Abstract
Short tandem repeats (STRs), or microsatellites, are genetic variants with repetitive 2–6 base pair motifs in many mammalian genomes. Using high-throughput sequencing and experimental validations, we systematically profiled STRs in five Holsteins. We identified a total of 60,106 microsatellites and generated the first high-resolution STR map, representing a substantial pool of polymorphism in dairy cattle. We observed significant STRs overlap with functional genes and quantitative trait loci (QTL). We performed evolutionary and population genetic analyses using over 20,000 common dinucleotide STRs. Besides corroborating the well-established positive correlation between allele size and variance in allele size, these analyses also identified dozens of outlier STRs based on two anomalous relationships that counter expected characteristics of neutral evolution. And one STR locus overlaps with a significant region of a summary statistic designed to detect STR-related selection. Additionally, our results showed that only 57.1% of STRs located within SNP-based linkage disequilibrium (LD) blocks whereas the other 42.9% were out of blocks. Therefore, a substantial number of STRs are not tagged by SNPs in the cattle genome, likely due to STR's distinct mutation mechanism and elevated polymorphism. This study provides the foundation for future STR-based studies of cattle genome evolution and selection.
Keywords: cattle genome, short tandem repeat, whole genome sequencing
Introduction
Short tandem repeats (STR) are highly variable genetic elements widely dispersed in mammalian genomes. Here, we focus on STRs with repetitive motifs of 2–6 base pairs, which are commonly referred to as microsatellites. The elevated level of polymorphism and mutability of STRs, due to the high incidence of replication slippage, has resulted in their application to the analysis of population differentiation, genetic diversity, and forensic identification (MacHugh et al. 1997; Chikhi et al. 2004; Li et al. 2007; Chambers et al. 2014).
An appreciable but unknown fraction of STRs contribute to gene regulation and have functional effects (Gemayel et al. 2010). Changes to the repeat length of STRs have been associated with gene function (Borel et al. 2012), transcriptional plasticity (Vinces et al. 2009), complex traits (Hammock and Young 2005; Queitsch et al. 2012; Gymrek et al. 2016), and morphological evolution (Wren et al. 2000; Fondon and Garner 2004). The potential for adaptive and deleterious STR mutation is considerable. For example, in human ∼17% of genes contain STRs in their open reading frames. Several human genetics disorders, including Huntington disease and Fragile X syndrome (Pearson et al. 2005), are caused by STR expansions.
In contrast to calling methods for single nucleotide polymorphisms (SNPs), insertions, deletions (indels) and copy number variations (CNVs), STRs are substantially more difficult to detect based on short reads produced by next generation sequencing (NGS); however, numerous methods have been recently developed to identify STR variants in human (Gymrek et al. 2012; Tae et al. 2013; Highnam et al. 2013; Anvar et al. 2014; Cao et al. 2014; Fungtammasan et al. 2015; Carlson et al. 2015). These programs enable STR detection in NGS data by, among other features, carefully adjusting for high mismatch/indel levels in STRs during the mapping step (lobSTR) and guiding genotyping of STRs using informed error profiles (RepeatSeq) (Gymrek et al. 2012; Highnam et al. 2013). Targeted STR region amplification, using capture array (Guilmatre et al. 2013) or single-molecule Molecular Inversion Probes (MIPSTR) (Carlson et al. 2015), followed by NGS has also been applied to identify human STRs. More recently, a flexible pipeline, STR-FM, was developed, which incorporates an error correction model into STR detection (Fungtammasan et al. 2015). We used the program lobSTR in our analysis, which has been used to reliably identify STRs in human (Gymrek et al. 2012), recover the surnames of individuals associated with putatively anonymous human genomes via profiling of STRs on the Y chromosome (Y-STRs) (Gymrek et al. 2013) and characterize the variation of nearly 700,000 STR loci across more than 1000 individuals in Phase 1 of the 1000 Genomes Project (Willems et al. 2014).
Numerous studies in cattle have generated several STR-based genetic maps using hundreds to thousands of STR markers at various resolutions (Stone et al. 1995; Barendse et al. 1997; Kappes et al. 1997; Ihara et al. 2004). Indeed, STR has become an essential marker for mapping quantitative trait loci (QTL) due to their high variability and easy amplification by PCR (Georges et al. 1993; Lipkin et al. 1998; Van Tassell et al. 2000; Ashwell et al. 2001; Schnabel et al. 2005). Furthermore, the exploration of the relationship between STRs with nearby SNPs can further help explain the results from SNP-based genome wide association studies (Brahmachary et al. 2014). Although previous studies have reported that most STRs can be tagged by SNPs (McClure et al. 2012, 2013), these conclusions were mainly based on a rather limited number of known microsatellites. Because it has been impossible to produce genome-wide profiles of STR variations, the majority of STRs in the cattle genome remain undetected and unexplored. Thus their population genetics and functional impacts on the cattle genome are poorly defined. The recent advent of next generation sequencing (NGS) has created a wealth of genomic data, offering an opportunity for profiling large numbers of STRs across the whole genome. We profiled the most comprehensive spectrum of STRs to date in the cattle genome, through the application of lobSTR to high coverage short-read sequencing data generated from five influential Holstein bulls.
In this study, we report the first large-scale STR study in livestock based on whole genome deep sequencing and generate a novel resource for the current research community. Furthermore, we perform an initial investigation of the population genetics and functional impacts of bovine STRs, and provide some new insights for further exploration of cattle genome.
Materials and Methods
Samples and Sequencing
The whole genome sequencing data was generated as previously described (Larkin et al. 2012). Five Holstein bulls (Bos taurus) were sequenced using the Illumina V3 PE 100 chemistry on a HiSeq 2000 platform to 30–50× coverage. The raw reads were first filtered using NGS QC toolkit to remove low-quality bases with quality score of 20 (−s 20) and the percentage of read length less than 75% of given quality (−l 75) (Patel and Jain, 2012). A summary of short read statistics was presented in supplementary table S1, Supplementary Material online.
Identification of STR in Cattle
We conducted a comprehensive survey of STR variants using high coverage NGS data in dairy cattle. The coverage of sequence data for each animal was around 30∼50× (supplementary table S1, Supplementary Material online), allowing sufficient power to detect STRs using lobSTR (version 2.0) (Gymrek et al. 2012). lobSTR was applied with default parameters for alignment and STR discovery as previously described (Gymrek et al. 2012). Briefly, we first created a lobSTR reference index based on Bovine UMD 3.1's STR data (retrieved from the UCSC genome browser) using lobstr_index.py script. Then we carried out lobSTR alignment to create the STR alignment bam files, and the final STR variants allelotypes were identified throughout all samples based on the merged alignment file. lobSTR employs an explicit model to enhance accuracy by avoiding stutter noise caused by PCR amplification of a STR locus. After filtering by quality of STR calling (QUAL > 30), we finally obtained 60,106 unique STRs across all analyzed animals.
STR-Overlapping Genes Annotation Using PANTHER and DAVID
The 60,106 STRs loci were intersected with RefSeq genes downloaded from the UCSC Genome Browser to obtain a total of 11,676 overlapped STRs. Based on 4,213 unique STR-containing genes, we tested the hypothesis that the PANTHER molecular function, biological process and pathway terms were under- or over-represented in genes regions after Bonferroni corrections (Mi et al. 2013). We also performed gene enrichment annotation and gene functional classification for these genes using the online tool DAVID (version 6.7) [9]. GO terms involved in molecular function, biological process and cellular component were selected as the functional annotation category in our studies. To explore the distribution of STR count in genes, we divided the total 4,213 genes into four groups (group 1 with more than 10 STRs, group 2 with 5–10 STRs, group 3 with 2–4 STRs, and group 4 with one STR), and performed enrichment tests on these groups separately using DAVID. To further explore the contributions of STRs involved in gene function, we overlapped the STRs with exon regions of RefSeq genes. We found 204 STRs were embedded in exon regions of 194 genes.
Validating lobSTR Accuracy Using Sanger Sequencing
To confirm lobSTR prediction, we randomly selected 18 STRs of different motif sizes (including di-, tri-, and tetranucleotides) and used Sanger sequencing to confirm whether the correct genotypes were derived after PCR and/or TA cloning. Primer information can be found in Table S9. DNA fragments obtained from five animals (Elevation, Blackstar, Starbuck, Chairman, and Ivanhoe) were sequenced by Sanger chemistry at Genewiz, Inc. according to standard procedures.
STRs Overlap with QTLs Associated with Important Traits
We downloaded QTL information from cattle QTLdb from http://www.animalgenome.org/cgi-bin/QTLdb/BT/index (last access November 2, 2016). Because previous QTL mapping studies have utilized both STR and SNP markers, and employed different design populations and mapping methods, we merged all QTLs into a set of unique non-redundant regions.
SNP-Based LD Block Estimation around STR
LD (R2 proxy for LD) was calculated and LD blocks were detected using Bovine HD SNPs datasets in Holstein population using PLINK v1.07 (http://pngu.mgh.harvard.edu/purcell/plink/; last access November 2, 2016) (Purcell et al. 2007). The SNPs within each block were overlapped with STRs. The STR overlapping with block region was considered as the potential candidate STRs which could be further tagged by proximate SNPs.
Selection and Population Genetic Analysis of STRs
A total of genotypes of 22,067 STRs were recovered in all five cattle, among which 20,059 (90.9%) were dinucleotides. Given their abundance, we restricted our analysis to this set of 20,059 dinucleotides STRs. We identified outlier loci based on two anomalous relationships that counter expected trends in microsatellite variation: (1) 100 invariant microsatellites with allele size ≥ 20 (supplementary table S10, Supplementary Material online) and (2) 44 dinucleotide loci that are diallelic with a maximum allele size at least two time greater than the size of the alternate allele (supplementary table S11, Supplementary Material online).
Principal Component Analysis of Genetic Relatedness
We used microsatellite calls for all dinculeotide loci where data were available for all five cattle and maximum allele size was ≤30 (n = 19,338). We carried out principal components analysis (PCA) using EIGENSOFT (Patterson et al. 2006) and each microsatellite allele was coded as a binary string of all zeroes except for the position corresponding to the size of allele, which was set to one.
STRs Overlap with Regions under Selection Predicted Based on SNPs
To identify STRs linked to potential regions under positive selection, we overlapped STRs with the 49 positive selection regions identified by the same Holstein sequencing samples as previously reported (Larkin et al. 2012). In addition, to further study the potential genome signals involved in selection for STRs in Holstein population, we utilized the integrated haplotype score (iHS) using Bovine HD SNP array in 44 unrelated Holstein, iHS was estimated using selscan program with default settings (Szpiech and Hernandez 2014). In this study, we considered 100-kb nonoverlapping windows; the density of signal in each region was measured by the proportion of SNPs with |iHS| > 2. Then the empirical cutoffs for the top 1% of signals were considered as candidate selection regions (Regions with SNP number <10 were dropped here) (Voight et al. 2006). To assess significance for the analyses of regions, we performed 10,000 times permutation tests to get empirical P-values for these overlaps using an R/Bioconductor package regioneR (Gel et al. 2016). To further explore the selection characteristics of STRs, we employed the recently developed ksk method; values were estimated with window size 100,000 and step size 5000 as described previously (Haasl et al. 2014).
Results and Discussion
Identification of STR Using Whole Genomics Sequencing
We used five sequenced Holstein bulls with coverage depth between 30 and 50× for STR identification (supplementary table S1, Supplementary Material online). Using lobSTR, we mapped 218,078 aligned reads and identified 37,997 STRs covered with low number of reads (Blackstar), whereas we mapped over 1 million aligned reads and identified 62,378 STRs with high number of reads (Starbuck with an average of 10.4× coverage). In total, we detected 72,615 STRs based on NGS data derived from the five genomes. On average, we obtained 52,186 STRs for a 7.99 × genome coverage (supplementary table S2, Supplementary Material online). After filtering with lobSTR calling quality (QUAL = 30), we obtained a final data set of 60,106 unique STRs with an average length of 36.4 bp, ranging from 25 to 188 bp, covering 21.9 Mb of polymorphic sequence, corresponding to 0.83% of the cattle genome (21.9/2,545.9 Mb). Among these STRs, 5624, 7517, 10,039, 14,859, and 22,067 STRs were identified in one, two, three, four, and five individual(s), respectively.
We observed a relatively uneven distribution of STRs with a maximum interval of 1,087,826 bp. The distribution of STRs shows large differences across chromosomes. For instance, we found 3683, 2926, and 2757 STRs on chromosome 1, 2, and 6, whereas only 931, 1071, and 1079 STRs on chromosome 25, 28, and 23. After normalizing by chromosome length, we observed that STR densities per Mb ranged from 20.54 to 23.26. Among 22,067 STRs found in all five sampled individuals, 20,059 (90.9%) were dinucleotide microsatellites, which were by far the most common, confirming the earlier result based on a small dataset (Stone et al. 1995). Two motifs, (AC)n and (AT)n, occupy a large proportion of our identified STRs, with counts of 35,402 and 14,697, respectively (supplementary fig. S1, Supplementary Material online). STRs with motifs longer than two nucleotides were rare in the cattle genome.
Because sequence and annotation of chrX and chrUn within the cattle genome are less satisfactory, we mainly focused on the high-confidence STRs on autosomes. Because lobSTR only has sufficient power to detect STRs with a motif size from 2 to 6 base pairs, we limited our analyses to these types of STRs on bovine autosomes. STRs with a motif size more than 6 bp were not covered in this study, which may require other detection programs such as VNTRseek (Gelfand et al. 2014).
STRs Overlap with Genes
In this study, we constructed the first STR map for five Holstein genomes. We observed 11,676 STRs overlapped with 4,213 unique annotated cattle RefSeq genes (fig. 1). Among these genes, we observed 188, 84, 45, 12 genes containing at least 10, 15, 20, 30 STRs, corresponding to total STR lengths of 11,7549, 72,981, 49,499, and 19,581 bp, respectively. Genes with over 30 STRs included RBFOX1, MACROD2, GALNTL6, CTNNA2, CA10, NRXN1, PRKG1, DPYD, NEGR1, CDH18, CTNNA3, and NRG3 (supplementary fig. S2, Supplementary Material online).
Fig. 1.—
Genomic landscape of STRs on autosomes in five Holsteins. Tracks from outside to inside are: STR frequencies across five Holsteins; chromosomes in different colors; frequencies of 11,676 STRs overlapped with genes; selected polymorphic genes; STR counts in each of 4,213 genes; Allele size plot for 100 invariant microsatellites with allele size ≥20; Variance plot in allele size for 44 dinucleotide loci that are diallelic with a maximum allele size at least two time greater than the size of the alternate allele.
Using the PANTHER classification system (Mi et al. 2010), STR-containing genes were enriched for the GO terms of synaptic vesicle exocytosis, heart development, visual perception, cellular amino acid metabolic process, and cell–cell adhesion (supplementary table S3, Supplementary Material online). We further divided the total 4,213 genes into four groups: group 1 with more than 10 STRs, group 2 with 5-10 STRs, group 3 with 2-4 STRs, and group 4 with one STR.
For group 1 genes with higher STR count, we observed that most of genes were enriched for heart development, nervous system development, ion transport and cell communication (with Enrichment Score >1.3, i.e., P-values < 0.05 after the Benjamini and Hochberg correction for the multiple testing in supplementary table S4, Supplementary Material online). Similarly, group 2 genes were involved in phosphate-containing compound metabolic process, regulation of catalytic activity, and voltage-gated calcium channel activity (supplementary table S5, Supplementary Material online). Group 3 genes were enriched for visual perception, blood coagulation, catabolic process and cell communication (supplementary table S6, Supplementary Material online), whereas group 4 genes were enriched for vesicle-mediated transport, protein transport, and primary metabolic process (supplementary table S7, Supplementary Material online). To evaluate STR's functional impacts, we further pinpointed 204 STRs located within the exons of 194 genes. DAVID results indicated that these genes are most enriched for neuron differentiation, lipid binding, and membrane-bounded vesicle (supplementary table S8, Supplementary Material online).
As an example, SLC11A1 overlaps with two STRs. This is a highly conserved gene across mammals and is associated with resistance and susceptibility to various intracellular pathogens in humans as well as in livestock species (Blackwell et al. 2001; Thomas and Joseph 2012). Previous studies have shown that microsatellite alleles localized in the 3′ UTR of the SLC11A1 are involved in macrophage function and resistance to Brucella abortus infections in both cattle and buffalo (Kumar et al. 2005; Borriello et al. 2006; Capparelli et al. 2007; Ganguly et al. 2008; Martinez et al. 2008; Kumar et al. 2011). Another gene ATP8B2 contains STR in its exon. This gene is involved in magnesium ion binding and cation-transporting ATPase activity. Notably, ATP8B2 has been specifically identified as a target of positive selection for the aquatic adaptation of dolphins by constructing whole-genome ortholog gene sets among five mammalian species, including dolphin, cow, dog, panda, and human (Sun et al. 2015).
Validating STR Predictions with Sanger Sequencing
To verify the STR detected using lobSTR in our cattle data, we performed PCR and Sanger sequencing. After sequencing these regions, we observed good concordance between lobSTR and the capillary sequencing results (table 1). We found that 16 (45.71%) regions were correctly genotyped by lobSTR and 14 (40%) regions were partly correctly genotyped (table 1). In only five instances, lobSTR called incorrect STR genotypes. We further performed two filters based on coverage and Q score: (1) After applying “DP ≥ 5” and “−LOG(1 − Q) ≥ 0.8” for each locus, 21 left validations showed similar results (42.86% completely correction rate); (2) After applying “DP ≥ 5” for each allele and “−LOG(1 − Q) ≥ 0.8” for each locus, 12 left validations showed a higher (58.33%) completely correction rate. It is noted that our lower validation rates might be related to the draft status of the cattle genome assembly, which is 95% complete and contains many gaps and unplaced contigs.
Table 1.
PCR Sanger Sequencing Results vs. lobSTR Results for Selected STRs
| UMD3.1 | PCR (bp) |
lobSTR (bp) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No | STR | Chr | Begin | End | Motif | (bp) | Animal | A1 | A2 | A1 | A2 | Call | Coverage | Q score |
| 1 | BM1818 | 23 | 39,294,224 | 39,294,249 | GT | 37 | Blackstar | 33 | 37 | 33 | 33 | P | 4 | 0.69 |
| 2 | Elevation | 35 | 37 | 37 | 37 | P | 10 | 2.39 | ||||||
| 3 | Ivanhoe | 33 | 37 | 37 | 37 | P | 2 | 0.30 | ||||||
| 4 | Chairman | 33 | 37 | 33 | 37 | Y | 30 | 0.00 | ||||||
| 5 | BM1824 | 1 | 132,498,006 | 132,498,034 | CA | 29 | Blackstar | 29 | 35 | 33 | 35 | P | 4 | 2.35 |
| 6 | Elevation | 29 | 35 | 27 | 35 | P | 10 | 2.39 | ||||||
| 7 | BM2113 | 2 | 127,591,877 | 127,591,917 | AC | 42 | Elevation | 26 | 48 | 28 | 38 | N | 14 | 5.52 |
| 8 | Blackstar | 28 | 36 | 28 | 40 | P | 6 | 3.88 | ||||||
| 9 | ETH10 | 5 | 56,657,954 | 56,657,996 | CA | 41 | Elevation | 39 | 43 | 37 | 43 | P | 6 | 4.09 |
| 10 | ETH152 | 5 | 114,885,382 | 114,885,416 | TG | 37 | Blackstar | 33 | 35 | 35 | 35 | P | 14 | 4.17 |
| 11 | Elevation | 33 | 35 | 35 | 35 | P | 6 | 1.79 | ||||||
| 12 | ETH225 | 9 | 10,858,165 | 10,858,199 | CA | 34 | Elevation | 32 | 34 | 30 | 32 | N | 8 | 0.69 |
| 13 | Blackstar | 26 | 38 | 38 | 38 | P | 2 | 0.25 | ||||||
| 14 | ETH3 | 19 | 56648417 | 56648461 | AC | 46 | Blackstar | 42 | 52 | 44 | 52 | P | 6 | 6.00 |
| 15 | HAUT27 | 26 | 29,127,336 | 29,127,370 | GT | 39 | Elevation | 29 | 41 | 31 | 31 | N | 2 | 0.37 |
| 16 | ILSTS006 | 7 | 96,709,240 | 96,709,279 | TG | 43 | Elevation | 39 | 43 | 35 | 41 | N | 6 | 4.12 |
| 17 | Blackstar | 39 | 43 | 43 | 43 | P | 2 | 0.37 | ||||||
| 18 | INRA023 | 3 | 33,011,005 | 33,011,044 | CA | 43 | Chairman | 31 | 37 | 31 | 31 | P | 28 | 3.62 |
| 19 | Blackstar | 31 | 35 | 31 | 35 | Y | 6 | 2.82 | ||||||
| 20 | Elevation | 35 | 39 | 35 | 39 | Y | 6 | 2.87 | ||||||
| 21 | Ivanhole | 31 | 39 | 31 | 39 | Y | 18 | 4.06 | ||||||
| 22 | INRA037 | 10 | 76,365,534 | 76,365,555 | TG | 32 | Blackstar | 31 | 38 | 31 | 31 | P | 8 | 1.94 |
| 23 | INRA063 | 18 | 40,699,867 | 40,699,892 | GT | 35 | Blackstar | 27 | 27 | 27 | 27 | Y | 4 | 0.80 |
| 24 | STR_chr8 | 8 | 38,693,251 | 38,693,297 | ACT | 46 | Blackstar | 39 | 39 | 39 | 39 | Y | 4 | 3.11 |
| 25 | Elevation | 39 | 39 | 39 | 39 | Y | 14 | 3.74 | ||||||
| 26 | STR_chr7 | 7 | 2,965,462 | 2,965,506 | ATCC | 44 | Blackstar | 44 | 44 | 44 | 44 | Y | 4 | 1.23 |
| 27 | Elevation | 44 | 48 | 44 | 48 | Y | 14 | 5.70 | ||||||
| 28 | STR_chr10 | 10 | 37,414,396 | 37,414,434 | ATCC | 38 | Blackstar | 38 | 38 | 38 | 38 | Y | 2 | 0.70 |
| 29 | Elevation | 38 | 38 | 38 | 38 | Y | 10 | 3.01 | ||||||
| 30 | STR_chr16 | 16 | 81,503,345 | 81,503,411 | ATCC | 66 | Blackstar | 30 | 66 | 30 | 66 | Y | 4 | big |
| 31 | Elevation | 30 | 30 | 30 | 30 | Y | 14 | 4.21 | ||||||
| 32 | STR_chr19 | 19 | 46,403,668 | 46,403,701 | ATCC | 33 | Blackstar | 33 | 37 | 33 | 37 | Y | 12 | big |
| 33 | Elevation | 33 | 33 | 33 | 33 | Y | 10 | 3.01 | ||||||
| 34 | STR_chr24 | 24 | 34,465,896 | 34,465,926 | AGAT | 30 | Blackstar | 30 | 30 | 30 | 30 | Y | 2 | 0.70 |
| 35 | Elevation | 26 | 26 | 30 | 30 | N | 8 | 2.41 | ||||||
Note.—Y: Both platforms agree. P: lobSTR reported only one allele out of two. N: lobSTR reported an allele that does not exist.
We observed lobSTR correctly called the homozygous STRs that were covered by low read coverage of ≤ 2×. For heterozygous STRs, lobSTR may correctly call one allele and miss the other allele due to insufficient sequencing coverage. We observed for homozygous loci 9/10 (90.00%) were correctly called whereas other heterozygous showed lower correct rates (7/25, 28.00%). Our results also revealed the allelotyping algorithm in lobSTR was not able to identify noisy reads and incorrectly assigned heterozygous genotypes to these loci. We also observed lobSTR made more correct callings for STRs of motif sizes of three and four (tri- and tetra-nucleotides) than dinucleotide STRs, although the dinucleotide STR is the most abundant STR type in cattle genome.
STRs Overlap with QTLs Associated with Important Traits
We found that 47.76% (28,705) of the STRs overlapped with the merged QTL regions (empirical P-value = 0.224). Most of overlapped QTLs were related to important milk and production traits. On the other hand, we also observed 52.24% of STRs did not overlap with any existing QTLs. These may represent some novel STRs, which may be used as new candidate markers to refine cattle QTLs after validation.
Selection and Population Genetic Analyses of Microsatellites
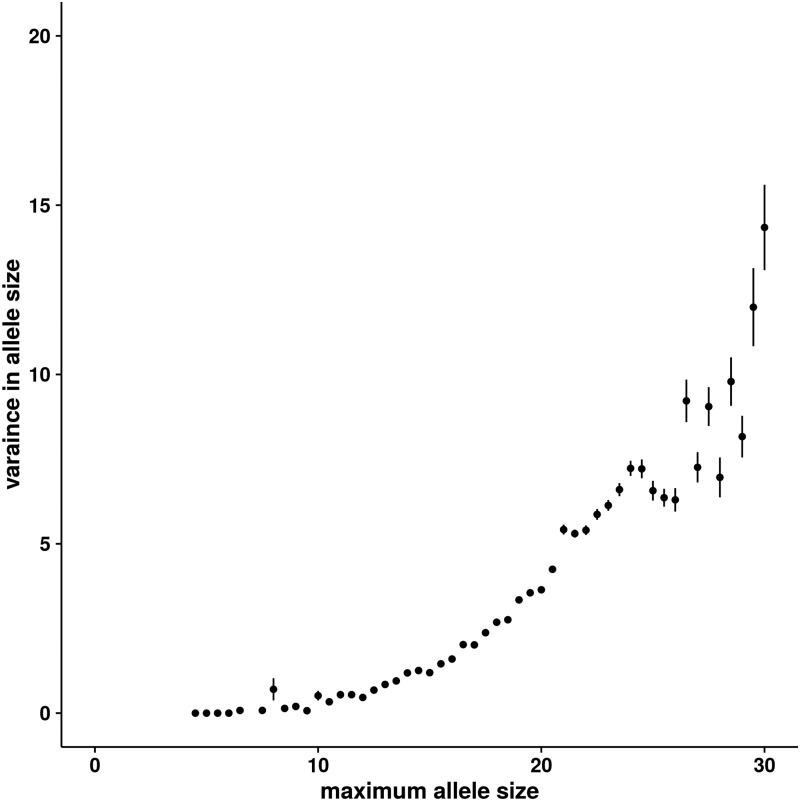
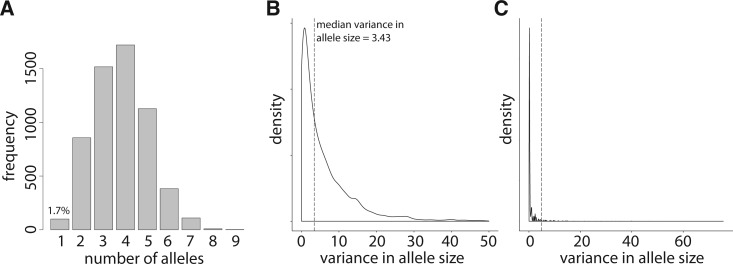
Of the 56,559 autosomal microsatellites, the genotypes of 22,067 microsatellites were recovered in all five cattle. Dinucleotide microsatellites were by far the most common (20,059 of 22,067 or 90.9%). Therefore, we restricted our analyses to diallelic STR loci with maximum allele size ≤30, to avoid spurious allele calls due to short read lengths. This results in a set of 19,338 dinucleotide STRs. We observed the plot of mean variance in allele size versus maximum allele size corroborates the well-established positive correlation between allele size and variance in allele size (fig. 2). Because sample size was limited (n = 5 diploid individuals, 10 chromosomes), we were unable to use the approximate Bayesian computation method for inferring natural selection on microsatellites (Haasl and Payseur 2013). However, we did identify outlier loci based on two anomalous relationships that countered expected trends in microsatellite variation. First, we identified 100 invariant microsatellites with allele size ≥20 (fig. 3A and B ; supplementary table S10, Supplementary Material online; empirical P-value = 0.017). These outlier loci are unusual because they demonstrate no variance despite allele size ≥20. Mutational studies (Marriage et al. 2009; Sun et al. 2012) as well as analyses of polymorphism data (Legendre et al. 2007; Brandstrom and Ellegren 2008; Kelkar et al. 2008; Payseur et al. 2011) have demonstrated that mutation rate of STRs increases with size. Thus, long STRs should be highly variable due to frequent mutation. The lack of variance at these loci therefore suggests that artificial selection (and perhaps genetic drift due to breed formation bottlenecks) has eliminated individuals that possessed mutated alleles at these loci. Second, we identified 44 diallelic microsatellites where the large allele was at least two times greater in size than the small allele (fig. 3C; supplementary table S11, Supplementary Material online; empirical P-value = 0.008). The second set of outlier loci is unusual for similar reasons. In each case, the longer allele is invariant, which defies the expectation that long alleles should be highly mutable. Furthermore, these loci are bimodal. Maintenance of two distinct allele sizes over time suggests elimination of mutated alleles by artificial and/or natural selection. A bimodal fitness surface, leading to the selection of two distinct alleles, might be especially common in regulatory regions, where specific STR sizes lead to the critical spacing patterns of promoter or enhancer sequence elements. Indeed, Elmore et al. (2012) identified promoter microsatellites in Aspergillus flavus where gene expression peaked at two distinct allele sizes, whereas intermediate allele sizes were associated with decreased expression (Elmore et al. 2012). An alternative possibility for the origin of both sets of outlier STRs is that the identified outlier loci are positioned in areas of reduced mutation. However, in the case of the bimodal outliers (supplementary table S11, Supplementary Material online) a low mutation rate begs the question of how the two alleles originated in the first place.
Fig. 2.—
Mean variance in allele size versus maximum allele size for 19,338 dinucleotide STRs with maximum allele size ≤30. Error bars are standard errors on the estimate of the mean variance in allele size.
Fig. 3.—
(A) The number of alleles for loci with maximum allele sizes on the interval [20,34]. Only 1.7% of these loci show no variation, which is unexpected for alleles of this size. (B) Kernel density estimate of variance in allele size of the same alleles summarized in (A). Median variance in allele size was 3.43 (vertical, dashed line). Only 1.7% of loci possessed variance in allele size of 0. (C) Kernel density estimate of variance in allele size for all 19,338 dinucleotide loci analysed. Of these, 44 (0.8%) were diallelic with a large alleles size at least two times as great as the small allele size. These loci show very high variance in allele size; all 44 loci possess variance in allele size >4.9, indicated by the vertical, dashed line.
In addition, we calculated ksk2 (20) values using 10-kb windows with a 5-kb step size as described previously (Haasl et al. 2014). ksk 2 (20) was developed to estimate selection on STRs by comparing the number of haplotypes (K) and segregating sites (S), representing a moving average for each 200 kb region. We obtained a total of 503,607 ksk 2 (20) values across the cattle genome, then chose the top 1% and top 5% highly negative values of ksk 2 (20). Our analysis revealed 25,182 top 5% positions and 5,035 top 1% positions. We overlapped the 25,182 top 5% positions with STR regions (60,106 regions), and found 25 STRs overlapping with top 5% positions.
Within these 25 STRs, we indeed found seven genes which may involve in selection signature: FAM171B, INHBB, TFCP2L1, RPRD2, KLHL1, SP2, and WRN. Three of these loci are of particular interest: INHBB has an important role of inhibin in reproduction (Chu et al. 2011; Lee et al. 2013); KLHL1 is involved in poor sperm motility in Holstein–Friesian bulls (Shin et al. 2014; Hering et al. 2014); and WRN gene is related to Werner syndrome (WS), also known as “adult progeria”, a rare and autosomal recessive progeroid syndrome (PS), which was characterized by the appearance of premature aging (Doan et al. 2012).
To investigate the potential selection involved in STRs, we next searched for all STRs that overlap with 1kb windows on either side of the midpoints of the top 5% of ksk 2 (20) values. We obtained 512 STRs in 97 genes which overlap with the identified ksk 2 (20) selection signature regions. Moreover, we found one STR located at 19.597 Mb on BTA21 overlapping with the detected outlier loci, indicating a strong selection signature with highly divergent alleles.
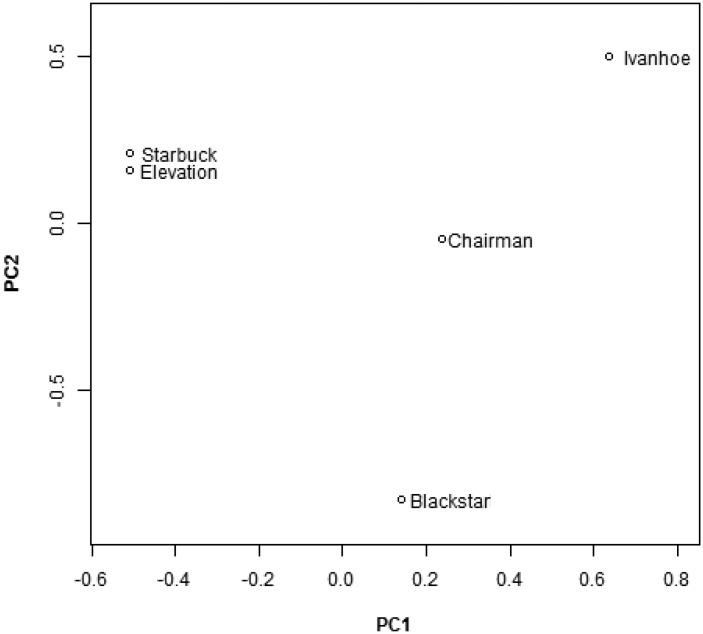
In order to estimate the genetic relatedness using identified STRs across five individual cattle, we used STR calls for all dinculeotide loci where data were available for all animals and maximum allele size was ≤30 (n = 19,338). Principal components analysis (PCA) was performed using EIGENSOFT (Patterson et al. 2006). The biplot of the first two principal components, which explained 63.5% of the variance, clearly showed that Starbuck and Elevation show little genetic differentiation (fig. 4). The other three individuals were divergent from each other as well as Starbuck and Elevation (fig. 4).
Fig. 4.—
Biplot of the first two principal components based on an analysis of genetic variation at 19,338 dincucleotide STR loci.
STRs and SNP-Based LD Blocks
STRs have been proposed as a major explanatory factor in explaining the heritability of complex traits in humans and model organisms (Press et al. 2014). Indeed, STRs are widely used for population genetics studies and linkage mapping complex traits due to their high variability. Although earlier studies based on a limited number of then known STRs suggested they are effectively tagged by SNPs (McClure et al. 2012, 2013), to fully explore the potential tagging relationship between STRs and SNPs, we investigated linkage disequilibrium (LD) pattern using SNPs around identified STRs, which is a simple method to understand their potential relationship without phasing STRs. We performed LD block estimation for each autosome using the 44 Holstein Bovine HD SNP array data retrieved from the Bovine HapMap panel.
We identified a total of 58,816 discrete LD blocks distributed across each chromosome with the maximum block size around 200 kb. Because genome regions characterized as LD blocks could imply the co-transmission of phenotype from parent to offspring, we overlapped the 60,106 STRs with the identified LD blocks. We found 57.1% of them (34,312 STRs) overlapped with 14,754 LD blocks, indicating the potential linkage characteristics between STRs and SNPs. The remaining 42.9% of STRs did not overlap with any SNP-base LD block, which suggests these STRs could serve as additional variants. Although SNP arrays and NGS have facilitated efficient SNP genotyping and QTL mapping, additional non-tagged STRs are likely to explain part of the heritability of complex trait missed by SNPs. Thus, a large proportion of the novel STRs reported here could contribute to STR-based fine mapping of hereditary traits of interest and characterization of meioses not tagged by SNPs. Given the LD pattern of STRs across the genome remains largely unknown, the exploration of LD between STRs and functional genes may help us identify novel candidate STR markers related to complex traits.
STRs Overlap with Regions under Selection Predicted Based on SNPs
Similar to SNPs, STRs are one type of the common variants in the cattle genome which may be under selection. Given their abundance in the genome, STRs represent a notable gap in our knowledge to interrogate genomes for selection signatures. The selective regime of a multi-allelic STR is potentially more complex than that of a di-allelic SNP. In conjunction with its complicated mutational properties, STRs therefore represent a substantially different selective target than SNPs (Putman and Carbone 2014).
We divided STRs into two types to explore their selection signatures. Type 1 included STRs that can be tagged by SNPs; these STRs can be simply explored by estimating the extent of homozygosity of haplotypes. Type 2 included STRs that cannot be tagged effectively by SNPs. Thus, they cannot be handled by currently existing methods directly but require additional research (Putman and Carbone 2014). Because our results were limited by a small sample size (five resequenced genomes), we focused on type 1 STRs. In the future, additional genome-wide STR will facilitate full genome scans for selection based on microsatellite. Here, we present our initial evidence for STR selection as a first step towards this future direction.
A previous study investigated selection signatures based on BovineSNP50 array and NGS data of Chief and Mark and a total of 49 regions were reported to be under recent selection (Larkin et al. 2012). To investigate the selection signature involved in STRs, we first checked if any STRs were embedded in those 49 regions. We observed 5243 STRs overlapping with 45 identified regions, where each of these regions contained variable numbers of STRs (empirical P-value = 0.014). The maximum and minimum counts of STRs contained in these regions were 1062 and 6, respectively, with an average of 118 per region.
To explore the recent selection of genome regions involved in STRs, we further utilized the Bovine HapMap HD SNP data, and then produced phased haplotypes for each Holstein individual. We then estimated the EHH and iHS in 44 resultant Holstein samples. In total, we found 249 identified regions at the top 1% level and 1194 regions at the top 5% level. Among these candidate regions, we observed 598 STRs within 54 genes overlapping with 220 regions at the top 1% level (empirical P-value = 0.037) and found 2700 STRs within 246 genes overlapping with 1025 regions at the top 5% level (empirical P-value = 0.003).
Notably, we identified some genes that may be as the potential targets of recent artificial selection for genetic improvement of milk production. For instance, gene SLCO2A1, encodes a prostaglandin transporter which is involved in maternal recognition of pregnancy (Bauersachs and Wolf 2015) and mammary development (Gao et al. 2013). We also identified genes related to milk production, fertility and milk fatty acid traits. For example, ATP1A2 was recently reported to be subject to artificial selection for milk production and fertility traits in multiple Holstein populations (Larkin et al. 2012; Lee et al. 2014). PRKG1 was identified as one of 20 genes associated with milk fatty acid traits in Holstein (Li et al. 2014), and its functions include calcium channel regulator activity and cGMP-dependent protein kinase activity. This study also identified ACSL1 associated with the milk fatty acid traits (Li et al. 2014) and supported by other studies (Widmann et al. 2011; Gao et al. 2013; Weber et al. 2013). Other identified genes in our current study are involved in somatic cell score and meat quality traits, such as GRIN2B (Wu et al. 2014) and PLXNA4 (Strillacci et al. 2014).
Conclusion
Short tandem repeats are highly mutable genetic elements that often reside in functional genomic regions and are involved in genome evolution. The advances of whole genome sequencing and STR genotype calling algorithms have made it possible to readily identify STR variants across the cattle genome. We concluded that STRs represent a significant source of polymorphism in the cattle genome. We proposed some novel candidate STRs which may be involved in important dairy traits. Our findings suggest that future studies of STRs using NGS could lead to many novel insights into their roles in contributing to complex trait heritability in farm animals. This study provides the foundation for future studies of STR's role in genome evolution and selection.
Data Accessibility
Cattle 60,106 STR genotypes predicted by lobSTR in the VCF format was uploaded under doi:10.5061/dryad.34v0d via Dryad. Raw sequencing data are available upon request (after a signed Material Transfer Agreement for exclusive research purpose).
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Acknowledgments
We thank Reuben Anderson and Alexandre Dimitriv for technical assistance. This work was supported in part by AFRI grant Nos. 2011-67015-30183 and 2013-67015-20951 from USDA NIFA. Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture. The USDA is an equal opportunity provider and employer. T.S.S. is an employee of Recombinetics, Inc. All other authors declare no potential conflict of interest.
Literature Cited
- Anvar SY, et al. 2014. TSSV: a tool for characterization of complex allelic variants in pure and mixed genomes. Bioinformatics 30:1651–1659. [DOI] [PubMed] [Google Scholar]
- Ashwell MS, Van Tassell CP, Sonstegard TS. 2001. A genome scan to identify quantitative trait loci affecting economically important traits in a US Holstein population. J Dairy Sci. 84:2535–2542. [DOI] [PubMed] [Google Scholar]
- Barendse W, et al. 1997. A medium-density genetic linkage map of the bovine genome. Mamm Genome 8:21–28. [DOI] [PubMed] [Google Scholar]
- Bauersachs S, Wolf E. 2015. Uterine responses to the preattachment embryo in domestic ungulates: recognition of pregnancy and preparation for implantation. Annu Rev Anim Biosci. 3:489–511. [DOI] [PubMed] [Google Scholar]
- Blackwell JM, et al. 2001. SLC11A1 (formerly NRAMP1) and disease resistance. Cell Microbiol. 3:773–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borel C, et al. 2012. Tandem repeat sequence variation as causative cis-eQTLs for protein-coding gene expression variation: the case of CSTB. Hum Mutat. 33:1302–1309. [DOI] [PubMed] [Google Scholar]
- Borriello G, et al. 2006. Genetic resistance to Brucella abortus in the water buffalo (Bubalus bubalis). Infect Immun. 74:2115–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahmachary M, et al. 2014. Digital genotyping of macrosatellites and multicopy genes reveals novel biological functions associated with copy number variation of large tandem repeats. PLoS Genet. 10:e1004418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandstrom M, Ellegren H. 2008. Genome-wide analysis of microsatellite polymorphism in chicken circumventing the ascertainment bias. Genome Res. 18:881–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao MD, et al. 2014. Inferring short tandem repeat variation from paired-end short reads. Nucleic Acids Res. 42:e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capparelli R, et al. 2007. The Nramp1AA genotype confers susceptibility to Brucella abortus in water buffalo. Mamm Genome 18:137–143. [DOI] [PubMed] [Google Scholar]
- Carlson KD, et al. 2015. MIPSTR: a method for multiplex genotyping of germline and somatic STR variation across many individuals. Genome Res. 25:750–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers GK, Curtis C, Millar CD, Huynen L, Lambert DM. 2014. DNA fingerprinting in zoology: past, present, future. Investig Genet. 5:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikhi L, Goossens B, Treanor A, Bruford MW. 2004. Population genetic structure of and inbreeding in an insular cattle breed, the Jersey, and its implications for genetic resource management. Heredity (Edinb.) 92:396–401. [DOI] [PubMed] [Google Scholar]
- Chu M, et al. 2011. Polymorphism of inhibin betaB gene and its relationship with litter size in sheep. Anim Sci J. 82:57–61. [DOI] [PubMed] [Google Scholar]
- Doan R, et al. 2012. Identification of copy number variants in horses. Genome Res. 22:899–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore MH, Gibbons JG, Rokas A. 2012. Assessing the genome-wide effect of promoter region tandem repeat natural variation on gene expression. G3 (Bethesda.) 2:1643–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fondon JW, III, Garner HR.x 2004. Molecular origins of rapid and continuous morphological evolution. Proc Natl Acad Sci U S A. 101:18058–18063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fungtammasan A, et al. 2015. Accurate typing of short tandem repeats from genome-wide sequencing data and its applications. Genome Res. 25:736–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly I, et al. 2008. Association of microsatellite (GT) n polymorphism at 3′ UTR of NRAMP1 with the macrophage function following challenge with Brucella LPS in buffalo (Bubalus bubalis). Vet Microbiol. 129:188–196. [DOI] [PubMed] [Google Scholar]
- Gao Y, Lin X, Shi K, Yan Z, Wang Z. 2013. Bovine mammary gene expression profiling during the onset of lactation. PLoS One 8:e70393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gel B, et al. 2016. regioneR: an R/Bioconductor package for the association analysis of genomic regions based on permutation tests. Bioinformatics 32:289–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelfand Y, Hernandez Y, Loving J, Benson G. 2014. VNTRseek-a computational tool to detect tandem repeat variants in high-throughput sequencing data. Nucleic Acids Res. 42:8884–8894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemayel R, Vinces MD, Legendre M, Verstrepen KJ. 2010. Variable tandem repeats accelerate evolution of coding and regulatory sequences. Annu Rev Genet. 44:445–477. [DOI] [PubMed] [Google Scholar]
- Georges M, et al. 1993. Microsatellite mapping of the gene causing weaver disease in cattle will allow the study of an associated quantitative trait locus. Proc Natl Acad Sci U S A. 90:1058–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilmatre A, Highnam G, Borel C, Mittelman D, Sharp AJ. 2013. Rapid multiplexed genotyping of simple tandem repeats using capture and high-throughput sequencing. Hum Mutat. 34:1304–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gymrek M, Golan D, Rosset S, Erlich Y. 2012. lobSTR: a short tandem repeat profiler for personal genomes. Genome Res. 22:1154–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gymrek M, McGuire AL, Golan D, Halperin E, Erlich Y. 2013. Identifying personal genomes by surname inference. Science 339:321–324. [DOI] [PubMed] [Google Scholar]
- Gymrek M, et al. 2016. Abundant contribution of short tandem repeats to gene expression variation in humans. Nat Genet. 48:22–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haasl RJ, Johnson RC, Payseur BA. 2014. The effects of microsatellite selection on linked sequence diversity. Genome Biol Evol. 6:1843–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haasl RJ, Payseur BA. 2013. Microsatellites as targets of natural selection. Mol Biol Evol. 30:285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammock EA, Young LJ. 2005. Microsatellite instability generates diversity in brain and sociobehavioral traits. Science 308:1630–1634. [DOI] [PubMed] [Google Scholar]
- Hering DM, Olenski K, Kaminski S. 2014. Genome-wide association study for poor sperm motility in Holstein-Friesian bulls. Anim Reprod. Sci. 146:89–97. [DOI] [PubMed] [Google Scholar]
- Highnam G, et al. 2013. Accurate human microsatellite genotypes from high-throughput resequencing data using informed error profiles. Nucleic Acids Res. 41:e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihara N, et al. 2004. A comprehensive genetic map of the cattle genome based on 3802 microsatellites. Genome Res. 14:1987–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappes SM, et al. 1997. A second-generation linkage map of the bovine genome. Genome Res. 7:235–249. [DOI] [PubMed] [Google Scholar]
- Kelkar YD, Tyekucheva S, Chiaromonte F, Makova KD. 2008. The genome-wide determinants of human and chimpanzee microsatellite evolution. Genome Res. 18:30–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar N, et al. 2011. DNA polymorphism in SLC11A1 gene and its association with brucellosis resistance in Indian zebu (Bos indicus) and crossbred (Bos indicus+ù Bos taurus) Cattle. Asian Aust J Anim Sci. 24:898–904. [Google Scholar]
- Kumar N, et al. 2005. Lack of association of brucellosis resistance with (GT) 13 microsatellite allele at 3ΓǦ UTR of Nramp1 gene in Indian zebu (Bos indicus) and crossbred (Bos indicus+ù Bos taurus) cattle. Vet Microbiol. 111:139–143. [DOI] [PubMed] [Google Scholar]
- Larkin DM, et al. 2012. Whole-genome resequencing of two elite sires for the detection of haplotypes under selection in dairy cattle. Proc Natl Acad Sci U S A. 109:7693–7698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, et al. 2014. Deciphering the genetic blueprint behind Holstein milk proteins and production. Genome Biol Evol. 6:1366–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T, et al. 2013. Genetic variants and signatures of selective sweep of Hanwoo population (Korean native cattle). BMB Rep. 46:346–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre M, Pochet N, Pak T, Verstrepen KJ. 2007. Sequence-based estimation of minisatellite and microsatellite repeat variability. Genome Res. 17:1787–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, et al. 2014. Genome wide association study identifies 20 novel promising genes associated with milk fatty acid traits in Chinese Holstein. PLoS One 9:e96186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MH, et al. 2007. The genetic structure of cattle populations (Bos taurus) in northern Eurasia and the neighbouring Near Eastern regions: implications for breeding strategies and conservation. Mol Ecol. 16:3839–3853. [DOI] [PubMed] [Google Scholar]
- Lipkin E, et al. 1998. Quantitative trait locus mapping in dairy cattle by means of selective milk DNA pooling using dinucleotide microsatellite markers: analysis of milk protein percentage. Genetics 149:1557–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacHugh DE, Shriver MD, Loftus RT, Cunningham P, Bradley DG. 1997. Microsatellite DNA variation and the evolution, domestication and phylogeography of taurine and zebu cattle (Bos taurus and Bos indicus). Genetics 146:1071–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marriage TN, et al. 2009. Direct estimation of the mutation rate at dinucleotide microsatellite loci in Arabidopsis thaliana (Brassicaceae). Heredity (Edinburgh) 103:310–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez R, et al. 2008. Bovine SLC11A1 3GǦ UTR SSCP genotype evaluated by a macrophage in vitro killing assay employing a Brucella abortus strain. J Anim Breed Genet. 125:271–279. [DOI] [PubMed] [Google Scholar]
- McClure M, Sonstegard T, Wiggans G, Van Tassell CP. 2012. Imputation of microsatellite alleles from dense SNP genotypes for parental verification. Front Genet. 3:140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure MC, et al. 2013. Imputation of microsatellite alleles from dense SNP genotypes for parentage verification across multiple Bos taurus and Bos indicus breeds. Front Genet. 4:176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Casagrande JT, Thomas PD. 2013. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc. 8:1551–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, et al. 2010. PANTHER version 7: improved phylogenetic trees, orthologs and collaboration with the Gene Ontology Consortium. Nucleic Acids Res. 38:D204–D210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel RK, Jain M. 2012. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One 7:e30619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N, Price AL, Reich D. 2006. Population structure and eigenanalysis. PLoS Genet. 2:e190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payseur BA, Jing P, Haasl RJ. 2011. A genomic portrait of human microsatellite variation. Mol Biol Evol. 28:303–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson CE, Nichol EK, Cleary JD. 2005. Repeat instability: mechanisms of dynamic mutations. Nat Rev Genet. 6:729–742. [DOI] [PubMed] [Google Scholar]
- Press MO, Carlson KD, Queitsch C. 2014. The overdue promise of short tandem repeat variation for heritability. Trends Genet. 30:504–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 81:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putman AI, Carbone I. 2014. Challenges in analysis and interpretation of microsatellite data for population genetic studies. Ecol Evol. 4:4399–4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queitsch C, Carlson KD, Girirajan S. 2012. Lessons from model organisms: phenotypic robustness and missing heritability in complex disease. PLoS Genet. 8:e1003041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabel RD, Sonstegard TS, Taylor JF, Ashwell MS. 2005. Whole-genome scan to detect QTL for milk production, conformation, fertility and functional traits in two US Holstein families. Anim Genet. 36:408–416. [DOI] [PubMed] [Google Scholar]
- Shin DH, et al. 2014. Deleted copy number variation of Hanwoo and Holstein using next generation sequencing at the population level. BMC Genomics 15:240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone RT, et al. 1995. A small-insert bovine genomic library highly enriched for microsatellite repeat sequences. Mamm Genome 6:714–724. [DOI] [PubMed] [Google Scholar]
- Strillacci MG, et al. 2014. Genome-wide association study for somatic cell score in Valdostana Red Pied cattle breed using pooled DNA. BMC Genet. 15:106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun JX, et al. 2012. A direct characterization of human mutation based on microsatellites. Nat Genet. 44:1161–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun YB, et al. 2015. Whole-genome sequence of the Tibetan frog Nanorana parkeri and the comparative evolution of tetrapod genomes. Proc Natl Acad Sci U S A. 112:E1257–E1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szpiech ZA, Hernandez RD. 2014. selscan: an efficient multithreaded program to perform EHH-based scans for positive selection. Mol Biol Evol. 31:2824–2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tae H, McMahon KW, Settlage RE, Bavarva JH, Garner HR. 2013. ReviSTER: an automated pipeline to revise misaligned reads to simple tandem repeats. Bioinformatics 29:1734–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas N, Joseph S. 2012. Role of SLC11A1 gene in disease resistance. Biotechnol Anim Husbandry 28:99–106. [Google Scholar]
- Van Tassell CP, Ashwell MS, Sonstegard TS. 2000. Detection of putative loci affecting milk, health, and conformation traits in a US Holstein population using 105 microsatellite markers. J Dairy Sci. 83:1865–1872. [DOI] [PubMed] [Google Scholar]
- Vinces MD, Legendre M, Caldara M, Hagihara M, Verstrepen KJ. 2009. Unstable tandem repeats in promoters confer transcriptional evolvability. Science 324:1213–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voight BF, Kudaravalli S, Wen X, Pritchard JK. 2006. A map of recent positive selection in the human genome. PLoS Biol. 4:e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber C, et al. 2013. Hepatic gene expression involved in glucose and lipid metabolism in transition cows: effects of fat mobilization during early lactation in relation to milk performance and metabolic changes. J Dairy Sci. 96:5670–5681. [DOI] [PubMed] [Google Scholar]
- Widmann P, Nuernberg K, Kuehn C, Weikard R. 2011. Association of an ACSL1 gene variant with polyunsaturated fatty acids in bovine skeletal muscle. BMC Genet. 12:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems T, Gymrek M, Highnam G, Mittelman D, Erlich Y. 2014. The landscape of human STR variation. Genome Res. 24:1894–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wren JD, et al. 2000. Repeat polymorphisms within gene regions: phenotypic and evolutionary implications. Am J Hum Genet. 67:345–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, et al. 2014. Genome-wide association studies using haplotypes and individual SNPs in Simmental cattle. PLoS One 9:e109330. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Cattle 60,106 STR genotypes predicted by lobSTR in the VCF format was uploaded under doi:10.5061/dryad.34v0d via Dryad. Raw sequencing data are available upon request (after a signed Material Transfer Agreement for exclusive research purpose).