Abstract
Although chromosome 1p36 deletion syndrome is considered clinically recognizable based on characteristic features, the clinical manifestations of patients during infancy are often not consistent with those observed later in life. We report a 4-month-old girl who showed multiple congenital anomalies and developmental delay, but no clinical signs of syndromic disease caused by a terminal deletion in 1p36.32-p36.33 that was first identified by targeted-exome sequencing for molecular diagnosis.
Chromosome 1p36 deletion syndrome (MIM 607872) is the most common terminal deletion syndrome in humans, occurring in ~1 in 5,000 individuals.1, 2 A pattern of characteristic functional deficits, congenital anomalies and physical features associated with 1p36 deletions has been observed in case reports, as well as clinical descriptions of large cohorts of individuals with 1p36 deletions.2–7 Defining this pattern has made chromosome 1p36 deletion syndrome a clinically recognizable entity. A characteristic facial appearance, hypotonia, psychomotor retardation and poor or absent speech are suggestive of chromosome 1p36 deletion syndrome, and the diagnosis is confirmed by detecting a deletion of the most distal band of the short arm of chromosome 1. However, a relatively high proportion of reported cases of chromosome 1p36 deletion syndrome have been diagnosed after the age of 1 year because these characteristics are often difficult to recognize during infancy, and significant phenotypic variability is observed among patients with this disease.2–7
Chromosomal microarray (CMA) is a powerful tool for detecting genomic copy number aberrations. In 2010, CMA testing was recommended as a first-tier examination for patients with unexplained developmental delay/intellectual disability (DD), autism spectrum disorders or multiple congenital anomalies.8 This diagnostic method is commonly used worldwide and can detect all types of chromosomal alterations associated with chromosome 1p36 deletion syndrome, such as terminal deletions, interstitial deletions, complex rearrangements and derivative chromosomes.3 CMA can be performed in the first-tier if patients are suspected of having this disease based on their clinical features. Recently, next-generation sequencing (NGS), particularly whole-exome sequencing (WES) or targeted-exome sequencing (TES), has become common worldwide as a clinical diagnostic tool for patients with suspected mutations, such as single nucleotide variations (SNVs) and small insertions/deletions (indels), that are responsible for human single gene disorders. Pathogenic copy number variations (CNVs), which frequently contain the genes that are functionally responsible for phenotypes of diseases,9 in targeted exons can also be simultaneously detected by WES and TES, leading to an appropriate clinical diagnosis.10,11 Therefore, it would be advantageous to screen genetic abnormalities using WES/TES data before performing CMA in undiagnosed syndromic patients suspected of having either single gene defects or cryptic chromosomal aberrations.
Here, we present a 4-month-old patient with chromosome 1p36 deletion syndrome, which was not suspected from clinical findings before genetic testing and was successfully diagnosed by the NGS-first approach using a disease-related exome panel.
The patient was a 4-month-old Japanese girl born to non-consanguineous healthy parents aged 20 (father) and 17 years (mother). Intrauterine growth retardation and ventricular septal defect (VSD) were indicated via a 28-week antenatal ultrasound. The child was born by artificial labor at 38 weeks and 3 days of gestation owing to oligohydramnios detected via a 38-week antenatal scan and had a birth weight of 1,718 g (−3.4 s.d.), a length of 41.5 cm (−3.2 s.d.) and an occipitofrontal circumference of 30 cm (−2.3 s.d.). Apgar scores were 9 and 10 at 1 and 5 min after birth, respectively. The neonate was admitted to the neonatal intensive care unit with polypnea that developed immediately after birth, and subsequently required oxygenation. The patient had multiple congenital heart defects, such as VSD, patent ductus arteriosus (PDA) and patent foramen ovale, as well as malrotation of the intestine. Although catecholamines and diuretics were used to treat decreased urine volume and cardiac dilation, pulmonary hypertension persisted, and increased cardiac dilation and decreased cardiac contractility were observed. At 16 days of age, the patient required artificial respiration with endotracheal intubation. After extubation due to stable breathing and circulatory dynamics at 40 days of age, nasal directional positive airway pressure (n-DPAP) was used. However, the patient’s respiratory condition worsened after discontinuing n-DPAP at 44 days of age, and artificial respiration with endotracheal intubation was again required at 50 days of age. Pulmonary artery banding and PDA ligation were performed at 67 days of age for severe cardiac hypofunction. After extubation at 84 days of age, she required high-flow nasal cannula for respiratory failure for 8 days and was transferred to the general ward at 109 days of age, although metabolic acidosis of uncertain cause was observed, and treatment for heart failure was required. The patient showed severe DD, generalized hypotonia and dysmorphic features, including a flat nasal bridge and low-set ears, and was suspected of having dilated cardiomyopathy (Table 1). Laboratory tests for various metabolic disorders were unremarkable. Conventional G-banded chromosome analysis showed a normal 46,XX karyotype. Her clinical features were not sufficient to diagnose a specific disease but suggested potential genetic disorders, including diseases caused by either a single gene or a chromosomal defect.
Table 1. Summary of clinical features frequently observed in patients with chromosome 1p36 deletion syndrome7 and the present case.
| Characteristics | Present case | Frequenciesa |
|---|---|---|
| Age at clinical evaluation | 4 months | 0–25 years |
|
Craniofacial and skeletal features | ||
| Characteristic craniofacial features | ||
| Microcephaly | − | 83 % (20/24) |
| Brachycephaly | − | 65 % (11/17) |
| Straight eyebrow | − | 84 % (32/38) |
| Deep-set eyes | − | 93 % (37/40) |
| Epicanthus | − | 86 % (30/35) |
| Broad nasal root/bridge | + | 97 % (32/33) |
| Long philtrum | ± | 63 % (22/35) |
| Low-set ears | + | 88 % (29/33) |
| Pointed chin | − | 89 % (34/38) |
| Late closure of the anterior fontanel | NA | 56 % (18/32) |
|
Neurological features (clinical) | ||
| Axial hypotonia | + | 92 % (44/48) |
| Poor sucking | ± | 70 % (32/46) |
| Difficulty in swallowing | − | 56 % (27/48) |
| Developmental delay | ||
| Intellectual disability | NA | 98 % (49/50) |
| Acquire independent gait | NA | 32 % (16/50) |
| Behavior disorders | ||
| Self-injury | NA | 30 % (11/37) |
| Temper tantrum | NA | 30 % (11/37) |
| Epilepsy | ||
| History of epilepsy | − | 70 % (35/50) |
|
Neurological features (radiological) | ||
| Cerebral white matter | ||
| Enlargement of lateral ventriclesb | + | 36 % (15/42) |
|
Cardiac abnormalities | ||
| Congenital heart defects | ||
| PDA | + | 37 % (18/49) |
| VSD | + | 37 % (18/49) |
|
Sensory organs | ||
| Ears | ||
| Hearing problems | Mild | 39 % (19/49) |
| Eyes | ||
| Strabismus | − | 33 % (15/46) |
Abbreviations: NA, not available; PDA, patent ductus arteriosus; VSD, ventricular septal defects.
Frequency of each clinical feature reported by Shimada et al.7 was shown. Only clinical features reported with high frequency (more than 30 %) were listed.
Enlargement of lateral ventricles was detected by computed tomography at 5 months of age and magnetic resonance imaging at 8 months of age.
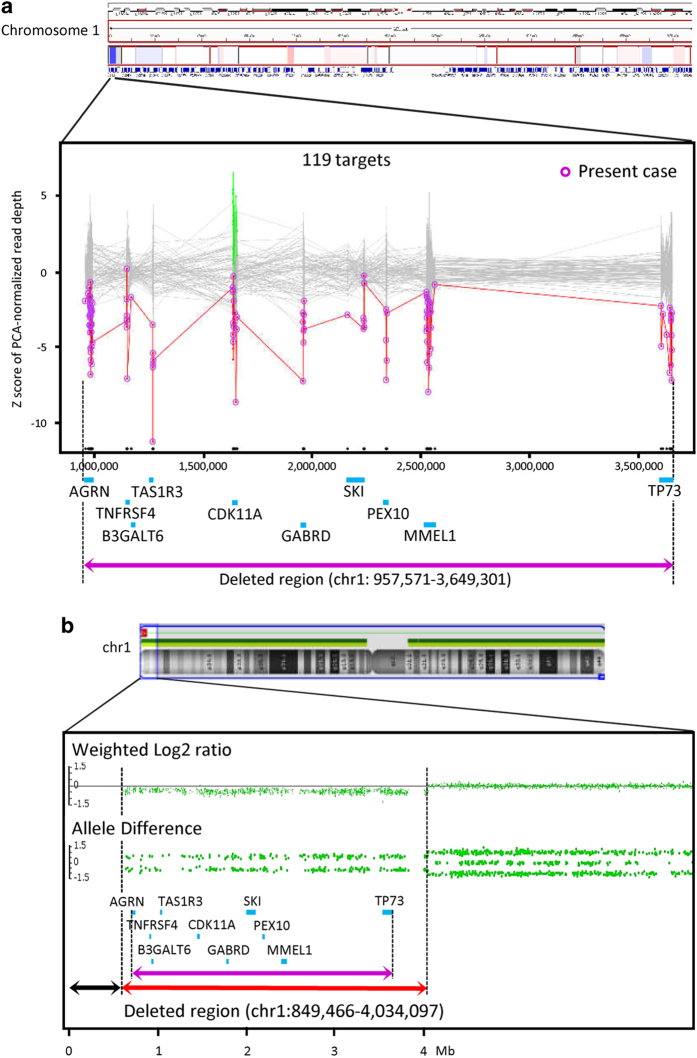
The ethics committees of Tokushima University approved this study. After obtaining written consent from the parents of the patient, molecular diagnosis was performed using genomic DNA extracted from patient whole blood. TES was performed using a TruSight One Sequencing Panel (Illumina, San Diego, CA, USA) and a MiSeq benchtop sequencer (Illumina). The alignments of sequencing reads to the human reference genome (GRCh37/hg19), duplicate read removal, local realignment around indels, base quality score recalibration, variant calling and annotation were performed as previously described.12 To identify SNVs, we excluded sequence variants with minor allele frequencies (>0.05) included in various human genome variation databases, as previously described,13 and used an integrative Japanese Genome Variation Database (https://ijgvd.megabank.tohoku.ac.jp/). Detection of CNVs using TES data with a resolution of a single exon to several exons, depending on the size of exons, was performed as previously described13 and using an eXome-Hidden Markov Model v1.0 (XHMM, https://atgu.mgh.harvard.edu/xhmm/). These analyses showed no disease-causing SNVs or indels, but detected monoallelic deletions of 10 genes (including the gene SKI (OMIM 164780), which may contribute to the 1p36 deletion phenotype)14 located from positions 957,571 to 3,649,301 on chromosome 1 (1p36.33-p36.32), suggesting the presence of a gross chromosomal deletion involving the 1p36.3 region (Figure 1a). Validation and fine mapping of the deleted region were performed by CMA using an Affymetrix CytoScan HD chromosome microarray platform (Affymetrix, Santa Clara, CA, USA), as previously described,15 and fluorescence in situ hybridization (FISH) using probes for the commonly deleted region (CDC2L1) and the subtelomeric region (CEB108/T7, maximal physical distance from telomere<300 kb). Although no probes were located within ~850 kb of the terminal region of 1p36.33 in the Affymetrix CytoScan HD array, CMA detected a monoallelic chromosomal deletion within 1p36.32-p36.33 as 46,XX.arr[hg19] 1p36.33-p36.32(849,466–4,034,097)x1 (Figure 1b). FISH detected the deletion of probes for CDC2L1 and CEB108/T7, suggesting that a pure terminal deletion occurred at 1p36.3-pter (data not shown). The FISH-based karyotype was 46,XX.ish del(1)(p36)(CEB108/T7-,CDC2L1-). The deletion was not confirmed as de novo because of the unavailability of parental DNA.
Figure 1.
(a) eXome-Hidden Markov Model (XHMM) analysis using targeted-exome sequencing (TES) data automatically detected the genomic copy number loss of 10 genes (blue bars) located within 1p36.33-p36.32, suggesting a 2.7-Mb deletion (purple closed arrow). The x axis shows the physical position, and the y axis shows the Z score of the principal component analysis (PCA) that was normalized to read depth. Purple circles connected by red lines represent values of the individual subjected to TES. Gray dots with gray connected lines indicate the results of normalized read depth obtained from in-house control data (N=106). Copy number gains (green dots) and losses (red dots) of target sites on the CDK11A gene were detected in three and four control samples, respectively. (b) Chromosomal microarray (CMA) using an Affymetrix Cytoscan HD array demonstrated a 3.2-Mb heterozygous deletion within 1p36.33-p36.32 (red closed arrow). The weighted-copy number log2 ratio, allele peak spots and genes included in Trusight One (blue bars) are shown. Because of the lack of probes in the 850-kb terminal region of 1p36.33 (black closed arrow), additional fluorescence in situ hybridization (FISH) analysis using a subtelomere probe was required to demonstrate the 1p36 terminal deletion. Compared with TES-based copy number variation (CNV) detection, the distal and proximal break points of the deleted region were shifted to the distal and proximal sides, respectively, in CMA (purple closed arrow).
Using the NGS-first approach, a disease-causing CNV in 1p36 was identified in the undiagnosed patient who lacked distinctive signs and symptoms, particularly typical craniofacial features, of chromosome 1p36 deletion syndrome (Table 1). At the time of clinical evaluation, the patient was likely too young to be recognized as having the clinical features of chromosome 1p36 deletion syndrome. The sequencing panel used in this study was constructed for clinical use to detect disease-causing SNVs, and targets in ~12-Mb regions covering ~5,000 genes associated with known clinical phenotypes based on Online Mendelian Inheritance in Man (OMIM; http://www.omim.org/). Therefore, the targeted regions of this panel represented ~20 % of the entire exome and 0.4 % of the genome. The potential clinical relevance of CNV increases with respect to the number of genes, particularly morbid OMIM genes, genes important in development or genes having a dosage phenotypic effect, within the region of genomic imbalance.9 Therefore, the TES data extracted from this sparse panel may be sufficient to simultaneously evaluate disease-causing SNVs and CNVs in first-choice diagnostic approaches for undiagnosed congenital diseases that are suspected of being genetic in origin. With respect to the interpretation of CNVs, the sequence-first approach using this panel has an advantage because those observed by TES using this panel may be designated as disease causing if CNVs are detected only in affected individuals. After WES or TES analysis, CMA with or without FISH is also necessary to accurately determine the range of the region of genomic imbalance for accurate cytogenetic diagnosis, estimation of prognosis, appropriate treatment and genetic counseling for patients, although no correlation between deletion size and the number of observed clinical features was reported in a large cohort of chromosome 1p36 deletion syndrome.16
Acknowledgments
Authors thank the patient and her family for participation in this study. This work was partly performed in the Cooperative Research Project Program of the Medical Institute of Bioregulation, Kyushu University. This work was supported by JSPS KAKENHI Grant Numbers 26293304 (II) and 15K19620 (TN) from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
Footnotes
The authors declare no conflict of interest.
References
- Shaffer LG, Lupski JR. Molecular mechanisms for constitutional chromosomal rearrangements in humans. Annu Rev Genet 2000; 34: 297–329. [DOI] [PubMed] [Google Scholar]
- Heilstedt HA, Ballif BC, Howard LA, Kashork CD, Shaffer LG. Population data suggest that deletions of 1p36 are a relatively common chromosome abnormality. Clin Genet 2003; 64: 310–316. [DOI] [PubMed] [Google Scholar]
- Battaglia A. 1p36 deletion syndrome. In:Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A (eds). GeneReviews [Internet]. University of Washington, Seattle: Seattle, WA, 1993–2015. [Google Scholar]
- Shapira SK, McCaskill C, Northrup H, Spikes AS, Elder FF, Sutton VR et al. Chromosome 1p36 deletions: the clinical phenotype and molecular characterization of a common newly delineated syndrome. Am J Hum Genet 1997; 61: 642–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilstedt HA, Ballif BC, Howard LA, Lewis RA, Stal S, Kashork CD et al. Physical map of 1p36, placement of breakpoints in monosomy 1p36, and clinical characterization of the syndrome. Am J Hum Genet 2003; 72: 1200–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battaglia A, Hoyme HE, Dallapiccola B, Zackai E, Hudgins L, McDonald-McGinn D et al. Further delineation of deletion 1p36 syndrome in 60 patients: a recognizable phenotype and common cause of developmental delay and mental retardation. Pediatrics 2008; 121: 404–410. [DOI] [PubMed] [Google Scholar]
- Shimada S, Shimojima K, Okamoto N, Sangu N, Hirasawa K, Matsuo M et al. Microarray analysis of 50 patients reveals the critical chromosomal regions responsible for 1p36 deletion syndrome-related complications. Brain Dev 2015; 37: 515–526. [DOI] [PubMed] [Google Scholar]
- Miller DT, Adam MP, Aradhya S, Biesecker LG, Brothman AR, Carter NP et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet 2010; 86: 749–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Iafrate AJ, Brothman AR. Copy number variations and clinical cytogenetic diagnosis of constitutional disorders. Nat Genet 2007; 39: S48–S54. [DOI] [PubMed] [Google Scholar]
- Miyatake S, Koshimizu E, Fujita A, Fukai R, Imagawa E, Ohba C et al. Detecting copy-number variations in whole-exome sequencing data using the eXome Hidden Markov Model: an 'exome-first' approach. J Hum Genet 2015; 60: 175–182. [DOI] [PubMed] [Google Scholar]
- Morita K, Naruto T, Tanimoto K, Yasukawa C, Oikawa Y, Masuda K et al. Simultaneous detection of both single nucleotide variations and copy number alterations by next-generation sequencing in Gorlin syndrome. PLoS ONE 2015; 10: e0140480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto N, Naruto T, Kohmoto T, Komori T, Imoto I. A novel PTCH1 mutation in a patient with Gorlin syndrome. Hum Genome Variat 2014; 1: 14022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromer M, Moran JL, Chambert K, Banks E, Bergen SE, Ruderfer DM et al. Discovery and statistical genotyping of copy-number variation from whole-exome sequencing depth. Am J Hum Genet 2012; 91: 597–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan VK, Zaveri HP, Scott DA. 1p36 deletion syndrome: an update. Appl Clin Genet 2015; 8: 189–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W, Higaki T, Eguchi-Ishimae M, Iwabuki H, Wu Z, Yamamoto E et al. DGCR6 at the proximal part of the DiGeorge critical region is involved in conotruncal heart defects. Hum Genome Variat 2015; 2: 15004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajecka M, Mackay KL, Shaffer LG. Monosomy 1p36 deletion syndrome. Am J Med Genet C Semin Med Genet 2007; 145C: 346–356. [DOI] [PubMed] [Google Scholar]
Data Citations
- Imoto Issei.HGV Database. 2016. 10.6084/m9.figshare.hgv.784. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Imoto Issei.HGV Database. 2016. 10.6084/m9.figshare.hgv.784. [DOI]