Abstract
The germline definition in metazoans was first based on few bilaterian models. As a result, gene function interpretations were often based on phenotypes observed in those models and led to the definition of a set of genes, considered as specific of the germline, named the “germline core”. However, some of these genes were shown to also be involved in somatic stem cells, thus leading to the notion of germline multipotency program (GMP). Because Porifera and Ctenophora are currently the best candidates to be the sister-group to all other animals, the comparative analysis of gene contents and functions between these phyla, Cnidaria and Bilateria is expected to provide clues on early animal evolution and on the links between somatic and germ lineages. Our present bioinformatic analyses at the metazoan scale show that a set of 18 GMP genes was already present in the last common ancestor of metazoans and indicate more precisely the evolution of some of them in the animal lineage. The expression patterns and levels of 11 of these genes in the homoscleromorph sponge Oscarella lobularis show that they are expressed throughout their life cycle, in pluri/multipotent progenitors, during gametogenesis, embryogenesis and during wound healing. This new study in a nonbilaterian species reinforces the hypothesis of an ancestral multipotency program.
Keywords: sponge, multi/pluripotency, GMP signature, stem cells, cell transdifferentiation, gametogenesis
Introduction
The majority of metazoans reproduce sexually, either exclusively like mammals, or alternating between sexual and asexual reproduction. In most bilaterians, gametes either differentiate from a well identified cell lineage, the germline, segregating early from somatic cells (maternal inheritance or preformation mode; Ewen-Campen et al. 2010) or are specified later (during or even after embryogenesis) by inductive signals (epigenesis or induction mode; Extavour and Akam 2003). In both cases, it has been shown that all or part of a conserved set of mRNAs and proteins is involved in their formation (such as Piwi, Nanos, Vasa, PL10, Pumilio, Tudor, Boule/Daz and Bruno/CELF) (Ewen-Campen et al. 2010; Voronina et al. 2011; Shukalyuk and Isaeva 2012) (supplementary table S1, Supplementary Material online). These determinants are usually organized in ribonucleoprotein complexes in a specific cytoplasmic region of the primordial germ cells (PGCs) called pole/germ plasm or nuage localized near nuclear pores and recognizable by various distinctive features (Eddy 1975), including electron-dense granules.
Interestingly in Porifera and Platyhelminthes, for which there is no distinction between the somatic and the germ lines, multipotent progenitor cells generate both somatic and germ cells throughout their lives, depending on environmental conditions (Simpson 1984; Ereskovsky 2010; Rink 2013). Both germ cells and multipotent progenitor cells express at least part of the previous set of genes, among them piwi, vasa, pl10 and nanos. This common set of genes was therefore named the “germline genes” (Juliano and Wessel 2010). These genes are mostly involved in the transcriptional repression of somatic programs, in chromatin reorganization, in meiosis control and in post-transcriptional regulation of several genes during gametogenesis and/or embryogenesis (for more information see supplementary table S1, Supplementary Material online).
However, because some of these genes have recently been shown to be expressed outside the gametogenesis process in some bilaterians, Juliano et al. (2011) have suggested the existence of a highly conserved germline multipotency program (GMP) that could operate in both pluri/multipotent somatic cells and germ cells. More recently, Solana put forward the hypothesis of primordial stem cells (PriSCs). These cells would appear during embryogenesis and their behavior would depend on the mode of reproduction and the regenerative capacities of each organism. In most sexually reproducing bilaterians, the PriSCs would give rise to the PGCs and some somatic stem cells; while in animals reproducing asexually and/or capable of regeneration, these cells might be present at the adult stage keeping a mixed germ/soma potential (Solana 2013).
The non-bilaterian phyla—sponges (Porifera), ctenophores (Ctenophora), placozoans (Placozoa) and cnidarians (Cnidaria)—are capable of both sexual and asexual reproduction, and of regeneration (Brusca and Brusca 2003). Unfortunately, studies on most of these phyla are scarce and thus our ability to propose scenarios on the early evolution of genetic mechanisms involved in metazoan gametogenesis is limited. In an attempt to answer this evolutionary question, our comparative study pays a particular attention to Ctenophora and Porifera: potential sister-groups of all other animals (Dunn et al. 2015).
In sponges, all of these processes seem to involve two principal cells types: choanocytes and/or archeocytes (Ereskovsky et al. 2013, 2015). Gametes are formed either by the differentiation of archeocytes and/or the transdifferentiation of choanocytes, depending on the species studied (Ereskovsky 2010). In this context, no somatic versus germline distinction exists in sponges, and according to the Solana’s hypothesis, both archeocytes and choanocytes might be considered as PriSCs. The recent comparative transcriptomic study of Ephydatia archeocytes and choanocytes (Alié et al. 2015) tends to confirm this hypothesis. Therefore, because there does not seem to be segregation of germline and soma in sponges, they are of particular interest for the present purpose. Unfortunately, expression studies of the GMP signature in sponges (Funayama et al. 2010; Leininger et al. 2014; Alié et al. 2015) remain too scarce to be compared with that of other animals. Notably expression data during all major events/processes of interest are not available to fully enable a comparison across animals: in particular expression patterns during spermatogenesis and regeneration have not been explored so far.
In a comparative and evolutionary perspective, we focus here on the study of the expression patterns of a larger set of GMP genes in the homoscleromorph sponge, Oscarella lobularis (fig. 1). Homoscleromorpha (Gazave et al. 2012) are of particular interest as they possess unique traits within Porifera (asynchronous spermatogenesis and the presence of an acrosome; Ereskovsky et al. 2009). No archeocytes were identified so far in this species in contrast to closely related species (Ereskovsky et al. 2015), and both oocytes and spermatocytes are formed by the transdifferentiation of choanocytes. The life cycle of this marine sponge is well known and moreover wound healing/regenerative experiments are managed in vitro (Ereskovsky and Tokina 2007; Ereskovsky et al. 2015) (supplementary fig. S1, Supplementary Material online). Therefore, we can access, for the first time in sponges, to all developmental stages/processes of interest to study GMP genes expression: asexual reproduction, oogenesis, spermatogenesis, embryogenesis and wound healing processes.
Fig. 1.—
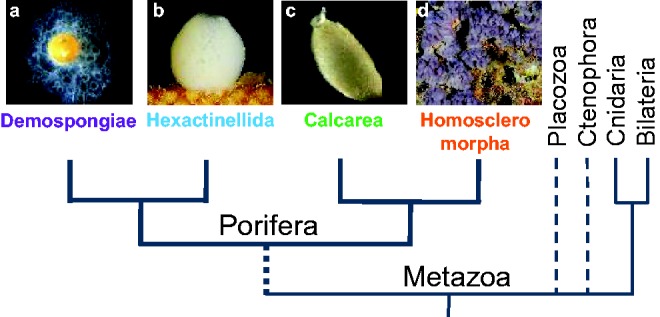
Phylogenetic relationships between the four Poriferan lineages (monophyly hypothesis) (a) Ephydatia muelleri, (b) Oopsacas minuta, (c) Sycon ciliatum, and (d) Oscarella lobularis (Gazave et al. 2012).
Because the GMP signature has already been thoroughly studied in diverse animal lineages and to enable a broad comparative approach, we chose to focus our study on genes for which the maximum amount of data is available in different taxa: piwi/argonaute, vasa/pl10, nanos, bruno, pumilio, tudor and boule. In the present study, we carry out an exhaustive bioinformatic analysis of this set of genes in both sponges and ctenophores. Supplementing previous incomplete data concerning these seven gene families in these two understudied non-bilaterian lineages is expected to provide a better view on their early evolution in metazoans. Moreover, we analyze the expression of 11 GMP genes throughout the life cycle of Oscarella lobularis: three distinct developmental stages, two types of reproduction and the beginning of the regenerative process. These expression data were compared and interpreted in the light of the large amount of data available in bilaterians and of the most complete data available so far in non-bilaterians: in ctenophorans (Alié et al. 2011; Reitzel et al. 2016) and cnidarians (Leclère et al. 2012). We then discuss the putative functional conservation of this genetic core and the hypothesis of a common ancestral stemness program shared by somatic and germ cell lineages (Solana 2013; Alié et al. 2015).
Material and Methods
Sampling and Fixation
Oscarella lobularis (Schmidt 1862) specimens were collected by SCUBA diving in the north-western Mediterranean Sea (Marseille Bay), for mRNA extraction, microscope observations and in situ hybridization (ISH) experiments. For microscopy observations and life cycle stage identification, samples were fixed (glutaraldehyde fixation) following published protocols (Ereskovsky et al. 2013) and semi-thin sections were observed under a light microscope (Leica DMLB—Evolution LC color). Ultra-thin sections were stained with uranyl acetate, contrasted with lead citrate and observed in a LEO 910 and LEO 912 transmission electron microscopes (TEM). To perform ISH, samples were fixed in 4% Paraformaldehyde (PFA) in sea water and conserved in 100% methanol at −20 °C for adult tissues and buds. Budding was triggered in vitro in Petri dishes from adult fragments by a mechanical (cut) or thermic stress (temperature variation).
Sequence Identification and Analyses
Oscarella lobularis orthologs of piwi, argonaute, boule, vasa, pl10, nanos, pumilio, tudor and bruno genes were searched in a personal transcriptomic database (Schenkelaars et al. 2015) by local TBALSTN using BioEdit software (Altschul et al. 1997) and in order to retrieve highly divergent sequences a threshold e-value of 1.0 was used. The relevance of the results was then tested by a reciprocal best hit approach (Wall et al. 2003) against NR NCBI databases. When required, sequences of interest were completed by PCR on cDNA according to previously published protocols described in Gazave et al. (2008) (primer choice in supplementary table S2, Supplementary Material online). Sequence assignations were confirmed and refined by protein domain analyses using InterProScan software (Mitchell et al. 2015) and phylogenetic analyses.
For this purpose, a dataset of each gene was constructed including the homologous genes of at least one representative species from each known taxa: either from already published data or by using default parameters of online software (BLASTP, TBLASTN/X) when sequences were not available (i.e. Choanoflagellata: Monosiga brevicollis and Salpingoeca rosetta; Filasterea: Capsaspora owczarzaki and Amoebozoa: Dictyostelium discoideum) (see supplementary tables S3, Supplementary Material online). Whatever the approach used, we always started the blast search by using as query the protein already annotated for the closest species. For comparative purposes, candidate genes were also retrieved in other sponge lineages—at least two species for each Porifera classes—from published data (Funayama et al. 2010; Leininger et al. 2014; Riesgo et al. 2014) and available databases: Oscarella carmela (Homoscleromorpha), Sycon coactum and S. ciliatum (Calcarea), Ephydatia muelleri, E. fluviatilis and Amphimedon queenslandica (Demospongiae); Aphrocallistes vastus (Hexactinellida), and Oopsacas minuta (Hexactinellida, personal transcriptome). All sequences, their origin and accession numbers identified and/or used for phylogenetic analyses are listed in supplementary tables S3, Supplementary Material online. The resulting dataset was aligned with ClustalW (Thompson et al. 1994) and/or MUSCLE (Edgar 2004) and refined manually using Bioedit Sequence Alignment Editor 5.0.9 (Hall 1999, 19). Highly divergent positions were removed using Gblock software (Castresana 2000) (alignments are available upon request). Maximum likelihood (ML) analysis were performed using ATGC-Montpellier online PhyML software (Guindon et al. 2010) and robustness was tested using aLRT SH-like method and bootstrap resampling (1000 replicates). Bayesian analyses were performed using MrBayes v3.2.3 with one cold chain and three heated chains until average deviation of split frequencies was <0.01 (100,000 generations). A Best-fit model of protein evolution was used for each protein family using ProtTest 3.2 (Abascal et al. 2005). Additional methodological details are available, for each phylogenetic analysis, in the legends of supplementary figures S3–S12, Supplementary Material online.
Wound Procedures
Wounds were inflicted manually by introducing a sterile needle (hole diameter 0.8 mm) in two perpendicular planes of each operated lobe, in order to observe the wound in every cut plane. By doing so, each cell layer of the specimen was injured in order to study the expression of the GMP genes in each cell type in the sponge during wound healing. The lobes were kept in Petri dishes with filtered natural sea water (0.22 µm) and wound healing was monitored and lobes were fixed at 0, 1, 3, 6, 10, 12, 24 and 30 h after injury. We prefer to use here the term “healing” here instead of regeneration because the process was not observed until after the complete reformation of structures.
ISHs were performed on whole mounted adult sponge lobes (whole-mount in situ hybridization, WMISH) and on serial 8 µm paraffin tissue sections of both adults and buds. These two different approaches are complementary and essential for the observation of all structures and cell types. WMISHs were performed using the InsituProVSi an instrument for automated in situ hybridization (Intavis AG) while sections ISHs were performed manually. ISH protocols were adapted and improved from Gazave et al. (2008). Probes of all candidate genes were synthesized using the DIG RNA labeling Roche kit and their length varied from 800 to 1,300 bp. Actin antisense probes were used as a positive control and as negative controls, experiments were conducted with sense probes and no probes.
Gene Expression Quantification by qPCR
mRNA extraction (IllustraTM QuickPrepTM micro mRNA Purification Kit GE Healthcare) was followed by cDNA synthesis on 150 ng mRNA (First-strand cDNA synthesis kit GE Healthcare) in order to study the expression level of each gene during different stages of the life cycle by quantitative PCR (qPCR). Five distinct individuals (biological replicates) were used for each stage: budding (asexual reproduction), gametogenesis, embryogenesis (embryos and prelarvae) and non-breeding/reproductive adults. qPCR experiments were run in triplicate (technical replicates) and performed in an Eppendorf Mastercycler® ep realplex Thermal Cycler (Platinium SYBRgreen qPCR super mix UDG Invitrogen), with 5 ng of cDNA, and 2 μM of primers in a final volume of 10 µl. Amplification parameters were 95 °C 2 min, 40 cycles with two steps: 95 °C 15 s/60 °C 1 min. Primers for candidate genes were designed using Primer3plus (Rozen and Skaletsky 1999). Primers were selected with amplification efficiency close to 100% and specificity was checked by melting curve analysis (primers sequences are given in supplementary table S2, Supplementary Material online). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was chosen as reference gene, because it showed the most stable expression (compared with cytochrome oxidase I (COI) and elongation factor 1 alpha (EF1a), data available upon request). Delta cycle threshold (dCt) values were calculated by subtracting the GAPDH Cts from genes of interest Ct. Delta delta Ct (ddCt) values were calculated by subtracting control dCts from dCts samples, and fold changes were calculated using the 2-ddCt method (Livak and Schmittgen 2001).
Statistical Analyses
Analyses were performed using the computing environment R (R-Studio under Windows®, version 3.2.3) for statistical computing and graphics (R Development Core Team 2009). Since 2-deltaCt results were not normally distributed, non-parametric analyses were performed using the Kruskal-Wallis and Mann-Whitney tests (Ihaka and Gentleman 1996).
Results and Discussion
GMP Genes Origin: An Old Toolkit with Ten Animal Innovations
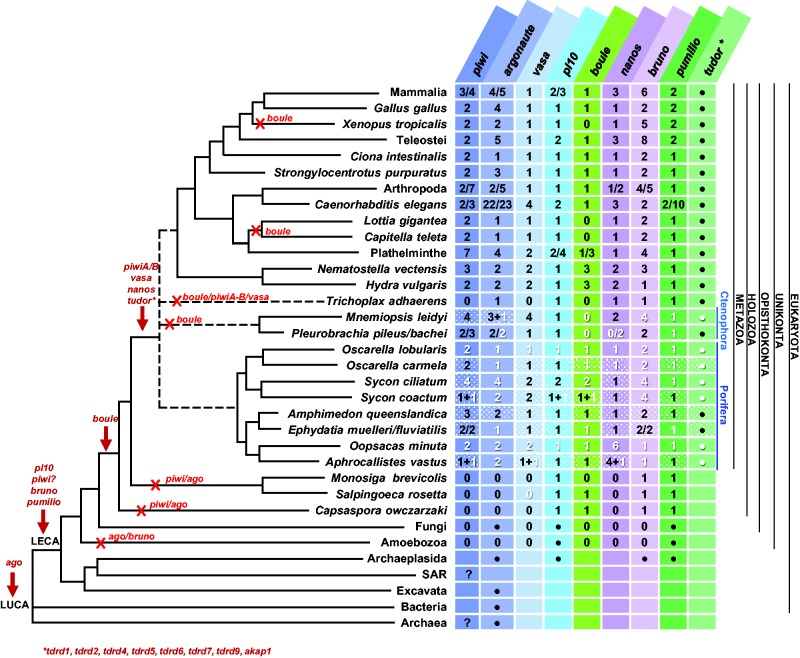
Figure 2 (completed by supplementary figs. S2–S12, Supplementary Material online) provides a synoptic view of the results available so far for the chosen set of genes (piwi, argonaute, vasa, pl10, nanos, boule, bruno, pumilio and tudor family), obtained here and by previous studies. While it seems that argonaute (ago) genes were already present in the last universal common ancestor (LUCA) (Parker et al. 2004; Ma et al. 2005), all the other genes have more recent histories. Pl10, bruno, pumilio, and members of the Tudor “royal family” were probably present in the last eukaryotic common ancestor (LECA) (for review, see references in fig. 2). Even if piwi genes were described in ciliates (Mochizuki et al. 2002) (questioning their emergence and calling for further studies in other unicellular eukaryotes), their orthology with metazoan genes is doubtful, therefore piwi is often considered as a metazoan innovation (Kerner et al. 2011; Alié et al. 2015).
Fig. 2.—
Distribution of studied genes within living organisms (phylogenetic relationships after Ren et al. 2016), information gathered from Ma et al. (2005), Cerutti and Casas-Mollano (2006), Makarova et al. (2009), Kerner et al. (2011), Gazave, Béhague et al. (2013), Suga et al. (2013), Alié et al. (2015) and completed in this study. Arrows show possible origins for each gene and crosses indicate secondary losses; 0 indicates that no paralogs were found and empty boxes indicate that, to our knowledge, no studies have been conducted so far; black/white dots indicate the presence/absence of at least one member of each family; white dotted boxes indicate genes already shown to be present but without any phylogenetic analyses or copy information, white numbers indicate genes found in this study while black numbers indicate previous published sequences and questions marks indicate that there is no clear affiliation. The asterisk in Tudor family indicate several protein members studied in detail in supplementary figure S2, Supplementary Material online. Only boule genes are shown here, therefore the paralogs DAZ/DAZLike genes were not included (see supplementary fig. S6, Supplementary Material online). Information about transcriptomic databases are detailed in supplementary tables S3, Supplementary Material online.
The latest innovations were nanos, vasa, boule (Kerner et al. 2011; Suga et al. 2013) and some members of the Tudor family. Among the ten germline specific Tudor proteins (Chen et al. 2009) Alié et al. (2015) suggested that four are metazoan innovations. Our enlarged analysis of these Tudor “germline specific” members, inside and outside metazoans, enabled us to show that a total of eight members are, in fact, metazoan innovations: Tdrd1, Tdrkh/Tdrd2, RNF17/Tdrd4, Tdrd5, Tdrd6, Tdrd7, Tdrd9, and Akap1 (supplementary fig. S2, Supplementary Material online).
For the other gene families, the complementary data we bring do not modify previously proposed evolutionary scenarios—except for vasa and boule. Indeed, the analyses of the sequence considered as vasa in Salpingoeca rosetta (Riesgo et al. 2014) showed that it is not a Vasa/Ddx4 ortholog (supplementary fig. S5, Supplementary Material online). Concerning boule, the ancestral founder of the DAZ family (Shah et al. 2010), our residue and phylogenetic analyses of the fungal sequences reported by Suga et al. (2013) showed that these RRM domain-containing proteins are not closely related to boule (see supplementary figs. S6 and S7, Supplementary Material online). Therefore, we have restored vasa as a metazoan innovation and restricted boule to the Holozoan lineage.
To summarize, we extend (from five to ten) the set of GMP genes that are metazoan innovations: vasa, nanos and eight tudor genes.
Evolution of the GMP across Animals
It has not been determined yet whether Ctenophora or Porifera are sister group of all other metazoans (Pick et al. 2010; Moroz et al. 2014). To trace back the early evolution of this set of genes in metazoans and to study their conservation (or otherwise) across metazoans, we established a more complete inventory in these two lineages. Unfortunately, previous studies in sponges are incomplete, for both taxa and genes studied (Funayama et al. 2010; Kerner et al. 2011; Suga et al. 2013; Leininger et al. 2014; Riesgo et al. 2014; Alié et al. 2015). The present study provides a more exhaustive view in terms of species studied and their genetic content (supplementary table S1, Supplementary Material online). Indeed, a unique species such as A. queenslandica, does not reflect the status of all demosponges or even sponges (Ereskovsky et al. 2009; Gazave et al. 2009). We thus complemented previous datasets with Ephydatia, two different Sycon species (Calcarea), two Oscarella species (Homoscleromorpha), and two glass sponges (Hexactinellida) from two different orders: Aphrocallistes vastus and Oopsacas minuta.
Our results show that all of the genes studied are conserved throughout the metazoan lineage, including all four Porifera classes (fig. 2 and supplementary fig. S2, Supplementary Material online). Nevertheless, we must point out two exceptions: as is often observed in Placozoa, several secondary losses were observed: piwi, vasa, boule (Kerner et al. 2011; Suga et al. 2013) and the majority of tudor (this study). Similarly, Ctenophora appeared to be missing boule (this study). Thus, boule seems to have been lost at least 4 times during animal evolution. This is quite astonishing as it is considered essential to gametogenesis in both vertebrates and invertebrates (Fu et al. 2015; Sekiné et al. 2015).
Gene Content: Sponges Are Not Less Rich than Others
Until now most data concerning sponges, presented only a presence/absence overview, thus preventing the comparison of gene-content diversity between animal lineages. Phylogenetic analyses of these copies enabled us to assign each gene copy to an orthology, which will be important for future functional comparative studies.
With some exceptions, pumilio, nanos, vasa, pl10 and boule were more often found in a single copy across metazoans (see fig. 2). In contrast, boule is absent in ctenophores and both boule and pl10 are present in two copies in calcareous sponges. Our attention was also drawn to the unusually high number of copies of nanos in glass sponges (five to six vs. one in other animals) (supplementary figs. S8, S9 and S17, Supplementary Material online). Concerning vasa, even though it was found in two or more copies (instead of one in most other animals) in calcareous sponges, glass sponges, cnidarians and in M. leidyi (Reitzel et al. 2016), a close look at the relationships among copies (supplementary fig. S5, Supplementary Material online) showed us that these copies are the result of duplication events that occurred independently in each of these lineages. Therefore, for these genes, it seemed clear that Urmetazoa possessed one copy of each, followed by multiple independent duplication/loss events.
Concerning the Argonaute family, it has been classified into three paralogous groups: Ago, Piwi and a worm-specific Wago subfamily (we focused on Piwi and Ago only) (Tolia and Joshua-Tor 2007). The number of argonaute (ago) genes ranged from one to four (fig. 2), and the topology of our phylogenetic analysis prevented us from proposing a reliable evolutionary scenario (supplementary fig. S4, Supplementary Material online). Nevertheless, it is interesting to note that many in-paralogous genes were found throughout the metazoan tree (i.e. in vertebrates, in S. ciliatum, O. minuta, H. vulgaris, S. purpuratus, P. pileus and P. bachei). However, deciphering if these genes are either specific ago duplications in each species or ancestral duplications undergoing concerted evolution, would require further investigation. In contrast, piwi was generally observed in at least two copies (additional lineage-specific paralogs Piwi X and Piwi P (Kerner et al. 2011) were not studied here). Our phylogenetic analyses supported the two PiwiA and PiwiB orthology groups proposed by Kerner et al. (2011) (supplementary fig. S3, Supplementary Material online). Surprisingly, ctenophores seem to be the only animal phylum lacking piwiB members. Indeed the two piwi copies reported in Pleurobrachia pileus by Alié et al. (2011) pertain to the A clade as well as the three copies we found in Pleurobrachia bachei and the four paralogs recently identified in M. leidyi (Reitzel et al. 2016) (in contrast of what was shown, this is probably due to the high divergence of ctenophoran sequences). According to the “Porifera first” hypothesis (Pick et al. 2010) our data confirm the scenario that an ancestral duplication may have occurred, just before metazoan radiation giving rise to A and B clades, and suggest that B copies could have been lost secondarily in ctenophores (or all copies homogenized by a concerted evolutionary process). In contrast, in the “Ctenophora first” hypothesis (Moroz et al. 2014) we might envisage that only one copy (more similar to piwiA) was present ancestrally, and that a duplication event occurred in the last common ancestor of all other metazoans, and gave rise to piwiA and piwiB (supplementary figs. S13 and S14, Supplementary Material online). Additional copies of piwiA are clearly resulting from several independent lineage-specific duplications (e.g. M. leidyi and in S. ciliatum).
As far as Bruno is concerned, it seems that the last common ancestor of Metazoa possessed two copies of Bruno (supplementary figs. S10 and S18, Supplementary Material online). According to our phylogenetic analyses one copy can be assigned to the B orthology group (node support > 85%) as defined by Kerner et al. (2011). The nature of the second copy remains uncertain because the position of sequences varies depending on the outgroup choice. Moreover, residue analyses (not shown but available on request) do not provide discriminating information. According to the currently favored hypothesis on the phylogenetic relationships among sponges, placing hexactinellids as sister group of demosponges (fig. 1), this second copy has been lost in glass sponges, while the genus Sycon underwent multiple bruno gene-duplication events.
Regarding the ten “germline” Tudor members, eight of them were retrieved in almost all metazoans including sponges, cnidarians and ctenophores, showing their presence ancestrally in animals, whereas tdrd8/stk31 and tdrd10 are innovations of bilaterians and chordates, respectively (supplementary figs. S2 and S12, Supplementary Material online).
Most of the observed exceptions to the “general rules” mentioned before are in accordance with generally available data: recurrent gene losses in Placozoa and Urochordata (Lanna 2015), gene copy-number polymorphism in C. elegans (Farslow et al. 2015) and in some plathyhelminth parasitic species (Cwiklinski et al. 2015), expansion of some gene families among cnidarians (Steele et al. 2011); whole genome duplication in vertebrates (Murphy et al. 2008). Finally, the combination of previous observations on various genes (Leininger et al. 2014; Schenkelaars et al. 2015) with our own current observations, strongly suggests that a polyploidization event occurred in the genus Sycon (Calcarea) followed, as often documented, by independent sorting-out of duplicated genes.
In conclusion, we show that Porifera and Ctenophora do not contain a reduced set of GMP genes compared with cnidarians or bilaterians.
Protein Domain Conservation Consistent with Functional Conservation
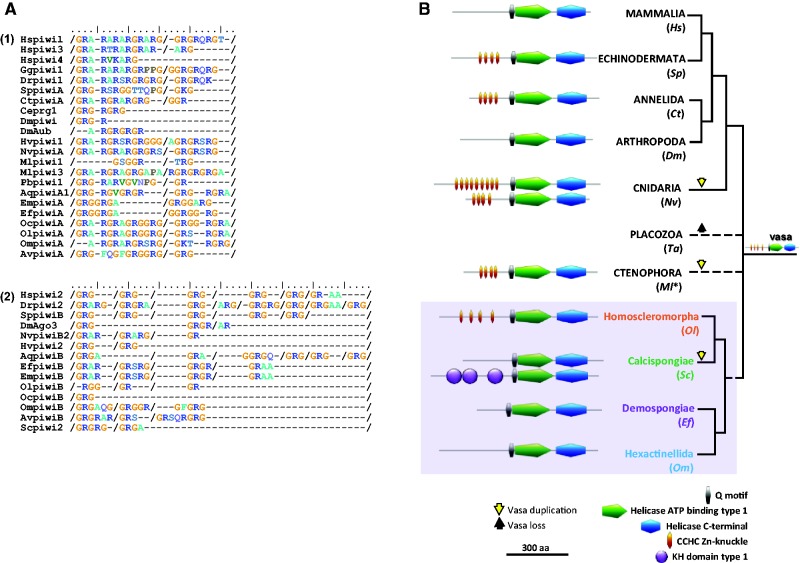
Our domain analyses led us to notice that ctenophoran and poriferan Piwi, Ago, Bruno, Nanos and Pumilio proteins harbor all the main characteristic features/domains of their cnidarian and bilaterian counterparts studied so far (supplementary figs. S13–S18, Supplementary Material online). For example, RG repeats noticed in the Piwi sequences of these two lineages (fig. 3A) are potential sites of dimethylation of Piwi, necessary for their interaction with Tudor (Kirino et al. 2009; Alié et al. 2015). This domain conservation may be compatible with a functional conservation.
Fig. 3.—
(A) Putative conservation of symmetrical dimethylation motifs (GRG, ARG/GRA) in the N terminus region of PiwiA (1) and PiwiB (2) metazoan proteins, essential for Tudor interaction according to Kirino et al. (2009). Slashes indicate that sequences have been cut in order to align motifs in each species studied. Complete alignment is available upon request. For a list of abbreviations used for species names, see supplementary tables S3, Supplementary Material online. (B) Schematic evolution of Vasa proteins in Metazoa (Protein domains analysis and copy numbers). *Only one of the five Mnemiopsis vasa paralogs is shown here (Mlvasa1) as they are highly conserved and branch together (Reitzel et al. 2016).
Concerning Vasa, our results showed that ctenophores, Nematostella vectensis (Anthozoa) and homoscleromorph sponges have a N-terminal Zn-knuckle motif repeats (fig. 3B) as found in Hydra and most bilaterians (Rebscher et al. 2007; Gustafson and Wessel 2010). We therefore suppose that this motif was present ancestrally and that its absence in some lineages, such as glass sponges and demosponges, is probably due to a secondarily loss, as previously observed in some bilaterians (Drosophila and vertebrates) by Gustafson and Wessel (2010). Surprisingly, the Zn-knuckle repeats are absent but are replaced by three KH domains in one of the Vasa copies of calcareous sponges. To the best of our knowledge, such lineage-specific domain shuffling has not been described so far in Vasa proteins. Interestingly, both CCHC Zn-knuckles and Type1 KH domains have DNA/RNA binding properties by providing helicase specificity to certain target mRNAs (Hollingworth et al. 2012). Therefore, is there a relation between Vasa protein interactions/specificity and their N-terminal motif variations? For example: (1) the absence of Zn-knuckle motif repeats in glass sponges, in demosponges (this study), and in some bilaterians (Gustafson and Wessel 2010); (2) the number of repeats in cnidarians; (3) the spacing of repeats in homoscleromorph sponges; or (4) their replacement by other binding domains in calcareous sponges (fig. 3B). Future functional experiments should pay particular attention to these N-terminal features in relation to expression patterns and molecular interactions.
Finally, concerning Boule, we did not find any evidence for the conservation of the DAZ domain as defined in vertebrates and much less convincingly in protostomes (Houston et al. 1998; Karashima et al. 2000; Xu et al. 2009; Kuales et al. 2011; Fu et al. 2015). Therefore, it remains uncertain whether or not a DAZ domain is conserved in Boule proteins across metazoans.
GMP Genes Are Expressed during Gametogenesis, Embryogenesis, Budding and Wound Healing in Oscarella lobularis
Oscarella lobularis (at both adult and bud stages) is characterized by a simple organization and few cell types (Ereskovsky and Tokina 2007; Ereskovsky et al. 2009), making expression patterns at the cellular level easy to observe. The external epithelium and the internal canals are formed by flattened cells, the pinacocytes (exo- and endopinacocytes, respectively). The water filtration takes place in the choanocyte chambers bordered by collar cells called choanocytes. The inner part, called the mesohyle, contains vacuolar cells (type 1 and 2) and symbiotic bacteria (supplementary fig. S1, Supplementary Material online).
The expression patterns of 11 genes are reported here (piwiA, piwiB, ago, vasa, pl10, boule, nanos, bruno, brunoB, pumilio and tudor1) by in situ hybridization (fig. 4A and supplementary fig. S19, Supplementary Material online) and their mRNA quantification by qPCR (fig. 4B). Different stages of the life cycle and developmental of O. lobularis were chosen to unravel the expression of these GMP genes during: (1) asexual reproduction, indeed O. lobularis is able to form buds throughout the year by external budding which is easy to trigger in vitro; (2) gametogenesis, sexual reproduction occurs naturally from May to September, then in the same individual, either entire choanocyte chambers transdifferentiate into spermatocysts or single choanocytes transdifferentiate into oocytes; (3) embryogenesis, after internal fertilization, embryos at various stages are observable simultaneously in the incubating adult; (4) wound healing/regeneration experiments performed in vitro. Regeneration is mainly performed by cell transdifferentiation and epithelial reorganization (Ereskovsky et al. 2015); and finally during (5) a non-reproductive stage: adults from December to February (supplementary fig. S1, Supplementary Material online).
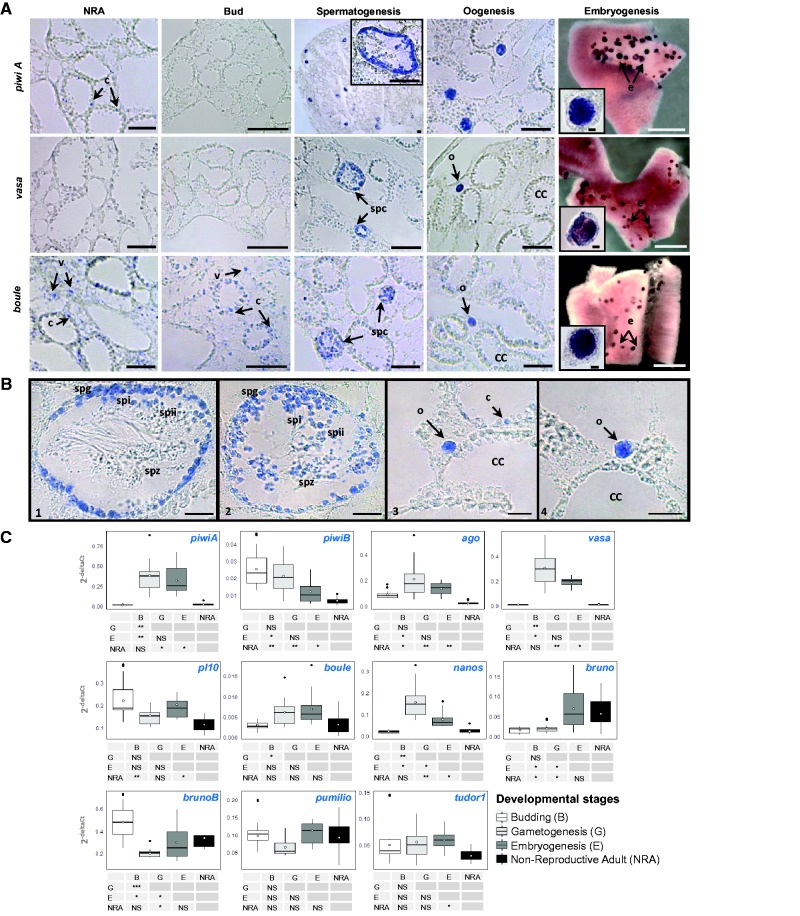
Fig. 4.—
(A) Expression of O. lobularis piwiA, vasa and boule genes during different developmental stages by serial sections ISH (in situ hybridization): non-reproductive adults (NRA), buds and gametogenesis; and WMISH (whole-mount in situ hybridization): embryogenesis. Black boxes represent zoomed figures. The remaining seven gene expressions are given in supplementary figure S19, Supplementary Material online. Black scale: 50 µm, grey scale: 1 mm. (B) Detailed expression in spermatocysts (1, 2) and in oocytes (3, 4) for piwiA (1, 3) and vasa (2, 4). Spg, spermatogonia; SpI, spermatocyte I; SpII, spermatocyte II; Spz, spermatids and/or spermatozoids; oo, oocytes; CC, choanocyte chamber. Scale: 20 µm. (C) Box plot representation of qPCR results measuring the relative RNA levels of OlpiwiA, OlpiwiB, Olago, Olvasa, Olpl10, Olboule, Olnanos, Olbruno, OlbrunoB, Olpumilio and Oltudor1 during the four developmental stages. All values were normalized against GAPDH RNA and are represented as 2exp-deltaCt, relative to the amount of RNA present in the tissues. Boxes denote median values with upper and lower quartiles, whiskers minimum and maximum extremes, black point outliers (which represent different stages of the buds) and white points mean values. Tables below show differences between expression levels of each gene at each stage (statistical significance P < 0.05*, <0.01**, <0.001***).
The results of our experiments evidenced, that the expression of the selected 11 genes was not at all restricted to the gametogenesis period. Indeed, all genes were expressed (at different levels and in different cell types) in embryos, buds and adults all year long whatever their reproductive status (fig. 5).
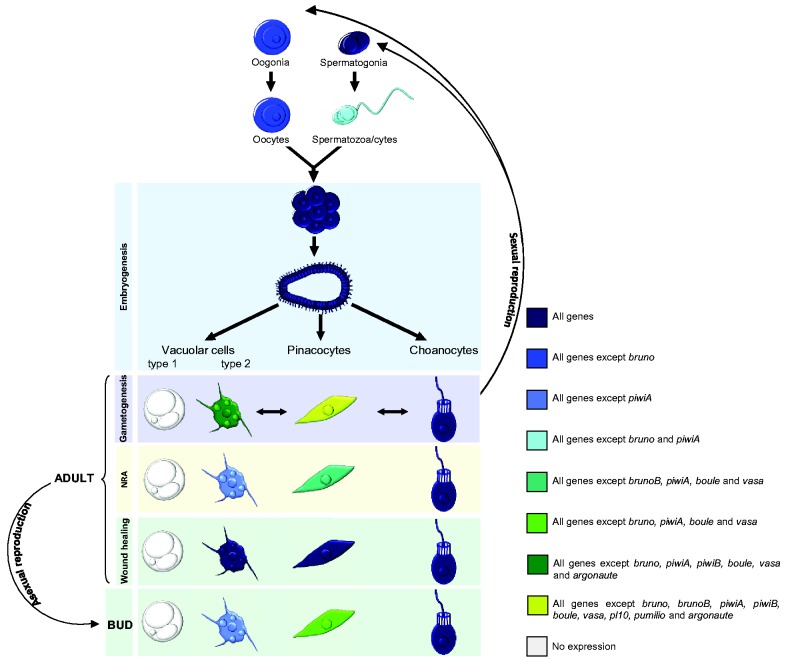
Fig. 5.—
Summary of the expression of the 11 genes studied in different cell types in O. lobularis: in the bud, the non-reproductive adult (NRA), during gametogenesis and embryogenesis represented in a sum up schematic synoptic view of gene expression in the different cell types.
In non-reproducing adults, these genes were expressed either in the specialized filtering cells, the choanocytes (piwiA), or in choanocytes and in type 2 vacuolar cells (vasa, boule, brunoB), or in both, as well as in pinacocytes (piwiB, ago, pl10, nanos, bruno, pumilio and tudor1). Note that when a certain cell type expressed the genes not all cells were concerned (fig. 4A and detailed in supplementary fig. S19, Supplementary Material online). We observed that cells expressing these genes in buds were generally the same as those in adults but at different levels. During gametogenesis, all the genes (except bruno) were expressed in both male and female gametes. Some genes appeared to be involved at the very beginning of gametogenesis (piwiA and bruno) because their expression was restricted to transdifferentiating choanocytes and spermatogonia, while most were expressed during the whole process (spermatogonia, spermatocytes and spermatozoa all clearly stained, and oocytes of different sizes). All genes were also expressed (at different levels) during embryogenesis. It is worth noting that nanos clearly showed a gradient of expression in blastula and prelarvae stages (supplementary fig. S21, Supplementary Material online).
Moreover, all genes were highly expressed in the healing zone in choanocytes, in some pinacocytes and a clear overexpression was observed in type 2 vacuolar cells (in comparison to non-affected zones and adult under standard conditions) (supplementary fig. S20, Supplementary Material online). This enhanced expression during healing or regeneration has already been observed for piwi in diverse metazoans (cnidarians, annelids and platyhelminthes) and for a larger number of GMP genes (e.g. argonaute, pl10, vasa, nanos) in annelids and platyhelminthes (Giani et al. 2011; van Wolfswinkel 2014; Kozin and Kostyuchenko 2015).
The expression of most of the genes investigated (except piwiA) in type 2 vacuolar cells, at different life stages, was of particular interest. These cells are localized in the mesohyle and present numerous vacuoles. They are moreover clearly amoeboid, the presence of long filopodes enable them to migrate to wounded zones during regeneration (Ereskovsky et al. 2015) (fig. 6A). These cytological features are commonly found in what was defined as archeocytes in most sponges (Ereskovsky 2010; Funayama 2010) including closely related Oscarella species (Gazave, Lavrov, et al. 2013). Together, both cytological and molecular aspects led us to propose that type 2 vacuolar cells might represent the equivalent of other sponge archeocytes in O. lobularis. In order to determine whether these cells represent either a stable stock of bona fide archeocytes-like cells or a transient stage during cell transdifferentiation, additional experiments are needed. These two hypotheses are equally reliable because: (1) transdifferentiation has been repeatedly suggested in sponges by histological observations (Volkova et al. 1981; Gaino et al. 1995; Korotkova 1997; Leys and Degnan 2002; Nakanishi et al. 2014; Borisenko et al. 2015; Degnan et al. 2015; Ereskovsky et al. 2015) and dedifferentiation was evidenced by cell tracking (Nakanishi et al. 2014) and (2) piwi and other GMP genes were shown to be expressed in E. fluviatilis archeocytes (Funayama et al. 2010; Alié et al. 2015). Either way, since the homology between archeocytes of different sponge species is not yet established, further studies will be necessary to test the proliferation capacities and pluripotency of type 2 vacuolar cells before renaming them.
Fig. 6.—
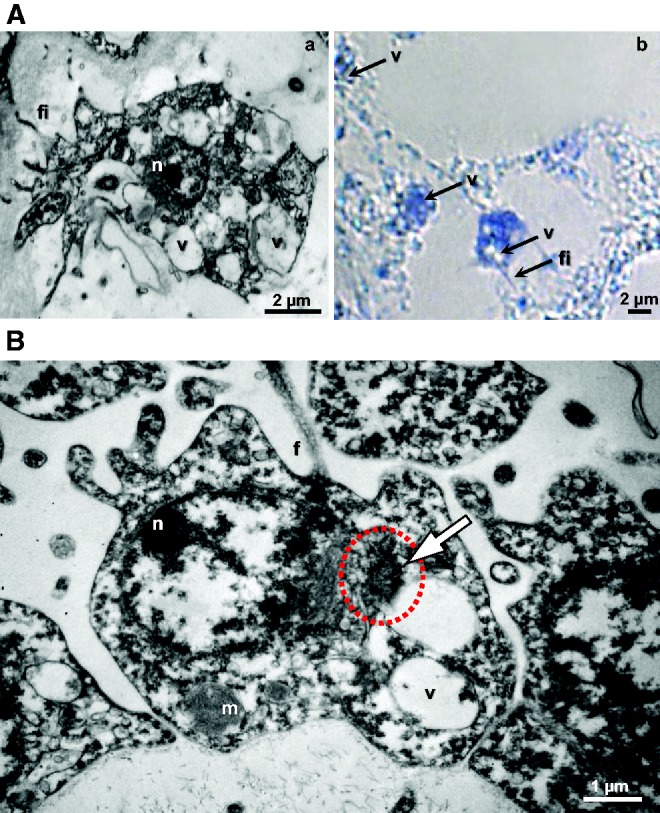
(A) Oscarella lobularis type 2 vacuolar cells during regeneration: (a) TEM transmission electron microscopy and (b) the expression of piwiA in type 2 vacuolar cells by optical microscopy after ISH. The same observation was made in the non-reproductive adult and the bud. (B) TEM of O. lobularis spermatogonia, nuage highlighted by the white arrow and red dotted circle, n, nucleus; m, mitochondria; f, flagella; v, vacuole; fi, filopodia; ep, exopinacocyte.
Other data on the expression of GMP genes in sponges are scarce thus limiting comparisons inside the poriferan lineage. GMP genes studied here were shown to be expressed in both choanocytes and archeocytes in Ephydatia fluviatilis (Funayama et al. 2010; Alié et al. 2015), but unfortunately no expression data are available during gametogenesis. Concerning in situs, as far as piwi is concerned, previous data for the adults of the fresh-water demosponge E. fluviatilis (Funayama et al. 2010), have shown that piwiA and piwiB have an overlapping expression in both choanocytes and archeocytes. Here, in O. lobularis adults the two genes had different expression patterns: piwiA in choanocytes only and piwiB in choanocytes, pinacocytes and type 2 vacuolar cells, thus advocating a non-redundant function of the two copies.
Concerning vasa and pl10, they are expressed in Ephydatia’s archeocytes (Alié et al. 2015) and in the calcareous sponge Sycon ciliatum (Leininger et al. 2014)—which possesses two copies of each gene—the two pl10 copies have similar expression patterns during oogenesis and embryogenesis, and only one of them is expressed in adult choanocytes (pl10B). In O. lobularis, this gene was shown to be additionally expressed during spermatogenesis and budding and we notice that type 2 vacuolar cells (and few pinacocytes nuclei) are also concerned. In O. lobularis, we observed that vasa was strongly expressed in embryos and during gametogenesis. Similarly, vasa in S. ciliatum is expressed at the same periods but with a “separation of duties” between the two copies (both in oocytes and embryos; but only vasaB in adult choanocytes). This could therefore, correspond to a subfunctionalization via the duplication degeneration complementation (DDC) model, (Force et al. 1999) as it is also observed for pl10 paralogs. Interestingly, Olvasa and ScvasaB expressions are not restricted to the germline, and both sequences are characterized by N-terminal binding motifs (Zn-knuckles and KH domains, respectively). Since the presence of Vasa Zn-knuckles is thought to be correlated, in many animals, with an expanded expression pattern outside of the germline (Gustafson and Wessel 2010), these N-terminal motifs may be of particular importance.
Interestingly, boule expression was more unexpected because, though intensively studied in bilaterians, it has very rarely been reported in other than gametogenetic cells (except for expression in Macrostomum lignano and Oryzias latipes embryos).
Finally, nanos seems to have different patterns in calcareous and homoscleromorph sponges: whereas it is expressed during oogenesis and embryogenesis in both studied species, it appeared to be expressed only in O. lobularis adults.
The Nuage, the GMP Genes and the Ancestral Reprograming Capacity
As we mentioned in the introduction section, the GMP products are frequently present in a structure called the germ plasm or nuage. In sponges, a germ plasm/nuage have so far been described in oocytes (Shukalyuk and Isaeva 2012). Our careful TEM (transmission electron microscopy) observations enabled us to observe one in O. lobularis spermatogonia (fig. 6B). The nuage has a roundish shape and is visible in the nucleus periphery, near nuclear pores, of spermatogonia within spermatocysts with different sizes (< 1 µm). This observation suggests that a germ plasm (therefore potentially GMP proteins) are a characteristic of sex cells shared by all animals.
Nevertheless, the observation of these cytoplasmic structures in other sponge cell types (i.e. archeocytes; Shukalyuk and Isaeva 2012) and in somatic stem cells of bilaterians (Voronina et al. 2011) is consistent with the increasingly obvious implication of GMP genes in other contexts than sexual reproduction.
Besides the expression of (all but one) GMP genes during both oogenesis and spermatogenesis in O. lobularis, suggesting a functional conservation of this toolkit in sexual reproduction, we showed additional evidence of the expression of this set of genes (fig. 5 and supplementary fig. S22, Supplementary Material online) in three other cell types: choanocytes, archeocyte-like type 2 vacuolar cells and, to a lesser extent, in pinacocytes. While archeocyte have been traditionally considered as sponge stem cells, choanocytes, in addition to their involvement in gamete formation (Simpson 1984; Ereskovsky 2010), seem to be able to (1) dedifferentiate into archeocytes (Funayama 2012; Nakanishi et al. 2014) and (2) transdifferentiate into pinacocytes during epithelial regeneration processes (Borisenko et al. 2015; Ereskovsky et al. 2015). Thus considering the multipotency of archeocytes and choanocytes, the expression of GMP genes in these two cell types supports the hypothesis of a primordial stem cell program (Solana 2013). Our findings (completing those of Leininger et al. 2014 and Alié et al. 2015) showed that an extended GMP program is not only present ancestrally in metazoans, but that it is also expressed in both pluri/multipotent cells and during gametogenesis just like in some bilaterians and cnidarians (fig. 7).
Fig. 7.—
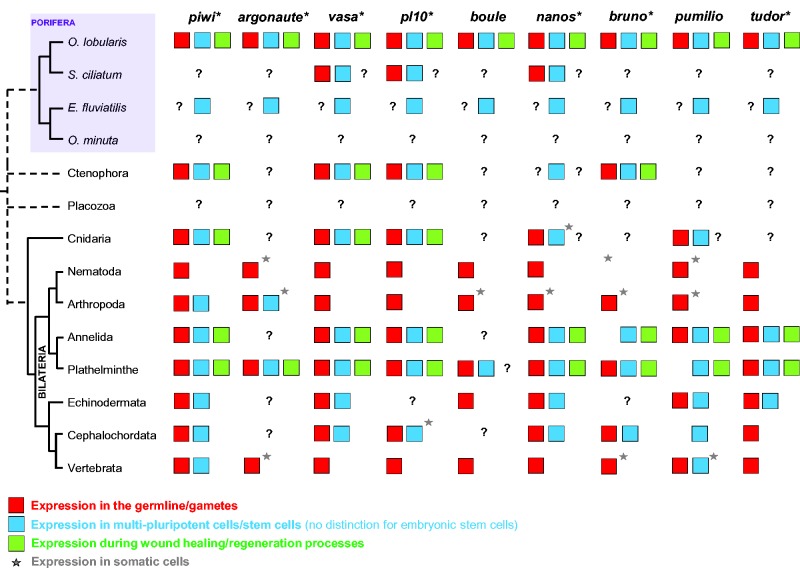
Summary of available data on each gene expression/function in different cell types and/or cell lineages across metazoans. White cases indicate “not found/not expressed”, questions marks indicate “not yet studied”. Asterisks indicate that several paralogs are concerned. Data gathered from Alié et al. (2011) and Gazave, Béhague, et al. (2013) and completed by the present study (additional references for each phylum are gathered in supplementary references, Supplementary Material online).
In contrast, the expression of these genes in pinacocytes was more surprising, even if the lability and transdifferentiation capacity of sponge cells was often reported (Simpson 1984; Lavrov and Kosevich 2014). Gaino et al. (1986) suggested that endopinacocytes were able to give rise to vacuolar cells in O. lobularis, but tracing experiments are needed to really measure the relative degree of versatility of sponge cells.
Nevertheless, when considered together, early observations and the expression of GMP genes observed here in all but one cell type (at some point in the life cycle), suggest that each cell type of O. lobularis may somehow keep a multi/pluripotency feature during its whole lifetime and that it may be capable of reprogramming in different contexts. We nevertheless avoid using the term “stem cell” because the capacity to self-renew remains unproven.
It now becomes clearer and clearer that in all metazoans studied so far, apart from the gametogenesis processes, cells that express GMP genes are stem cells or multipotent progenitors: choanocytes and archeocytes (and in less extent pinacocytes) in sponges (Funayama et al. 2010; Leininger et al. 2014; Alié et al. 2015; this study), i-cells in hydrozoans (Rebscher et al. 2008), tentacle root cells or related to the tentacle bulb in ctenophores (Alié et al. 2011; Reitzel et al. 2016), neoblasts of plathyhelminths (Handberg-Thorsager and Saló 2007), posterior multipotent progenitors or teloblasts in annelids (Gazave, Béhague, et al. 2013) and cell islands in Botryllus schlosseri (Rinkevich et al. 2013). This observation at such an extended phylogenetic scale (at least for piwi, vasa and nanos) has led few authors to envisage that either a RNA/transposon regulation toolkit has been co-opted in somatic and germlines, or that somatic and germline stem cells (GSCs) share an ancestral set of genes inherited from a common origin (Juliano and Wessel 2010; Alié et al. 2011; Leclère et al. 2012). On a limited set of genes, either the cooption or the sharing of an inherited ancestral stemness program in somatic and germlines may, equally and rationally, account for this observation. Nevertheless, the more recent and more complete results in an annelid (Gazave, Béhague, et al. 2013), in ctenophores (Alié et al. 2011; Reitzel et al. 2016) and in sponges (Alié et al. 2015; this study) strongly support the ancestral hypothesis. Indeed, even if the cooption of a few genes, or even of a whole network have been reported (Glassford et al. 2015), to our knowledge, there is no clear evidence so far, that the aforementioned 11 genes form a unique interaction network, then making co-option of several networks less probable (but not to be fully rejected). Therefore, these results reinforce the hypothesis of ancestry of a complex RNA metabolizing toolkit necessary for reprogramming properties and then multi/pluripotency capabilities.
Conclusions
To retrace the early evolution of animal GMP content and functions, we largely complete the inventory of GMP genes in four sponge lineages and two ctenophoran lineages, and provide the expression patterns of 11 of them at different stages of the life cycle of a sponge.
Briefly, 18 GMP genes (piwiA, piwiB, argonaute, vasa, pl10, boule, two bruno, nanos, pumilio, Tud1, Tudkh/Tdrd2, RNF17/Tdrd4, Tud5, Tud6, Tud7, Tud9 and Akap1) were already present in the last common ancestor of metazoans, Urmetazoa. We show that most of these genes are expressed during the whole life cycle of Oscarella lobularis and during wound healing. Thus, none of the 11 genes studied are germ specific in this sponge species. Furthermore, all genes are expressed in type 2 vacuolar cells, very similar to archeocytes of other sponges. We therefore suggest that this cell type might be the equivalent of archeocyte-like cells in O. lobularis.
Expression studies in other sponges are necessary for comparative interpretation. Based on our current knowledge, we note the following facts: (1) piwi, vasa, nanos and pl10 are not devoted only to gametogenesis, in at least three of the four sponge classes; (2) the cells expressing these genes are not always the same in adults, possibly reflecting different capacities depending on the sponge; (3) comparing expression patterns of different copies of a same family suggests either subfunctionalization or functional redundancy depending on the gene/species considered, as observed in other species (Kuramochi-Miyagawa et al. 2001; Extavour et al. 2005; Funayama 2010; Giani et al. 2011); and (4) nanos shows a polarized expression in embryos (Sycon and Oscarella) as observed in various animals even if its role in axis patterning was recently challenged (Kanska and Frank 2013).
Our study shows that the GMP toolkit is involved in sexual reproduction in sponges, as it is in all animals studied so far. Furthermore, the GMP is also expressed in other sponge cell types not involved in sexual reproduction. Therefore not only the well documented—in a plethora of bilaterians—piwi, vasa and nanos genes (reviewed in fig. 7) have to be abandoned as germline specific markers, but rather a wider GMP set (for which expression data were sparser until now) (Juliano et al. 2011; Leclère et al. 2012; Alié et al. 2015). Together with recent studies on the subject, our findings make the hypothesis of the co-option of RNA metabolizing toolkit during gametogenesis and development less reliable and strikingly support that the GMP is rather an ancestral program. Our results and recent ones (Alié et al. 2015) enable to extend Solana’s hypothesis (2013) to all animal lineages: the unlimited PriSCs model (uPriSCs) proposes that PriSCs (primordial stem cells) can continuously self-renew and retain a mixed germ/soma potential. In demosponges, both choanocytes and archeocytes fulfill both criteria (Funayama et al. 2010; Nakanishi et al. 2014; Sogabe et al. 2016). In this context, it is worth noting that in both mammals and insects oogonia taken from gonads and grown in particular conditions, can revert back to an embryonic stem cell-like state (Reik and Surani 2015). Therefore, the bilaterian germline seem to retain a pluripotency, and that their storage in a particular niche (gonad tissues) would be responsible for their limited cell fate. If we assume that all animal multi/pluripotent cells share the same origin and the same ancestral genetic toolkit—whatever their more or less restricted potency—we need to reconsider the exact role of the GMP by further exploring non-conventional animal models. Future studies should pay particular attention to the fact that germ and stem cells share the property of reprogramming their genome by erasing epigenetic imprints (Reik and Surani 2015).
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank the molecular biology and scleronochrology support services of the Institut méditerranéen de Biodiversité et d’Ecologie marine et continentale (IMBE) and the diving staff of the OSU Pythéas. We thank Mr Olivier Chabrol for performing transcriptome partial annotation of O. lobularis that enabled us to initiate this study. We also thank Mr Thomas Smith, a native English speaker, for providing proofreading services, Mr Federico Fierro for the help editing the figures and Adam Reitzel for providing Mnemiopsis piwi3 sequence. This work has been carried out thanks to the doctoral fellowship Provence-Alpes-Côte-d'Azur region and the support of the A*MIDEX project (n° ANR-11-IDEX-0001-02) funded by the “investissements d'Avenir” French Government program, managed by the French National Research Agency (ANR), the CNRS (UMR7288) and the Aix-Marseille University.
Literature Cited
- Abascal F, Zardoya R, Posada D. 2005. ProtTest: selection of best-fit models of protein evolution. Bioinforma. 21:2104–2105. [DOI] [PubMed] [Google Scholar]
- Alié A, et al. 2011. Somatic stem cells express Piwi and Vasa genes in an adult ctenophore: ancient association of ‘germline genes’ with stemness. Dev Biol. 350:183–197. [DOI] [PubMed] [Google Scholar]
- Alié A, et al. 2015. The ancestral gene repertoire of animal stem cells. Proc Natl Acad Sci. 201514789: E7093–E71100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borisenko IE, Adamska M, Tokina DB, Ereskovsky AV. 2015. Transdifferentiation is a driving force of regeneration in Halisarca dujardini (Demospongiae, Porifera). Peer J. 3:e1211.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brusca RC, Brusca GJ. 2003. Invertebrates second edition. Sinauer Associates Sunderland, Massachusetts. [Google Scholar]
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17:540–552. [DOI] [PubMed] [Google Scholar]
- Cerutti H, Casas-Mollano JA. 2006. On the origin and functions of RNA-mediated silencing: from protists to man. Curr Genet. 50:81–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, et al. 2009. Mouse Piwi interactome identifies binding mechanism of Tdrkh Tudor domain to arginine methylated Miwi. Proc Natl Acad Sci U S A. 106:20336–20341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cwiklinski K, et al. 2015. The extracellular vesicles of the helminth pathogen, Fasciola hepatica: biogenesis pathways and cargo molecules involved in parasite pathogenesis. Mol Cell Proteomics MCP 14:3258–3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan BM, et al. 2015. Porifera In: Wanninger A, editor. Evolutionary developmental biology of invertebrates 1. Vienna: Springer; p. 65–106. [Google Scholar]
- Dunn CW, Leys SP, Haddock SHD. 2015. The hidden biology of sponges and ctenophores. Trends Ecol Evol. 30:282–291. [DOI] [PubMed] [Google Scholar]
- Eddy EM. 1975. Germ plasm and the differentiation of the germ cell line. Int Rev Cytol. 43:229–280. [DOI] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ereskovsky AV. 2010. The comparative embryology of sponges. Springer Netherlands: Dordrecht. [Google Scholar]
- Ereskovsky AV, et al. 2009. The Homoscleromorph sponge Oscarella lobularis, a promising sponge model in evolutionary and developmental biology. BioEssays 31:89–97. [DOI] [PubMed] [Google Scholar]
- Ereskovsky AV, et al. 2013. Pluri-annual study of the reproduction of two Mediterranean Oscarella species (Porifera, Homoscleromorpha): cycle, sex-ratio, reproductive effort and phenology. Mar Biol. 160:423–438. [Google Scholar]
- Ereskovsky AV, et al. 2015. Oscarella lobularis (Homoscleromorpha, Porifera) regeneration: epithelial morphogenesis and metaplasia, Singh, SR, editor. PLoS One 10:e0134566.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ereskovsky A, Renard E, Borchiellini C. 2013. Cellular and molecular processes leading to embryo formation in sponges: evidences for high conservation of processes throughout animal evolution. Dev Genes Evol. 223:5–22. [DOI] [PubMed] [Google Scholar]
- Ereskovsky AV, Tokina DB. 2007. Asexual reproduction in homoscleromorph sponges (Porifera; Homoscleromorpha). Mar Biol. 151:425–434. [Google Scholar]
- Ewen-Campen B, Schwager EE, Extavour CGM. 2010. The molecular machinery of germ line specification. Mol Reprod Dev. 77:3–18. [DOI] [PubMed] [Google Scholar]
- Extavour CG, Akam M. 2003. Mechanisms of germ cell specification across the metazoans: epigenesis and preformation. Development 130:5869–5884. [DOI] [PubMed] [Google Scholar]
- Extavour CG, Pang K, Matus DQ, Martindale MQ. 2005. vasa and nanos expression patterns in a sea anemone and the evolution of bilaterian germ cell specification mechanisms. Evol Dev. 7:201–215. [DOI] [PubMed] [Google Scholar]
- Farslow JC, et al. 2015. Rapid Increase in frequency of gene copy-number variants during experimental evolution in Caenorhabditis elegans. BMC Genomics 16:1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Force A, et al. 1999. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151:1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X-F, et al. 2015. DAZ family proteins, key players for germ cell development. Int J Biol Sci. 11:1226–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funayama N. 2010. The stem cell system in demosponges: insights into the origin of somatic stem cells. Dev Growth Differ. 52:1–14. [DOI] [PubMed] [Google Scholar]
- Funayama N. 2012. The stem cell system in demosponges: suggested involvement of two types of cells: archeocytes (active stem cells) and choanocytes (food-entrapping flagellated cells). Dev Genes Evol. 223:23–38. [DOI] [PubMed] [Google Scholar]
- Funayama N, Nakatsukasa M, Mohri K, Masuda Y, Agata K. 2010. Piwi expression in archeocytes and choanocytes in demosponges: insights into the stem cell system in demosponges. Evol Dev. 12:275–287. [DOI] [PubMed] [Google Scholar]
- Gaino E, Burlando B, Buffa P. 1986. The vacuolar cells of Oscarella lobularis (Porifera, Demospongiae): ulatrastructural organization, origin, and function. J Morphol. 188:29–37. [DOI] [PubMed] [Google Scholar]
- Gaino E, Manconi R, Pronzato R. 1995. Organizational plasticity as a successful conservative tactics in sponges. Anim Biol. 4:31–43. [Google Scholar]
- Gazave E, Béhague J, et al. 2013. Posterior elongation in the annelid Platynereis dumerilii involves stem cells molecularly related to primordial germ cells. Dev Biol. 382:246–267. [DOI] [PubMed] [Google Scholar]
- Gazave E, et al. 2008. NK homeobox genes with choanocyte-specific expression in homoscleromorph sponges. Dev Genes Evol. 218:479–489. [DOI] [PubMed] [Google Scholar]
- Gazave E, et al. 2009. Origin and evolution of the Notch signalling pathway: an overview from eukaryotic genomes. BMC Evol Biol. 9:249.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazave E, et al. 2012. No longer Demospongiae: Homoscleromorpha formal nomination as a fourth class of Porifera. Hydrobiologia 687:3–10. [Google Scholar]
- Gazave E, Lavrov DV, et al. 2013. Systematics and molecular phylogeny of the family Oscarellidae (Homoscleromorpha) with description of two new oscarella species. PLoS One 8:doi: 10.1371/journal.pone.0063976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giani VC, Yamaguchi E, Boyle MJ, Seaver EC. 2011. Somatic and germline expression of piwi during development and regeneration in the marine polychaete annelid Capitella teleta. Evo Devo. 2:10.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glassford WJ, et al. 2015. Co-option of an ancestral hox-regulated network underlies a recently evolved morphological novelty. Dev Cell. 34:520–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, et al. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59:307–321. [DOI] [PubMed] [Google Scholar]
- Gustafson EA, Wessel GM. 2010. Vasa genes: emerging roles in the germ line and in multipotent cells. BioEssays News Rev Mol Cell Dev Biol. 32:626–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 41:95–98. [Google Scholar]
- Handberg-Thorsager M, Saló E. 2007. The planarian nanos-like gene Smednos is expressed in germline and eye precursor cells during development and regeneration. Dev Genes Evol. 217:403–411. [DOI] [PubMed] [Google Scholar]
- Hollingworth D, et al. 2012. KH domains with impaired nucleic acid binding as a tool for functional analysis. Nucleic Acids Res. 40:6873–6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houston DW, Zhang J, Maines JZ, Wasserman SA, King ML. 1998. A Xenopus DAZ-like gene encodes an RNA component of germ plasm and is a functional homologue of Drosophila boule. Development 125:171–180. [DOI] [PubMed] [Google Scholar]
- Ihaka R, Gentleman R. 1996. R: a language for data analysis and graphics. J Comput Graph Stat. 5:299–314. [Google Scholar]
- Juliano C, Wang J, Lin H. 2011. Uniting germline and stem cells: the function of Piwi proteins and the piRNA pathway in diverse organisms. Annu Rev Genet. 45:447–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juliano C, Wessel G. 2010. Versatile germline genes. Science 329:640–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanska J, Frank U. 2013. New roles for Nanos in neural cell fate determination revealed by studies in a cnidarian. J Cell Sci. 126:3192–3203. [DOI] [PubMed] [Google Scholar]
- Karashima T, Sugimoto A, Yamamoto M. 2000. Caenorhabditis elegans homologue of the human azoospermia factor DAZ is required for oogenesis but not for spermatogenesis. Development 127:1069–1079. [DOI] [PubMed] [Google Scholar]
- Kerner P, Degnan SM, Marchand L, Degnan BM, Vervoort M. 2011. Evolution of RNA-binding proteins in animals: insights from genome-wide analysis in the sponge Amphimedon queenslandica. Mol Biol Evol. 28:2289–2303. [DOI] [PubMed] [Google Scholar]
- Kirino Y, et al. 2009. Arginine methylation of Piwi proteins catalysed by dPRMT5 is required for Ago3 and Aub stability. Nat Cell Biol. 11:652–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korotkova GP. 1997. Regeneration in animals. St Petersburg: St Petersburg Univ. Press. [Google Scholar]
- Kozin VV, Kostyuchenko RP. 2015. Vasa, PL10, and Piwi gene expression during caudal regeneration of the polychaete annelid Alitta virens. Dev Genes Evol. 225:129–138. [DOI] [PubMed] [Google Scholar]
- Kuales G, et al. 2011. Boule-like genes regulate male and female gametogenesis in the flatworm Macrostomum lignano. Dev Biol. 357:117–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, et al. 2001. Two mouse piwi-related genes: miwi and mili. Mech Dev. 108:121–133. [DOI] [PubMed] [Google Scholar]
- Lanna E. 2015. Evo-devo of non-bilaterian animals. Genet Mol Biol. 38:284–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavrov AI, Kosevich IA. 2014. Sponge cell reaggregation: mechanisms and dynamics of the process. Russ J Dev Biol. 45:205–223. [PubMed] [Google Scholar]
- Leclère L, et al. 2012. Maternally localized germ plasm mRNAs and germ cell/stem cell formation in the cnidarian Clytia. Dev Biol. 364:236–248. [DOI] [PubMed] [Google Scholar]
- Leininger S, et al. 2014. Developmental gene expression provides clues to relationships between sponge and eumetazoan body plans. Nat Commun. 5:3905. [DOI] [PubMed] [Google Scholar]
- Leys SP, Degnan BM. 2002. Embryogenesis and metamorphosis in a haplosclerid demosponge: gastrulation and transdifferentiation of larval ciliated cells to choanocytes. Invertebr Biol. 121:171–189. [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. [DOI] [PubMed] [Google Scholar]
- Ma J-B, et al. 2005. Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature 434:666–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, van der Oost J, Koonin EV. 2009. Prokaryotic homologs of Argonaute proteins are predicted to function as key components of a novel system of defense against mobile genetic elements. Biol Direct. 4:29.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell A, et al. 2015. The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res. 43:D213–D221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki K, Nishimiya-Fujisawa C, Fujisawa T. 2001. Universal occurrence of the vasa-related genes among metazoans and their germline expression in Hydra. Dev. Genes Evol. 211:299–308. [DOI] [PubMed] [Google Scholar]
- Moroz LL, et al. 2014. The ctenophore genome and the evolutionary origins of neural systems. Nature 510:109–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy D, Dancis B, Brown JR. 2008. The evolution of core proteins involved in microRNA biogenesis. BMC Evol Biol. 8:92.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi N, Sogabe S, Degnan BM. 2014. Evolutionary origin of gastrulation: insights from sponge development. BMC Biol. 12:26.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JS, Roe SM, Barford D. 2004. Crystal structure of a PIWI protein suggests mechanisms for siRNA recognition and slicer activity. EMBO J. 23:4727–4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pick KS, et al. 2010. Improved phylogenomic taxon sampling noticeably affects nonbilaterian relationships. Mol Biol Evol. 27:1983–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. 2009. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing. [Google Scholar]
- Rebscher N, et al. 2007. Vasa unveils a common origin of germ cells and of somatic stem cells from the posterior growth zone in the polychaete Platynereis dumerilii. Dev Biol. 306:599–611. [DOI] [PubMed] [Google Scholar]
- Rebscher N, Volk C, Teo R, Plickert G. 2008. The germ plasm component vasa allows tracing of the interstitial stem cells in the cnidarian Hydractinia echinata. Dev Dyn. 237:1736–1745. [DOI] [PubMed] [Google Scholar]
- Reik W, Surani MA. 2015. Germline and pluripotent stem cells. Cold Spring Harb Perspect Biol. 7:a019422.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitzel AM, Pang K, Martindale MQ. 2016. Developmental expression of ‘germline’- and ‘sex determination’-related genes in the ctenophore Mnemiopsis leidyi. Evo Devo. 7:17.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren R, et al. 2016. Phylogenetic resolution of deep eukaryotic and fungal relationships using highly conserved low-copy nuclear genes. Genome Biol Evol. 8:2683–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riesgo A, Farrar N, Windsor PJ, Giribet G, Leys SP. 2014. The analysis of eight transcriptomes from all Porifera classes reveals surprising genetic complexity in sponges. Mol Biol Evol. 1102–1120. [DOI] [PubMed] [Google Scholar]
- Rink JC. 2013. Stem cell systems and regeneration in planaria. Dev Genes Evol.. 223:67–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinkevich Y, et al. 2013. Repeated, long-term cycling of putative stem cells between niches in a basal chordate. Dev Cell. 24:76–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. Clifton NJ. 132:365–386. [DOI] [PubMed] [Google Scholar]
- Schenkelaars Q, Fierro-Constain L, Renard E, Hill AL, Borchiellini C. 2015. Insights into Frizzled evolution and new perspectives. Evol Dev. 17:160–169. [DOI] [PubMed] [Google Scholar]
- Sekiné K, Furusawa T, Hatakeyama M. 2015. The boule gene is essential for spermatogenesis of haploid insect male. Dev Biol. 399:154–163. [DOI] [PubMed] [Google Scholar]
- Shah C, et al. 2010. Widespread Presence of Human BOULE Homologs among Animals and Conservation of Their Ancient Reproductive Function. PLoS Genet. 6:e1001022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukayuk AL, Isaeva W. 2012. Molecular and sub-cellular gametogenic machinery of stem and germline cells across metazoa. In: Najman S, editor. Current Frontiers and Perspectives in Cell Biology. p. 279–314. [Google Scholar]
- Simpson TL. 1984. The cell biology of sponges. New York, NY: Springer New York. [Google Scholar]
- Sogabe S, Nakanishi N, Degnan BM. 2016. The ontogeny of choanocyte chambers during metamorphosis in the demosponge Amphimedon queenslandica. Evo Devo. 7:016.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solana J. 2013. Closing the circle of germline and stem cells: the primordial stem cell hypothesis. Evo Devo. 4:2.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele RE, David CN, Technau U. 2011. A genomic view of 500 million years of cnidarian evolution. Trends Genet. 27:7–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suga H, et al. 2013. The Capsaspora genome reveals a complex unicellular prehistory of animals. Nat Commun. 4:2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolia NH, Joshua-Tor L. 2007. Slicer and the Argonautes. Nat Chem Biol. 3:36–43. [DOI] [PubMed] [Google Scholar]
- van Wolfswinkel JC. 2014. Piwi and potency: PIWI proteins in animal stem cells and regeneration. Integr Comp Biol. 54:700–713. [DOI] [PubMed] [Google Scholar]
- Volkova MA, Zolotoreva GA, Korotkova GP. 1981. Halisarca dujardini Jonhston development from somatic cell conglomerates. In: Korotkhova GP. (ed) Morphogenesis in sponges. Leningrad University Press, Leiningrad, 74–93. [Google Scholar]
- Voronina E, Seydoux G, Sassone-Corsi P, Nagamori I. 2011. RNA granules in germ cells. Cold Spring Harb Perspect Biol. 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall DP, Fraser HB, Hirsh AE. 2003. Detecting putative orthologs. Bioinformatics 19:1710–1711. [DOI] [PubMed] [Google Scholar]
- Xu H, Li Z, Li M, Wang L, Hong Y. 2009. Boule is present in fish and bisexually expressed in adult and embryonic germ cells of medaka, Rusche, L, editor. PLoS One 4:e6097.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.