Abstract
Widespread species spanning strong environmental (e.g., climatic) gradients frequently display morphological and physiological adaptations to local conditions. Some adaptations are common to different species that occupy similar environments. However, the genomic architecture underlying such convergent traits may not be the same between species. Using genomic data from previous studies of three widespread eucalypt species that grow along rainfall gradients in southern Australia, our probabilistic approach provides evidence that adaptation to aridity is a genome-wide phenomenon, likely to involve multiple and diverse genes, gene families and regulatory regions that affect a multitude of complex genetic and biochemical processes.
Keywords: adaptation, genetic architecture, outlier markers, Eucalyptus, parallel evolution, convergent evolution, climate
Introduction
Widespread species spanning strong environmental gradients frequently display morphological and physiological adaptations to local conditions. For example, local adaptation to high temperature extremes and aridity in many plants appears to involve multiple genetically independent phenotypic traits encompassing facets of ontogeny, resource allocation, defense (Gauli et al. 2015), phenology, water relations (Alberto et al. 2013), and morphology (Steane et al. 2014, 2017). Populations of Eucalyptus camaldulensis are differentially adapted to aridity, and Dillon et al. (2014) identified five putatively adaptive SNPs in three candidate genes (each on a different chromosome) involved just in plant water relations. It is logical, therefore, that the genetic basis of adaptation is complex and probably involves numerous genes, distributed across the genome, that affect a broad range of genetically complex phenotypic traits.
There is mounting evidence to support this hypothesis (Turner et al. 2008; Pujolar et al. 2014; Nicotra et al. 2015). Similar adaptive phenotypes may arise through complex genetic and biochemical networks (Marazzi et al. 2012) via (i) shared ancestry (lineage sorting of an ancestral polymorphism or reversion to an ancestral genotype; Eckert et al. 2013) or the presence of “evolutionary precursors” (Marazzi et al. 2012), (ii) parallel de novo mutation or (iii) completely different genetic and/or biochemical pathways [see Martin and Orgogozo (2013) for a review]. To avoid the confusion surrounding the terms “convergent” and “parallel” evolution (where usage is generally dependent on the degree of relatedness of the organisms being compared; Scotland 2011; Martin and Orgogozo 2013; Nouhaud et al. 2014), we here adopt the terminology sensu Scotland (2011), whereby homoplasy (i.e., a character shared by different species but not present in their most recent common ancestor) at the genetic level is referred to as “parallelism” and homoplasy at the phenotypic level is “convergence”.
There are numerous examples in which phenotypic convergence has come about through different underlying genetic/biochemical pathways (Bigham et al. 2010; Turner et al. 2010; Pascoal et al. 2014; Renaut et al. 2014). There are also many examples of “gene re-use” in adaptation (i.e., changes to the same gene for adaptation by different populations; see Conte et al. 2012; Nouhaud et al. 2014), but only rarely are parallel mutations involved (e.g., Projecto-Garcia et al. 2013). More often, mutations within a gene may have similar biochemical/phenotypic effects, but the mutations themselves are not identical (Conte et al. 2012; Renaut et al. 2014; Hodgins et al. 2015). Well-studied examples of this are domestication genes in cereal and pulse crops (Sang 2009; Weller et al. 2012).
Recent analyses of adaptation in three keystone eucalypt species using directly comparable methodology (Steane et al. 2014, 2015, 2017) provide an ideal opportunity to investigate parallelism in adaptation in this important tree genus. In these species, genomic scans and outlier marker analysis were used to identify signals of adaptation. On a probabilistic level, the identification of outlier markers associated with a specific trait or variable (e.g., aridity) common to two or more species could indicate a shared adaptive pathway (i.e., through shared ancestry or parallelism, e.g., Nouhaud et al. 2014). On the other hand, if two or more species do not share any putatively adaptive markers, it may suggest that adaptation in each taxon is occurring through different genetic/biochemical pathways (or, simply, that the screening method is not comprehensive enough to detect shared pathways).
Using DArTseq technology (Sansaloni et al. 2011), we previously conducted population-level genetic studies of three widespread eucalypt species (family Myrtaceae) that grow across rainfall gradients in southern Australia: Eucalyptus tricarpa (L.A.S. Johnson) L.A.S. Johnson & K.D. Hill in south-eastern Australia, and Eucalyptus salubris F. Muell. and Eucalyptus loxophleba ssp. lissophloia L.A.S. Johnson & K.D. Hill (hereon referred to as E. loxophleba) in south-western Australia. We detected genomic signals of environmental adaptation across climate gradients in both E. tricarpa (Steane et al. 2014) and E. loxophleba (Steane et al. 2017). However, in E. salubris we found evidence for two discrete lineages and, while there may be climate-adaptation within each lineage, the small sample size and distribution of each lineage made confirmation difficult (Steane et al. 2015). In both E. loxophleba and E. salubris, we found indications that soil type may influence patterns of adaptation. In this paper, we aim to assess genomic data from these three keystone species to evaluate evidence for (i) genome-wide adaptation to aridity, and (ii) parallelism/common ancestry in patterns of adaptation. If the emerging evidence of complex adaptive networks (Eckert et al. 2013; Martin and Orgogozo 2013) applies in these species, we hypothesize that putatively adaptive genetic markers will be distributed across the genome with little commonality among species. Alternatively, if species have genetic adaptations derived from parallelism or common ancestry we would expect to see evidence of shared adaptive genetic markers associated with specific traits or variables.
Materials and Methods
Study Species
The three study species, E. tricarpa (Steane et al. 2014), E. loxophleba (Steane et al. 2017) and E. salubris (Steane et al. 2015) were selected for genomic analysis of adaptation because their natural distributions span aridity (rainfall and temperature) gradients (table 1). Eucalyptus tricarpa (subgenus Symphyomyrtus, section Adnataria) is a tree that grows to 35 m in height, occurring in open forest throughout south-eastern Australia, with a distribution spanning a mean annual precipitation (MAP) range of 450–1,200 mm. Eucalyptus loxophleba [subgenus Symphyomyrtus, section Bisectae sensu Brooker (2000)] is a mallee that grows up to 8 m tall. Its range in south-western Australia spans a MAP range from 230 mm in the Goldfields region to 360 mm in the eastern Wheatbelt region. Eucalyptus salubris [subgenus Symphyomyrtus, section Bisectae sensu Brooker (2000)] is a small tree that grows to 15 m tall; it is widespread in south-western Australia, distributed across a MAP range from 200 mm in the Goldfields region to 440 mm in the Wheatbelt region. The two south-western Australian species grow across a variety of soil types. For each study species, we sampled leaves from approximately 30 individuals from each of nine populations across the climatic gradient, with a minimum of two tree heights between each sampled individual. A serendipitous discovery of two cryptic lineages within E. salubris (Steane et al. 2015) necessitated that, for genomic analyses (see below), the E. salubris data be divided into one set (Sal1) of five and a second set (Sal2) of four populations (see table 1).
Table 1.
Environmental Parameters at Each Population of Three Species of Eucalyptus Included in This Study
| Species/Population | Lat (°N) | Long (°E) | Mean Annual P/PE | T ann (°C) | R ann (mm) | TMXWM (°C) | TMNCM (°C) |
|---|---|---|---|---|---|---|---|
| E. tricarpa | |||||||
| Tarnagulla | −36.76 | 143.85 | 0.598 | 13.84 | 480.9 | 29.48 | 2.47 |
| Mt Bealiba | −36.81 | 143.65 | 0.659 | 13.28 | 543.6 | 28.64 | 2.33 |
| Craigie | −37.08 | 143.77 | 0.683 | 13.27 | 534.3 | 28.48 | 2.48 |
| Heyfield | −37.94 | 146.73 | 0.786 | 13.31 | 713.2 | 25.84 | 2.94 |
| Heathcote | −36.98 | 144.75 | 0.816 | 13.14 | 622.8 | 28.14 | 2.19 |
| Mt Nowa Nowa | −37.7 | 148.11 | 0.908 | 13.21 | 846.6 | 24.72 | 2.22 |
| Tuckerbox | −37.63 | 148.24 | 1.026 | 12.89 | 892.5 | 24.75 | 1.78 |
| Christmas Hills | −37.69 | 145.31 | 1.141 | 13.23 | 897.1 | 25.89 | 4.12 |
| Martins Creek | −37.47 | 148.58 | 1.231 | 12.6 | 1,059 | 25.04 | 1.21 |
| E. loxophleba ssp. lissophloia | |||||||
| Pianko Rd | −29.92 | 121.67 | 0.114 | 19.6 | 252 | 35.88 | 5.03 |
| Goongarrie | −29.97 | 121.06 | 0.118 | 19.8 | 266 | 35.97 | 5.27 |
| Karonie | −31.02 | 123.06 | 0.138 | 18.3 | 264 | 34.28 | 4.24 |
| Quairnie Rock | −31.27 | 121.09 | 0.17 | 17.9 | 299 | 33.69 | 4.81 |
| Yellowdine | −31.29 | 119.68 | 0.192 | 17.9 | 301 | 34.53 | 4.07 |
| Hines Hill | −31.54 | 118.06 | 0.242 | 18.1 | 314 | 34.36 | 5.34 |
| Burracoppin | −31.38 | 118.46 | 0.252 | 18.1 | 325 | 34.52 | 4.99 |
| Narembeen | −32.02 | 118.54 | 0.263 | 17.8 | 325 | 34.33 | 4.83 |
| Graham Rock | −32.46 | 119.06 | 0.287 | 16.7 | 360 | 32.99 | 4.09 |
| E. salubris Lineage 1 | |||||||
| Queen Victoria Spring Reserve | −30.15 | 123.32 | 0.112 | 18.3 | 218 | 34.29 | 3.89 |
| Bullock Holes Reserve | −30.52 | 121.79 | 0.127 | 18.1 | 229 | 33.6 | 4.29 |
| Credo Station | −30.19 | 120.65 | 0.141 | 18.3 | 244 | 34.3 | 4.16 |
| Lake Johnston | −32.03 | 120.82 | 0.189 | 16.9 | 269 | 32.38 | 4.38 |
| Bruce Rock | −31.87 | 118.17 | 0.27 | 17.2 | 324 | 33.75 | 4.43 |
| E. salubris Lineage 2 | |||||||
| Kangaroo Hills | −30.99 | 121.12 | 0.156 | 17.5 | 263 | 33.01 | 4.22 |
| Dunn Rock | −33.24 | 119.55 | 0.291 | 15.7 | 345 | 29.82 | 5.01 |
| Ravensthorpe | −33.45 | 120.03 | 0.297 | 15.9 | 368 | 29.2 | 5.73 |
| Lockhart Rd (Newdegate) | −33.3 | 119.02 | 0.323 | 15.6 | 354 | 30.04 | 4.85 |
Note.—All populations of E. tricarpa were in the Australian state of Victoria. All populations of E. loxophleba ssp. lissophloia and E. salubris were in the Australian state of Western Australia. Climate data were downloaded from Atlas of Living Australia (P/PE, ratio of precipitation to pan evaporation) and ANUCLIM (Xu and Hutchinson 2011). Lat, latitude; long, longitude; T ann, mean annual temperature; R ann, mean annual rainfall; TMXWM, mean maximum temperature of the warmest period (week); TMNCM, mean minimum temperature of the coldest period (week).
Genome Scans
Tissue collection, DNA extraction and genome scan (DArTseq) procedures have been described previously for E. tricarpa (Steane et al. 2014), E. loxophleba (Steane et al. 2017) and E. salubris (Steane et al. 2015). Briefly, DNA was extracted from leaves of each individual, standardized to approximately 50 ng µl − 1 and sent to Diversity Arrays Technology Pty. Ltd. (Canberra, Australia) for genotyping using DArTseq technology (Sansaloni et al. 2011). These studies were some of the earliest to use the DArTseq technology and dominant (presence/absence) data from DArTseq were used for the analyses; however, the advantage of using DArTseq was that each marker was tagged by approximately 60 bp of DNA sequence data that allowed it to be placed on the Eucalyptus grandis reference genome. The working data sets for each species (6,544 markers in E. tricarpa, 4,851 in E. loxophleba and 16,122 in E. salubris) included only markers for which (i) there was ≤5% missing data, and (ii) the “Q value” (i.e., average read depth/standard deviation) was >2.5. In each data set the proportion of missing data ranged from 1.6% to 4.6%.
Identification of Putatively Adaptive Markers
BAYESCAN Ver. 2.1 (Foll and Gaggiotti 2008) was used, as described previously (Steane et al. 2014), to identify outlier markers that differentiated the sampled populations of each species (or of each lineage, in the case of E. salubris) more than would be expected solely from stochastic processes (such as drift), and would therefore signal that selective processes may have been involved in the differentiation of the populations within each species. BAYESCAN is one of the most robust differentiation methods for outlier detection (Perez-Figueroa et al. 2010; Narum and Hess 2011; Vilas et al. 2012; Savolainen et al. 2013), even when the island model of allele frequency correlation, upon which the software is based, is violated (e.g., in a population with a hierarchical structure; De Mita et al. 2013). Although Lotterhos and Whitlock (2014) found that recent demographic history, particularly expansion from refugia, influenced false discovery rate (FDR) in BAYESCAN, this was not considered likely to be problematic for E. loxophleba, because this species is known to have persisted throughout the range during the Pleistocene climatic fluctuations (Byrne and Hines 2004). This is common in many species that have been studied in the ancient landscape of south-western Australia (Byrne 2008), so we consider it is also likely to be the case in E. salubris. For Eucalyptus tricarpa, where recent demographic history might confound outlier detection, our routine two-pronged approach (Manel et al. 2009; Funk et al. 2012; De Mita et al. 2013) increased the likelihood that our final set of “outlier” markers represented regions of the genome that were under selection. This approach (see Steane et al. 2014) comprised a differentiation-based outlier detection method (BAYESCAN) followed by further filtering of the outlier markers on the basis of linear regression, saving only markers whose population-level allele frequencies were significantly (P ≤ 0.05) correlated with population-level variation in (i) climate and/or (ii) soil variables. Linear regressions were done with the PROC REG procedure of SAS, correcting for multiple testing within each set of environmental variables using a conservative “dependent” FDR of 0.05 (DFDR; Benjamini and Yekateuli 2001) that allowed for correlation between tests within each set of variables (i.e., climate or soil).
To determine the impact of different BAYESCAN settings on outlier detection in our data sets, we conducted a sensitivity analysis. In the original studies of E. tricarpa (Steane et al. 2014) and E. loxophleba (Steane et al. 2017), prior settings for BAYESCAN followed the recommendations of the manual: (i) prior odds for the neutral model of selection were higher for larger data sets than for smaller data sets; (ii) the inbreeding coefficients (F IS) were set according to specific data available for E. loxophleba (F IS uniform between 0.01 and 0.08; Byrne 2008), and a more general value (F IS uniform between 0.01 and 0.3) for E. salubris and E. tricarpa, estimated from a range of values from several Eucalyptus species (Byrne 2008). Here, we tested the effects on the BAYESCAN output of using (i) high prior odds (as recommended by Lotterhos and Whitlock (2014), (ii) uniform F IS settings and (iii) the BAYESCAN default values. The various analyses were rated as low, medium, medium–high and high stringency (see table 2).
Table 2.
Parameters and Output Summary of BAYESCAN Sensitivity Test
| Species | BAYESCAN Model Parameters | Total No. Dominant DArT Markers | No. Markers Included in BS Analysis | F IS Prior (Uniform between… ) | Prior Odds | No. Outliers Detected by BS Analysis | Max Outlier Q Value a | % of Input | Outlier FST Mean | Nonoutlier FST Mean | Stringency |
|---|---|---|---|---|---|---|---|---|---|---|---|
| E. tricarpa | Default | 6,544 | 3,684 | 0–1 | 10 | 131 | 0.049 | 3.50 | 0.3312 | 0.1368 | Low |
| E. tricarpa | BS-recommended PO + generic eucalypt FIS ( Byrne 2008 ) | 6,544 | 3,684 | 0.01–0.3 b | 100 | 94 | 0.050 | 2.60 | 0.3210 | 0.1200 | Medium |
| E. tricarpa | Relatively high BS-recommended PO + generic eucalypt F IS (Byrne 2008) | 6,544 | 3,684 | 0.01–0.3 b | 200 | 80 | 0.0571 | 2.22 | 0.3305 | 0.1206 | Medium |
| E. tricarpa | High PO (Lotterhos and Whitlock 2014) + generic eucalypt F IS (Byrne 2008) | 6,544 | 3,684 | 0.01–0.3 b | 3,685 | 41 | 0.044 | 1.10 | 0.3210 | 0.1238 | High |
| E. loxophleba | Default | 4,851 | 3,530 | 0–1 | 10 | 62 | 0.047 | 1.80 | 0.2456 | 0.0663 | Low |
| E. loxophleba | BS-recommended PO + species-specific F IS (Byrne 2008) | 4,851 | 3,530 | 0.01–0.08 c | 100 | 50 | 0.047 | 1.40 | 0.2159 | 0.0443 | Medium |
| E. loxophleba | Relatively high BS-recommended + species-specific F IS (Byrne 2008) | 4,851 | 3,530 | 0.01–0.08 c | 200 | 45 | 0.044 | 1.28 | 0.2236 | 0.0444 | Medium |
| E. loxophleba | High PO (Lotterhos and Whitlock 2014) + species-specific FIS | 4,851 | 3,530 | 0.01–0.08 c | 3,531 | 50 | 0.050 | 1.40 | 0.2160 | 0.0440 | Medium–high |
| E. loxophleba | BS-recommended PO + generic eucalypt F IS (Byrne 2008) | 4,851 | 3,530 | 0.01–0.3 | 100 | 49 | 0.047 | 1.39 | 0.2286 | 0.0480 | Medium–high |
| E. loxophleba | Relatively high BS-recommended + species-specific F IS (Byrne 2008) | 4,851 | 3,530 | 0.01–0.3 | 200 | 44 | 0.043 | 1.25 | 0.2365 | 0.0482 | Medium–high |
| E. loxophleba | High PO (Lotterhos and Whitlock 2014) + generic eucalypt F IS (Byrne 2008) | 4,851 | 3,530 | 0.01–0.3 | 3,531 | 32 | 0.034 | 0.90 | 0.2616 | 0.0487 | High |
| E. salubris lineage 1 | Default | 16,122 | 14,949 | 0–1 | 10 | 43 | 0.048 | 0.20 | 0.2141 | 0.0517 | Low |
| E. salubris lineage 1 | Low PO + generic eucalypt F IS (Byrne 2008) | 16,122 | 14,949 | 0.01–0.3 b | 100 | 22 | 0.046 | 0.15 | 0.1933 | 0.0341 | Medium |
| E. salubris lineage 1 | BS-recommended PO + generic eucalypt FIS ( Byrne 2008 ) | 16,122 | 14,949 | 0.01–0.3 b | 200 | 18 | 0.043 | 0.12 | 0.2164 | 0.0371 | Medium |
| E. salubris lineage 1 | High PO (Lotterhos and Whitlock 2014) + generic eucalypt F IS (Byrne 2008) | 16,122 | 14,949 | 0.01–0.3 b | 14,950 | 7 | 0.007 | 0.04 | 0.2730 | 0.0370 | High |
| E. salubris lineage 2 | Default | 16,122 | 8,089 | 0–1 | 10 | 33 | 0.049 | 0.40 | 0.3196 | 0.0874 | Low |
| E. salubris lineage 2 | BS-recommended PO + generic eucalypt FIS ( Byrne 2008 ) | 16,122 | 8,089 | 0.01–0.3 b | 100 | 17 | 0.047 | 0.20 | 0.3284 | 0.0672 | Medium |
| E. salubris lineage 2 | Relatively high BS-recommended PO + generic eucalypt F IS (Byrne 2008) | 16,122 | 8,089 | 0.01–0.3 b | 200 | 13 | 0.0517 | 0.16 | 0.3507 | 0.0673 | Medium–high |
| E. salubris lineage 2 | High PO (Lotterhos and Whitlock 2014) + generic eucalypt F IS (Byrne 2008) | 16,122 | 8,089 | 0.01–0.3 b | 8,090 | 8 | 0.015 | 0.10 | 0.3997 | 0.0677 | High |
Note.—Bold rows represent the sets of outlier markers that were used in the mapping and colocation analyses.
The Q value is the false discovery rate (FDR) analog of a P-value; it is the minimum FDR at which a locus may become significant. A Q value of 0.05 means that 5% of outliers (i.e., those having a Q value ≤0.05) are expected to be falsely positive. A 5% threshold for Q values is much more stringent than a 5% threshold for P values in classical statistics (Foll 2012).
No species-specific information available—F IS prior based on estimated average value for eucalypts (Byrne 2008).
F IS for E. loxophleba ca. 0.046 (Byrne 2008).
For each marker in both outlier and “nonoutlier” data sets from E. tricarpa, E. loxophleba and E. salubris lineage 1, linear regression was used to identify significant associations between population-level allele frequencies and climate and soil variables. Regressions were not done for E. salubris lineage 2 because it comprised only four populations, which was considered too few for meaningful statistical correlations. Only outlier markers that were also correlated with at least one environmental variable (climate and/or soil) were included in further analyses (all outliers detected for E. salubris lineage 2 were included).
To quantify the rate of false outlier identification within each species’ data set, we conducted a randomization analysis with 100 replicates. For each species, R (R Core Team 2014) was used to randomize individuals across populations and a new BAYESCAN file was created with the same population sizes as the original data set. Each new BAYESCAN file was analyzed using the parameters of the medium-stringency analysis shown in bold in table 2. The number of outliers detected in each bootstrap replicate (allowing for an FDR of 0.05) was recorded.
Context of DArTseq Markers in the E. grandis Genome
BLAST searches were used to compare all DArTseq marker sequences to the complete genome sequence of Eucalyptus grandis (v. 1.1) (http://www.phytozome.net/; last accessed January 10, 2017; Goodstein et al. 2012) and, where possible, locations on the 11 main eucalypt chromosomes were determined. When more than one alignment was obtained, the alignment that had the lowest “expect value” (E) (i.e., the number of DNA sequence matches one could expect to see by chance) was used to assign a position to the marker.
Data for all markers (from all species/lineages) that could be linked to the eleven chromosomes of E. grandis were combined in a text file. Data that related to outlying markers from each species were copied into a second text file. All E. grandis gene positions and their associated gene ontology (GO) terms were downloaded from the Phytozome database, using the built-in BioMart application, and saved as a third text file. A Perl script was used to connect the three text documents (marker data vs. E. grandis gene data), thereby providing for each mapped marker a “context”, that is, whether it was contained “In” a gene or “Near” a gene (i.e., within 5,000 bp upstream or downstream of, but not In, the gene; Bierne et al. 2011). Overall gene length, rather than transcript length, was used so that In a gene also included 3′ and 5′ untranslated regions. The GO information for the designated gene was tied to the marker. All results were entered into a MySQL database which permitted sorting on the basis of: (i) species from which the marker originally came, (ii) context (In or Near gene) and (iii) whether the marker was in the outlier set.
Enrichment Analysis
Blast2Go (Conesa et al. 2005) was used to test whether any GO terms were more common among the Outliers than would be expected from a random sample of genes from the “All Markers” reference data set. The GO terms for “All Markers” were loaded into Blast2Go. The test set of Outliers was compared with the “All markers” reference data set. The search was narrowed using search terms relating to the environmental gradients in the study: “Water”, “Osmotic stress”, “Temperature”, “Heat”, “Radiation”, “Phosphorus” and “Nitrogen”. The Blast2Go enrichment analysis used Fisher’s exact test in a two-sided analysis (i.e., looking for over-represented and under-represented GO terms); the P-value Filter was set to 0.05. Analyses were conducted with and without correcting for a FDR of 5%.
Identification of Outliers Common to Two or More Species
The complete catalogue of outliers from all species, their positions on the E. grandis genome and associated GO terms (supplementary material S1, Supplementary Material online) was searched for colocations, using as criteria: (i) exact start position, (ii) In the same gene or (iii) Near (i.e., within 5,000 bp upstream or downstream of, but not In) the same gene.
The probability of discovering outlier markers that colocated to the same region on the E. grandis genome in two species was calculated in R (R Core Team 2014), using the scaffold number and either the start position or the gene name (see below) as the “location” of each marker. Each mapped marker was tagged as either “neutral” or outlier, and the total set of colocating markers in a species pair was determined using the “merge” command in R. This procedure excluded markers that did not occur in both species, and produced a data set comprising (i) the marker position, (ii) whether the marker was neutral or an outlier in species A and (iii) whether the marker was neutral or an outlier in species B. The number of colocations was recorded. One hundred thousand randomizations of the neutral/outlier status of the markers in species B were carried out and the number of times (N) a marker was deemed to be an outlier in both species was counted. This provided an estimate (N × 10− 5) of the probability that outliers (i) with the same start position, (ii) falling within the same gene and (iii) falling within 5,000 bp of the same gene (but not In the gene), would be found in two species by chance alone.
Results and Discussion
The number of DArTseq markers for each species varied considerably [from 4,851 in E. loxophleba to 16,122 for both lineages (combined) of E. salubris], and seemed to correspond to the quality and/or quantity of the starting DNA; extractions conducted “in house” using a protocol optimized for Eucalyptus yielded more markers than extractions that had been done by a commercial provider using a generic DNA extraction procedure. Although all samples of DNA were fully digested with restriction enzymes and all DNA samples of each species were standardized to a uniform (within each species) concentration between 30 and 100 ng µl− 1, it appeared that higher concentrations of starting DNA yielded more DArTseq markers.
The proportion of outlier loci differentiating populations within E. tricarpa and E. loxophleba were 2.6% and 1.4%, respectively. The number of outliers detected within each lineage of E. salubris represented only 0.12% and 0.2% of the BAYESCAN input. The lower proportion of outlier loci in each E. salubris lineage is likely due to the small sample size within each lineage (outlier analysis of all nine populations of E. salubris yielded 438 outlier loci, approximately 5% of the Bayescan input file; data not shown). Up to 53% of all markers—and a similar proportion of outliers—could be located on the E. grandis reference genome version 1.1, and just over a third of these were associated with (i.e., in or within 5,000 bp of) genes (table 3). In E. tricarpa and E. loxophleba there were more mappable markers in the Near gene than In gene category. This may be a result of surveying regions on either side of genes that were bigger than the genes themselves (e.g., we surveyed a total of 10,000 bp outside genes, while genes average 3,500 bp in length [in E. grandis; Myburg et al. 2014)]. The E. grandis reference genome is relatively new (Myburg et al. 2014) and little study of the synteny, collinearity and gene sequence homology with genomes of other eucalypt species has yet been done. However, high synteny and colinearity has been found between E. grandis (section Latoangulatae) and E. globulus (section Maidenaria, which is closely related to section Latoangulatae) (Hudson et al. 2012). The three species in the present study come from two more-distantly related sections of subgenus Symphyomyrtus, sections Adnataria and Bisectae (Steane et al. 2002), and the extent of synteny and colinearity with E. grandis is unknown.
Table 3.
Numbers and Proportions of DArTseq Markers in the Three Eucalypt Species that Were Mappable to the 11 Main Chromosomes of the Eucalyptus grandis Reference Genome, In or Near (i.e., within 5,000 bp of but not Inside) a Coding Region
| E. tricarpa | E. loxophleba ssp. lissophloia | E. salubris Lineage 1 (Sal1) | E. salubris Lineage 2 (Sal2) | |
|---|---|---|---|---|
| No. Pops | 9 | 9 | 5 | 4 |
| Total No. markers | 6,544 | 4,851 | 15,147 | 14,428 |
| No. outlier markers | 94 | 50 | 19 | 17 |
| Percentage of Bayescan input that were outliers (FDR = 0.05) | 2.60% | 1.40% | 0.12% | 0.20% |
| Total No. mappable markers (%) | 3,489 (53%) | 1,644 (34%) | 6,962 (46%) | 7,417 (51%) a |
| No. mappable outlier markers (%) | 48 (51%) | 15 (30%) | 9 (50%) | 7 (41%) |
| No. mappable markers In genes (%) | 1,319 (38%) | 557 (34%) | 2,152 (31%) | 2,055 (28%) |
| No. mappable outliers In genes (%) | 17 (35%) | 5 (33%) | 4 (44%) | 4 (57%) |
| No. mappable markers Near genes (%) | 1,352 (39%) | 619 (38%) | 2,787 (40%) | 2,645 (36%) |
| No. mappable outliers Near genes (%) | 21 (44%) | 6 (40%) | 5 (44%) | 2 (29%) |
Plus four markers that mapped to the chloroplast genome.
Outlier detection methods for dominant markers are few. BAYESCAN is widely thought to be reasonably conservative in the detection of outliers, even though simulation studies by Lotterhos and Whitlock (2014) indicated some problems. Three of the four randomization tests (where individuals of each species/lineage were shuffled among populations to create 100 new data sets, each of which was analyzed using BAYESCAN) did not find any outlier markers; one of them (E. salubris lineage 2) identified one outlier marker in one of the 100 BAYESCAN runs when the FDR was set to 0.05 (but this became nonsignificant when FDR was set to 0.01). These results indicated that it was unlikely that outliers would be detected by chance alone.
BAYESCAN sensitivity tests in each species indicated that using the high stringency prior odds recommended by Lotterhos and Whitlock (2014) resulted in a reduction of approximately 36–61% in the number of outliers compared with the model settings suggested by the BAYESCAN manual (i.e., medium and medium–high stringency in table 2), and a reduction of 48–84% relative to the default settings of the program. In order to avoid the risk of a high rate of false nondiscovery of outliers, we used the outliers detected using the medium settings (suggested by the BAYESCAN manual) and, to increase stringency, markers that were not correlated with at least one environmental variable (supplementary material S2, Supplementary Material online) were discarded from further analyses (see below). In E. loxophleba, where we had specific prior information for F IS values, it appeared that increasing the value/range of the F IS prior setting (to a more generic range) reduced the number of outliers detected by BAYESCAN (table 2) when using the very high prior odds recommended by Lotterhos and Whitlock (2014), but did not make a marked difference when using the moderate prior odds recommended by the BAYESCAN manual. In general, higher priors for F IS and the neutral model resulted in fewer outliers.
As previously detailed (Steane, et al. 2014, 2017), population-level allele frequencies of outlier and neutral markers were regressed against site-specific climate and/or soil data and/or population-level functional traits (fig. 1; supplementary material S2, Supplementary Material online). The majority of outlier loci were significantly associated (P < 0.05) with at least one climate or soil variable (all 94 outlier markers in E. tricarpa; 40/50 outliers in E. loxophleba; and 16/18 in E. salubris lineage 1; correlations were not done for E. salubris lineage 2 because we felt the correlations derived from only four populations would not be statistically reliable). Those outliers that were not correlated with any environmental variables were removed from the set of outliers.
Fig. 1.—
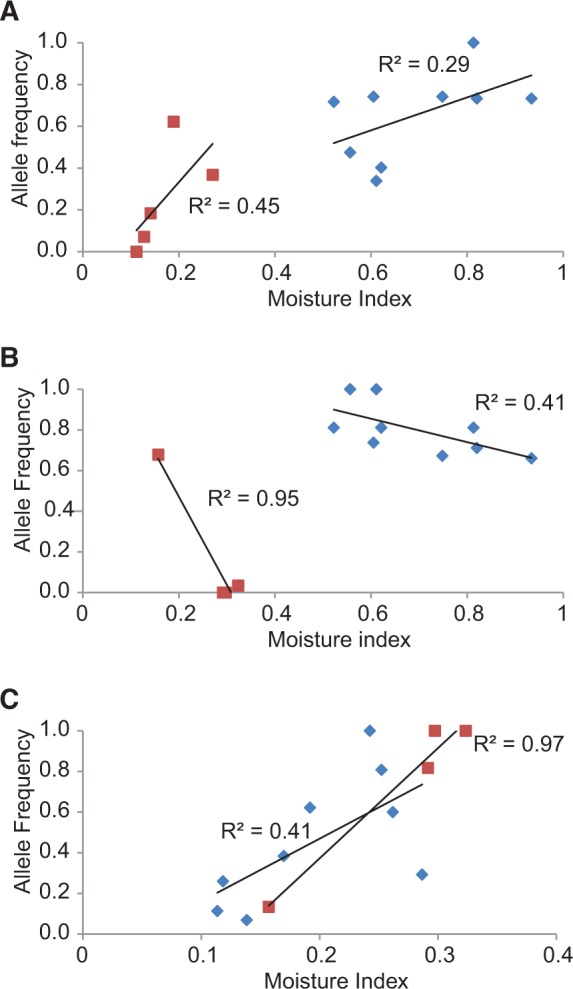
Linear regression of population-level allele frequencies of outlier markers that were common to two of the three studied Eucalyptus species plotted against a moisture index (“Aridity Index” from the Atlas of Living Australia = precipitation/pan evaporation). The markers were also correlated with other environmental variables (supplementary material S2, Supplementary Material online). (A) TriDArTseq 1174 (E. tricarpa, blue diamond) and SalDArTseq 9159 (E. salubris lineage 1, brown square) were both In Eucgr.A02872 (Chromosome 1), K13458—a disease resistance gene; (B) TriDArTseq 1079 (E. tricarpa, blue diamond) and SalDArTseq 3803 (E. salubris lineage 2, brown square) were both In Eucgr.F00208 (Chromosome 6), a proprotein convertase subtilisin/kexin gene; (C) LoxDArTseq 1012 (E. loxophleba, blue diamond) and SalDArTseq 11926 (E. salubris lineage 2, brown square) were both Near Eucgr.G00352 (Chromosome 7), an ATP phosphoribosyl-transferase gene. See text for definitions of In versus Near genes.
In previous studies, significantly higher proportions of such marker-environment and marker-trait correlations were found for outlier markers compared with neutral markers in E. loxophleba (Steane et al. 2017) and E. tricarpa (Steane et al. 2014), supporting the hypothesis that the population-level differences in allele frequencies of outlier markers were due to selective influences at the site of origin, rather than merely stochastic processes. Such comparisons of neutral versus outlier marker associations with climatic variables and functional traits were not made for the two E. salubris lineages because of the small number of populations within each.
Are the Same Biochemical/Physiological Pathways Involved in Adaptation?
There was no obvious enrichment of GO terms in the outlier marker data set relative to the neutral marker data set (e.g., approximately 5.5% of markers in both data sets were involved in trans-membrane transport; supplementary material S1, Supplementary Material online). Tests for enrichment using Blast2GO and Revigo confirmed this. The Blast2GO analysis with no FDR correction yielded 25 enrichment results (data not shown), the majority of which involved three markers that were common to two of our species (see below). However, implementation of an FDR correction of 0.05 indicated that the level of enrichment was not significant. The lack of enrichment may be due to the lack of power arising from the small number of outlier loci included in the analyses; alternatively, it may mean that the selective forces acting on populations of the three species were not acting on the same cellular processes, so that no strong signal of selection was detected for any particular GO term. Linkage disequilibrium could be a confounding factor in the GO analysis. In this study, only genes within 5 kb of a putatively adaptive marker were considered, as this appears to be the average size of linkage blocks in Eucalyptus (see below). However, if the region of the genome in LD were greater than 5 kb, it could be another factor reducing the power of the GO analysis.
Are the Same Genes Involved in Adaptation?
There were three instances where outlier markers from two species colocated on the E. grandis genome (table 4), each of which was unlikely to have occurred by chance (P ranged from 0.013 to 0.062; table 5). Population-level allele frequencies of each marker were correlated with at least one climatic variable, including moisture availability (fig. 1). Of course, the fact that the three species in the study all belong to subgenus Symphyomyrtus [E. loxophleba and E. salubris belong to different series within section Bisectae sensu Brooker (2000)] may increase the likelihood of the same genes/markers being involved in convergent phenotypes if an ancestral genotype or “precursor” (Marazzi et al. 2012) persisted from the most recent common ancestor. The marker-linked genes that were common to two species included an ATP phosphoribosyltransferase (involved in the biosynthesis of the amino acid, histidine), a subtilisin-like protease (cleaves proteins where there is a serine amino acid) and a disease resistance protein. A subtilisin-like protease has been reported to be associated with climate adaptation in pines (Nadeau 2014) and such proteases have been reported widely to be influenced by drought (Vaseva et al. 2012 and references therein). The suggestion that selection may be acting on disease resistance proteins is not surprising, because the risk of disease varies with climate and there is coadaptation between forest trees and disease. For example, in E. globulus, the level of genetic resistance to Teratosphaeria spp. (syn. Mycosphaerella) leaf disease is correlated to the predicted risk of the disease at the site of origin, with the risk greater in areas of higher temperature and, to a lesser degree, increased humidity (Hamilton et al. 2013).
Table 4.
Gene Ontogeny (GO) Terms Associated with Outlier DArTseq Markers from Three Eucalyptus Species that Colocate on the Eucalyptus grandis Reference Genome (Ver2)
| Marker 1 | Marker 2 | Context | E. grandis Gene | Description | Linkage Group | Gene Start Position | Gene Stop Position | GO Term ID | Description |
|---|---|---|---|---|---|---|---|---|---|
| LoxDArTseq1012 | SalDArTseq 11926 (Sal2) | Near | Eucgr.G00352 | ATP phosphoribosyl-transferase | 7 | 5,749,137 | 5,754,442 | 105 | Histidine biosynthesis process |
| 287 | Magnesium ion binding | ||||||||
| 3,879 | ATP phophoribosyl transferase activity | ||||||||
| 5,737 | Cytoplasm | ||||||||
| TriDArTseq1079 | SalDArTseq 3803 (Sal2) | In | Eucgr.F00208 | Proprotein convertase subtilisin/kexin | 6 | 2,435,562 | 2,438,350 | 4,252 | Serine type endopeptidase activity |
| 42,802 | Identical protein binding | ||||||||
| 43,086 | Negative regulation of catalytic activity | ||||||||
| 6,508 | Proteolysis | ||||||||
| TriDArTseq 1174 | SalDArTseq 9159 (Sal1) | In | Eucgr.A02872 | K13458—disease resistance protein | 1 | 43,795,662 | 43,798,891 | NA |
Note.—“Context” refers to whether markers are located within the same gene (In) or are located within 5,000 bp of (i.e., Near but not In) the same gene.
Table 5.
Number of Markers that Could be Mapped to Eucalyptus grandis Chromosomes 1–11, and the Probability (P) of Finding (by Chance Alone) Outlier Markers Common to Two of the Three Eucalypt Species in this Study that: (i) Share the Same Start Position in the E. grandis Reference Genome; (ii) Are Located In the Same Gene; or (iii) Are Located Within 5,000 bp of (i.e., Near but not In) the Same Gene
| Species 1 (Total No. Mappable Markers) | Species 2 (Total No. Mappable Markers) | No. Mappable Markers/Outliers in Common that have Exactly the Same Start Position (P) | No. Mappable Markers/Outliers in Common In Gene (P) | No. Mappable Markers/Outliers in Common Near Gene (P) |
|---|---|---|---|---|
| E. loxophleba ssp. lissophloia (1,644) | E. tricarpa (3,489) | 238/0 (0.008) | 133/0 (0) | 81/0 (0.050) |
| E. loxophleba ssp. lissophloia (1,644) | E. salubris lineage 1 (6,962) | 460/0 (0.007) | 196/0 (0) | 113/0 (0.036) |
| E. loxophleba ssp. lissophloia (1,644) | E. salubris lineage 2 (7,417) | 472/1 (0.013) a | 194/0 (0.005) | 112/1 a (0.035) |
| E. tricarpa (3,489) | E. salubris lineage 1 (6,962) | 662/0 (0.003) | 379/1 (0.062) a | 726/0 (0.017) |
| E. tricarpa (3,489) | E. salubris lineage 2 (7,417) | 681/0 (0) | 369/1 (0.037) a | 709/0 (0) |
| E. salubris lineage 1 (6,962) | E. salubris lineage 2 (7,417) | 6,962/0 (0.006) | 1,653/0 (0.008) | 2,937/0 (0.003) |
The colocations detailed in table 2. See main text for details regarding the calculation of P.
Is Adaptation a Genome Wide Phenomenon?
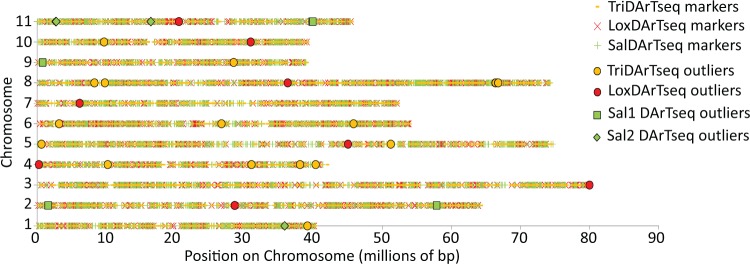
The outlier DArTseq markers were spread across the genome (fig. 2) and there were outliers from at least two (usually three) species on each chromosome. There was little evidence of clustering of the putatively adaptive outlier markers except for the six E. tricarpa markers within a 3 Mb stretch at the end of chromosome 8 as identified previously by Steane et al. (2014). However, the overall density of DArTseq markers across each genome may not have been sufficient to detect actual genetic linkage (cf. a general broadscale “clustering”) of outlier loci. For example, a conservative estimate of DArTseq marker density in E. tricarpa would be one marker every 98 kb (based on 6,544 markers over a genome of 640 Mb); comparable estimates for E. loxophleba and E. salubris are one marker per 133 kb and one marker per 40 kb, respectively. While early studies suggested that linkage disequilibrium in Eucalyptus decayed rapidly relative to other tree species, more recent research (Silva-Junior and Grattapaglia 2015) has indicated that the linkage blocks are, on average, larger than previously thought. For example, at the genome-wide level, linkage disequilibrium decays within ca. 4–6 kb, but there is a lot of variation in rate across the genome (Silva-Junior and Grattapaglia 2015; Gion et al. 2016), ranging from absence to complete linkage disequilibrium up to 50 kb.
Fig. 2.—
Approximate positions of DArTseq markers from Eucalyptus tricarpa, E. loxophleba and E. salubris on the 11 main chromosomes of the Eucalyptus grandis reference genome. Line symbols represent all markers in the study. Filled shapes represent markers that were identified as outliers in each species and were correlated with at least one environmental variable.
The results of this survey add to the growing body of evidence supporting the notion that phenotypic adaptation to the environment (in particular, aridity) is not controlled by a restricted set of key genes or key mutations, but is more likely to involve a wide range of genes that do not necessarily affect common developmental or metabolic pathways (Pritchard and Di Rienzo 2010; Neale and Kremer 2011; Prunier et al. 2011; Berg and Coop 2014; Hudson et al. 2015) that lead to convergent adaptive phenotypes. Genetic differences may include point-mutations or insertions/deletions in genes or regulatory regions. Epigenetic changes such as methylation may affect acclimation via changes in gene expression (e.g., Ahuja et al. 2010; Nicotra et al. 2015; Shaar-Moshe et al. 2015) and, if such changes are heritable, they may facilitate adaptation of subsequent generations to environmental change (Brautigam et al. 2013; Burton and Metcalfe 2014; Kinoshita and Seki 2014; Meyer 2015).
The idea of genomic “hot spots” of adaptation—where adaptive genes cluster into a relatively small region of the genome—may apply to individual traits, such as wing patterning in butterflies (Supple et al. 2013), drought tolerance in chick peas (Varshney et al. 2014) or disease resistance in sorghum (Wang et al. 2014), but frequently there are numerous such hot-spots distributed across a genome. For example, disease resistance may involve few genes of large effect or multiple genes of small effect that cluster in particular regions of the genome across a number of chromosomes (Wang et al. 2001, 2014; Chu et al. 2004). Such clustering of genes could result from tandem duplication events, as seen in Eucalyptus (Myburg et al. 2014). Duplicated regions often include stress-response genes (Hanada et al. 2008), suggesting that tandem duplication may be important for adaptation in dynamically changing environments (e.g., through dosage effects or through redundant paralogs developing new functions; Lynch and Conery 2000; Flagel and Wendel 2009; Kondrashov 2012). Genetic hitchhiking (Barton 2000) is another potential explanation for apparent hot spots of adaptation (e.g., the cluster of markers at the end of chromosome 8 of E. tricarpa; Steane et al. 2014), where population-level allele frequencies of an outlier marker changes because it is near (i.e., in linkage disequilibrium with) another marker that is linked to a gene that is under selection. For example, Kubota et al. (2015) found around 500 genomic islands, across the genome, postulated to be involved in adaptation to altitude in populations of Arabidoposis hallii, growing on the slopes of two mountains in Japan.
Other analyses of genomic architecture of adaptation have also found genome-wide signatures of selection (e.g., Loblolly pine (Eckert et al. 2010), Arabidopsis (Lee and Mitchell-Olds 2012; Kubota et al. 2015) and various animals (Hohenlohe et al. 2010; Deagle et al. 2011; Fan et al. 2014; Pujolar et al. 2014; and see Nosil et al. 2009 and references therein)). Conifers, in particular, are providing many examples of genome-wide adaptation, where convergent adaptations do not come from parallel mutations. In a comparative study of environmental adaptation in two pine species (Pinus monticola and P. strobus), Nadeau (2014) found six genes in common that were correlated with similar environmental variables. However, in that study, the vast majority of genes postulated to be under selection were not common to both species, and the number of genes in common did not differ from random expectation. Similar to our findings, Nadeau (2014) concluded that although a small number of common outliers were detected, generally the two species were adapted to climate via different suites of genes.
Other conifer studies have yielded similar results (Grivet et al. 2011; Prunier et al. 2011; Mosca et al. 2012). Mosca et al. (2012), comparing climate adaptation in four alpine conifer species, detected seven climate-associated genes that were shared between two or more species, although in most cases the mutations were not homologous. There are many examples of such “gene reuse” in adaptation (Kubota et al. 2015, and see Nouhaud et al. 2014). Some genes might contribute to adaptation more often than others because they have more standing allelic variation, higher mutation rates, larger effect sizes, more numerous beneficial mutations, fewer pleiotropic constraints, particular linkage relationships, or because they are involved in vital epistatic interactions (Conte, et al. 2012). Occasionally the mutations might be the same; for example, among hummingbird species, two epistatic substitutions in the βA globin gene, related to altitude adaptation, have occurred independently at least 17 times (Projecto-Garcia et al. 2013). More often, however, specific mutations within a gene may be similar, but not identical (Conte et al. 2012; Renaut et al. 2014; Hodgins et al. 2015) despite phenotypic convergence.
Conclusion
Our research adds to a growing body of evidence suggesting that within-species population-level adaptation to contrasting environments is a genome-wide phenomenon involving a range of mutations in multiple and diverse genes, gene families and regulatory regions that affect a multitude of complex genetic and biochemical processes. Screening candidate genes (e.g., those known to be involved in stomatal conductance) for adaptive mutations may be informative, but such targeted studies are likely to overlook other equally important mutations elsewhere in the genome. Genomic scans have the potential to flag situations in which adaptation has occurred, and this information may be applied to environmental management (e.g., seed transfer guidelines; Prober et al. 2015) or to launch more detailed genetic studies into the functions of particular adaptive genes. In this context, a key challenge for geneticists and physiologists is to link variants of candidate genes, gene families or regulatory regions with functional adaptation.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgment
We thank Michael Charleston (University of Tasmania) for providing computing resources to allow completion of multiple BAYESCAN runs on shuffled data. This work was funded by a grant from the Australian National Climate Change Adaptation Research Facility (TB11 03) with additional support through the Great Western Woodlands Supersite of Australia’s Terrestrial Ecosystem Research Network and ARC Discovery Grant DP130104220.
Literature Cited
- Ahuja I, de Vos RCH, Bones AM, Hall RD. 2010. Plant molecular stress responses face climate change. Trends Plant Sci. 15:664–674. [DOI] [PubMed] [Google Scholar]
- Alberto FJ, et al. 2013. Potential for evolutionary responses to climate change: evidence from tree populations. Global Change Biol. 19:1645–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton NH. 2000. Genetic hitchhiking. Philos Trans R Soc Lond Ser B Biol Sci. 355:1553–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Yekateuli D. 2001. The control of the false discovery rate in multiple testing under dependency. Ann Stat. 29:1165–1188. [Google Scholar]
- Berg JJ, Coop G. 2014. A population genetic signal of polygenic adaptation. PLoS Genet. 10:e1004412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierne N, Welch J, Loire E, Bonhomme F, David P. 2011. The coupling hypothesis: why genome scans may fail to map local adaptation genes. Mol Ecol. 20:2044–2072. [DOI] [PubMed] [Google Scholar]
- Bigham A, et al. 2010. Identifying signatures of natural selection in Tibetan and Andean populations using dense genome scan data. PLoS Genet. 6:e1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brautigam K, et al. 2013. Epigenetic regulation of adaptive responses of forest tree species to the environment. Ecol Evol. 3:399–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooker MIH. 2000. A new classification of the genus Eucalyptus L'Her. (Myrtaceae). Aust Syst Bot. 13:79–148. [Google Scholar]
- Burton T, Metcalfe NB. 2014. Can environmental conditions experienced in early life influence future generations? Proc R Soc B Biol Sci. 281:20140311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne M. 2008. Phylogeny, diversity and evolution of eucalypts In: Sharma A, Sharma A, editors. Plant genome biodiversity and evolution. Part E. Phanerogams - angiosperms. Berlin: Science Publishers; p. 303–346. [Google Scholar]
- Byrne M, Hines B. 2004. Phylogeographical analysis of cpDNA variation in Eucalyptus loxophleba (Myrtaceae). Aust J Bot. 52:459–470. [Google Scholar]
- Chu Z, Ouyang Y, Zhang J, Yang H, Wang S. 2004. Genome-wide analysis of defense-responsive genes in bacterial blight resistance of rice mediated by the recessive R gene xa13. Mol Genet Genomics. 271:111–120. [DOI] [PubMed] [Google Scholar]
- Conesa A, et al. 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676. [DOI] [PubMed] [Google Scholar]
- Conte GL, Arnegard ME, Peichel CL, Schluter D. 2012. The probability of genetic parallelism and convergence in natural populations. Proc R Soc B Biol Sci. 279:5039–5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Mita S, et al. 2013. Detecting selection along environmental gradients: analysis of eight methods and their effectiveness for outbreeding and selfing populations. Mol Ecol. 22:1383–1399. [DOI] [PubMed] [Google Scholar]
- Deagle BE, et al. 2011. Population genomics of parallel phenotypic evolution in stickleback across stream-lake ecological transitions. Proc R Soc B Biol Sci. 279(2012):1277–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon S, et al. 2014. Characterisation of adaptive genetic diversity in environmentally contrasted populations of Eucalyptus camaldulensis Dehnh. (River Red Gum). PLoS ONE 9:e103515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert AJ, et al. 2013. Multilocus analyses reveal little evidence for lineage-wide adaptive evolution within major clades of soft pines (Pinus subgenus Strobus). Mol Ecol. 22:5635–5650. [DOI] [PubMed] [Google Scholar]
- Eckert AJ, et al. 2010. Patterns of population structure and environmental associations to aridity across the range of loblolly pine (Pinus taeda L., Pinaceae). Genetics 185:969–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan HZ, et al. 2014. Genome-wide detection of selective signatures in Simmental cattle . J Appl Genet. 55:343–351. [DOI] [PubMed] [Google Scholar]
- Flagel LE, Wendel JF. 2009. Gene duplication and evolutionary novelty in plants. N Phytol. 183:557–564. [DOI] [PubMed] [Google Scholar]
- Foll M. 2012. Bayescan v2.1 User Manual. http://cmpg.unibe.ch/software/BayeScan/files/BayeScan2.0_manual.pdf.
- Foll M, Gaggiotti O. 2008. A genome scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics 180:977–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk WC, McKay JK, Hohenlohe PA, Allendorf FW. 2012. Harnessing genomics for delineating conservation units. Trends Ecol Evol. 27:489–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauli A, Vaillancourt RE, Bailey TG, Steane DA, Potts BM. 2015. Evidence for climate adaptation in early-life traits of Tasmanian populations of Eucalyptus pauciflora . Tree Genet Genom. 11:104. [Google Scholar]
- Gion J-M, et al. 2016. Genome-wide variation in recombination rate in Eucalyptus . BMC Genom. 17:590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstein DM, et al. 2012. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40:D1178–D1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivet D, et al. 2011. Molecular footprints of local adaptation in two Mediterranean conifers. Mol Biol Evol. 28:101–116. [DOI] [PubMed] [Google Scholar]
- Hamilton MG, et al. 2013. A latitudinal cline in disease resistance of a host tree. Heredity 110:372–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K, Zou C, Lehti-Shiu MD, Shinozaki K, Shiu S-H. 2008. Importance of lineage-specific expansion of plant tandem duplicates in the adaptive response to environmental stimuli. Plant Physiol. 148:993–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgins KA, et al. 2015. Comparative genomics in the Asteraceae reveals little evidence for parallel evolutionary change in invasive taxa. Mol Ecol. 24:2226–2240. [DOI] [PubMed] [Google Scholar]
- Hohenlohe PA, et al. 2010. Population genomics of parallel adaptation in Threespine Stickleback using sequenced RAD tags. PLoS Genet. 6:e1000862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson CJ, Freeman JS, Myburg AA, Potts BM, Vaillancourt RE. 2015. Genomic patterns of species diversity and divergence in Eucalyptus . New Phytol. 206:1378–1390. [DOI] [PubMed] [Google Scholar]
- Hudson CJ, et al. 2012. High synteny and colinearity among Eucalyptus genomes revealed by high-density comparative genetic mapping. Tree Genet Genomes 8:339–352. [Google Scholar]
- Kinoshita T, Seki M. 2014. Epigenetic memory for stress response and adaptation in plants. Plant Cell Physiol. 55:1859–1863. [DOI] [PubMed] [Google Scholar]
- Kondrashov FA. 2012. Gene duplication as a mechanism of genomic adaptation to a changing environment. Proc R Soc B Biol Sci. 279:5048–5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota S, et al. 2015. A genome scan for genes underlying microgeographic-scale local adaptation in a wild Arabidopsis species. PLOS Genet. 11:e1005361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CR, Mitchell-Olds T. 2012. Environmental adaptation contributes to gene polymorphism across the Arabidopsis thaliana genome. Mol Biol Evol. 29:3721–3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotterhos KE, Whitlock MC. 2014. Evaluation of demographic history and neutral parameterization on the performance of FST outlier tests. Mol Ecol. 23:2178–2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Conery JS. 2000. The evolutionary fate and consequences of duplicate genes. Science 290:1151–1155. [DOI] [PubMed] [Google Scholar]
- Manel S, Conord C, Despres L. 2009. Genome scan to assess the respective role of host-plant and environmental constraints on the adaptation of a widespread insect. BMC Evol Biol. 9:288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marazzi B, et al. 2012. Locating evolutionary precursors on a phylogenetic tree. Evolution 66:3918–3930. [DOI] [PubMed] [Google Scholar]
- Martin A, Orgogozo V. 2013. The loci of repeated evolution: a catalog of genetic hotspots of phenotypic variation. Evolution 67:1235–1250. [DOI] [PubMed] [Google Scholar]
- Meyer P. 2015. Epigenetic variation and environmental change. J Exp Bot. 66:3541–3548. [DOI] [PubMed] [Google Scholar]
- Mosca E, et al. 2012. The geographical and environmental determinants of genetic diversity for four alpine conifers of the European Alps. Mol Ecol. 21:5530–5545. [DOI] [PubMed] [Google Scholar]
- Myburg AA, et al. 2014. The genome of Eucalyptus grandis . Nature 510:356–362. [DOI] [PubMed] [Google Scholar]
- Nadeau S. 2014. Genetic population structure and adaptation to climate across the range of eastern white pine (Pinus strobus L.) and western white pine (Pinus monticola Douglas ex. D. Don) [MSc Thesis]. [Vancouver]: The University of British Columbia.
- Narum SR, Hess JE. 2011. Comparison of FST outlier tests for SNP loci under selection. Mol Ecol Resour. 11:184–194. [DOI] [PubMed] [Google Scholar]
- Neale DB, Kremer A. 2011. Forest tree genomics: growing resources and applications. Nat Rev Genet. 12:111–122. [DOI] [PubMed] [Google Scholar]
- Nicotra AB, et al. 2015. Adaptive plasticity and epigenetic variation in response to warming in an alpine plant. Ecol Evol. 5:634–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosil P, Funk DJ, Ortiz-Barrientos D. 2009. Divergent selection and heterogeneous genomic divergence. Mol Ecol. 18:375–402. [DOI] [PubMed] [Google Scholar]
- Nouhaud P, et al. 2014. Genomic regions repeatedly involved in divergence among plant-specialized pea aphid biotypes. J Evol Biol. 27:2013–2020. [DOI] [PubMed] [Google Scholar]
- Pascoal S, et al. 2014. Rapid convergent evolution in wild crickets. Curr Biol. 24:1369–1374. [DOI] [PubMed] [Google Scholar]
- Perez-Figueroa A, Garcia-Pereira MJ, Saura M, Rolan-Alvarez E, Caballero A. 2010. Comparing three different methods to detect selective loci using dominant markers. J Evol Biol. 23:2267–2276. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Di Rienzo A. 2010. Adaptation - not by sweeps alone. Nat Rev Genet. 11:665–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prober SM, et al. 2015. Climate-adjusted provenancing: a strategy for climate-resilient ecological restoration. Front Ecol Evol. 3:65. [Google Scholar]
- Projecto-Garcia J, et al. 2013. Repeated elevational transitions in hemoglobin function during the evolution of Andean hummingbirds. Proc Natl Acad Sci U S A. 110:20669–20674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunier J, Laroche J, Beaulieu J, Bousquet J. 2011. Scanning the genome for gene SNPs related to climate adaptation and estimating selection at the molecular level in boreal black spruce. Mol Ecol. 20:1702–1716. [DOI] [PubMed] [Google Scholar]
- Pujolar JM, et al. 2014. Genome-wide single-generation signatures of local selection in the panmictic European eel. Mol Ecol. 23:2514–2528. [DOI] [PubMed] [Google Scholar]
- R Core Team. 2014. Vienna, Austria: R Foundation for Statistical Computing.
- Renaut S, Owens GL, Rieseberg LH. 2014. Shared selective pressure and local genomic landscape lead to repeatable patterns of genomic divergence in sunflowers. Mol Ecol. 23:311–324. [DOI] [PubMed] [Google Scholar]
- Sang T. 2009. Genes and mutations underlying domestication transitions in grasses. Plant Physiol. 149:63–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansaloni CP, et al. 2011. Diversity Arrays Technology (DArT) and next-generation sequencing combined: genome-wide, high throughput, highly informative genotyping for molecular breeding of Eucalyptus . BMC Proc. 5:54. [Google Scholar]
- Savolainen O, Lascoux M, Merila J. 2013. Ecological genomics of local adaptation. Nat Rev Genet. 14:807–820. [DOI] [PubMed] [Google Scholar]
- Scotland RW. 2011. What is parallelism? Evol Dev. 13:214–227. [DOI] [PubMed] [Google Scholar]
- Shaar-Moshe L, Hubner S, Peleg Z. 2015. Identification of conserved drought-adaptive genets using a cross-species meta-analysis approach. BMC Plant Biol. 15:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva-Junior OB, Grattapaglia D. 2015. Genome-wide patterns of recombination, linkage disequilibrium and nucleotide diversity from pooled resequencing and single nucleotide polymorphism genotyping unlock the evolutionary history of Eucalyptus grandis . N Phytol. 208:830–845. [DOI] [PubMed] [Google Scholar]
- Steane DA, et al. 2017. Evidence for adaptation and acclimation in a widespread eucalypt of semi-arid Australia. Biol J Linn Soc in press.
- Steane DA, Nicolle D, McKinnon GE, Vaillancourt RE, Potts BM. 2002. Higher-level relationships among the eucalypts are resolved by ITS-sequence data. Aust Syst Bot. 15:49–62. [Google Scholar]
- Steane DA, et al. 2014. Genome-wide scans detect adaptation to aridity in a widespread forest tree species. Mol Ecol. 23:2500–2513. [DOI] [PubMed] [Google Scholar]
- Steane DA, et al. 2015. Genome-wide scans reveal cryptic population structure in a dry-adapted eucalypt. Tree Genet Genomes 11:33. [Google Scholar]
- Supple MA, et al. 2013. Genomic architecture of adaptive color pattern divergence and convergence in Heliconius butterflies. Genome Res. 23:1248–1257. doi: 10.1101/gr.150615.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner TL, Bourne EC, Von Wettberg EJ, Hu TT, Nuzhdin SV. 2010. Population resequencing reveals local adaptation of Arabidopsis lyrata to serpentine soils. Nat Genet. 42:260. [DOI] [PubMed] [Google Scholar]
- Turner TL, Von Wettberg EJ, Nuzhdin SV. 2008. Genomic analysis of differentiation between soil types reveals candidate genes for local adaptation in Arabidopsis lyrata . PLoS ONE 3:e3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney RK, et al. 2014. Genetic dissection of drought tolerance in chickpea (Cicer arietinum L.). Theor Appl Genet. 127:445–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaseva I, et al. 2012. The response of plants to drought stress: the role of dehydrins, chaperones, proteases and protease inhibitors in maintaining cellular protein function In: Neves DF, Sanz JD, editors. Droughts: new research. Hauppauge (NY): Nova Science Publishers. [Google Scholar]
- Vilas A, Perez-Figueroa A, Caballero A. 2012. A simulation study on the performance of differentiation-based methods to detect selected loci using linked neutral markers. J Evol Biol. 25:1364–1376. [DOI] [PubMed] [Google Scholar]
- Wang X, et al. 2014. Two distinct classes of QTL determine rust resistance in sorghum. BMC Plant Biol. 14. doi:36610.1186/s12870-014-0366-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, et al. 2001. Rice ESTs with disease-resistance gene- or defense-response gene-like sequences mapped to regions containing major resistance genes or QTLs. Mol Genet Genomics 265:302–310. [DOI] [PubMed] [Google Scholar]
- Weller JL, et al. 2012. A conserved molecular basis for photoperiod adaptation in two temperate legumes. Proc Natl Acad Sci U S A. 109:21158–21163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Hutchinson M. 2011. ANUCLIM Version 6.1 User Guide. The Australian National University, Fenner School of Environment and Society, Canberra.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.