Abstract
Recent studies of interactions between hosts and their resident microbes have revealed important ecological and evolutionary consequences that emerge from these complex interspecies relationships, including diseases that occur when the interactions go awry. Given the preponderance of these interactions, we hypothesized that effects of the microbiota on gene expression in the developing gut—an important aspect of host biology—would be pervasive, and that these effects would be both comparable in magnitude to and contingent on effects of the host genetic background. To evaluate the effects of the microbiota, host genotype, and their interaction on gene expression in the gut of a genetically diverse, gnotobiotic host model, the threespine stickleback (Gasterosteus aculeatus), we compared RNA-seq data among 84 larval fish. Surprisingly, we found that stickleback population and family differences explained substantially more gene expression variation than the presence of microbes. Expression levels of 72 genes, however, were affected by our microbiota treatment. These genes, including many associated with innate immunity, comprise a tractable subset of host genetic factors for precise, systems-level study of host–microbe interactions in the future. Importantly, our data also suggest subtle signatures of a statistical interaction between host genotype and the microbiota on expression patterns of genetic pathways associated with innate immunity, coagulation and complement cascades, focal adhesion, cancer, and peroxisomes. These genotype-by-environment interactions may prove to be important leads to the understanding of host genetic mechanisms commonly at the root of sometimes complex molecular relationships between hosts and their resident microbes.
Keywords: host–microbe systems, gnotobiology, fish model, genotype-by-environment interaction, RNA-seq
Introduction
Over the past two decades, the importance of the microbiota to the biology of their associated hosts, including humans, has become abundantly clear (Dethlefsen et al. 2007; McFall-Ngai et al. 2013). However, causal relationships between microbial variation and host traits like gene expression and disease are difficult to infer in descriptive human microbiome studies due to the presence of covariates such as host genetic variation and a number of environmental variables (Mai et al. 2016). Controlled gnotobiology experiments using inbred animal host models have been the most recent answer to these limitations. For example, a comparison of larval zebrafish with mouse transcriptional responses to microbes in the gut revealed molecular patterns shared among vertebrates (Rawls et al. 2004). Specifically, microbe-induced changes in expression for dozens of genes related to innate immune function, metabolism of nutrients, epithelial tissue proliferation, and xenobiotic metabolism are consistent in presence and direction between zebrafish and mouse (Hooper et al. 2001; Backhed et al. 2004; Rawls et al. 2004; Semova et al. 2012; Kanther et al. 2014). Unfortunately, inbred models like mouse and zebrafish are in turn limited in their ability to evaluate the sensitivity of microbiota-induced gene expression across the mélange of ecologically relevant host genetic variation observed in nature. Indeed, understanding how genetically heterogeneous hosts such as humans respond to their gut microbiota is key to one major objective of host–microbe systems biology: identification of complex host genetic regulatory programs that variably shape the interaction landscape, in the context of disease or otherwise (Ko et al. 2009; Goodrich et al. 2014; Huttenhower et al. 2014). For example, knowing whether host responses are dependent on structured genetic variation among families or populations, and whether key gene expression phenotypes sensitive to microbes have a simple or polygenic basis, may influence perspectives on disease treatment and/or host–microbe co-evolutionary dynamics. Despite these motivations, to our knowledge, no controlled experiment has addressed the extent to which transcriptional responses to microbes in the digestive tract vary among animal hosts with naturally heterogeneous genetic backgrounds. Such studies are necessary for the reasons above and to provide perspective on the relevance of inbred model systems to the genetic complexity within and among human populations.
We surveyed transcriptional responses to the microbiota across large scale (population-level) and small scale (family-level) genetic variation using an especially promising alternative host model, the threespine stickleback fish (Gasterosteus aculeatus). Evolved genetic diversity among natural populations, often a product of adaptation to diverse environments, has successfully been leveraged to map the genetic basis of many complex traits in stickleback (Colosimo et al. 2004; Cresko et al. 2004; Kimmel et al. 2012; Glazer et al. 2015). This naturally occurring form of genetic variation, in contrast to induced variation among laboratory animal lines, commonly bears a historical signature of environmental influences, both abiotic and biotic. In addition to a rich assortment of genetic and phenotypic variation among countless populations throughout the Northern Hemisphere (Bell and Foster 1994), stickleback, like other popular fish models such as zebrafish and medaka, are amenable to well-replicated, controlled studies in the laboratory. The term “evolutionary mutant model” was coined to describe systems like stickleback, in which naturally occurring genetic variation may be linked to phenotypes that mirror aspects of human disease (Albertson et al. 2009). These natural systems have advantages over traditional, induced mutant models, including phenotypes that are less severe, are later onset, or are products of complex genetic and environmental contributions, properties shared with many human diseases. Evolutionary mutant models are especially compatible with systems biology approaches, which translate the diffuse effects of many genomic differences among individuals to phenotypic differences through gene expression variance and covariance patterns. High levels of standing genetic variation within and among stickleback populations, the ability to cross individuals from diverse populations, and amenability to manipulative studies in the laboratory all enable stickleback as promising models for systems biology. In addition, the recent development of a protocol to generate gnotobiotic individuals (Milligan-Myhre et al. 2016) and identification of significant variation in the gut microbiota among stickleback populations in the wild (Smith et al. 2015), places the stickleback model in a rare position to address systems-level questions about host genetic variation and host–microbe relationships.
Global transcriptional profiles offer comprehensive, systematic insights into multi-level processes such as microbe-sensitive gene regulatory cascades (Morgan et al. 2015) and are therefore useful for evaluating the relative contributions of host and environmental factors to variation in the behavior of gene regulatory networks. Also, because a transcriptomic profile is itself a composite of co-varying complex traits, it is ultimately a useful framework for establishing connections between gene regulatory networks and quantitative traits such as morphological, physiological, and disease variants (Hodgins-Davis and Townsend 2009; van Nas et al. 2010). Indeed, in many cases, only through systems biology approaches are we able to understand how nucleotide-level variation among individuals mechanistically affects these traits, because measurements of gene regulatory variation provide missing pieces of predictive information about the structure of gene-gene and gene-environment interactions (Civelek and Lusis 2014; Albert and Kruglyak 2015). This framework has been applied successfully to both confirm the involvement of known developmental pathways and reveal novel gene regulation patterns that explain how genetic variation translates to host-specific responses. For example, Orozco et al. (2012) revealed macrophage-specific biology during inflammation by measuring genome-wide transcriptional responses of macrophages from 92 genetically diverse mouse strains to chemical stimuli associated with acute and chronic inflammation (Orozco et al. 2012). Through the mapping of genetic variation among the mouse lines to variation in transcriptional responses, the authors were able to identify novel regulatory connections between cholesterol transport and Toll-like receptor signaling during acute inflammation, among other insights.
To understand the robustness of microbiota-mediated variation in gene expression across different host genotypes and to identify specific microbe-associated genes and gene pathways in stickleback, we measured transcript abundance in the guts of 84 larval stickleback using RNA-seq. The study fish, derived from two genetically divergent source populations separated in nature for as long as 15,000 years (Cresko et al. 2004) were experimentally exposed or unexposed to microbes in an otherwise uniform environment. We also endeavored to identify genes that demonstrate conserved microbe-sensitive gene expression across stickleback and other vertebrates, as these may be of importance in a biomedical context. To this end, we identified dozens of stickleback genes differentially expressed by microbial treatment, and confirmed their overlap with microbe-sensitive orthologs in zebrafish. To contrast the magnitude of microbiota effects on gene expression with those of host genetic background, we compared two populations evolutionarily adapted to very different environments: a freshwater lake and the ocean. Because our previous work documented innate immune responses to the microbiota contingent on these two genetic backgrounds (Milligan-Myhre et al. 2016), we also tested for a statistical interaction between these two experimental factors in the current study. We expected pervasive Genotype-by-Environment (G-by-E) interactions, though the transcriptome-wide evidence for G-by-E interaction was unexpectedly subtle. Our findings point to host-genotype-specific transcriptional sensitivities to the presence of microbes across a small subset of the gastrointestinal transcriptome, including genes associated with focal adhesion, coagulation and the complement system, and other innate immune system components. The relatively small number of affected genes points to the tractability of this experimental system for deciphering the effects of natural host genetic variation on the microbiota and associated traits. Broadly, this work sets the stage for future studies of the genetic underpinnings of variation in the host response to microbes, and it makes the case for careful consideration regarding design of gnotobiology experiments and fundamental sources of phenotypic variance in these studies.
Materials and Methods
Generation of Gnotobiotic Stickleback Larvae and Experimental Design
We derived germ-free threespine stickleback embryos from multiple laboratory crosses using methods described by Milligan-Myhre et al. (2016). Briefly, we fertilized ova from each lab-reared female in vitro using sperm from dissected testes of a lab-reared adult male from the same source population. Fertilized eggs were incubated in sterile stickleback embryo medium (SBEM, 4 g/L Instant Ocean, sodium-bicarbonate-buffered to pH 7) with ampicillin (100 ug/ml), kanamycin (5 µg/ml) and amphotericin (250 ng/ml). Four hours post fertilization, we surface-sterilized all embryos using filter-sterilized 0.2% Polyvinylpyrrolidone-iodine (PVP-I, diluted in sterile SBEM and filter sterilized) and 0.003% bleach (diluted in SBEM and filter sterilized), followed by three rinses with filter-sterilized SBEM. At this point we transferred subsets of 20 embryos to 250 mL sterile polystyrene flasks with filter caps (Techno Plastic Products AG) containing 50 mL of sterile SBEM.
We included three crosses (different families) derived from a freshwater (FW) Alaskan population stock (Boot Lake), and two crosses (also different families) derived from an oceanic (OC) Alaskan stock (Rabbit Slough). These stocks have been maintained in the laboratory for at least eight generations, effectively controlling for maternal and paternal effects related to the original environments in which the populations evolved. Initially we inoculated two flasks for each family with 500 µL of water from our recirculating stickleback system to approximate a “conventional” (CV) microbiota and left another two flasks for each family “germ-free” (GF). In all cases, rearing flasks were maintained at a constant temperature of 18 °C until 14 days post fertilization (dpf), at which point all surviving larvae were counted, euthanized with a lethal dose of tricaine, and either dissected immediately to obtain the gut for bacteria plating, or tail-clipped for sex genotyping and fixed individually in RNAlater (Life Technologies). We also collected water samples on filter disks for PCR with bacteria-specific 16S rRNA gene primers to detect bacterial contamination.
To ensure balanced sampling of males and females for the RNA-seq analysis, we amplified a sex-specific region of the genome using the “GA1” PCR primer pair described by Griffiths (2000). Briefly, we isolated DNA from each tail clip using a 5% chelex solution, exactly as described in Rose et al. (2014) and amplified the sex-specific marker for each sample using the following PCR reagent cocktail: 1 µL DNA isolate, 7.5 µL 2X PCR buffer, 0.015 µL GA1 primer mix (50 µM), 0.078 µL Taq polymerase (2U/µL), 6.27 µL nuclease-free water. Each 15 µL PCR reaction underwent an initial denaturation at 94 °C for 5 min, 44 cycles of denaturation, primer annealing, and extension (50 s at 94 °C, 50 s at 44 °C, 80 s at 72 °C, respectively), and a final extension at 72 °C for 10 min. We determined sex by visualizing the presence or absence of the male-specific amplicon on a 2% agarose gel (0.5X Tris-Borate-EDTA running buffer) stained with SafeView (abm).
Validation of Conventional Microbiota and Germ-Free Treatments
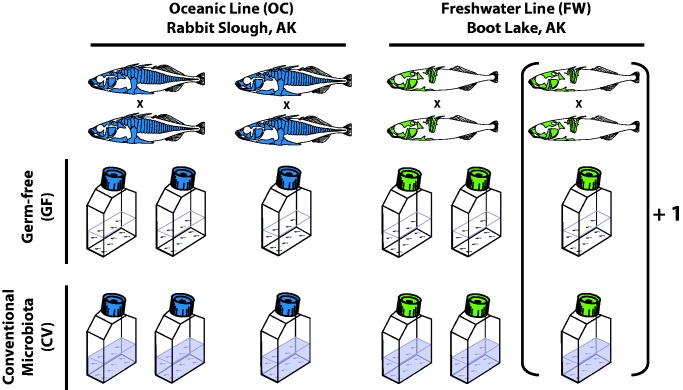
We confirmed the effectiveness of the microbial treatments in our experiments with three methods. (1) Flasks were directly visualized for microbial contamination using phase optics at 40X magnification. (2) Water from rearing flasks and the contents of individual, homogenized stickleback guts were plated on standard LB-agarose media in petri dishes and incubated at 20°C. Bacterial and fungal growth was noted 1 day and 4 days post plating. (3) Water from each flask was collected on a filter, DNA was isolated from the filter, and the 16S ribosomal RNA gene was amplified by PCR via the method described in Bates et al. (2006). Briefly, 0.1 mm zirconia beads were added to the tube containing the filter and SDS lysis solution (10% SDS, 0.5M Tris/HCl pH 8.0, 0.1M NaCl), followed by bead beating for 2 min. The supernatant was treated with lysozyme for 30 min at 37°C, and the DNA was precipitated with ammonium acetate and isopropanol and washed with ethanol. The 16S ribosomal RNA gene was amplified from 5 µl of DNA template per sample, following the details described in Bates et al. (2006). Negative controls included sterile SBEM, and positive controls contained DNA isolated from several CV larval intestines. Three of the ten GF flasks tested positive for bacteria via plating or PCR and were excluded from the study. To maintain balance with respect to the microbial treatment, we randomly excluded one of the CV flasks from each of the families for which a contaminated germ-free flask existed. In total, six individuals were sampled from each of the 14 remaining flasks, for a total of 84 individuals. Figure 1 depicts these flasks and their treatments in our experimental design, upon which all analyses for the study were based.
Fig. 1.—
Experimental design. An illustration of the factors and their levels included in our experiment. In total we generated 84 RNA-seq libraries (6 fish each from 14 flasks), each using mRNA isolated from the entire gut—posterior esophagus to anus—of individual fish. The two stickleback lines (“OC” and “FW“) were derived from natural Alaskan populations, as indicated. Two OC families and three FW families (FW family 3 not shown, but referenced by brackets), were represented in the study. Note that for one OC family and one FW family, conventional and germ-free treatments were duplicated to examine housing (“flask“) effects.
To assess effects of the experimental factors on normal development within flasks included in the study, we counted the number of dead and abnormally developed fish per flask. We used log-linear modeling to test for dependence of abnormal development on microbiota treatment and host genotype (family and population levels).
Preparation of RNA-Seq Libraries from Individual Stickleback Guts
We dissected whole guts from three male and three female, normally developed larvae per flask and flash-froze in liquid nitrogen each gut in 200 µl TRIzol reagent (Life Technologies). We thawed and homogenized each sample by bead beating with a 0.5/1.0 mm mix of zirconia oxide beads in a Bullet Blender Storm homogenizer (Next Advance) for three minutes at maximum speed. After homogenization we added an additional 800 µl TRIzol reagent, froze again at −80 C, then isolated total RNA using a protocol adapted from Leung and Dowling (2005). Briefly, homogenate was spun through a Qiashredder centrifuge column (Qiagen), two rounds of phase separation after the addition of chloroform were performed with the assistance of 2.0 mL phase-lock gel tubes (5prime), and total RNA was washed and eluted by centrifuge column using the RNeasy Minelute Kit (Qiagen). We quantified each RNA sample using a Qubit fluorometer (Life Technologies) and confirmed high RNA integrity for a subset of samples using a Fragment Analyzer (Advanced Analytical Technologies).
To construct RNA-seq libraries we used the TruSeq mRNA v2 Kit (Illumina) according to manufacturer recommendations, but with several modifications. We scaled reaction volumes down to one third of the suggested value, used 200 ng of total RNA as starting material, incubated reactions for 5 min at 94°C during the RNA fragmentation step, and applied 10 cycles of PCR during the library amplification step. We quantified all samples by fluorometry and multiplexed between 12 and 22 libraries per Illumina HiSeq 2500 lane to achieve raw sequencing coverage of approximately 8.9 million 100 nt single-end reads per sample. To avoid confounding “batch” effects with experimental variables of interest, we adopted a stratified library preparation and sequencing scheme, in which each bout of library preparation and sequencing included balanced representation from experimental factor level combinations. All sequencing was performed by the University of Oregon Genomics and Cell Characterization Core Facility (GC3F).
RNA-Seq Data Processing, Normalization, and Differential Expression Analysis
For all sequencing reads we demultiplexed, trimmed Illumina adapters, and performed base quality filtering using process_shortreads from the Stacks software suite (Catchen et al. 2013). The splice-aware aligner GSNAP (Wu and Nacu 2010) was used to align processed reads to the Ensembl threespine stickleback reference genome and annotation (version 80). We used default GSNAP parameters, except that we set the proportion of each read length allowed to mismatch the reference at 0.1, we enabled novel spice site detection, and we used the split_output flag to produce different alignment files for different alignment types. For each sample we counted the number of uniquely-mapped reads per exonic region of each annotated gene using HTSeq-count (Wu and Nacu 2010). Counts of uniquely aligned reads per gene model for each sample were merged into a single text file for all subsequent analyses, which were performed using the R statistical language and programing environment (R Core Team 2015).
We limited differential expression analysis to only those genes represented by at least two reads per million mapped (“copies per million,” CPM) in at least 12 of the 84 libraries (see supplementary fig. S1, Supplementary Material online). We normalized read counts for these 15,847 genes using TMM normalization (Robinson and Oshlack 2010) as implemented by the calcNormFactors function of the R/Bioconductor package edgeR (Robinson et al. 2010). In order to perform gene-wise differential expression analyses in a general linear model framework (Law et al. 2014), we supplied the TMM normalization factors to the voom function of the R/Bioconductor package limma (Ritchie et al. 2015), which generated appropriately weighted log2CPM expression values for all observations. We then fit a linear model for each gene including the fixed effects of factor levels for host population, host family (nested within host population), sex, and microbiota treatment using the limma lmFit function. We did not include a library “batch” effect in the model because initial nMDS ordination did not suggest batch as a major source of transcriptional variation, and our stratified assignment of samples to batches controlled for any confounding effect of batch with respect to other factors of interest. To account for variation between replicate flasks we incorporated flask as a random effect in the model using the limma duplicateCorrelation function. Each hypothesis of interest was tested, for each gene, using one or more contrasts via moderated t-tests applied by the limma function eBayes. To evaluate the effect of our microbiota treatment we performed a within-OC contrast, a within-FW contrast, and an overall contrast. Genes expressed differentially in any of these three contrasts were interpreted as being associated with the presence of microbes. We performed a single contrast to test for an overall effect of host population, and a single contrast to test for an interaction between host population and microbiota, both of these accounting for family differences nested within population. Finally, we performed contrasts to test for an effect of sex and a sex-by-microbiota interaction. For each of these seven contrasts, we controlled the false discovery rate (FDR) at 0.1 using the approach of Benjamini and Hochberg (1995), as implemented by the limma topTable function.
Given the reduced power to detect real interaction effects relative to main term effects in general linear models, and given that effects of the microbiota alone were subtle, we adopted a second, more sensitive regression-based approach to identify genes and regulatory pathways likely subject to an interaction between host population and microbiota treatment. We identified extreme residuals based on deviations from a null linear model (y-intercept = 0, slope = 1) of identical CV-GF log2 fold changes between genes in OC and FW groups. Genes falling in the upper and lower 1.25% of the distribution of residuals were considered to deviate substantially from OC-FW unity regarding the effects of the microbiota treatment and were therefore likely subject to population-specific effects of microbial presence. This approach was based on the distribution of effect sizes (log2 fold changes) alone, and not on individual hypothesis tests for differential expression, enabling the identification of G-by-E interaction effects across the transcriptome without relying on thousands of under-powered, individual tests.
To characterize multivariate patterns of gene expression among the 84 gut transcriptomes, and subsets when appropriate, we performed non-metric multidimensional scaling (nMDS) using TMM-normalized CPM expression values for all 15,847 gut-expressed genes. We calculated Bray-Curtis dissimilarity, performed the nMDS ordinations, and tested multivariate differences for the fixed effects of microbiota treatment, population, family, and relevant interactions, using permutation-based multivariate analysis of variance (perMANOVA), as implemented by the R package vegan (Oksanen et al. 2016),
Comparison of Microbiota-Specific Gene Expression between Zebrafish and Stickleback
Using Ensembl’s BioMart data mining tool we endeavored to assign Ensembl IDs (release 80) to 212 EST-derived microarray probes identified by Rawls et al. (2004) as differentially expressed between germ-free and conventional-microbiota zebrafish. For the non-redundant, Ensembl zebrafish sequences from this set we attempted to find putative threespine stickleback orthologs using Ensembl orthology assignments, or BLASTp. Specifically, in the infrequent event that no or multiple orthologs were identified based on the Ensembl database, we considered any top BLASTp hit (with sequence identity ≥ 60%) as a putative ortholog. We used these stickleback orthologs, and their expression patterns with respect to the microbiota treatment, as a basis for comparisons between the two fish species. We assessed the strength of the relationship between zebrafish and stickleback CV-GF log2 fold changes via Kendall’s Tau nonparametric tests of correlation. We tested whether the number of ortholog pairs demonstrating the same CV-GF log2 fold change direction in stickleback and zebrafish (CV-enriched in both species, or GF-enriched in both species) was greater than that expected by chance using chi-squared tests.
Gene Ontology and KEGG Pathway Analyses
We obtained Gene Ontology descriptions for stickleback genes via Ensembl BioMart, formatted the information using the R/Bioconductor databases AnnotationDbi (Pages et al. 2016) and GSEABase (Morgan et al. 2016) and conducted GO term overrepresentation tests using hypergeometric tests implemented by the R/Bioconductor package GOstats (Falcon and Gentleman 2007). We tested for GO terms enriched among the 72 genes differentially expressed between GF and CV microbiota treatments, relative to all gut-expressed genes. We also tested for GO terms enriched among the 398 genes most likely subject to genotype-by-microbiota interaction, relative to all gut-expressed genes.
To associate KEGG Pathways with stickleback genes we first identified putative Ensembl human orthologs using the approach described above for zebrafish. We then extracted KEGG Pathway assignments via the human orthologs using the R databases AnnotationDbi (Pages et al. 2016) and org.Hs.eg.db (Carlson 2016). We identified KEGG Pathways enriched for stickleback genes with extreme CV-GF log2 fold change values using the general gene set enrichment analysis framework of the R/Bioconductor package GAGE (Luo et al. 2009). We performed these KEGG Pathway enrichment analyses separately for the OC and FW data sets, and as previously we controlled the false discovery rate at 0.1 in both cases. To identify KEGG pathways most likely subject to genotype-by-microbiota interaction, we performed a similar enrichment test based on residual values from the regression approach described above. To visualize stickleback gene members of the coagulation and complement cascade KEGG pathway, and their scaled CV-GF log2 fold change values in both OC and FW data sets, we used the R/Bioconductor package Pathview (Luo and Brouwer 2013).
Results
Microbial Influences on the Gastrointestinal Transcriptome of Larval Stickleback Are Subtle Relative to the Effects of Host Genotype
One concern regarding our experimental design was whether the microbial treatment, host genotype (family and population), or both, might affect normal fish development and survival, potentially limiting our inferences about direct effects on gene expression. We first fitted and compared full and reduced log-linear models to evaluate the dependence of whether fish developed normally on microbial treatment and family. The proportion of surviving, normally developed fish depended on family (Analysis of Deviance: G2 = 24.32, P = 0.002), but neither on microbiota treatment (Analysis of Deviance: G2 = 7.59, P = 0.18) nor their interaction (Analysis of Deviance: G2 = 7.24, P = 0.12). Importantly, we evaluated similar models at the population level and found no evidence for dependence of normal development on population (Analysis of Deviance: G2 = 0.66, P = 0.72), treatment (Analysis of deviance: G2 = 0.49, P = 0.78), or their interaction (Analysis of Deviance: G2 = 0.16, P = 0.68). Therefore, while some families demonstrated significantly higher or lower rates of normal development, there was no systematic effect of treatment or population of origin on normal development. This suggests that neither the microbiota treatment, nor population-specific sensitivity to a rearing salinity of 4 g/L, affected normal development in our experiment.
To measure the relative contributions of the microbiota and host genotype to gene expression, we generated RNA-seq data from 84 larval stickleback guts collected from families of freshwater and oceanic fish reared in the presence or absence of microbes (fig. 1). On average we obtained 8,932,868 high-quality reads per individual, of which nearly 5.5 million per individual could be aligned uniquely to exons annotated in the stickleback reference genome. Over 70% (15,847 out of 22,456) of genes annotated in the stickleback reference genome were expressed consistently among gut libraries, based on a threshold of 2 reads per one million aligned (to exons) or greater, in at least 12 of 84 libraries. For a summary of the proportion of annotated genes expressed across various coverage thresholds, see supplementary figure S1, Supplementary Material online.
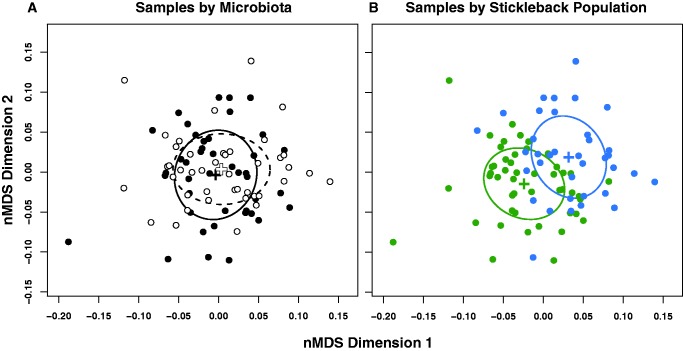
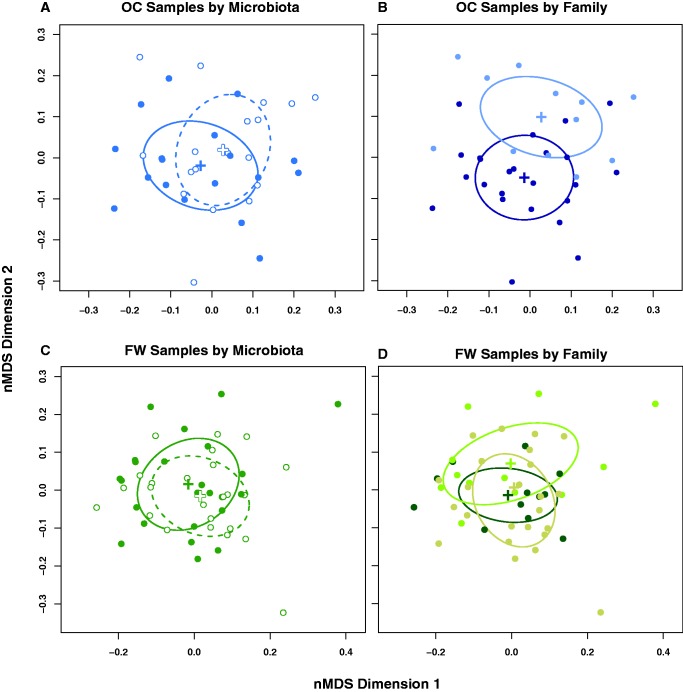
Contrary to expectations, host population and family differences explained substantially more gene expression variation than the presence of the microbiota. The germ-free state did not induce gross, global transcriptional changes in the gut, as overall transcriptome composition did not differ significantly between CV and GF fish in a multivariate sense (fig. 2A; perMANOVA: F1,80 = 1.36, P = 0.17), and the treatment explained only 1.45% of the total dissimilarity among fish. In contrast to effects of the microbiota, host population significantly differentiated individual guts in transcript space, explaining 11.59% of the total dissimilarity (fig. 2B; perMANOVA: F1,80 = 10.84, P = 0.001). Restricting inferences to oceanic (OC) fish, microbiota explained 3.93% of the total dissimilarity (fig. 3A; perMANOVA: F1,32 = 1.48, P = 0.12), while family explained 9.04% of the total dissimilarity, a significant effect (fig. 3B; perMANOVA: F1,32 = 3.41, P = 0.001). Likewise, within freshwater (FW) fish, microbiota explained 2.56% of the total dissimilarity (fig. 3C; perMANOVA: F1,42 = 1.32, P = 0.20), while family explained 9.61% of the total dissimilarity, a significant effect (fig. 3D; perMANOVA: F2,42 = 2.44, P = 0.009).
Fig. 2.—
Effects of the microbiota on global gene expression in the 14 dpf stickleback gut are weak relative to effects of host population. An nMDS ordination of the 84 stickleback guts in multivariate transcript space labeled by (A) microbiota treatment and (B) stickleback population. The “plus” symbols denote group centroids, and ellipses mark 95% confidence intervals about the centroids. In panel (A), open circles represent germ-free individuals and closed circles represent conventional individuals. In panel (B), blue circles represent oceanic (OC) individuals and green circles represent freshwater (FW) individuals. Note the clearer separation of population groups in panel (B), relative to microbiota treatment groups in panel (A).
Fig. 3.—
Effects of the microbiota on global gene expression in the 14 dpf stickleback gut are weak relative to effects of host family. (A and B): An nMDS ordination of the 36 stickleback guts from OC fish in transcript space labeled by (A) microbiota treatment and (B) stickleback family. (C and D): An nMDS ordination of the 48 stickleback guts from FW fish, labeled by (C) microbiota and (D) stickleback family. In panels (A) and (C) open circles represent germ-free individuals and closed circles represent conventional individuals. In panels (B) and (D), different colors represent the two different OC families and three different FW families, respectively, included in the study. Note the clearer separation of transcriptomes by family, relative to separation by microbiota treatment.
Because previous work in mice has shown that cage effects on the gut microbiota are large relative to other sources of variation (Hildebrand et al. 2013), we thought rearing flask might be a significant factor explaining stickleback gene expression, especially for flasks with a conventional microbiota. For instance, if microbial effects on gastrointestinal gene expression were contingent on the immediate housing environment, it might suggest stochastic variation between flasks in microbial community structure and/or host colonization dynamics. We included replicate rearing flasks for both CV and GF treatments in the case of one oceanic and one freshwater family (fig. 1), so we made an effort to assess relative contributions of flask and microbial effects to variation in transcriptome dissimilarity among individual fish. Flask-level replication was low, so the following results should be interpreted with this caveat. In general, effects of co-housing environment on global transcriptional variation were weak, but greater than those of the microbiota. In a multivariate sense, flask explained more overall gut transcriptome dissimilarity among fish than did microbiota, with neither factor explaining a statistically significant proportion. (FW perMANOVA: R2flask(microbiota) = 0.094, R2microbiota = 0.066; OC perMANOVA: R2flask(microbiota) = 0.110, R2microbiota = 0.051; also see supplementary fig. S2C and F, Supplementary Material online). Individual transcriptomes from duplicate flasks were as dissimilar as those compared between CV and GF flasks (supplementary fig. S2A, B, and D–E, Supplementary Material online). Fish across duplicate CV flasks were not significantly more dissimilar than fish across duplicate GF flasks (FW dispersion test: F1,22 = 0.0001, P = 0.991; OC dispersion test: F1,22 = 0.099, P = 0.756), although there was a trend in this direction within the OC family.
We did observe significant effects of the microbiota on expression for a targeted subset of genes, particularly those belonging to innate immunity pathways. Seventy-two genes were differentially expressed between CV and GF fish, based on our linear model contrasts (see Methods) and a false discovery rate controlled at 0.1 (see supplementary table S3, Supplementary Material online). Table 1 includes information regarding the top ten (by fold change) CV-enriched and top ten GF-enriched genes. Of the dozens of stickleback genes subject to a pronounced transcriptional effect of the microbiota, many were associated with innate immune processes, including a number of granulocyte-specific genes. The adaptive immune system is unlikely operative in larval fish of this age, based on knowledge from zebrafish (Lam et al. 2004), so we expected few, if any, adaptive immune responses. The group of 72 differentially expressed genes demonstrated overrepresentation of a number of Biological Process GO terms, relative to all genes expressed in the 14-dpf stickleback gut, including “response to external biotic stimulus,” “innate immune response,” “arginine metabolic process,” “endopeptidase activity,” “lipid binding,” and others (see supplementary table S4, Supplementary Material online). In contrast to the effects of the microbiota, and consistent with the multivariate transcriptome analysis (see above), influences of host population on gene-wise expression patterns in the gut were much more extensive; we identified 3,451 genes differentially expressed between OC and FW fish, nearly 22% of the annotated genes expressed at appreciable levels in the gut (see supplementary table S5, Supplementary Material online).
Table 1.
Top Genes Expressed Differentially by Microbiota Treatment
| Ensembl Gene ID | Gene Description | Direction of Diff. Expr. | Fold Change |
|---|---|---|---|
| ENSGACG00000008429 | myeloperoxidase-like protein* | CV-enriched | 5.35 |
| ENSGACG00000010234 | cathepsin Bb | CV-enriched | 3.99 |
| ENSGACG00000017166 | myeloid-specific peroxidase | CV-enriched | 3.52 |
| ENSGACG00000011676 | BPI fold containing family C | CV-enriched | 3.47 |
| ENSGACG00000006045 | zymogen granule protein 16B | CV-enriched | 3.23 |
| ENSGACG00000006706 | tumor necrosis factor receptor superfamily, member 11b | CV-enriched | 2.99 |
| ENSGACG00000010404 | interleukin 22 receptor, alpha 2 | CV-enriched | 2.65 |
| ENSGACG00000010912 | deleted in malignant brain tumors 1 protein* | CV-enriched | 2.18 |
| ENSGACG00000001729 | interleukin 8* | CV-enriched | 2.13 |
| ENSGACG00000014415 | low choriolytic enzyme precursor* | CV-enriched | 2.11 |
| ENSGACG00000001881 | leukocyte elastase inhibitor* | GF-enriched | 1.67 |
| ENSGACG00000014112 | developing brain homeobox 1 | GF-enriched | 1.65 |
| ENSGACG00000012888 | heat shock protein 90, alpha (cytosolic), class A member 1, tandem duplicate 2 | GF-enriched | 1.50 |
| ENSGACG00000006375 | serpin peptidase inhibitor, clade H (heat shock protein 47), member 1b | GF-enriched | 1.47 |
| ENSGACG00000014099 | mucin 5f | GF-enriched | 1.34 |
| ENSGACG00000020294 | V-set and immunoglobulin domain containing 1 | GF-enriched | 1.33 |
| ENSGACG00000018013 | protease, serine, 12 (neurotrypsin, motopsin) | GF-enriched | 1.31 |
| ENSGACG00000010026 | ankyrin repeat and death domain containing 1B | GF-enriched | 1.28 |
| ENSGACG00000008795 | inositol-trisphosphate 3-kinase A | GF-enriched | 1.25 |
| ENSGACG00000011569 | transient receptor potential cation channel, subfamily M, member 6 | GF-enriched | 1.24 |
Note.—Ten top-ranked (by fold change) conventional-enriched and germ-free-enriched stickleback genes, of 72 total differentially expressed. In cases where no Ensembl gene description was available (marked by an asterisk), descriptions were obtained from top BLASTp hits of searches against the NCBI nr database.
To identify known gene regulatory pathways responsive to the microbiota in stickleback, we tested whether certain KEGG pathways were enriched for stickleback orthologs with high or low CV-GF log2 fold changes. Ten KEGG pathways were significantly enriched (FDR = 0.1) for genes transcriptionally sensitive to microbes in both FW and OC stickleback populations, including those associated with JAK-STAT signaling, chemokine signaling, Toll-like receptor signaling, leukocyte transendothelial migration, and rheumatoid arthritis (see supplementary table S6, Supplementary Material online). An additional 34 KEGG pathways were significantly enriched for microbiota-sensitive transcription in either, but not both stickleback populations (see supplementary table S6, Supplementary Material online).
Microbiota-Sensitive Gene Expression Patterns Are Conserved between Zebrafish and Stickleback
We identified unique zebrafish Ensembl gene annotations for 191 of the microarray features considered differentially expressed between germ-free and conventional-microbiota 6-dpf zebrafish by Rawls et al. (2004). Of these we were able to assign putative stickleback orthologs with Ensembl annotations to 173. Of these stickleback orthologs, 166 were expressed at detectable levels based on our RNA-seq data. Fifteen of these 166 (9.04%) were characterized by at least one GF-CV contrast p-value less than 0.05 from our RNA-seq experiment (see supplementary table S7, Supplementary Material online), including the neutrophil-specific gene myeloperoxidase, inflammatory cytokine-responsive genes, and metabolism-related genes such as cell death-inducing DFFA-like effector c and carnitine palmitoyltransferase 1Ab.
The magnitude of CV/GF log2 fold change co-varied significantly between zebrafish and FW stickleback (see supplementary fig. S8A, Supplementary Material online, Kendall T = 0.133, z = 2.497, P = 0.013), as well as between zebrafish and OC stickleback (see supplementary fig. S8B, Supplementary Material online, Kendall T = 0.192, z = 3.596, P < 0.001). Furthermore, the direction of fold change for orthologs was consistent between zebrafish and each stickleback line, more so than expected by chance. Specifically, 102 of 166 genes were consistent in log2 fold change sign between zebrafish and FW stickleback (X21 = 8.699, P = 0.003), and 111 of 166 were consistent between zebrafish and OC stickleback (X21 = 18.892, P < 0.001).
In both zebrafish and stickleback data sets a greater proportion of the microbiota-sensitive genes were expressed at higher levels in CV relative to GF fish. In zebrafish, for example, approximately 63% of differentially expressed genes were expressed at higher levels in CV fish, relative to GF fish (Rawls et al. 2004). In stickleback, 52 (72.22%) were expressed at higher levels in CV fish, and 20 (27.78%) were over-expressed in GF fish. This cross-species pattern suggests a directional bias toward positive, as opposed to negative, microbe-induced gene regulation in fish hosts.
Microbe-Associated Transcriptional Responses of Innate Immunity Pathways and NLRC3-like NOD-like Receptors Depend on the Genetic Background of the Host
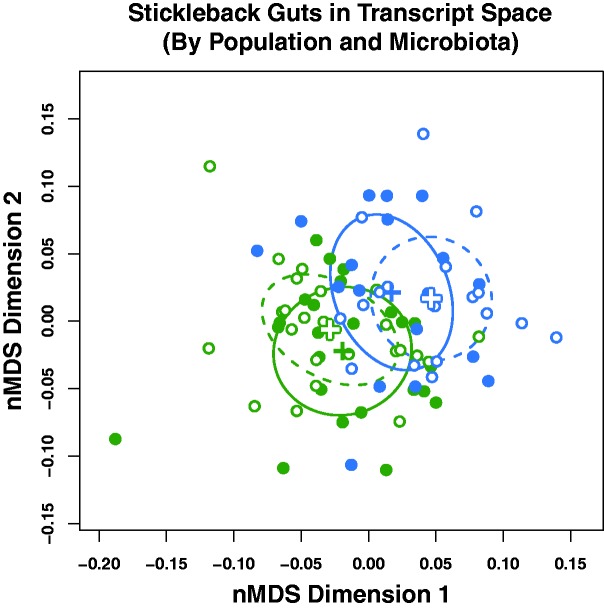
Considering global, multivariate patterns of gene expression, the interaction between host population and microbiota treatment was not statistically significant (fig. 4; perMANOVA: F1,80 = 1.28, P = 0.21), meaning transcriptome disparity between CV and GF fish did not differ by host population. Furthermore, we detected no individual genes subject to a statistical interaction between stickleback population and microbiota treatment based on the gene-wise limma-voom tests for differential expression (FDR = 0.1, see supplementary table S9, Supplementary Material online). Using an alternative analysis of log2 fold change residuals (see Methods), we identified 398 genes in the extreme upper and lower quantiles (1.25% per tail) of the residual distribution. The 40 genes in the outer most tails of the residual distribution may be found in supplementary table S10, Supplementary Material online. Among those more strongly CV-enriched in FW fish relative to OC fish were genes associated with innate immunity: a paralog of myeloperoxidase, complement component C3, and toll-like receptor 5. Those that were more strongly CV-enriched in OC fish relative to FW also included immune response genes: the T-cell-expressed mal, and five genes that encode NLRC3-like (NOD-like receptor family CARD domain-containing 3) proteins. We analyzed this subset of 398 genes and found overrepresented GO terms including “regulation of body fluid levels,” “blood coagulation and fibrin clot formation,” “response to wounding,” transmembrane transport,” and more (see supplementary table S11, Supplementary Material online).
Fig. 4.—
Global transcriptional patterns in the 14 dpf stickleback gut are not strongly shaped by an interaction between microbiota treatment and host population. The same ordination from figure 2, but with groups labeled by population-treatment combination. Again, open circles denote germ-free fish, closed circles denote conventional fish, blue circles denote OC fish, and green circles denote FW fish. Outlined “plus” signs denote germ-free centroids, and solid "plus" signs denote conventional centroids. Dashed ellipses reflect 95% confidence intervals about germ-free centroids, and solid ellipses reflect 95% confidence intervals about conventional centroids. Note the lack of a clear difference in the orientation of CV vs. GF individuals between the two populations, which would indicate a statistical interaction.
We also tested whether certain KEGG pathways were enriched for genes with extreme residual values from the above analysis. Using this approach, we identified four significantly enriched KEGG pathways (FDR = 0.1): “complement and coagulation cascades” (P = 0.0001), “pathways in cancer” (P = 0.0003), “focal adhesion” (P = 0.0007), and “peroxisome” (P = 0.005). For an illustration of the population-specific responsiveness of components of the “complement and coagulation cascades” pathway to the microbiota, see supplementary figure S12, Supplementary Material online. Specifically, FW fish responded to the presence of microbes mostly via increased transcript abundance, while the OC fish transcriptional response was minimal.
Sex Biases in Gene Expression Are Concentrated on Stickleback Chromosome 19 and Do Not Depend on the Presence of Microbes
We identified 703 genes differentially expressed between females and males, based on our linear model contrasts (see Methods) and a false discovery rate controlled at 0.1 (see supplementary table S13, Supplementary Material online), of which 530 (75.39%) corresponded to annotations on stickleback linkage group 19. This chromosome corresponds to the stickleback sex chromosomes (Peichel et al. 2004). Nearly 68% of these sex-biased genes were female-enriched, and over 32% were male-enriched. A previous study of sex-biased gene expression in threespine stickleback reported 1268 sex-biased genes, of which 23% localized to chromosome 19 (Leder et al. 2010). Leder et al., however, examined adult liver tissue gene expression, so our finding of fewer overall sex-biased genes, but a larger chromosome 19 bias, suggests that sex-biased gene expression in undifferentiated larval stickleback guts most likely reflects lack of dosage compensation for X and Y-specific gene expression.
Ward clustering of normalized expression values for a subset of 314 genes on chromosome 19—located between 6 and 12 Mb and corresponding to high sex-biased gene expression according to Leder et al. (2010)— revealed two major clusters of individuals that corresponded perfectly with our PCR-based sex genotypes (supplementary fig. S14A, Supplementary Material online). A randomly selected subset of 314 genes from the genome at large showed no such clustering by sex (supplementary fig. S14B, Supplementary Material online), confirming the high incidence of sex-biased gene expression on chromosome 19 and the reliability of our PCR sexing assay.
We also tested for a statistical interaction between sex and microbiota treatment with respect to gene-wise differential expression but found no strong evidence for this after controlling the false discovery rate at 0.1. Other specific details regarding patterns of sex-biased gene expression are beyond the scope of this study and will be addressed, using these and additional data, in a future report.
Discussion
Understanding the landscape of molecular interactions from which healthy and dysbiotic relationships between hosts and their resident microbes emerge is a major, ongoing research endeavor (Morgan et al. 2015). Several valuable studies have addressed host-specific components of this landscape by measuring cellular and transcriptional responses to microbes using gnotobiotic vertebrate host models (Rawls et al. 2004; Kanther et al. 2011; Camp et al. 2014; Sommer et al. 2015; Milligan-Myhre et al. 2016). We advanced this fundamental objective of host–microbe systems research using RNA-seq in larval threespine stickleback, a powerful new model system for the influences of host genetic diversity. By incorporating population- and family-level genetic variation as well as housing environment replicates into our experimental design, we were able to contrast the relative influence of these factors with that of the microbiota on gene expression in the gut, putting into context the potential significance of host genetic variation in human-microbe interactions.
Microbe-Sensitive Gene Expression Patterns in Stickleback Align with Inferences from Other Vertebrate Host–Microbe Model Systems
The first major objective of our study was to characterize the transcriptional response of larval stickleback hosts to the gut microbiota. We found that while over 70% of the 22,456 genes annotated in the stickleback genome were expressed in the guts of 14 dpf fish, a surprisingly small number—72—responded robustly to the presence of microbes. Global multivariate transcriptome dissimilarity between CV and GF fish was also very low, perhaps reflecting a select group of modular regulatory networks influenced by the microbiota. A previous study of genome-wide transcription patterns in the whole guts of larval zebrafish reported 212 genes (of 16,228 tested) differentially expressed between CV and GF treatments (Rawls et al. 2004). Recent studies in mouse considered CV versus GF gene expression differences for various intestinal cell types separately, and reported 261 (of 26,900 tested) (Camp et al. 2014) and 2,256 (of 33,648 tested) (Sommer et al. 2015) microbe-sensitive genes, regardless of cell type.
Direct, between-study comparisons of the number of microbe-sensitive genes are made difficult by fundamental differences among the studies with respect to technology (i.e., microarray vs. RNA-seq), tissue type, experimental design, statistical analysis, and host genetic diversity (see below). For example, cell type-specific expression differences in the presence versus absence of microbes may be numerous, but undetectable when averaged across an entire gastrointestinal tract. Host developmental stage may also explain discrepancies among studies, as there is evidence that gut transcriptional effects of the microbiota become more pronounced with age of Drosophila melanogaster hosts (Broderick et al. 2014). The age of fish in our experiment was limited to a maximum of 14 dpf, because beyond this age the yolk sac no longer provides nutrition, feeding is necessary, and it is not currently practical to maintain a quality, germ-free food source for juvenile stickleback. The 14 dpf fish in our study were not likely starving, or affected differently between treatments from a nutritional standpoint, given our tests for developmental differences (see Results). It is possible, however, that sampling fish at such an early stage of microbial colonization reflects more variation in community structure (and possibly less stable microbe-related gene expression) than would be observed at later stages. Future stickleback studies in which later developmental stages can be included, and in which both metagenomic and host transcriptomic data are collected from the same individuals, will address this potential source of gut community and transcriptome variability.
Despite these technical differences across studies, we found significant similarities regarding identity and general expression direction of microbe-sensitive genes in the zebrafish study (Rawls et al. 2004) and our stickleback data set (see supplementary fig. S8, Supplementary Material online). For instance, as in zebrafish, we observed a marked signature of the innate immune response based on the identity of genes differentially expressed between GF and CV stickleback, such as the granulocyte-specific gene mpx (myeloid-specific peroxidase). Genes involved in the recruitment of these cells such as Il8 (interleukin-8) and stat4 (signal transducer and activator of transcription 4) were also among those differentially expressed. Gene Ontology terms associated with innate immunity were overrepresented in the subset of stickleback genes sensitive to the microbiota, and KEGG pathway analysis revealed that pathways associated with innate immunity were enriched for genes transcriptionally responsive to the microbiota in stickleback (see supplementary table S6, Supplementary Material online). Also similar to zebrafish, genes associated with nutrient metabolism were differentially expressed between CV and GF stickleback. Examples from our study include dao.3 (D-amino-acid oxidase, tandem duplicate 3), the fatty acid transporter slc27a2, and the sulfotransferase sult5a1. These concordances between zebrafish and stickleback confirm a conserved system of responses to microbes among vertebrates, as inferred from past comparisons between zebrafish and mouse (Rawls et al. 2004). Furthermore, our relatively small list of stickleback genes robustly influenced by the microbiota provides a manageable subset of host genetic factors for future, manipulative studies aimed at perturbing host–microbe interactions in gnotobiotic stickleback.
A number of recent studies have identified regions of the host genome at which genetic variation maps to variation in microbial community structure and/or microbe-related disease traits (reviewed in Goodrich et al. 2016). Candidate genes near the genomic regions identified by this work, mostly inferred using quantitative trait locus (QTL) mapping and genome-wide association (GWA) approaches in humans and inbred mouse lines, align with some genes and pathways we identified as microbe-sensitive in stickleback. For example, Benson et al. (2010) identified Ifng (interferon-gamma) and Il22 (interleukin-22) as belonging to a genomic interval strongly associated with the fraction of Gram-positive bacteria in the mouse gut microbiome. As mentioned, stickleback stat4, whose mammalian ortholog is a known regulator of Ifng (Nguyen et al. 2002) and the inflammatory cascade in general (Good et al. 2009), was expressed at higher levels in CV stickleback. Also, isoform-specific expression patterns of human STAT4 are associated with Inflammatory Bowel Disease (Jabeen et al. 2015). There is currently no annotation for an Il22 ortholog in stickleback, but one of its receptors (il22ra2), which is regulated by the inflammasome and controls colon epithelial cell proliferation in mammals (Huber et al. 2012), was expressed 2 to 3.5 times higher in CV relative to GF stickleback.
Several mouse studies based on QTL and/or eQTL data (Benson et al. 2010; McKnite et al. 2012; Leamy et al. 2014; Org et al. 2015) have reported that genetic variants near genes in the Toll-like receptor (TLR) and T cell receptor signaling pathways are associated with microbiota traits, with gene expression patterns within the pathways, or with both. Of the 72 stickleback genes that we found most influenced transcriptionally by the microbiota in the larval gut, at least 13 have likely human orthologs represented in the TLR signaling pathway or those directly linked (see supplementary table S3, Supplementary Material online). These fundamental similarities justify use of the gnotobiotic stickleback model in research aimed at understanding how genetic variation maps to human disease states related to host–microbe interactions. Studies using stickleback offer advantages because experiments in which control over environmental conditions such as the microbiota, diet, housing conditions (and others) are possible. To illustrate this potential, we evaluated multiple sources of variation in stickleback gene expression by incorporating several additional factors into our experimental design.
Extensive Population-Specific Gene Expression in the Stickleback Gut Reflects Evolutionary Divergence among Hosts
The experimental factor that explained most of the global variation and co-variation in transcriptional patterns of the 14 dpf stickleback gut was host population (fig. 2). Our finding that 22% of the genes expressed in the larval stickleback gut are differentially expressed between the two populations is perhaps not surprising, given that the populations used in our study have been isolated from one another for at least 9,000, and perhaps as many as 15,000 years (Cresko et al. 2004). These populations also show substantial genetic divergence across the genome (Hohenlohe et al. 2010), and they differ with respect to a number of morphological phenotypes (Cresko et al. 2004; Kimmel et al. 2005). It should be noted, however, that our study was restricted to three FW and two OC families, largely due to logistical constraints. Although we accounted for family-level variation in our analyses of population differences in gene expression, larger samples of genetic variation within populations (i.e. inclusion of many, unrelated families) in future studies will provide less biased, more general estimates of population divergence.
Recent studies of gene expression variation among other threespine stickleback populations have focused on non-gastrointestinal tissues such as the liver (Nikinmaa et al. 2013; Leder et al. 2015), and those associated with immune function, particularly the head kidney and spleen (Lenz et al. 2013; Stutz et al. 2015; Huang et al. 2016). In these studies, the magnitude of population-specific gene expression, and explanations for its occurrence, has varied. Nikinmaa et al. (2013) identified 1698 liver genes differentially expressed among populations of F2 individuals reared under controlled laboratory settings, and Leder et al. (2015) inferred from similar data that 1411 liver transcripts have diverged in expression phenotype among populations owing to directional selection alone. Lenz et al. (2013) found a significant effect of population on global head kidney gene expression in adult fish, but only for those fish exposed to helminth parasites. Stutz et al. (2015) provided strong evidence for environmental effects on head kidney expression via qPCR for seven immunity-related genes, by finding that fish experimentally transplanted to waters inhabited by genetically distinct populations resembled the unrelated resident fish more than related fish reared in their native waters. Huang et al. (2016) demonstrated extensive population differences in head kidney and spleen gene expression among wild-caught fish, while highlighting similarity among different populations occurring in the same type of environment (i.e. river versus lake).
More generally, few well-replicated, controlled laboratory studies have measured population-level, genome-wide divergence in gene expression. In those that have, insights regarding the fraction of a transcriptome differentially expressed range extensively, for example 0.41% for two hybridizing crow species (Poelstra et al. 2014), 7.64% for two Coregonus whitefish populations (Nolte et al. 2009), and 31.7% between African and European Drosophila melanogaster populations (Hutter et al. 2008). The neutral and/or adaptive accumulation of genetic differences in cis- and trans-regulatory regions of genomes in diverging populations could explain the expression disparities we observed in larval stickleback, but the relative contributions from these different mechanisms depend on time since divergence and on the taxa being studied (Coolon et al. 2014). Future studies, including assessments of allele-specific expression in between-population crosses, for example, should address some of these outstanding questions.
Developmental differences at 14 dpf between FW and OC fish may at least partially explain the expression differences, as both body size and the degree of gut epithelial fold complexity are known to differ subtly in stickleback at this age (Milligan-Myhre et al. 2016). Future studies that measure transcriptional divergence at multiple developmental time points will be necessary to address this possibility. For example, a valuable comparison would be to measure gene expression in adult stickleback guts from multiple populations raised in the same environment, as others have shown that microbial community structure in wild-caught adult guts differs substantially among stickleback populations from Vancouver Island (Smith et al. 2015). Assuming these differences in the gut microbiota are at least in part explained by genetic variation, which has been demonstrated in other vertebrate hosts (Goodrich et al. 2014; Davenport et al. 2015; Org et al. 2015; Sullam et al. 2015), widespread, evolved differences in digestive tract gene expression among populations are a potential mechanism connecting host genotype to microbial phenotype in stickleback.
Sources of Global Transcriptional Variation in the Gut Highlight Considerations for Future Host–Microbe Systems Research
Given our conclusion that substantial divergence in host transcription within the digestive tract can evolve on relatively short timescales, studies of host–microbe interactions in genetically heterogeneous host species deserve special consideration. On the one hand, systems like stickleback are extremely promising with respect to genetic mapping studies in which variation for microbe-relevant traits such as gut transcription or the microbiota are the focus. Especially diverse recombinant phenotypes are expected in F2 progeny owing to the marked genetic and phenotypic differences between parents in initial between-population crosses. In mapping studies that use such diverse populations, statistical power to detect QTLs should be high. On the other hand, experimental inferences about host or microbial biology made using a single host population may not be extendable to other populations. In general, studies of host–microbe relationships should include multiple host populations if possible, or at least be consistent in using the same host population over multiple experiments, if results from the experiments are to be interpreted together.
Variation in gut transcriptome dissimilarity among fish in our experiment was also explained by family-level effects nested within each population (fig. 3). Effects of family explained more than 9% of the total transcriptome dissimilarity within each population, suggesting that segregating genetic variation contributes significantly to gene expression patterns in the stickleback gut. Authors of a recent quantitative genetic study of transcriptional variation among livers of adult threespine stickleback (Leder et al. 2015) reported that narrow-sense heritability for transcript abundance was, on average, quite high (h2 = 0.23). The detectable family-level effects we observed are consistent with larval gut gene expression patterns being heritable, although our experimental design was not intended to estimate quantitative genetic parameters like heritability. This insight is again relevant to experimental design for studies of host–microbe biology, as experimenters should take care not to confound family with treatments of interest. Furthermore, to understand host population- or species-level effects on host–microbe traits, it is necessary to adequately sample the genetic variation within these higher levels by including individuals from multiple, unrelated families when possible. Any treatment factors should be applied in a randomized block framework, allowing for treatment levels to be compared within groups of varying genetic structure.
We attempted to evaluate the effect of housing environment, as compared to the microbiota, on transcriptome dissimilarity for a very limited subset (one OC and one FW family) of individuals in our experiment. Unlike the clear influences of host population and family on transcriptional patterns in the full data set, the effects of housing and microbiota were weak within these two families (see supplementary fig. S2, Supplementary Material online), although our sample was small. Rearing flask explained 10.98% and 9.37% of the variation in gut transcriptome dissimilarity within the OC and FW families, respectively, but we could not reject the null hypothesis of no average flask effect in either case. The effect of microbiota, for which we also could not reject the null hypothesis, explained 5.15% and 6.57% of the variation, respectively. One unanswered question regarding gnotobiology experiments is whether conventional or experimentally controlled microbiota treatments lead to more variance in host traits relative to germ-free treatments. Such a pattern seems likely for cases in which environmental or within-host community structure is expected to be largely stochastic, for example when communities are in early stages of assembly (Gillilland et al. 2012), when host selective agents like the immune system are ablated via mutation (Kubinak et al. 2015), or when the immune system is immature (Burns et al. 2016). Although this pattern would seem especially likely in very young stickleback, we did not observe statistically significant flask effects or greater transcriptome dissimilarity in CV relative to GF flasks in our study. However, trends from the data suggest that the effect of flask is greater than the effect of the microbiota, and that transcriptome dissimilarity may be greater among CV flasks. Given these tentative results and other work demonstrating strong housing effects on the microbiota (Hildebrand et al. 2013; McCafferty et al. 2013), we strongly recommend that multiple rearing vessel replicates - more extensive than those included in this study—should be included for each factor or factor level combination in gnotobiology experiments to account for this potentially important source of variation.
Interaction between Host Population and the Microbiota Influences Specific Transcriptional Patterns in the Larval Stickleback Gut
Stickleback have advantages over other experimental vertebrate models such as mouse and zebrafish, given the ability to sample a vast reservoir of host genetic variation in a tractable, gnotobiotic experimental system (Milligan-Myhre et al. 2016). In fact, the intersection of genetic variation and control over key environmental variables is where stickleback promise to offer novel insights into non-additive effects of host genotype and environmental factors on traits related to microbes and host health, a feature directly relevant to the notion of “personalized medicine.” Motivated largely by this potential for the stickleback system to reveal statistical interaction between host genotype and the microbial environment, we assessed the phenomenon using our transcriptional data. The aforementioned study by Lenz et al. (2013) provided evidence for effects of an interaction between stickleback population and parasite infection status on gene expression. In our previous study (Milligan-Myhre et al. 2016), we documented a clear G-by-E interaction, wherein the host response to microbes, as measured by neutrophil abundance in the gut, was greater in magnitude for stickleback from the OC population, relative to fish from the FW population. We predicted that this host population-specific innate immune response to microbes might be reflected in a parallel, widespread pattern across the larval gut transcriptome.
We did not observe a significant, population-specific shift in treatment-associated, global transcription patterns (fig. 4), perhaps for the same reasons that we did not detect an effect of the microbiota on multivariate gene expression patterns in general (discussed above). Furthermore, after controlling the false discovery rate across gene-by-gene analyses, we did not reject the null hypothesis of no interaction for any gene. In general, statistical power of linear models is lower for hypothesis tests of interaction terms, relative to those of main effect terms, so higher replication at the family or individual level may have changed this outcome.
We adopted a second, regression-based approach to identify genes and regulatory pathways most likely subject to an interaction between host population and microbiota treatment. This approach has the advantage of revealing patterns based on effect size (i.e. log2 fold change), as opposed to FDR-adjusted interaction test p-values from many individual differential expression tests, which are more likely to yield false negative hypothesis tests. Both GO term overrepresentation and KEGG pathway enrichment analyses based on this approach revealed that coagulation and complement cascades, two closely connected pathways that underlie hemostasis and innate immune responses, and are responsive to bacterial infection in primates (Lupu et al. 2014), were positively regulated by the microbiota in FW fish, but not in OC fish (see supplementary fig. S12, Supplementary Material online). Conversely, a suite of NOD-like receptors similar to mammalian NLRC3, a negative regulator of inflammation in mice (Schneider et al. 2012; Coutermarsh-Ott et al. 2016) and in synovium from rheumatoid arthritis patients (Park et al. 2015), showed the opposite pattern, as these NLRC3s were more positively responsive to microbes in OC fish relative to FW fish (see supplementary table S10, Supplementary Material online). In mammals NLRC3 may reduce the activity of NLRP3 inflammasomes—protein complexes that initiate inflammatory cascades in response to both microbial and endogenous signals (Levy et al. 2015) - by suppressing pro-caspase-1 cleavage and interleukin 1ß maturation (Gultekin et al. 2015). Inflammasomes have been shown to mediate important host–microbe interactions (Wlodarska et al. 2014; Levy et al. 2015), and inflammasome dysfunction is associated with inflammatory bowel disease (Levy et al. 2015; Opipari and Franchi 2015), so genotype-dependent, microbe-sensitive gene expression of NLRC3 orthologs in stickleback may be relevant in an evolutionary mutant model context. Given the expression patterns for complement and NLRC3 genes, it is likely that multiple components of stickleback innate immunity react to microbes in fundamentally different ways depending on the genetic background of the host.
One potential explanation for population-specific expression responses to the microbiota treatment in our study is that the assemblage of microbes in the CV inoculum more closely resembled the historical environmental microbiota of wild OC ancestors relative to FW ancestors, or vice versa, resulting in an asymmetric response. Currently we lack sufficient information about the relative dissimilarities between the laboratory microbiota, and the historical environmental microbiota of the two populations, to address this. Another possibility is that, owing to genetic differences among host populations, individuals select for different gut microbial assemblages, in turn affecting gene expression differently. Exploring this explanation will require the characterization of both microbial community structure and host transcription from the same individuals, a strategy we were unable to apply to the current study given limitations in the amount of material from larval guts. Although the direct causes and ecological and evolutionary implications of these “Genotype-by-Microbiota” interactions are unclear at this stage, their existence warrants further investigation in stickleback and consideration in other gnotobiotic models.
Because genetic variation among stickleback hosts influences the manner in which regulatory systems such as the complement cascade and NOD-like receptor signaling respond to the presence of microbes, and these conserved systems are relevant to disease in humans (e.g. Rittirsch et al. 2008; Saxena and Yeretssian 2014), stickleback may offer a promising model for the study of microbiota-associated disease. Recent in vitro studies of human cellular responses to microbes have revealed a clear importance of host genetic variation (Quach et al. 2016; Richards et al. 2016). In vivo systems like gnotobiotic stickleback, however, promise to help inform on the genetic architecture of complex traits like chronic inflammatory diseases, but in the context of the entire host organism and its microbiota. Along these lines, the work described here has set the stage for critical follow-up studies that aim to identify specific regions of the genome underlying variation in traits related to the immune response to and the structure of microbial communities in the stickleback gut, including GWAS, QTL, and eQTL mapping studies, approached from a systems-level perspective. Through these approaches, host models like stickleback, that leverage naturally occurring, ecologically meaningful genetic variation, will make fundamental contributions to our understanding of interaction landscapes in host–microbe systems.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We are grateful to M. Currey for advice and help with fish husbandry. We also thank Dr D. Turnbull of the University of Oregon Genomics and Cell Characterization Core Facility (GC3F) for assistance with Illumina sequencing logistics. This work was funded by National Institutes of Health grants P50GM098911 (to KG, WAC et al.) and RR032670 (to WAC et al.). KM-M was supported by a National Institutes of Health NRSA fellowship (F32DK096753).
Literature Cited
- Albert FW, Kruglyak L. 2015. The role of regulatory variation in complex traits and disease. Nat Rev Genet. 16:197–212. [DOI] [PubMed] [Google Scholar]
- Albertson RC, Cresko W, Detrich HW, 3rd, Postlethwait JH. 2009. Evolutionary mutant models for human disease. Trends Genet. 25:74–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backhed F, et al. 2004. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci U S A. 101:15718–15723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates JM, et al. 2006. Distinct signals from the microbiota promote different aspects of zebrafish gut differentiation. Dev Biol. 297:374–386. [DOI] [PubMed] [Google Scholar]
- Bell MA, Foster SA. 1994. The evolutionary biology of the threespine stickleback. Oxford: University Press. [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate—a practical and powerful approach to multiple testing. J Roy Stat Soc Ser B-Methodol. 57:289–300. [Google Scholar]
- Benson AK, et al. 2010. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc Natl Acad Sci U S A. 107:18933–18938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broderick NA, Buchon N, Lemaitre B. 2014. Microbiota-induced changes in drosophila melanogaster host gene expression and gut morphology. MBio 5:e01117–e01114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns AR, et al. 2016. Contribution of neutral processes to the assembly of gut microbial communities in the zebrafish over host development. ISME J. 10:655–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp JG, et al. 2014. Microbiota modulate transcription in the intestinal epithelium without remodeling the accessible chromatin landscape. Genome Res. 24:1504–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M. 2016. org.Hs.eg.db: Genome wide annotation for Human. R package version 3.2.3.
- Catchen J, Hohenlohe PA, Bassham S, Amores A, Cresko WA. 2013. Stacks: an analysis tool set for population genomics. Mol Ecol. 22:3124–3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civelek M, Lusis AJ. 2014. Systems genetics approaches to understand complex traits. Nat Rev Genet. 15:34–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colosimo PF, et al. 2004. The genetic architecture of parallel armor plate reduction in threespine sticklebacks. PLoS Biol. 2:E109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coolon JD, McManus CJ, Stevenson KR, Graveley BR, Wittkopp PJ. 2014. Tempo and mode of regulatory evolution in Drosophila. Genome Res. 24:797–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutermarsh-Ott S, Eden K, Allen IC. 2016. Beyond the inflammasome: regulatory NOD-like receptor modulation of the host immune response following virus exposure. J Gen Virol. 97:825–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cresko WA, et al. 2004. Parallel genetic basis for repeated evolution of armor loss in Alaskan threespine stickleback populations. Proc Natl Acad Sci U S A. 101:6050–6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport ER, et al. 2015. Genome-wide association studies of the human gut microbiota. PLoS One 10:e0140301.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L, McFall-Ngai M, Relman DA. 2007. An ecological and evolutionary perspective on human–microbe mutualism and disease. Nature 449:811–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcon S, Gentleman R. 2007. Using GOstats to test gene lists for GO term association. Bioinformatics 23:257–258. [DOI] [PubMed] [Google Scholar]
- Gillilland MG, 3rd, et al. 2012. Ecological succession of bacterial communities during conventionalization of germ-free mice. Appl Environ Microbiol. 78:2359–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazer AM, Killingbeck EE, Mitros T, Rokhsar DS, Miller CT. 2015. Genome assembly improvement and mapping convergently evolved skeletal traits in sticklebacks with genotyping-by-sequencing. G3 (Bethesda) 5:1463–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good SR, et al. 2009. Temporal induction pattern of STAT4 target genes defines potential for Th1 lineage-specific programming. J Immunol. 183:3839–3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich JK, et al. 2014. Human genetics shape the gut microbiome. Cell 159:789–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich JK, Davenport ER, Waters JL, Clark AG, Ley RE. 2016. Cross-species comparisons of host genetic associations with the microbiome. Science 352:532–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths R. 2000. DNA sex identification in the three-spined stickleback. J Fish Biol. 57:1331–1334. [Google Scholar]
- Gultekin Y, Eren E, Ozoren N. 2015. Overexpressed NLRC3 acts as an anti-inflammatory cytosolic protein. J Innate Immun. 7:25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand F, et al. 2013. Inflammation-associated enterotypes, host genotype, cage and inter-individual effects drive gut microbiota variation in common laboratory mice. Genome Biol. 14:R4.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgins-Davis A, Townsend JP. 2009. Evolving gene expression: from G to E to GxE. Trends Ecol Evol. 24:649–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohenlohe PA, et al. 2010. Population genomics of parallel adaptation in threespine stickleback using sequenced RAD tags. PLoS Genet. 6:e1000862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper LV, et al. 2001. Molecular analysis of commensal host-microbial relationships in the intestine. Science 291:881–884. [DOI] [PubMed] [Google Scholar]
- Huang Y, et al. 2016. Transcriptome profiling of immune tissues reveals habitat-specific gene expression between lake and river sticklebacks. Mol Ecol. 25:943–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber S, et al. 2012. IL-22BP is regulated by the inflammasome and modulates tumorigenesis in the intestine. Nature 491:259–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenhower C, Kostic AD, Xavier RJ. 2014. Inflammatory bowel disease as a model for translating the microbiome. Immunity 40:843–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutter S, Saminadin-Peter SS, Stephan W, Parsch J. 2008. Gene expression variation in African and European populations of Drosophila melanogaster. Genome Biol. 9:R12.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabeen R, et al. 2015. Altered STAT4 Isoform Expression in Patients with Inflammatory Bowel Disease. Inflamm Bowel Dis 21:2383–2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanther M, et al. 2011. Microbial colonization induces dynamic temporal and spatial patterns of NF-kappaB activation in the zebrafish digestive tract. Gastroenterology 141:197–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanther M, et al. 2014. Commensal microbiota stimulate systemic neutrophil migration through induction of serum amyloid A. Cell Microbiol. 16:1053–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, et al. 2005. Evolution and development of facial bone morphology in threespine sticklebacks. Proc Natl Acad Sci U S A. 102:5791–5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Hohenlohe PA, Ullmann B, Currey M, Cresko WA. 2012. Developmental dissociation in morphological evolution of the stickleback opercle. Evol Dev. 14:326–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko DC, et al. 2009. A genome-wide in vitro bacterial-infection screen reveals human variation in the host response associated with inflammatory disease. Am J Hum Genet. 85:214–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubinak JL, et al. 2015. MyD88 signaling in T cells directs IgA-mediated control of the microbiota to promote health. Cell Host Microb. 17:153–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam SH, Chua HL, Gong Z, Lam TJ, Sin YM. 2004. Development and maturation of the immune system in zebrafish, Danio rerio: a gene expression profiling, in situ hybridization and immunological study. Develop Comp Immunol 28:9–28. [DOI] [PubMed] [Google Scholar]
- Law CW, Chen Y, Shi W, Smyth GK. 2014. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15:R29.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leamy LJ, et al. 2014. Host genetics and diet, but not immunoglobulin A expression, converge to shape compositional features of the gut microbiome in an advanced intercross population of mice. Genome Biol. 15:552.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leder EH, et al. 2010. Female-biased expression on the X chromosome as a key step in chromosome evolution in threespine sticklebacks. Mol Biol Evol. 27:1495–1503. [DOI] [PubMed] [Google Scholar]
- Leder EH, et al. 2015. The evolution and adaptive potential of transcriptional variation in sticklebacks–signatures of selection and widespread heritability. Mol Biol Evol. 32:674–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz TL, Eizaguirre C, Rotter B, Kalbe M, Milinski M. 2013. Exploring local immunological adaptation of two stickleback ecotypes by experimental infection and transcriptome-wide digital gene expression analysis. Mol Ecol. 22:774–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung Y, Dowling J. 2005. Gene expression profiling of zebrafish embryonic retina. Zebrafish 2:269.. [DOI] [PubMed] [Google Scholar]
- Levy M, Thaiss CA, Katz MN, Suez J, Elinav E. 2015. Inflammasomes and the microbiota–partners in the preservation of mucosal homeostasis. Semin Immunopathol 37:39–46. [DOI] [PubMed] [Google Scholar]
- Levy M, et al. 2015. Microbiota-modulated metabolites shape the intestinal microenvironment by regulating NLRP6 inflammasome signaling. Cell 163:1428–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Brouwer C. 2013. Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 29:1830–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Friedman MS, Shedden K, Hankenson KD, Woolf PJ. 2009. GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinformatics 10:161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupu F, Keshari RS, Lambris JD, Coggeshall KM. 2014. Crosstalk between the coagulation and complement systems in sepsis. Thromb Res. 133(Suppl 1):S28–S31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai V, Prosperi M, Yaghjyan L. 2016. Moving microbiota research toward establishing causal associations that represent viable targets for effective public health interventions. Ann Epidemiol. 26:306–310. [DOI] [PubMed] [Google Scholar]
- McCafferty J, et al. 2013. Stochastic changes over time and not founder effects drive cage effects in microbial community assembly in a mouse model. ISME J. 7:2116–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFall-Ngai M, et al. 2013. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci U S A. 110:3229–3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnite AM, et al. 2012. Murine gut microbiota is defined by host genetics and modulates variation of metabolic traits. PLoS One 7:e39191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan-Myhre K, et al. 2016. Innate immune responses to gut microbiota differ between oceanic and freshwater threespine stickleback populations. Dis Model Mech. 9:187–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan XC, et al. (Morgan2015 co-authors). 2015. Associations between host gene expression, the mucosal microbiome, and clinical outcome in the pelvic pouch of patients with inflammatory bowel disease. Genome Biol. 16:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan M, Falcon S, Gentleman R. 2016. GSEABase: gene set enrichment data structures and methods. R package version 1.34.1.
- Nguyen KB, et al. 2002. Critical role for STAT4 activation by type 1 interferons in the interferon-gamma response to viral infection. Science 297:2063–2066. [DOI] [PubMed] [Google Scholar]
- Nikinmaa M, et al. 2013. Transcription and redox enzyme activities: comparison of equilibrium and disequilibrium levels in the three-spined stickleback. Proc R Soc B. 280:20122974.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolte AW, Renaut S, Bernatchez L. 2009. Divergence in gene regulation at young life history stages of whitefish (Coregonus sp.) and the emergence of genomic isolation. BMC Evol Biol. 9:59.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, et al. 2016. vegan: Community Ecology Package. R package version 2.3-5.
- Opipari A, Franchi L. 2015. Role of inflammasomes in intestinal inflammation and Crohn's disease. Inflamm Bowel Dis. 21:173–181. [DOI] [PubMed] [Google Scholar]
- Org E, et al. 2015. Genetic and environmental control of host-gut microbiota interactions. Genome Res. 25:1558–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco LD, et al. 2012. Unraveling inflammatory responses using systems genetics and gene-environment interactions in macrophages. Cell 151:658–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pages H, Carlson M, Falcon S, Li N. 2016. AnnotationDbi: Annotation Database Interface. R package version 1.34.4.
- Park MC, Lee SW, Park YB, Lee SK. 2015. AB0128 expression patterns of nod-like receptors in rheumatoid synovium and its effect on inflammatory responses of rheumatoid fibroblast-like synoviocytes. Ann Rheumat Dise. 74:933.932-933. [Google Scholar]
- Peichel CL, et al. 2004. The master sex-determination locus in threespine sticklebacks is on a nascent Y chromosome. Curr Biol. 14:1416–1424. [DOI] [PubMed] [Google Scholar]
- Poelstra JW, et al. 2014. The genomic landscape underlying phenotypic integrity in the face of gene flow in crows. Science 344:1410–1414. [DOI] [PubMed] [Google Scholar]
- Quach H, et al. 2016. Genetic adaptation and neandertal admixture shaped the immune system of human populations. Cell 167:643–656 e617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2015. R: A language and environment for statistical computing R Foundation for Statistical Computing, Vienna, Austria. URL http://www.R-project.org/. [Google Scholar]
- Rawls JF, Samuel BS, Gordon JI. 2004. Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. Proc Natl Acad Sci U S A. 101:4596–4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards AL, et al. 2016. Genetic and transcriptional analysis of human host response to healthy gut microbiota. mSystems. 1: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, et al. 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43:e47.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittirsch D, Flierl MA, Ward PA. 2008. Harmful molecular mechanisms in sepsis. Nat Rev Immunol. 8:776–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, Oshlack A. 2010. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11:R25.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose E, Small CM, Saucedo HA, Harper C, Jones AG. 2014. Genetic evidence for monogamy in the dwarf seahorse, Hippocampus zosterae. J Hered. 105:828–833. [DOI] [PubMed] [Google Scholar]
- Saxena M, Yeretssian G. 2014. NOD-like receptors: master regulators of inflammation and cancer. Front Immunol. 5:327.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider M, et al. 2012. The innate immune sensor NLRC3 attenuates toll-like receptor signaling via modification of the signaling adaptor TRAF6 and transcription factor NF-kappaB. Nat Immunol. 13:823–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semova I, et al. 2012. Microbiota regulate intestinal absorption and metabolism of fatty acids in the zebrafish. Cell Host Microb. 12:277–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CC, Snowberg LK, Gregory Caporaso J, Knight R, Bolnick DI. 2015. Dietary input of microbes and host genetic variation shape among-population differences in stickleback gut microbiota. isme J. 9:2515–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer F, Nookaew I, Sommer N, Fogelstrand P, Backhed F. 2015. Site-specific programming of the host epithelial transcriptome by the gut microbiota. Genome Biol. 16:62.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutz WE, Schmerer M, Coates JL, Bolnick DI. 2015. Among-lake reciprocal transplants induce convergent expression of immune genes in threespine stickleback. Mol Ecol. 24:4629–4646. [DOI] [PubMed] [Google Scholar]
- Sullam KE, et al. 2015. Divergence across diet, time and populations rules out parallel evolution in the gut microbiomes of Trinidadian guppies. ISME J. 9:1508–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nas A, et al. 2010. Expression quantitative trait loci: replication, tissue- and sex-specificity in mice. Genetics 185:1059–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodarska M, et al. 2014. NLRP6 inflammasome orchestrates the colonic host-microbial interface by regulating goblet cell mucus secretion. Cell 156:1045–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TD, Nacu S. 2010. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 26:873–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.