Abstract
Conifers’ exceptionally large genome (20–30 Gb) is scattered with 60% retrotransposon (RT) components and we have little knowledge on their origin and evolutionary implications. RTs may impede the expression of flanking genes and provide sources of the formation of novel small RNA (sRNAs) populations to constrain events of transposon (TE) proliferation/transposition. Here we show a declining expression of 24-nt-long sRNAs and low expression levels of their key processing gene, pgRTL2 (RNASE THREE LIKE 2) at seed set in Picea glauca. The sRNAs in 24-nt size class are significantly less enriched in type and read number than 21-nt sRNAs and have not been documented in other species. The architecture of MIR loci generating highly expressed 24-/21-nt sRNAs is featured by long terminal repeat—retrotransposons (LTR-RTs) in families of Ty3/Gypsy and Ty1/Copia elements. This implies that the production of sRNAs may be predominantly originated from TE fragments on chromosomes. Furthermore, a large proportion of highly expressed 24-nt sRNAs does not have predictable targets against unique genes in Picea, suggestive of their potential pathway in DNA methylation modifications on, for instance, TEs. Additionally, the classification of computationally predicted sRNAs suggests that 24-nt sRNA targets may bear particular functions in metabolic processes while 21-nt sRNAs target genes involved in many different biological processes. This study, therefore, directs our attention to a possible extrapolation that lacking of 24-nt sRNAs at the late conifer seed developmental phase may result in less constraints in TE activities, thus contributing to the massive expansion of genome size.
Keywords: 21- and 24-nt-long small RNAs, MIR loci, long terminal repeat-retrotransposons (LTR-RTs), seed set, conifers, Picea glauca
Introduction
Two distinctive categories of plant regulatory small RNAs (sRNAs) are generated from single- or double-stranded RNA precursors; the former can form self-complementary foldback structures in hairpin shapes, primarily known as microRNAs (miRNAs), and the latter often refers to small interfering RNAs (siRNAs). The sRNA duplexes range in size from 18 to 24 nucleotides (nt) owing to diverse DICER-LIKE ribonucleases (DCLs) (Henderson et al. 2006). Vascular plant sRNAs are typically found in two predominant size classes, 21- and 24-nt (Chávez Montes et al. 2014). Canonical plant miRNAs are 20- to 22-nt-long and mediate target gene cleavage or protein translation inhibition, while 24-nt sRNAs classically comprise siRNAs (Ghildiyal and Zamore 2009). Yet, some miRNAs in Arabidopsis thaliana and Oryza sativa are 24-nt-long and after biogenesis, enter into the heterochromatic siRNA effector pathway (Vazquez et al. 2008; Chellappan et al. 2010; Wu et al. 2010). In angiosperms, the majority of endogenous sRNAs are 24-nt-long siRNAs (Chen et al. 2010), which guide target chromatin remodeling via the RNA-directed DNA methylation (RdDM) pathway (Du et al. 2015; Matzke et al. 2015), but other seed plant lineages have inconsistent patterns. For instance, conifers (e.g., Picea abies, Pinus contorta) do not yield conspicuous 24-nt-long sRNA mass (Dolgosheina et al. 2008; Morin et al. 2008; Källman et al. 2013; Nystedt et al. 2013), while a 24-nt size class is prevalent in Cycas rumphii and has an elevated production in Ginkgo biloba and Marsilea quadrifolia (Chávez Montes et al. 2014). This suggests that the 24-nt sRNAs have originated prior to the gymnosperm diversification. Ribonuclease III (RNase III) is essential to the processing and maturation of sRNAs (Comella et al. 2008). A previous presumption was that conifers have a novel DCL (a type of RNase III), while lack of the DCL3 enzyme that matures 24-nt long RNAs in angiosperms (Dolgosheina et al. 2008). However, DCL3 has been found in all representative plant groups (Ma et al. 2015) and multiple DCL3 homologs have recently been identified in conifers (Wan et al. 2012; Zhang et al. 2013; Niu et al. 2015). The enigma of lacking 24-nt sRNA populations in conifers, therefore, remains an intriguing ambiguity particularly from the perspective of biological significances.
Evolutionary forces drive gymnosperms to produce many haploid eggs within an ovule, increasing the chance for selection among female gametophytes (Lenormand and Dutheil 2005), while to reduce frequencies of whole genome duplications (i.e., polyploidization) in all but Ephedra (Gnetales) (Leitch and Leitch 2012). This indicates alternative mechanisms to constrain genetic divergence (e.g., epigenetics and activities of transposable elements (TEs)) in gymnosperms. The conifer genomes are elusively large (20–30 gigabases) (De La Torre et al. 2014) and abundant and diverse retrotransposons (RTs) are the main component of non-genic portions (Hamberger et al. 2009; Kovach et al. 2010). It is estimated that 62% of Pinus taeda genome is composed of RTs, of which 70% are long terminal repeats (LTRs), mainly Pseudoviridae (also known as Ty1/Copia elements) and Metaviridae (Ty3/Gypsy) (Nystedt et al. 2013; Neale et al. 2014; Wegrzyn et al. 2014). Excessive TEs can destroy eukaryotic genome and many organisms have developed diverse mechanisms to inhibit TE activities, including RNA-based silencing pathways (Almeida and Allshire 2005; Nosaka et al. 2012). Some TEs fold into stem-loop secondary structures and thus potentially contribute to the formation of sRNAs (Piriyapongsa and Jordan 2008; Sun et al. 2012). Endogenous 24-nt-long siRNAs are enriched in intergenic and repetitive genomic regions (Kasschau et al. 2007; Zhang et al. 2007), and more generally, most plant MIR loci are mapped to intergenic segments (Piriyapongsa and Jordan 2008). The feature of exceptionally large amount of excessive genomic DNA in conifers implies a rich source of TE-derived sRNAs, thus building a distinct landscape of sRNA populations targeting TEs and genes involved in various biological processes.
In this study, we chose three Picea glauca populations with contrasting seed set patterns in terms of developmental durations (supplementary fig. S1, Supplementary Material online) and sequenced sRNA molecules for developing seeds at four phases. We focused on hairpin-derived small RNAs (i.e., hpRNAs, containing miRNAs and hairpin-siRNAs), and hypothesized that those sRNAs orchestrate reproductive programs and coevolve with the genome. We aimed to delineate the landscape of sRNA and hpRNA populations by reads and abundances across seed set with special emphasis on two size classes, 21- and 24-nt, to computationally predict the origin of mature sRNAs (i.e., endogenous loci of MIR genes), and to classify putative target genes. We expected to extrapolate what features of MIR loci are evidenced for conifer genome evolution and in what role that mature hpRNAs may be at play in the evolution of genome size and architecture. Addressing these questions reveals meaningful insights into the joint evolution of the genome with sRNA modulators.
Materials and Methods
Plant Material, Growing Conditions, and Sample Collection
Three populations of white spruce (Picea glauca), which bear different timing of fertilization and seed set duration, were selected and 20 developing cones for each population were collected at early, middle, and late seed set for a total of four time points in the Kalamalka seed orchard (50°–51°37'N, 119°16'–120°29'W), British Columbia, Canada (supplementary fig. S1, Supplementary Material online). After dissection from white spruce cones, developing seeds were immediately frozen in liquid nitrogen and stored at − 80 °C until further use.
RNA Isolation, Library Construction, and sRNA Sequencing
Total RNAs were extracted from developing seed samples using PureLink Plant RNA Reagent (Ambion) according to the instruction from the manufacturer. The integrity and the quantity of the RNAs were assessed by BioAnalyser 2100 (Agilent Technologies) and Nanodrop ND-1000 spectrophotometer (Thermo Fisher Scientific). Methods for sRNA library construction followed that previously described (Chu et al. 2015) with minor modifications. Briefly, sRNA-seq libraries were constructed using a strand-specific and plate-based protocol. To enrich sRNAs, total RNA samples underwent polyA selection using Miltenyi MultiMACS mRNA isolation kit (cat. 130-092-519) following the manufacturer’s protocol and the flowthrough (i.e., containing sRNA species without mRNA) was used for plate-based sRNA construction. A 3′ adapter that is an adenylated single-stranded DNA was selectively ligated to the sRNA template using a truncated T4 RNA ligase 2 (NEB Canada, cat. M0242L). A 5′ adapter was then added using a T4 RNA ligase (Ambion USA, cat. AM2141) and ATPs. After ligation, first-strand cDNA was synthesized using a Superscript II Reverse Transcriptase (Invitrogen, cat. 18064 014) and one RT primer. This was the template for the final library PCR, into which 6-nt mers index was introduced to identify libraries (i.e., demultiplexed) from a sequenced pool. Constructed libraries were pooled in one 31 base SET lane and sequencing (Illumina HiSeqTM 2500) was performed using one short SET indexed lane in pool (BC Cancer Agency Genome Sciences Centre, Vancouver, Canada).
Small RNA Dataset Analysis
The sequence data are separated into individual libraries based on the index read sequences, and the reads underwent an initial QC assessment (Chu et al. 2015). After being preprocessed to clean reads by trimming adapters and barcode sequences using an internal matching algorithm (BC Cancer Agency), the raw sequencing data (bam format) were parsed into fastq and fasta formats under Linux in a command-line environment for subsequent use. The sRNA toolbox was used to profile sRNAs and size distribution (Rueda et al. 2015). Highly enriched miRNAs in sRNA sequencing libraries (≥ 1,000 copies in at least one phase) were computationally predicted against the P. glauca genome assemblies (PG29 v3, 20Gb divided into 30Mb per file) (Warren et al. 2015) using miRPlant (An et al. 2014) and their mRNA targets were predicted using transcripts without miRNA genes on psRNATarget (Dai and Zhao 2011) with default options for search parameters. As the conifer genome massively accumulates transposable elements [43.4% of loblolly pine genome is composed of long terminal repeats (LTRs)] (De La Torre et al. 2014) and they are potential sources for sRNAs generation and targets, the predicted MIR loci of hairpin structure (∼160 nt) for highly abundant miRNAs and target genes were used for the identification of retroelements with autonomous LTRs, including Ty1/Copia, Ty3/Gypsy, Bel/Pao, Retroviridae, and Caulimoviruses (Llorens et al. 2011). Due to most conifer genes unannotated, homologs for miRNA-targeted genes in P. glauca were retrieved via a BLASTN search against Arabidopsis genome on EnsemblPlants (http://plants.ensembl.org). To annotate target mRNA functions, top one predicted target gene for each miRNA of high abundance was aligned against the Gene Ontology (GO) protein database for GO term classification and KEGG pathway enrichment (Ashburner et al. 2000; Kanehisa and Goto 2000).
Gene Expression Analysis
AtDCL3 gene expression map during Arabidopsis seed set was browsed by Arabidopsis eFP 2.0 (Schmid et al. 2005). To retrieve ‘pgDCL3’, tBLASTN was executed through AtDCL3 protein with reference to conserved domains against translated (six frames) nucleotide using P. glauca PlantGDB Putative Unique Transcripts (PUTs) database on ConGenIE (http://congenie.org/). The relative expression of putative ‘pgDCL3’ was assayed using quantitative RT-PCR (qRT-PCR) as follows. Two µg of the other aliquot of total RNAs was reverse-transcribed into cDNA using the EasyScript Plus™ kit (abmGood, cat. G235) with oligo-dT primers following the instructions of the manufacturer and first-strand cDNA synthesis products were diluted fivefold as qRT-PCR template. qRT-PCR was run in 15 µl reaction volumes on an ABI StepOnePlus™ machine (Life Technologies) using the PerfeCTa® SYBR® Green SuperMix with ROX (Quanta Biosciences, cat. 101414 152). The reaction components and procedure were carried out as previously described (Liu et al. 2015). Three technical replicates of each of three biological replicates were used. Reference genes were used as previously described (Czechowski et al. 2005; Liu et al. 2015). In addition, the primer pair used for ‘pgDCL3’ amplication was 5'-GGAAGCAGAGGAGACAAAGG and 3'-CCGCCCTCACTTATACACCT.
Results
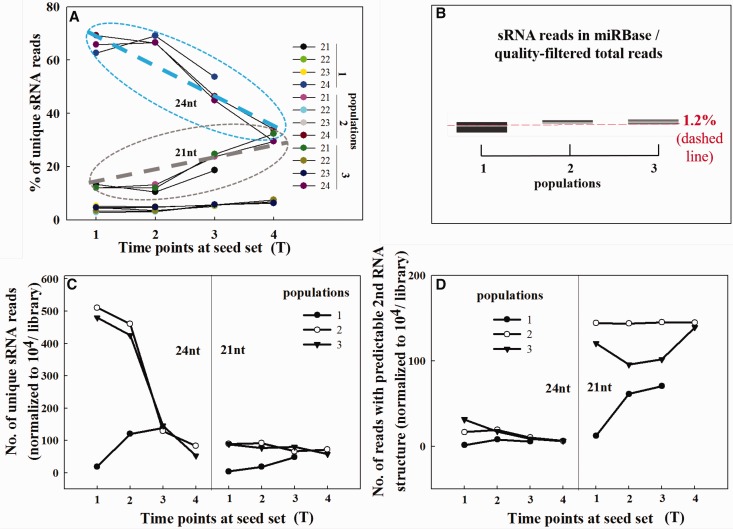
Prior to maturation at seed set (T1-2), sRNA repertoire was enriched in 24-nt-long reads across the three populations (∼66.59%) (fig. 1A and supplementary fig. S2, Supplementary Material online), while at maturation, the proportion declined from an average of 48.28 at T3 to 31.72% at T4 (fig. 1A ). By contrast, the percentage of 21-nt sRNAs pronouncedly increased, on average, from 12.23% at T1∼2 to 22.35 and 30.93% at T3 and T4, respectively (fig. 1A ). These findings are in agreement with previous investigations (Dolgosheina et al. 2008; Morin et al. 2008; Wan et al. 2012) and observations in genome sequencing of Picea abies (Nystedt et al. 2013) concerning the highly specific expression of 24-nt sRNAs in reproductive tissues. The sRNA abundance of studied species across the plant kingdom ranged from 0.14 to 41% of total sRNAs identified in libraries, in which Picea abies is 25% (Chávez Montes et al. 2014). In the repertoires for quality-filtered sRNAs, only an average of 1.2% sRNA families has been curated in miRBase across the studied three populations (fig. 1B , reads curated in supplementary table S2, Supplementary Material online). This percentage is rather small compared with 5% counterpart in Arabidopsis thaliana flowers (Mosher et al. 2009). Interestingly, of the detected sRNAs, only some 21∼22-nt families have been archived in miRBase and no 24-nt reads from this study had records in miRBase (supplementary table S1, Supplementary Material online). This indicates that a small portion of coniferous short sRNAs (19–22-nt-long) belongs to documented conserved sRNAs in plant lineages and conifers may produce unique long sRNAs (23–25-nt-long) at seed set to execute unknown functions. In general, the absolute number of unique sRNAs in 24-nt-long outnumbered that in the size of 21-nt across populations over time (fig. 1C ), particularly, at early phases in population 2 and 3 (T1–2), the burst of diversified 24-nt sRNA production was 4–5 times more than in other phases (fig. 1C ). However, the number of 24-nt sRNAs (i.e., sRNAs mapped to the genome with predictable secondary RNA structures) did not exceed that of 21-nt sRNAs (fig. 1D ). The large portion of 24-nt sRNAs that was not hpRNAs (left panels in fig. 1C and D ) may belong to siRNA categories (e.g., heterochromatic siRNA class). Moreover, numerous low abundant 21-nt sRNAs were synthesized across time at seed set (compare right panels in fig. 1C and D ).
Fig. 1.—
Distribution of sRNAs by size (A) and by populations (B), and absolute read numbers of 24- and 21-nt sRNAs (C and D). Note: The long thick dashed lines in ellipses represent the relative change on read numbers of 24nt (in blue) and 21nt (in grey); changes on absolute number of unique sRNA reads (C) and on that of reads mapped to miRBase hairpins (D) in each population over time during seed set of P. glauca; sequencing statistics was presented in supplementary table S1 (Supplementary Material online).
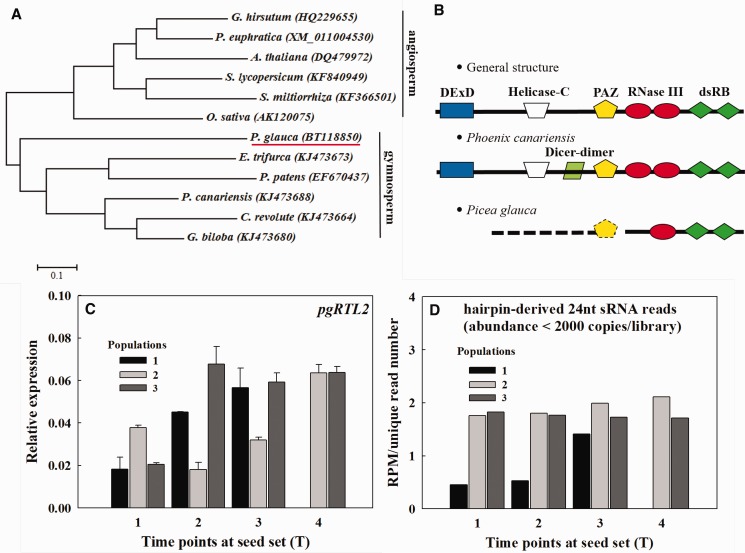
The gene tree for DCL3 homologs was divided into two major clades corresponding to two spermatophytes: angiosperm and gymnosperm (fig. 2A ). Compared with the linear arrangement of domains typically found in DCL3 proteins, DCL3 in Picea glauca lacks two domain types (i.e., DExD-helicase and helicase-C) (fig. 2B ). PAZ, RNaseIII and dsRB domains are thought to be crucial for specific recognition and spatial cleavage of dsRNAs into sRNAs (Zhang et al. 2004). Recent reports showed that RNASE THREE-LIKE 2 (RTL2, another type of RNase III) only contains one RNase III and two dsRB domains and cleaves dsRNAs into longer molecules (e.g., 24-nt) (Shamandi et al. 2015; Elvira-Matelot et al. 2016). Along with this evidence, our investigation, therefore, provides an indication that Picea glauca produces 24-nt sRNAs possibly via RTL2 instead of DCL3. The relative expression of RTL2 in P. glauca was significantly lower than that in Arabidopsis (compare the range of vertical axes in fig. 2C and supplementary S3, Supplementary Material online). Moreover, the relative expression of pgRTL2 had tendency to increase during seed ripening across the three populations (fig. 2C ); while RPM scaled by unique read counts increased significantly and slightly for population 1 and 2, respectively, and they kept in a constant high level for population 3 (fig. 2D ). This indicates that pgRTL2 expression level is in agreement with the abundance of 24-nt reads (more than 2,000 copies by library excluded).
Fig. 2.—
Comparisons of DCL3 homologs (A and B), pgRTL2 relative expression (C) and RPM scaled by the number of unique reads (D). Note: gene trees for DCL3 homologs in gymnosperms and several model angiosperms (A); components of DCL3 domains in Arabidopsis (general), Phoenix canariensis and Picea glauca (B); PAZ and dsRB represent Piwi–Argonaute–Zwille and double stranded RNA-binding domains, respectively; thick dashed line in (B) means the upstream of incomplete pgRTL2 mRNA may contain PAZ domain after mapped to its genome (PG29-v.4); RPM represents reads per million and if the absolute expression of the 24-nt sRNA exceeds 2,000 copies in a single library, it is not used for the calculation of RPM and RPM per unique read number. We excluded around 4-5 sRNA reads per library (see supplementary fig. S4 C, Supplementary Material online).
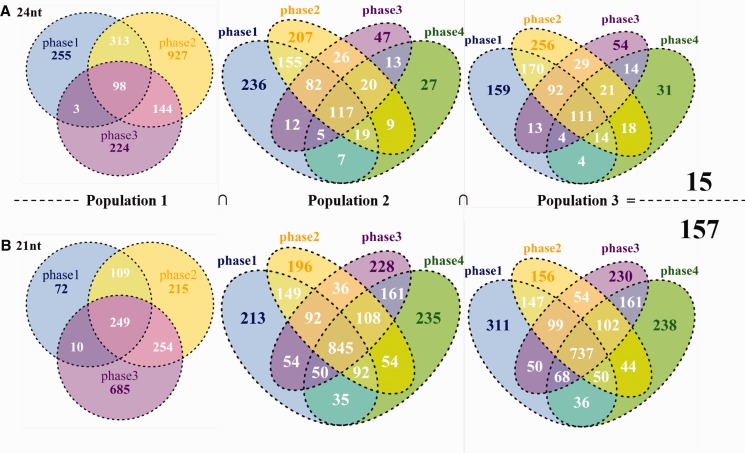
Approximately 100 hpRNAs in 24-nt-long were identified throughout seed set phases in each of the three studied populations (fig. 3A ), but in which only 15 were shared across populations (fig. 3A and listed in supplementary table S3, Supplementary Material online). These 15 hpRNA reads were enriched in medium (copies bounded between [200, 2,000] per sRNA read in at least one phase) to high (>2,000 copies per sRNA read) levels (supplementary table S3, Supplementary Material online). The distribution of 24-nt hpRNA reads in time and population (supplementary fig. S4, Supplementary Material online) showed that highly expressed hpRNAs greatly affected changes on the total sRNA reads over time in populations (fig. 1D ). By contrast, ∼250, 845, and 740 hpRNAs in 21-nt-long were detected across phases in population 1–3 (fig. 3B ), in which, 157 were in common across populations with medium to high expression levels (fig. 3B and supplementary table S3, Supplementary Material online). In addition to more variants over time (fig. 3B ), 21-nt hpRNA reads were also featured in production at high (copies ranged from 10,000 to 50,000 per sRNA read in a single phase) and extremely high (up to 800,000 copies per sRNA) levels (supplementary fig. S5, Supplementary Material online). These discrepant features between the two sRNA classes imply that 24-nt hpRNAs in conifers may target genes with a specific function fine-tuning developmental programs, while 21-nt sRNAs regulate genes covering a spectrum of functions.
Fig. 3.—
Numbers of sRNA reads of 24- (A) and 21-nt (B) by seed set phases in populations. Note: the number of reads detected throughout phases and populations respectively provided in black and detailed information listed in supplementary table S3, Supplementary Material online.
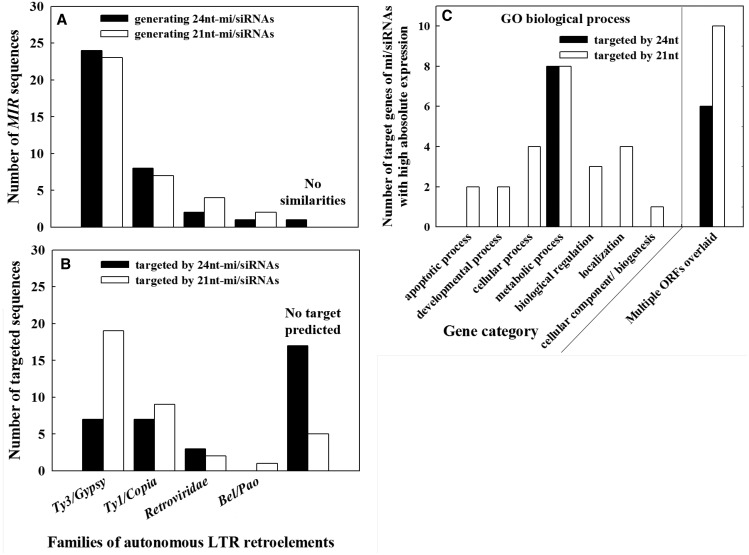
In light of the principle that ‘conserved’ sRNAs are usually highly and constantly expressed (Chávez Montes et al. 2014), we selected the 36 most highly and/or constantly expressed sRNAs in 24- and 21-nt populations for subsequent comparisons (all miRs in supplementary table S4, Supplementary Material online and all 24-nt and highlighted 21-nt miRs in supplementary table S3, Supplementary Material online). An overview of typical secondary MIR structures is available in supplementary fig. S6 (Supplementary Material online). The architecture of MIR loci had characteristics of autonomous long terminal repeat-retrotransposons (LTR-RTs), particularly in families of Ty3/Gypsy and Ty1/Copia (fig. 4A ). Ty1/Copia RTs are often scattered on the chromosomes, whereas Ty3/Gypsy elements preferentially accumulate in specific chromosomal locations and structures (e.g., (peri-)centromeric regions) (Neumann et al. 2011). More Ty3/Gypsy elements (fig. 4A ) thus suggest that the biogenesis of 21- or 24-nt sRNAs may have site bias on chromosomes. A large portion of 24-nt sRNAs did not target genes (note that we used Picea unigene libraries for target search) (fig. 4B ), supporting their roles in TE silencing. The classification of sRNA target genes showed that genes targeted by 24-nt sRNAs were specifically involved in metabolic processes, while 21-nt sRNAs targeted genes encompassing seven gene categories, also evidenced by the fact that the functionality of 21-nt sRNAs is mainly associated with genes (Dolgosheina et al. 2008; Morin et al. 2008; Wan et al. 2012; Nystedt et al. 2013) (fig. 4C and targets curated in supplementary tables S3 and 4, Supplementary Material online). These findings reinforce the notion that 24-nt sRNAs may target genes with specific functions at reproductive programs in conifers. In addition, homologs for some putative target mRNAs in P. glauca were related to multiple ORFs for overlapped genes in Arabidopsis (fig. 4C ).
Fig. 4.—
Analyses of abundant sRNA populations in 24-/21-nt size classes via their biogenesis sequences (A) and target genes (B and C). Note: the highly expressed sRNA reads provided in supplementary table S4 (Supplementary Material online) with details on computational predictions; one gene may be classified in more than one gene category or none of the categories; gene category and GO code: apoptotic process (GO:0006915), developmental process (GO:0032502), cellular process (GO:0009987), metabolic process (GO:0008152), biological regulation (GO:0065007), localization (GO:0051179), and cellular component organization or biogenesis (GO:0071840).
Discussion
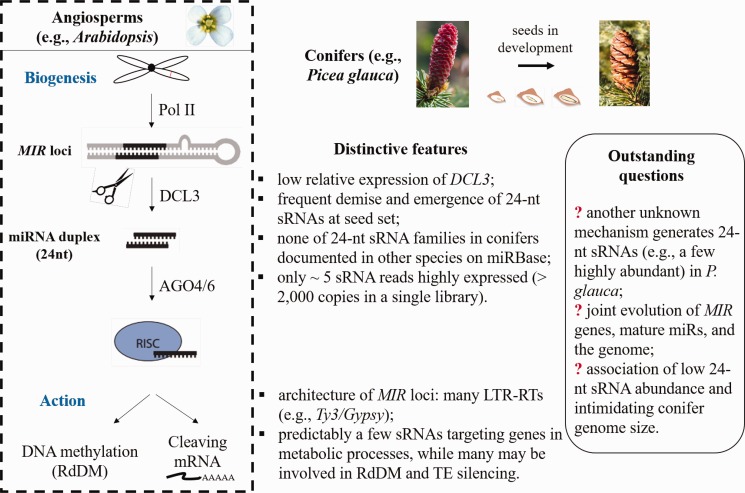
In this study, we unraveled the spatiotemporal expression pattern of sRNAs and computationally predicted loci of MIR genes and putative targets using sieved 21- and 24-nt-long hpRNAs in Picea glauca. The landscape of sRNA production at seed set manifested the burst of 24-nt-long sRNAs particularly in early and middle stages, and none of these sRNA families has been documented in other species in miRBase (summarized in fig. 5). The low pgRTL2 abundance and the frequent demise and emergence of different 24-nt sRNAs at different seed set stages in different populations indicate distinctive production mechanism and function of 24-nt sRNAs in conifers. Moreover, the origin and feature of MIR loci may direct our attention to a possible rationale concerning the joint evolution of MIR loci, mature sRNAs, and target genes, which may be coupled in time with genome evolution.
Fig. 5.—
Summary of distinctive features of 24-nt sRNAs and outstanding questions regarding their production in conifers. Note: Pol II, DCL3, and AGO4/6 are abbreviations of polymerase II, Dicerlike 3, and argonaute 1/4/7, respectively, which are key effectors in 24-nt sRNA biogenesis; RdDM represents the RNA-directed DNA methylation pathway.
It has become clear that pervasive non-coding RNA transcripts are involved in the regulation of genome functions (Ponting et al. 2009; Wilusz et al. 2009). Epigenetic variation (e.g., methylation sites) is a key player in the evolution of biological diversity (Diez et al. 2014; Turck and Coupland 2014) and DNA methylation changes between parents and their progeny in Arabidopsis are characterized by the abundance of 24-nt siRNAs through the RNA-directed DNA methylation (RdDM) pathway (Zhang et al. 2016). Such changes are correlated with altered genetic variation within the genome, suggesting that they may play an important role in genome evolution. In addition, small non-coding RNAs are involved in global cytosine methylation (Finnegan 2010; Hirsch et al. 2012), which contributes to genome size evolution via silencing redundant genes and regulating the activity of TEs (e.g., polyploidization) (Song and Chen 2015; Alonso et al. 2016; Springer et al. 2016). As such, the substantial decrease of 24-nt sRNAs at late seed set in conifers may have correlation with the insufficient suppression of replicative mechanisms of TEs that may result in invasive genome expansion.
TEs have been mostly regarded as parasitic DNA (Doolittle and Sapienza 1980; Orgel and Crick 1980) and allow the evolutionary increase in size and complexity of the plant genome (Fedoroff 2012). The TE insertion may trigger epigenetic modifications in surrounding genome sequences, suggesting that important epigenetic mechanisms originally evolved to constrain the activity of TEs and thus maintain genome integrity, such as RNA-based silencing pathways (Almeida and Allshire 2005; Nosaka et al. 2012), histone modifications (Gao et al. 2008; Huda et al. 2011), or methylations (Yoder et al. 1997). The functionality of TEs depends on the TE chromosomal context (e.g., near a gene, within a gene, in a pericentromere/TE island, and at the centromere core) (Sigman and Slotkin 2016). As 24-nt sRNAs are largely implicated in TE silencing (Henderson and Jacobsen 2007), the dynamic changes on the expression of 21- and 24-nt sRNAs (figs. 1 and 3) indicate that the epigenetic landscape was differentially regulated throughout phases at seed set of P. glauca. On the other hand, TEs contribute to the generation of species-specific MIR genes (Nozawa et al. 2012) and both 21- and 24-nt-long silencing RNAs can be produced from TEs (Kasschau et al. 2007). Small RNA-generating loci, especially those spawning predominantly 24-nt siRNAs, were highly correlated with repetitive elements across the genome (Kasschau et al. 2007). Our results conclusively showed that TEs constitute a vital source of conifer-specific sRNA populations (fig. 4A ).
The number of MIR genes has increased substantially from green algae to land plant lineages but altered in a lineage-specific manner after the divergence of eudicots and monocots (Nozawa et al. 2012). Similar to protein-coding genes, the evolution of MIR genes is affected by genomic features (Maher et al. 2006; Axtell and Bowman 2008; Zhao et al. 2015) and correlated with the utility of certain paralogs of DCLs resulting in specific size classes of sRNAs (Vazquez et al. 2008). However, our tBLASTN search and conserved domain analysis corroborate the availability of RTL2 instead of DCL3 homolog in P. glauca (fig. 2) and RTL2 is likely to play a dual role in modulating the accumulation of 24-nt sRNAs in the RdDM pathway (Elvira-Matelot et al. 2016). Positive and negative regulation of sRNAs via RTL2 may explain conspicuous changes in the net production of 24-nt sRNAs between reproductive and vegetative life stages in P. glauca. DCL3-dependent siRNA production system associated with transposons and other non-genic regions of the genome is an ancestral feature within land plants, although the size of relevant siRNAs differs between lineages (Cho et al. 2008). The existence of DCL3 homologs in other conifers (Wan et al. 2012; Zhang et al. 2013; Niu et al. 2015) remains supportive to DCL3 as a common feature in plant lineages, while the lack of DCL3 in P. glauca reports an exception in the gene phylogeny and may indicate a possible bifurcation in gene evolution. In general, ancient MIR genes give rise predominantly to 21-nt-long RNAs, generated by DCL1 and regulating targets via mRNA cleavage or translation inhibition, while recently evolved MIR genes can be processed by any member of the DCL family proteins into diverse lengths with great tendency toward longer sRNAs (e.g., 24-nt-long), consequently repressing targets via various modes (e.g., mRNA cleavage, chromatin remodeling) (Vazquez et al. 2008; Nozawa et al. 2012). As such, production and enrichment of 24-nt-long sRNAs may embody sRNA evolution in coordination with genomic features. Moreover, duplicated MIR genes (again similar to protein-coding genes) exhibit higher expression than singletons (Liu et al. 2016), in agreement with our observation of the extremely high expression of TE-derived sRNAs, particularly represented by ancient sRNAs in 21-nt-long (supplementary table S4 and figs. S4 and S5, Supplementary Material online).
In general, MIR genes have higher evolutionary rates than small RNA targets (Liu et al. 2016), while MIR genes that are conserved across species have a relatively slow evolutionary rate (Abrouk et al. 2012; Smith et al. 2015). In contrast, conserved sRNAs experience stronger purifying selection (Ehrenreich and Purugganan 2008; Takuno and Innan 2011). This opinion is favored by our findings, i.e., conifer-specific ‘conserved’ sRNAs (supplementary table S3, Supplementary Material online). To maintain regulatory functionality and base-pairing ability, sRNAs have undergone joint evolution with their target genes when targets are subject to functional divergence (Felippes et al. 2008). Functions of conserved sRNAs in different plants are scarcely diverse due to long-term evolutionary selection of sRNA-target interplays (Voinnet 2009; Axtell 2013).
Regarding the two major sRNA classes, 21-nt sRNAs are more enriched in gymnosperms; while in angiosperms, 24-nt sRNAs are predominant in number (Chen et al. 2010; Chávez Montes et al. 2014) and primarily involved in properties of DNA methylation and heterochromatinization (Law and Jacobsen 2010). This indicates that in gymnosperms, the 21-nt sRNAs may replace heterochromatin siRNAs or the number of sRNA genes plays a more important role (Lelandais-Brière et al. 2010). At early-to-middle reproductive phases, the conifers are able to spawn millions of 24-nt sRNAs (fig. 1A and C ) comprising a significant amount of 24-nt sRNAs (fig. 1D ), indicating that 24-nt sRNAs are not an angiosperm-specific innovation and may restrain TE activities (fig. 4B ) or harbor particular functions specifically at seed set (fig. 4C ). Methylation of TEs in developing seeds seems to be driven by maternally inherited 24-nt-long siRNAs (Calarco et al. 2012), and due to their mobility between cells (Molnar et al. 2010), transmitting RNA silencing signal into developing seed or pollen (Mosher et al. 2009; Slotkin et al. 2009) may induce epigenetic changes that ultimately initiate transgenerational effects. Moreover, the basal angiosperm, Amborella trichopoda, lacks evidence of recent genome duplications and transposon insertions but is coincident with enriched 23/24-nt-long-miRNAs (78%) ( Amborella Genome Project 2013). The long-lived forest tree, populus, has 1/24 size of conifer genome and produces abundant 24-nt sRNAs (Klevebring et al. 2009). We, therefore, postulate that failure to maintain 24-nt sRNA-mediated silencing of TE activities at late developmental programs (fig. 1D ) and/or lacking 24-nt sRNA transport from maternal tissues (Dolgosheina et al. 2008; Morin et al. 2008; Källman et al. 2013; Nystedt et al. 2013) may contribute to the massive proliferation of mobile elements in conifers. Each species possesses unique sets of sRNA families in an organ-specific manner with broad predicted targets also as crucial determinants of various biological processes. These non-universally conserved sRNAs are tailored to individual species, involved in specific functions in response to environmental settings, and may evolve in coalition with genome evolution, as observed in this study (fig. 3 and supplementary table S3, Supplementary Material online). Species-specific conserved sRNAs should occupy a crucial position to exert adapted functions.
In C. Darwin’s eyes, the obscure origin of angiosperms in fossil records is a ‘perplexing phenomenon’ and an ‘abominable mystery’. As living fossils, extant subgroups within gymnosperms rarely have undergone polyploidization but are quite diverse in terms of genome size while large genome size variation in gymnosperms reflects more or less the lack of effective mechanisms to govern TE activities, thus likely linking to different rates at which new species occur. 24-nt-long sRNAs act as genome ‘guardians’, providing multigenerational protections against invasive TEs. Through this study, we postulate that lacking the regulation of 24-nt-long sRNAs that occurs at late seed set in conifer alters developmental programs and may lead to increased TE activities and exceptionally large genome size. The balance between large amount of TEs and novel sRNAs originated from foldback TEs to in turn restrain TE expansion may steer the direction and speed of genome evolution. This study, therefore, provides important evolutionary insights in a possible mechanism on genome evolution. Further understanding of whether such a connection is true in the plant kingdom requires experimental validation assays that motivate future studies for the correlation of the decrease in 24-nt sRNAs, DNA methylation changes, and TE invasion in the genome.
Data Accessibility
The sRNA sequencing data has been deposited at the Sequence Read Archive (SRA) in the National Center for Biotechnology Information (NCBI) under the accession number, SRP096198. (N.B. only populations 1, 2, and 4 used in this study.)
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Author Contributions
Y.L. conceived this study, performed data analyses, and wrote the manuscript; Y.A.E. coordinated the project.
Supplementary Material
Acknowledgments
We would like to extend our sincere gratitude to B. Jaquish (Ministry of Forestry) for sampling developing cones of white spruce (P. glauca) and to R. Hamelin (UBC) for providing ABI StepOnePlusTM machine. We are equally thankful to the constructive comments from three anonymous referees. This project was supported by the Johnson’s Family Forest Biotechnology Endowment and the National Science and Engineering Research Council (NSERC) of Canada Discovery and Industrial Research Chair to Y.A.E.
Literature Cited
- Abrouk M, et al. 2012. Grass microRNA gene paleohistory unveils new insights into gene dosage balance in subgenome partitioning after whole-genome duplication. Plant Cell. 24:1776–1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida R, Allshire RC. 2005. RNA silencing and genome regulation. Trends Cell Biol. 15:251–258. [DOI] [PubMed] [Google Scholar]
- Alonso C, Balao F, Bazaga P, Pérez R. 2016. Epigenetic contribution to successful polyploidizations: variation in global cytosine methylation along an extensive ploidy series in Dianthus broteri (Caryophyllaceae). New Phytol. 212:571–576. [DOI] [PubMed] [Google Scholar]
- Amborella Genome Project. 2013. The Amborella genome and the evolution of flowering plants. Science 342:1241089. [DOI] [PubMed] [Google Scholar]
- An J, Lai J, Sajjanhar A, Lehman ML, Nelson CC. 2014. miRPlant: an integrated tool for identification of plant miRNA from RNA sequencing data. BMC Bioinformatics 15:275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, et al. 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ. 2013. Classification and comparison of small RNAs from plants. Annu Rev Plant Biol. 64:137–159. [DOI] [PubMed] [Google Scholar]
- Axtell MJ, Bowman JL. 2008. Evolution of plant microRNAs and their targets. Trends Plant Sci. 13:343–349. [DOI] [PubMed] [Google Scholar]
- Calarco JP, et al. 2012. Reprogramming of DNA methylation in pollen guides epigenetic inheritance via small RNA. Cell 151:194–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chávez Montes RA, et al. 2014. Sample sequencing of vascular plants demonstrates widespread conservation and divergence of microRNAs. Nat Commun. 5:3722. [DOI] [PubMed] [Google Scholar]
- Chellappan P, et al. 2010. siRNAs from miRNA sites mediate DNA methylation of target genes. Nucleic Acids Res. 38:6883–6894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, et al. 2010. Small RNAs in angiosperms: sequence characteristics, distribution and generation. Bioinformatics 26:1391–1394. [DOI] [PubMed] [Google Scholar]
- Cho SH, et al. 2008. Physcomitrella patens DCL3 is required for 22-24 nt siRNA accumulation, suppression of retrotransposon-derived transcripts, and normal development. PLoS Genet. 4:e1000314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu A, et al. 2015. Large-scale profiling of microRNAs for the cancer genome atlas. Nucleic Acids Res. 44:e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comella P, et al. 2008. Characterization of a ribonuclease III-like protein required for cleavage of the pre-rRNA in the 3' ETS in Arabidopsis . Nucleic Acids Res. 36:1163–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. 2005. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139:5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Zhao PX. 2011. psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res. 39:W155–W159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Torre AR, et al. 2014. Insights into conifer giga-genomes. Plant Physiol. 166:1724–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez CM, Roessler K, Gaut BS. 2014. Epigenetics and plant genome evolution. Curr Opin Plant Biol. 18:1–8. [DOI] [PubMed] [Google Scholar]
- Dolgosheina EV, et al. 2008. Conifers have a unique small RNA silencing signature. RNA 14:1508–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle WF, Sapienza C. 1980. Selfish genes, the phenotype paradigm and genome evolution. Nature 284:601–603. [DOI] [PubMed] [Google Scholar]
- Du JM, Johnson LM, Jacobsen SE, Patel DJ. 2015. DNA methylation pathways and their crosstalk with histone methylation. Nat Rev Mol Cell Biol. 16:519–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenreich IM, Purugganan MD. 2008. Sequence variation of MicroRNAs and their binding sites in Arabidopsis . Plant Physiol. 146:1974–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elvira-Matelot E, et al. 2016. Arabidopsis RNASE THREE LIKE2 modulates the expression of protein-coding genes via 24-nucleotide small interfering RNA-directed DNA methylation. Plant Cell 28:406–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedoroff NV. 2012. Transposable elements, epigenetics, and genome evolution. Science 338:758–767. [DOI] [PubMed] [Google Scholar]
- Felippes FF, Schneeberger K, Dezulian T, Huson DH, Weigel D. 2008. Evolution of Arabidopsis thaliana microRNAs from random sequences. RNA 14:2455–2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan EJ. 2010. DNA methylation: a dynamic regulator of genome organization and gene expression in plants In: Pua EC, Davey MR, editors. Plant developmental biology—biotechnological perspectives. Vol. 2 Berlin, Germany: Springer-Verlag. [Google Scholar]
- Gao X, Hou Y, Ebina H, Levin HL, Voytas DF. 2008. Chromodomains direct integration of retrotransposons to heterochromatin. Genome Res. 18:359–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD. 2009. Small silencing RNAs: an expanding universe. Nat Rev Genet. 10:94–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamberger B, et al. 2009. Targeted isolation, sequence assembly and characterization of two white spruce (Picea glauca) BAC clones for terpenoid synthase and cytochrome P450 genes involved in conifer defence reveal insights into a conifer genome. BMC Plant Biol. 9:106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson IR, Jacobsen SE. 2007. Epigenetic inheritance in plants. Nature 447:418–424. [DOI] [PubMed] [Google Scholar]
- Henderson IR, et al. 2006. Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat Genet. 38:721–725. [DOI] [PubMed] [Google Scholar]
- Hirsch S, Baumberger R, Grossniklaus U. 2012. Epigenetic variation, inheritance, and selection in plant populations. Cold Spring Harbor Symp Quant Biol. 77:97–104. [DOI] [PubMed] [Google Scholar]
- Huda A, Bowen NJ, Conley AB, Jordan IK. 2011. Epigenetic regulation of transposable element derived human gene promoters. Gene 475:39–48. [DOI] [PubMed] [Google Scholar]
- Källman T, Chen J, Gyllenstrand N, Lagercrantz U. 2013. A significant fraction of 21-nucleotide small RNA originates from phased degradation of resistance genes in several perennial species. Plant Physiol. 162:741–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. 2000. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasschau KD, et al. 2007. Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol. 5:e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klevebring D, et al. 2009. Genome-wide profiling of populus small RNAs. BMC Genomics 10:620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovach A, et al. 2010. The Pinus taeda genome is characterized by diverse and highly diverged repetitive sequences. BMC Genomics 11:420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE. 2010. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 11:204–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch AR, Leitch IJ. 2012. Ecological and genetic factors linked to contrasting genome dynamics in seed plants. New Phytol. 194:629–646. [DOI] [PubMed] [Google Scholar]
- Lelandais-Brière C, et al. 2010. Small RNA diversity in plants and its impact in development. Curr Genomics 11:14–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenormand T, Dutheil J. 2005. Recombination difference between sexes: a role for haploid selection. PLoS Biol. 3:e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, et al. 2016. Global investigation of the co-evolution of MIRNA genes and microRNA targets during soybean domestication. Plant J. 85:396–409. [DOI] [PubMed] [Google Scholar]
- Liu Y, Müller K, El-Kassaby YA, Kermode AR. 2015. Changes in hormone flux and signaling in white spruce (Picea glauca) seeds during the transition from dormancy to germination in response to temperature cues. BMC Plant Biol. 15:292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorens C, et al. 2011. The Gypsy Database (GyDB) of mobile genetic elements: release 2.0. Nucleic Acids Res. 39:D70–D74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, et al. 2015. Angiosperms are unique among land plant lineages in the occurrence of key genes in the RNA-directed DNA methylation (RdDM) pathway. Genome Biol Evol. 7:2648–2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher C, Stein L, Ware D. 2006. Evolution of Arabidopsis microRNA families through duplication events. Genome Res. 16:510–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke MA, Kanno T, Matzke AJ. 2015. RNA-directed DNA methylation: the evolution of a complex epigenetic pathway in flowering plants. Annu Rev Plant Biol. 66:243–267. [DOI] [PubMed] [Google Scholar]
- Molnar A, et al. 2010. Small silencing RNAs in plants are mobile and direct epigenetic modification in recipient cells. Science 328:872–875. [DOI] [PubMed] [Google Scholar]
- Morin RD, et al. 2008. Comparative analysis of the small RNA transcriptomes of Pinus contorta and Oryza sativa . Genome Res. 18:571–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosher RA, et al. 2009. Uniparental expression of PolIV-dependent siRNAs in developing endosperm of Arabidopsis . Nature 460:283–286. [DOI] [PubMed] [Google Scholar]
- Neale DB, et al. 2014. Decoding the massive genome of loblolly pine using haploid DNA and novel assembly strategies. Genome Biol. 15:R59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann P, et al. 2011. Plant centromeric retrotransposons: a structural and cytogenetic perspective. Mobil DNA 2:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu SH, et al. 2015. Identification and expression profiles of sRNAs and their biogenesis and action-related genes in male and female cones of Pinus tabuliformis . BMC Genomics 16:693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosaka M, et al. 2012. Role of transposon-derived small RNAs in the interplay between genomes and parasitic DNA in rice. PLoS Genet. 8:e1002953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa M, Miura S, Nei M. 2012. Origins and evolution of MicroRNA genes in plant species. Genome Biol Evol. 4:230–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nystedt B, et al. 2013. The Norway spruce genome sequence and conifer genome evolution. Nature 497:579–584. [DOI] [PubMed] [Google Scholar]
- Orgel LE, Crick FH. 1980. Selfish DNA: the ultimate parasite. Nature 284:604–607. [DOI] [PubMed] [Google Scholar]
- Piriyapongsa J, Jordan IK. 2008. Dual coding of siRNAs and miRNAs by plant transposable elements. RNA 14:814–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP, Oliver PL, Reik W. 2009. Evolution and functions of long noncoding RNAs. Cell 136:629–641. [DOI] [PubMed] [Google Scholar]
- Rueda A, et al. 2015. sRNAtoolbox: an integrated collection of small RNA research tools. Nucleic Acids Res. 43:W467–W473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, et al. 2005. A gene expression map of Arabidopsis thaliana development. Nat Genet. 37:501–506. [DOI] [PubMed] [Google Scholar]
- Shamandi N, et al. 2015. Plants encode a general siRNA suppressor that is induced and suppressed by viruses. PLoS Biol. 13:e1002326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigman MJ, Slotkin RK. 2016. The first rule of plant transposable element silencing: location, location, location. Plant Cell 28:304–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin RK, et al. 2009. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136:461–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LM, et al. 2015. Rapid divergence and high diversity of miRNAs and miRNA targets in the Camelineae. Plant J. 81:597–610. [DOI] [PubMed] [Google Scholar]
- Song Q, Chen ZJ. 2015. Epigenetic and developmental regulation in plant polyploids. Curr Opin Plant Biol. 24:101–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer NM, Lisch D, Li Q. 2016. Creating order from chaos: epigenome dynamics in plants with complex genomes. Plant Cell 28:314–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Zhou M, Mao ZT, Li CX. 2012. Characterization and evolution of microRNA genes derived from repetitive elements and duplication events in plants. PLoS One 7:e34092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takuno S, Innan H. 2011. Selection fine-tunes the expression of microRNA target genes in Arabidopsis thaliana . Mol Biol Evol. 28:2429–2434. [DOI] [PubMed] [Google Scholar]
- Turck F, Coupland G. 2014. Natural variation in epigenetic gene regulation and its effects on plant developmental traits. Evolution 68:620–631. [DOI] [PubMed] [Google Scholar]
- Vazquez F, Blevins T, Ailhas J, Boller T, Meins F., Jr. 2008. Evolution of Arabidopsis MIR genes generates novel microRNA classes. Nucleic Acids Res. 36:6429–6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O. 2009. Origin, biogenesis, and activity of plant microRNAs. Cell 136:669–687. [DOI] [PubMed] [Google Scholar]
- Wan LC, et al. 2012. Identification and characterization of small non-coding RNAs from Chinese fir by high throughput sequencing. BMC Plant Biol. 12:146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren RL, et al. 2015. Improved white spruce (Picea glauca) genome assemblies and annotation of large gene families of conifer terpenoid and phenolic defense metabolism. Plant J. 83:189–212. [DOI] [PubMed] [Google Scholar]
- Wegrzyn JL, et al. 2014. Unique features of the loblolly pine (Pinus taeda L.) megagenome revealed through sequence annotation. Genetics 196:891–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, Sunwoo H, Spector DL. 2009. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 23:1494–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, et al. 2010. DNA methylation mediated by a microRNA pathway. Mol Cell 38:465–475. [DOI] [PubMed] [Google Scholar]
- Yoder JA, Walsh CP, Bestor TH. 1997. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 13:335–340. [DOI] [PubMed] [Google Scholar]
- Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W. 2004. Single processing center models for human Dicer and bacterial RNase III. Cell 118:57–68. [DOI] [PubMed] [Google Scholar]
- Zhang JH, et al. 2013. Dynamic expression of small RNA populations in larch (Larix leptolepis). Planta 237:89–101. [DOI] [PubMed] [Google Scholar]
- Zhang QZ, et al. 2016. Methylation interactions in Arabidopsis hybrids require RNA-directed DNA methylation and are influenced by genetic variation. Proc Natl Acad Sci U S A. 113:E4248–E4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Henderson IR, Lu C, Green PJ, Jacobsen SE. 2007. Role of RNA polymerase IV in plant small RNA metabolism. Proc Natl Acad Sci U S A. 104:4536–4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Meyers BC, Cai C, Xu W, Ma J. 2015. Evolutionary patterns and coevolutionary consequences of MIRNA genes and microRNA targets triggered by multiple mechanisms of genomic duplications in soybean. Plant Cell 27:546–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sRNA sequencing data has been deposited at the Sequence Read Archive (SRA) in the National Center for Biotechnology Information (NCBI) under the accession number, SRP096198. (N.B. only populations 1, 2, and 4 used in this study.)