMed] [Google Scholar]
- Jansen RK, et al. 2007. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci U S A. 104:19369–19374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RK, Palmer JD. 1987. A chloroplast DNA inversion marks an ancient evolutionary split in the sunflower family (Asteraceae). Proc Natl Acad Sci U S A. 84:5818–5822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RK, Ruhlman TA. 2012. Plastid genomes of seed plants In: Bock R, Knoop V, editors. Genomics of chloroplasts and mitochondria, Vol. 35. Dordrecht: Springer; p. 103–126. [Google Scholar]
- Jones CS, Bakker FT, Schlichting CD, Nicotra AB. 2009. Leaf shape evolution in the South African genus Pelargonium L’Her. (Geraniaceae). Evol Int J Org Evol. 63:479–497. [DOI] [PubMed] [Google Scholar]
- Kim K-J, Choi K-S, Jansen RK. 2005. Two chloroplast DNA inversions originated simultaneously during the early evolution of the sunflower family (Asteraceae). Mol Biol Evol. 22:1783–1792. [DOI] [PubMed] [Google Scholar]
- Knox EB. 2014. The dynamic history of plastid genomes in the Campanulaceae sensu lato is unique among angiosperms. Proc Natl Acad Sci U S A. 111:11097–11102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kode V, Mudd EA, Iamtham S, Day A. 2005. The tobacco plastid accD gene is essential and is required for leaf development. Plant J. 44:237–244. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Frost SDW, Muse SV. 2005. HyPhy: hypothesis testing using phylogenies. Bioinformatics 21:676–679. [DOI] [PubMed] [Google Scholar]
- Lee H-L, Jansen RK, Chumley TW, Kim K-J. 2007. Gene relocations within chloroplast genomes of Jasminum and Menodora (Oleaceae) are due to multiple, overlapping inversions. Mol Biol Evol. 24:1161–1180. [DOI] [PubMed] [Google Scholar]
- Löytynoja A, Goldman N. 2005. An algorithm for progressive multiple alignment of sequences with insertions. Proc Natl Acad Sci U S A. 102:10557–10562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee AM, et al. 2010. Localized hypermutation and associated gene losses in legume chloroplast genomes. Genome Res. 20:1700–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maréchal A, Brisson N. 2010. Recombination and the maintenance of plant organelle genome stability. New Phytol. 186:299–317. [DOI] [PubMed] [Google Scholar]
- Metzlaff M, Börner T, Hagemann R. 1981. Variations of chloroplast DNAs in the genus Pelargonium and their biparental inheritance. Theor Appl Genet. 60:37–41. [DOI] [PubMed] [Google Scholar]
- Müller KF. 2005. SeqState: primer design and sequence statistics for phylogenetic DNA datasets. Appl Bioinfo. 4:65–69. [DOI] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinforma Oxf Engl, 20:289–290. [DOI] [PubMed] [Google Scholar]
- Röschenbleck J. 2015. Die Gattung Pelargonium (Geraniaceae): Taxonomie, Biogeography und Plastomevolution. Ph.D. Thesis, Westfälische Wilhelms-Universität Münster: Münster, Germany.
- Röschenbleck J, Albers F, Müller KF, Weinl S, Kudla J. 2014. Phylogenetics, character evolution and a subgeneric revision of the genus Pelargonium (Geraniaceae). Phytotaxa 159:31–76. [Google Scholar]
- Rousseau-Gueutin M, et al. 2013. Potential functional replacement of the plastidic accD gene by recent transfers to the nucleus in some angiosperm lineages. Plant Physiol. 16:1918–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schliep KP. 2011. phangorn: phylogenetic analysis in R. Bioinforma Oxf Engl. 27:592–593. [21169378] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons MP, Ochoterena H. 2000. Gaps as characters in sequence-based phylogenetic analyses. Syst Biol. 49:369–381. [PubMed] [Google Scholar]
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. [DOI] [PubMed] [Google Scholar]
- Stothard P. 2000. The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. BioTechniques 28:1102–1104. [DOI] [PubMed] [Google Scholar]
- Sveinsson S, Cronk Q. 2014. Evolutionary origin of highly repetitive plastid genomes within the clover genus (Trifolium). BMC Evol Biol. 14:228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sytsma KJ, Spalink D, Berger B. 2014. Calibrated chronograms, fossils, outgroup relationships, and root priors: re-examining the historical biogeography of Geraniales: re-examining historical biogeography of Geraniales. Biol J Linn Soc. 113:29–49. [Google Scholar]
- Temnykh S, et al. 2001. Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential. Genome Res. 11:1441–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weihe A, Apitz J, Pohlheim F, Salinas-Hartwig A, Börner T. 2009. Biparental inheritance of plastidial and mitochondrial DNA and hybrid variegation in Pelargonium . Mol Genet Genomics MGG 282:587–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng M-L, Blazier JC, Govindu M, Jansen RK. 2014. Reconstruction of the ancestral plastid genome in Geraniaceae reveals a correlation between genome rearrangements, repeats, and nucleotide substitution rates. Mol Biol Evol. 31:645–659. [DOI] [PubMed] [Google Scholar]
- Weng M-L, Ruhlman TA, Gibby M, Jansen RK. 2012. Phylogeny, rate variation, and genome size evolution of Pelargonium (Geraniaceae). Mol Phylogenet Evol. 64:654–670. [DOI] [PubMed] [Google Scholar]
- Wicke S, et al. 2013. Mechanisms of functional and physical genome reduction in photosynthetic and non-photosynthetic parasitic plants of the broomrape family. Plant Cell 25:3711–3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicke S, et al. 2016. Mechanistic model of evolutionary rate variation en route to a nonphotosynthetic lifestyle in plants. Proc Natl Acad Sci U S A. 113:9045–9050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicke S, Schäferhoff B, dePamphilis CW, Müller KF. 2014. Disproportional plastome-wide increase of substitution rates and relaxed purifying selection in genes of carnivorous Lentibulariaceae. Mol Biol Evol. 31:529–545. [DOI] [PubMed] [Google Scholar]
- Wicke S, Schneeweiss GM, de Pamphilis CW, Müller KF, Quandt D. 2011. The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol Biol. 76:273–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman SK, Boore JL, Jansen RK. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20:3252–3255. [DOI] [PubMed] [Google Scholar]
- Zhang J, et al. 2016. Coevolution between nuclear-encoded DNA replication, recombination, and repair genes and plastid genome complexity. Genome Biol Evol. 8:622–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.