Abstract
The objective of this study was to examine five different sources of RNA, namely mammary gland tissue (MGT), milk somatic cells (SC), laser microdissected mammary epithelial cells (LCMEC), milk fat globules (MFG) and antibody-captured milk mammary epithelial cells (mMEC) to analyze the bovine mammary gland transcriptome using RNA-Sequencing. Our results provide a comparison between different sampling methods (invasive and non-invasive) to define the transcriptome of mammary gland tissue and milk cells. This information will be of value to investigators in choosing the most appropriate sampling method for different research applications to study specific physiological states during lactation. One of the simplest procedures to study the transcriptome associated with milk appears to be the isolation of total RNA directly from SC or MFG released into milk during lactation. Our results indicate that the SC and MFG transcriptome are representative of MGT and LCMEC and can be used as effective and alternative samples to study mammary gland expression without the need to perform a tissue biopsy.
The mammary gland is a secretory skin gland unique to the class Mammalia. Mammary epithelial cells are unique in that they synthesize and secrete milk. They also form part of a complex and heterogeneous tissue along with multiple cell types including myoepithelial, stromal and immune cells, making it difficult to precisely assign gene expression to the different cell types. Thus, it is important to distinguish which cell populations play specific functions, and to obtain relevant expression profile data reflecting the in vivo state in order to recognize differences in these cell fractions and to obtain reliable transcriptomic results1. Mammary epithelial cells are normally subject to turnover and are replaced by new cells2. These exfoliated cells are shed into the milk and represent a proportion of the somatic cells found in milk. Separately, a portion of the cytoplasm of the mammary epithelial cells is included within the milk fat globules during the secretion process3.
Total RNA extracted from these different fractions such as milk epithelial cells and milk fat globules has been used to assess the transcriptional activity of the secretory mammary epithelium in livestock4. Alternative sampling approaches include isolating RNA directly from biopsies of mammary gland tissue5, from somatic cells in milk that are naturally released during lactation6,7, from antibody-captured epithelial cells in milk8,9,10 and from milk fat globules11. Also, laser-capture microdissection has been used to selectively isolate epithelial cells from frozen tissue sections of the mammary gland1. Some of these sampling procedures were validated in an earlier study using qPCR for specific genes and/or using microarray11. With the advent of Next Generation sequencing, RNA-Sequencing presents the opportunity to examine gene expression on a global basis, thereby providing numerous advantages over microarray analyses because of its wide dynamic range and its ability to quantify all genes present and not only those targets found on the array12. Only SC have been validated in relation to MGT using RNA-Sequencing5. Hence, here we compared the transcriptome for cells isolated by all the above different sampling methods to document the representation of genes and their levels of expression. We collected samples from the same cows and examined the transcriptome of mammary gland tissue (MGT), milk somatic cells (SC), laser microdissected mammary epithelial cells (LCMEC), milk fat globules (MFG) and antibody-captured mammary epithelial cells from milk (mMEC) using RNA-Sequencing.
Results
RNA preparation and amplification
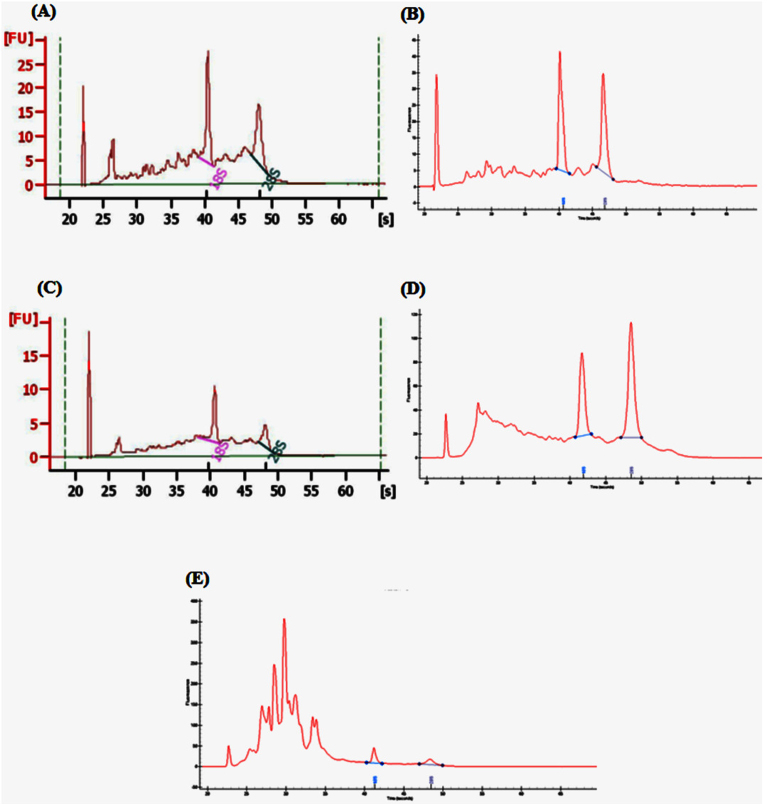
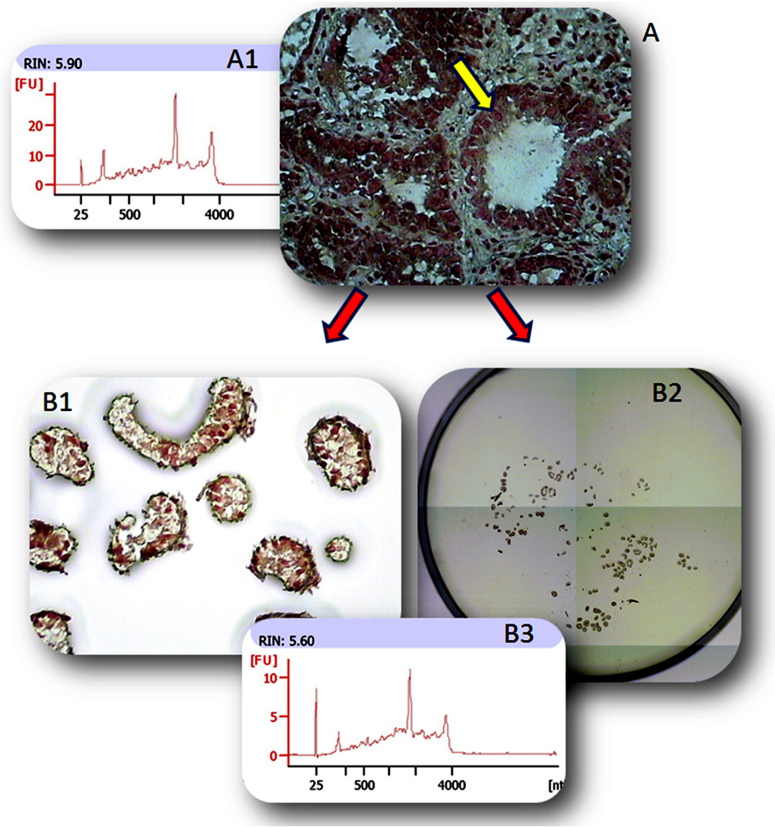
A total of 14 bovine samples (three MGT, three LCMEC, three MFG, two SC and three mMEC from two Jersey and one Normande cows were analyzed by RNA-Sequencing. Figure 1 shows the chromatogram traces profiling the quality of the RNA obtained for each fraction. The spectrophotometric absorbance profile for RNA from the MGT (Figure 1A) and LCMEC (Figure 1C) were similar (RIN = 6.5–7.0). The quality of the RNA may be affected by time taken to obtain the tissue biopsy until it is frozen, and the time to cut the sections to freeze in the OCT blocks used for LCMEC. Comparison between mammary gland epithelial cells before and after laser microdissection was performed (Figure 2). Alveolar (acinar) structures (Figure 2A) lined by mammary epithelial cells (yellow arrows) could be easily distinguished after staining mammary tissue sections with Mayer's hematoxylin (Sigma). The LCMEC were dissected and collected on a cap at 60× (Figure 2B1) and 4× (Figure 2B2) magnifications. The RIN was similar (6.5 to 7.0) between MGT (Figure 2A1) and LCMEC (Figure 2B3), as illustrated in the chromatogram profiles. The total RNA yield for one laser capture cap ranged between 5 to 10 ng. In addtion, to assess a possible contamination of microdissected MEC, qPCR systems were carried out to quantify specific markers for mammary myoepithelial cells (KRT14), lymphocytes (CD3e), neutrophils (CD18), macrophages (CD68 and CD18) and MEC (CSN3). In general, LCMEC showed a similar profile for each sample, with over-expression of MEC specific markers (LALBA and CSN2S2) and under-expression of mammary myoepithelial cells markers (KRT14) and immune cells markers (CD3e and CD68). Also, the same profile was observed in MFG and mMEC fractions with an over-expression of marker CD68, suggesting the presence of macrophages (Supplementary File 2).
Figure 1. Total RNA capillary electrophoresis chromatograms from mammary and milk fractions.
(A) Mammary tissue (MGT), (B) Somatic cells (SC), (C) Laser capture mammary epithelial cells (LCMEC), (D) Antibody-captured milk mammary epithelial cells (mMEC), (E) Milk fat globule (MFG). Traces (A) and (C) (Agilent Bioanalyzer), traces (B), (D), (E) Experion Analyzer (BioRad). These diagrams show the differences in quality of RNA obtained from the different fractions.
Figure 2.
Mammary gland tissue (MGT) before microdissection (A) and mammary epithelial cells after laser microdissection (LCMEC) (B1 and B2). Chromatograms from total RNA capillary electrophoresis; MGT before microdissection (A1) LCMEC after microdissection (B3).
High RIN values (around 8.0) were consistently observed for total RNA from SC (Figure 1B) despite the fact that the milk sample was kept on ice for three hour until transported to the laboratory for RNA extraction. The RNA from MFG contains low molecular weight fragments and a very low amount of ribosomal RNA (RIN = 6.0) (Figure 1E).
The membrane of MFG contains specific components such as phospholipids, proteins and glycophospholipids that are known to have an affinity for the surface of bacterial cells. By analyzing all the MFG reads not mapping to the bovine genome (~11 million reads), we identified the presence of 618 different bacteria. In contrast, approximately 9 million not mapped reads were found in SC as being associated just with E. coli.
The mMEC samples purified with the antibody method presented a pattern similar to the MFG RNA, with a large fraction of low molecular weight RNA (Figure 1D) significantly impacting the RIN value (RIN = 6.0). The quality of RNA obtained from the mMEC in this study was not representative of the quality generally obtained with this method8.
RNA-Seq expression analyses
An average of 32 million short-sequence reads was obtained for each individual sample; these were assembled and mapped to the annotated UMD3.1 bovine genome assembly (27,368 genes). In all the samples, 60–75% of the reads were categorized as mapped to the bovine reference sequence (Table 1).
Table 1. Total reads and mapped reads obtained in each sample analyzed.
| RNA Fraction | Sample number | Breed | Reads (n) | *Mapped reads (n) | % mapped reads |
|---|---|---|---|---|---|
| MGT | 653 | Jersey | 40,147,740 | 24,732,197 | 61.6 |
| 671 | Jersey | 38,508,068 | 23,163,085 | 60.2 | |
| 1816 | Normande | 26,480,391 | 19,732,280 | 74.5 | |
| SC | 653 | Jersey | 40,209,699 | 28,036,287 | 69.7 |
| 671 | Jersey | 39,507,994 | 26,136,100 | 66.2 | |
| LCMEC | 653 | Jersey | 27,335,082 | 17,121,159 | 62.6 |
| 671 | Jersey | 24,355,141 | 14,345,582 | 58.9 | |
| 1816 | Normande | 23,173,807 | 16,973,772 | 73.3 | |
| MFG | 653 | Jersey | 23,918,861 | 14,588,022 | 60.9 |
| 671 | Jersey | 24,334,219 | 14,475,382 | 59.5 | |
| 1816 | Normande | 34,023,717 | 22,407,126 | 65.9 | |
| mMEC | 653 | Jersey | 38,899,626 | 29,284,686 | 75.3 |
| 671 | Jersey | 38,495,029 | 28,292,867 | 73.5 | |
| 1816 | Normande | 29,191,918 | 22,056,958 | 75.6 | |
| Average | 32,041,521 | 21,524,679 | 66.9 |
*Reads categorized as mapped to the annotated UMD3.1 bovine genome assembly (27,368 genes). MGT: mammary gland tissue; SC: milk somatic cells; LCMEC: laser microdissected mammary epithelial cells; MFG: milk fat globules; mMEC: antibody- captured milk mammary epithelial cells.
RPKM values13 were used to establish the total number of genes expressed in the MGT, SC, LCMEC, MFG and mMEC transcriptome. In this context, approximately 90% of the total annotated genes in the UMD3.1 bovine genome assembly were expressed in the samples under study (24,616 genes out of a total of 27,368).
Correlations between RPKM expression levels for the MGT, SC, LCMEC, MFG and mMEC transcriptomes
The different sampling methods were compared by calculating a Pearson correlation between the RPKM expression levels of the ~24,616 expressed genes in samples of MGT, SC, LCMEC, MFG and mMEC. Our results indicate that gene expression is highly correlated among LCMEC, MGT, SC and MFG. These sampling methods showed the highest correlation values in gene expression with LCMEC, ranging from r = 0.80 to r = 0.99 (MGT), from r = 0.80 to r = 0.95 (SC) and from r = 0.81 to r = 0.93 (MFG) in Jersey and Normande cows, respectively (Table 2). Interestingly, gene expression in SC was significantly correlated with that in MGT in Jersey and Normande cows (r = 0.98 and r = 0.92, respectively). By contrast, a low correlation existed between mMEC and LCMEC in both Jersey and Normande cows (correlation ranged from r = 0.15 to r = 0.39, Table 2). In addition, independent correlation analysis were performed using the list of highly expressed (≥500 RPKM), normally/medium expressed (ranged from 500 to 10 RPKM) and lowly expressed (<10 RPKM) genes14. A homogeneous distribution of expressed genes was obtained across LCMEC, MGT, SC, MFG and mMEC. All fractions had a higher number of lowly expressed genes. The high correlation among LCMEC, MGT, SC and MFG persist in highly-expressed, normally/medium-expressed and lowly-expressed genes (Additional files 3, 4 and 5).
Table 2. Correlations between RPKM expression levels of MGT, SC, LCMEC, MFG and mMEC transcriptome in Jersey and Normande cows.
| MGT | SC | LCMEC | MFG | mMEC | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Jersey | Normande | Jersey | Jersey | Normande | Jersey | Normande | Jersey | Normande | ||
| MGT | Jersey | 1.00 | - | - | - | - | - | - | - | - |
| Normande | 0.97 | 1.00 | - | - | - | - | - | - | - | |
| SC | Jersey | 0.98 | 0.92 | 1.00 | - | - | - | - | - | - |
| LCMEC | Jersey | 0.80 | 0.90 | 0.79 | 1.00 | - | - | - | - | - |
| Normande | 0.90 | 0.99 | 0.95 | 0.80 | 1.00 | - | - | - | - | |
| MFG | Jersey | 0.90 | 0.88 | 0.93 | 0.81 | 0.85 | 1.00 | -- | - | - |
| Normande | 0.88 | 0.92 | 0.95 | 0.85 | 0.93 | 0.93 | 1.00 | - | - | |
| mMEC | Jersey | 0.18 | 0.15 | 0.26 | 0.39 | 0.23 | 0.50 | 0.37 | 1.00 | - |
| Normande | 0.15 | 0.16 | 0.22 | 0.23 | 0.15 | 0.37 | 0.32 | 0.67 | 1.00 | |
MGT: mammary gland tissue; SC: milk somatic cells; LCMEC: laser microdissected mammary epithelial cells; MFG: milk fat globules; mMEC: antibody- captured milk mammary epithelial cells.
A correlation was also calculated using the ranking/position of the RPKM gene expression values. The ranking/position stretched from the most highly expressed to the lowest expressed gene for each sample analyzed. Consistently, the highest correlated values were between LCMEC and MGT, MFG and SC samples in both breeds (correlation ranged from r = 0.90 to r = 0.94; p < 0.0001). In this analysis, relatively high correlations (ranging from r = 0.79 to r = 0.90) were observed between LCMEC and mMEC (Supplementary File 6).
Most highly expressed genes in MGT, SC, LCMEC, MFG and MEC
We also compared the genes having the greatest expression in MGT, SC, LCMEC, MFG and mMEC (Table 3). As reported by Wickramasinghe et al.14 for milk SC, the genes having the greatest expression in all MGT, SC, LCMEC and MFG transcriptomes were the caseins (i.e. CSN2, CSN3, CSN1S1 and CSN1S2), the main two whey proteins (BLG and LALBA) and GLYCAM1 (glycosylation-dependent cell adhesion molecule 1). Among these, CSN2 and CSN3 showed the highest RPKM values ranging from 207,305 to 1,877 depending on the sample (Table 3). BLG, LALBA and GLYCAM1 were abundantly expressed in MGT, SC, LCMEC and MFG. Surprisingly BLG, LALBA and GLYCAM1 were not highly expressed in mMEC. In general the overall transcriptome expression level in mMEC was several orders of magnitude lower than in the other samples. In cattle, mMEC only account for no more than 5% of the somatic cells15. Also, the additional step in the purification of mMEC from SC may explain the observed different expression profile.
Table 3. Most highly expressed genes in MGT, SC, LCMEC, MFG and mMEC transcriptome in Jersey and Normande cows expressed in RPKM values (reads per kilo base per million mapped reads). Green color represents the most highly expressed genes, red color represents the lowest expressed genes and the different color tones are intermediate values.
| MGT | SC | LCMEC | MFG | mMEC | |||||
|---|---|---|---|---|---|---|---|---|---|
| Gene | Jersey | Normande | Jersey | Jersey | Normande | Jersey | Normande | Jersey | Normande |
| CSN2 | 207305 | 145263 | 156130 | 105065 | 179820 | 76891 | 45571 | 13651 | 2591 |
| CSN3 | 72233 | 87335 | 30208 | 51838 | 84031 | 15602 | 18689 | 1877 | 1991 |
| CSN1S1 | 39736 | 45340 | 24903 | 25982 | 50297 | 11528 | 7652 | 956 | 710 |
| CSN1S2 | 12056 | 32973 | 4010 | 9093 | 29740 | 2156 | 4592 | 206 | 551 |
| LALBA | 5619 | 6537 | 3056 | 2706 | 11139 | 2693 | 1719 | 43 | 42 |
| GLYCAM1 | 4035 | 8346 | 3579 | 2668 | 10633 | 1758 | 2995 | 119 | 90 |
| BLG | 3231 | 2407 | 6835 | 2697 | 5925 | 3854 | 707 | 121 | 84 |
| SPP1 | 1406 | 1036 | 6975 | 845 | 1301 | 927 | 6250 | 10680 | 992 |
| RPS17 | 1374 | 366 | 3164 | 120 | 90 | 2183 | 916 | 118 | 69 |
| TCTP | 937 | 1098 | 2724 | 974 | 917 | 2451 | 2656 | 1320 | 42 |
| PTMA | 893 | 123 | 2876 | 401 | 104 | 1843 | 392 | 3345 | 28 |
| MFGE8 | 705 | 186 | 576 | 75 | 84 | 95 | 49 | 177 | 2 |
| CD36 | 486 | 691 | 264 | 322 | 673 | 56 | 112 | 65 | 27 |
| SCD | 265 | 459 | 44 | 222 | 570 | 39 | 98 | 4 | 6 |
| FABP4 | 171 | 527 | 138 | 182 | 103 | 90 | 123 | 5 | 4 |
| FRIH | 127 | 134 | 1898 | 121 | 87 | 779 | 3400 | 2165 | 53 |
| RL19 | 519 | 358 | 1351 | 348 | 405 | 1142 | 717 | 255 | 16 |
| RS12 | 412 | 152 | 1172 | 516 | 186 | 66 | 66 | 1379 | 84 |
| CD68 | 3 | 3 | 1108 | 7 | 4 | 84 | 62 | 1755 | 46 |
| RS17 | 157 | 117 | 117 | 1577 | 433 | 87 | 119 | 1499 | 33 |
| B2MG | 171 | 156 | 766 | 482 | 265 | 97 | 1055 | 84 | 21 |
| PLIN2 | 57 | 65 | 77 | 75 | 73 | 994 | 8195 | 38 | 23 |
| TUBA1B | 46 | 39 | 38 | 35 | 37 | 59 | 46 | 1625 | 53 |
| HMOX1 | 4 | 0.2 | 1 | 0.09 | 0.04 | 7 | 3 | 1073 | 54 |
| S100A11 | 33 | 4 | 2 | 10 | 5 | 51 | 10 | 936 | 57 |
| RL10 | 36 | 4 | 34 | 32 | 35 | 45 | 33 | 668 | 53 |
MGT: mammary gland tissue; SC: milk somatic cells; LCMEC: laser microdissected mammary epithelial cells; MFG: milk fat globules; mMEC: antibody- captured milk mammary epithelial cells. Jersey data provided correspond to mean values of the two analyzed Jersey cows.
A comparison of the list of the most highly expressed genes revealed that approximately 15 out of the 17 genes with greatest levels of expression in LCMEC were also expressed abundantly in both MGT and SC. In contrast, only 9 out of these 17 were abundantly expressed in mMEC (Table 3). The transcriptome for MGT, SC, LCMEC and MFG had a very similar expression pattern. Both SC and MFG showed highly expressed genes, such as SPP1 (secreted phosphoprotein 1 or osteopontin), RPS17 (ribosomal protein S17), TCTP (translationally controlled tumor protein) and PTMA (prothymosin alpha). Among these, SPP1 cooperates with other pro-inflammatory cytokines such as TNFα to participate in the overall inhibition of mammary epithelial cell viability that is required for tissue remodeling in the bovine mammary gland during the course of involution16. Furthermore, MFG showed high levels of gene expression for perilipin 2 (PLIN2) that is related to lipid accumulation and secretion17. In contrast, mMEC showed a different profile for all caseins and major whey proteins, and also uniquely expressed high levels of the TUBA1B (tubulin alpha 1B), HMOX1 (heme oxygenase (decycling) 1), S100A11 (S100 calcium binding protein A11) and RL10 (ribosomal protein L10) genes in Jersey cows. These genes represent markers of cell development and immunity. Also, expression of CD68 (CD68 molecule), a specific marker for macrophages, was elevated in the SC and mMEC transcriptomes in Jersey cows, perhaps due to the presence of a higher proportion of macrophages.
Discussion
The transcriptome for mammary gland was studied using five different sampling methods. Total RNA from MGT, SC, LCMEC, MFG and mMEC were compared using RNA-Sequencing to examine the representation of genes and their levels of expression.
The integrity and type of the RNA varied considerably between samples depending on the sampling method, which in turn had a bearing on the RNA-Sequencing results (Table 1). The RNA from the MGT, LCMEC and SC showed similar electrophoresis chromatograms profiles. The RNA from MFG contained low molecular weight fragments and a very low amount of ribosomal RNA due to the presence of small amounts of cytoplasmic material in the bovine milk fat globules during the MFG secretory process3. This likely explains the lower RIN value (RIN = 6.0) of this tissue. The low MW RNA may be due to the presence of bacteria and also very small RNAs that are abundant in this fat fraction18. The MFG membrane contains specific components such as phospholipids, proteins and glycophospholipids that are unique in this cell fraction. Interestingly, these components are known to have an affinity for the cell surface of bacteria19. Considering this, we analyzed all the MFG reads not mapping to the bovine genome (~11 million reads) for the presence of bacterial genes. From this analysis we found that 618 different kinds of bacteria were identified with MFG. Comparatively, approximately 9 million reads were found in SC as being associated just with E. coli. The mMEC samples purified with the antibody method presented a pattern similar to the MFG RNA, with a large fraction of low molecular weight RNA significantly impacting the RIN value (RIN = 6.0).
The transcriptome for bovine LCMEC was highly correlated with that for MGT, SC and MFG, and showed a high correlation with MGT in Jersey (r = 0.80) and Normande (r = 0.99) cows (p < 0.0001). A significant correlation was found between MGT and SC in Jersey (r = 0.98) and Normande (r = 0.92) cows. By contrast, a low correlation existed between mMEC and LCMEC in both Jersey and Normande cows (correlation ranged from r = 0.15 to r = 0.39). The MGT is complex and heterogeneous, with multiple cell types including myoepithelial, stromal and immune cells. In terms of immune cells, SC is more similar to MGT than mMEC due to the high proportion of immune cells such as neutrophils, lymphocytes and macrophages which account for ca. 95% of SC in milk from healthy cows15. It is important to note that the isolation of mMEC utilized the binding of bovine MEC to a monoclonal antibody recognizing cytokeratins8. Different antibody reactions both with not much high RNA quality level can alter and affect the final product, which may explain why these samples had a low correlation with all the cellular fractions analyzed (range from r = 0.13 to r = 0.50).
In general, the highly expressed genes such as caseins (CSN1S1, CSN1S2, CSN2, CSN3) and whey proteins (BLG, LALBA) showed a similar expression profile in MGT, LCMEC, SC and MFG. However, MFG had a unique abundant expression of the PLIN2 gene that is implicated in lipid accumulation.
The majority of the lipid produced during lactation is secreted into milk by a process of membrane envelopment of cytoplasmic lipid droplets (CLDs). Others have hypothesized that PLIN2 plays a pivotal role in both formation and secretion of milk lipids, where it functions as an adaptor to couple CLDs to the cytoplasmic leaflet of the apical plasma membrane of MEC20. This gene may also be involved in development and maintenance of adipose tissue. Furthermore, PLIN2 is expressed by a wide range of cultured cell lines, including fibroblasts, endothelial and epithelial cells, and tissues, suggesting that it may serve as a marker of lipid accumulation in diverse cell types and diseases21.
Expression of CD68 (CD68 molecule), a specific marker for macrophages, was elevated in the SC and mMEC transcriptomes in Jersey cows, perhaps due to the presence of a higher proportion of macrophages. This finding would suggest that mMEC purification was not totally effective. Although the antibody-directed mMEC purification method allowed the specific selection of epithelial cells, which account for less than 5% of total milk cells (SC), the mMEC transcriptome was different from the rest of the transcriptomes analyzed. In our results, immunomagnetic mMEC purification method showed that a conjoint of reactions was affecting the proportion and/or the function of white cells present in milk, showing a unique set of highly expressed genes such as TUBA1B, HMOX1, S100A11 and RL10 involved in anti-inflammatory processes, cell death or apoptosis, specifically in Jersey mMEC samples.
Up to this point, the majority of gene expression analyses related to bovine lactation have been developed using tissues obtained by biopsy22,23. This biopsy approach is invasive during a stage when vascularization of the gland is at a maximum, which presents significant technical challenges and management issues in the recovery of the animals.
An alternative sampling procedure has been proposed via isolating mRNA directly from mMEC8,9, MFG11 and SC6,7 that are released into milk during lactation. Using RNA extracted from mMEC, MFG or SC represents a more accessible method for assessing gene expression in the mammary epithelium4, particularly when dynamic studies require the repeated analysis of samples from the same animal. Considering the cost and labor involved to obtain a sample of mammary parenchyma for RNA extraction, analyzing milk SC and MFG appears to be an alternative, direct and simple approach that provides a high correlation with gene expression in MGT and LCMEC without the need to perform a biopsy of the mammary gland tissue. Our results present the similarities between different sampling methods by examining the transcriptome of mammary gland tissue and different RNA sources from milk. This information can guide investigators in choosing the most appropriate sampling method to examine specific physiological states from different sources of RNA during lactation for different research applications. One of the simplest procedures to examine the transcriptome associated with milk appears to be to isolate mRNA directly from MFG and SC that are released into milk during lactation. Our analyses suggest that the SC and MFG transcriptome are the most representative of MGT and LCMEC and can be used as an effective and easy way to study gene expression using RNA-Sequencing, without the need to perform a mammary gland biopsy.
Methods
Animal material
Mammary gland tissue and milk samples were obtained from two Jerseys and one Normande cow at the University of California – Davis (UCD) and the Institut National de la Recherche Agronomique (INRA) herd (Le Pin-au-Haras, France), respectively. Jersey cows were in their 3rd and 4th lactation around 100 days in milk whereas Normande cow was in its 2nd lactation around 180 days in milk. Mammary tissue biopsies were performed using a 9-gauge, 12-cm probe with a 12-mm aperture as described by VanKlompenberg et al.23 in Jersey cows and 70 × 4 mm rotating stainless steel cannula in Normande cow24. A 5-mm incision was made through the skin and connective tissue approximately half-way up the rear udder quarter using a scalpel blade before the parenchyma was penetrated with a Hologic ATEC Biopsy Console Handpiece for excision of a tissue core (Hologic Inc., Bedford, MA) and Farr's Instrument. Triplicate milk samples were collected before performing tissue biopsies and were held on ice. Sample collections and procedures were performed in accordance with the approved guidelines of the UC Davis Institutional Animal Care and Use Committee (IACUC).
RNA extraction
Biopsied mammary gland tissue (MGT) was divided in two: one part was immediately put in a cryomold with OCT compound and frozen with a mix of dry ice and isopropanol for future microdissection. The other portion was frozen in liquid nitrogen for subsequent extraction of RNA. Total RNA from MGT was extracted using Trizol (Invitrogen).
Laser capture microdissection of epithelial cells (LCMEC) from the mammary tissue was performed using an Arcturus Veritas Microdissection system and software (Applied Biosystems). Cryosectioning, staining, dehydration and harvesting of microdissected cells was performed as described Bevilacqua et al.1. Total RNA was extracted from captured cells using the PicoPure® RNA Isolation Kit (Arcturus, Applied Biosystems) according to the manufacturer's instructions, including an on-column DNase I treatment (Qiagen). An aliquot of RNA was reverse transcribed to validate the selectivity and specificity of laser capture microdissection by qPCR for markers of myoepithelial cells, MEC and immune cells. To assess a possible contamination of microdissected MEC, qPCR systems designed to quantify specific markers for mammary myoepithelial cells (Krt14), lymphocytes (CD3e), neutrophils (CD18), macrophages (CD68 and CD18) and MEC (CSN3) were implemented. Sequences were from GenBank (Supplementary File 1).
Milk was centrifuged at 2,000 g for 10 min to pellet somatic cells (SC) and to separate the upper milk fat globule (MFG) fraction. The MFG fraction was mixed with Trizol LS and heated briefly at 30°C while shaking to emulsify the fat. Total RNA was extracted from milk somatic cells (SC) and from milk fat using Trizol (Invitrogen) following the protocol from the manufacturer as described in Cánovas et al.25 and Brenaut et al.11, respectively. In addition, MEC from milk (mMEC) were obtained according to the procedure described by Boutinaud et al.8 using a protocol based on the binding of mMEC to a monoclonal antibody recognizing cytokeratin 8 (clone 34BE12, Sigma-Aldrich), except that milk was kept on ice for approximately 3 h until it was transported to the laboratory. In all cases the purified total RNA was treated with Turbo DNase (Invitrogen, Carlsbad, CA). Quality of the total RNA was evaluated using the RNA Integrity Number (RIN) value in the Agilent Bioanalyzer 2100 or in the Experion Analyzer (BioRad).
RNA-Sequencing
RNA-Sequencing was used to measure gene expression in the MGT, SC, LCMEC, MFG and mMEC transcriptome. Total RNA (DNAse-treated) was used to synthesize amplified cDNA following the protocol in the Ovation RNA-seq system and the Encore NGS library system (NuGEN, San Carlos, CA). Total RNA (100 ng DNase treated) was subjected to an RNA amplification process using the Ribo-SPIA technology from the Ovation RNA-seq System (NuGEN, San Carlos, CA). The amplified cDNA (200 ng) was fragmented using the Bioruptor sonicator (Diagenode) 30 sec ON, 30 sec OFF cycles for 10 min to produce fragments with 150–200 bp size distribution. Adaptor molecules were ligated to the ends of each fragment and amplified by PCR to produce the final library using the Encore NGS library system (NuGEN, San Carlos, CA). A single library was made for each sample.
Fragmented DNA was sequenced on a GAII sequencer analyzer (Illumina, San Diego, CA).
Quality control (QC) analysis was performed using the application NGS quality control tool of CLC Genomics workbench software (CLC Bio, Aarhus, Denmark). This tool assess sequence quality indicators based on the FastQC-project (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Quality was measured taking into account sequence-read lengths and base-coverage, nucleotide contributions and base ambiguities, quality scores as emitted by the base caller and over-represented sequences. All the samples analyzed passed all the QC parameters having the same length (36 bp), 100% coverage in all bases, 25% of A, T, G and C nucleotide contributions, 50% GC on base content and less than 0.1% over-represented sequences, indicating a very good quality.
Short sequence single reads (36 bp) were mapped to the annotated UMD3.1 bovine reference genome (http://www.ncbi.nlm.nih.gov/genome/?term=bos+taurus). Analysis of RNA-Seq expression data was performed using the transcriptomics analysis tool of CLC Genomics workbench software. Data were normalized by calculating the ‘reads per kilobase per million mapped reads' (RPKM) for each gene13. To select expressed genes a threshold of RPKM ≥ 0.2 was used14.
Large gap mapping analysis was performed to identify those reads not mapping to the bovine genome. Then, de novo assembly analysis was performed from unmapped reads to generate contigs using CLC Genomics workbench software (CLC Bio, Aarhus, Denmark). Blast2GO software (http://www.blast2go.com) was used for contig annotation and bacteria identification (Gotz et al., 2008). Blats2Go performs the Blast step by communicating directly to NCBI (National Center for Biotechnology Information) blast service. The Blastn algorithm was used to compare a nucleotide query sequence against all the nucleotide sequence database. A statistical significance threshold of 1.0 E-3 (e-value) was established for reporting matches against the database sequences. Also a minimal length (30 bp) of a blast hit and 90% of similarity was used to exclude hits with only small local alignments from the blast results.
Statistical analysis
To examine the similarities between the different sampling procedures a correlation between the RPKM expression levels of MGT, SC, LCMEC, MFG and mMEC transcriptomes was performed using the CORR procedure of SAS (SAS Institute Inc.).
Supplementary Material
Supplementary file
Acknowledgments
The authors would like to thank Doug Gisi from U.C. Davis Dairy Facility and S. Leurent and Y. Gallard from INRA Experimental Farm of Le Pin-au-Haras, France and S. Marion from UMR PEGASE for their technical help. We gratefully acknowledge the support for this collaborative project from the France-Berkeley Fund.
Footnotes
This project was partially funded by the France Berkeley Fund (http://fbf.berkeley.edu/), collaboration between JFM and PM and by Incentive credits from the Animal Genetics Department of INRA. Angela Cánovas received a contractual Postdoctoral grant from ME/Fulbright.
Author Contributions A.C. analyzed the data and wrote the manuscript, G.R. design, analysis and sample collection, C.B. laser capture experiments and sample collection, A.I.T. sample collection and construction of sequencing libraries, P.B. study design and sample collection, R.C.H. sample collection, M.B. sample collection and processing, C.M. sample collection, M.K.V. sample collection, P.M. and J.F.M. conceived, designed and funded the study. All authors reviewed and contributed to the final manuscript.
References
- Bevilacqua C., Makhzami S., Helbling J. C., Defrenaix P. & Martin P. Maintaining RNA integrity in a homogeneous population of mammary epithelial cells isolated by Laser Capture Microdissection. BMC Cell Biol 11, 95 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capuco A. V., Wood D. L., Baldwin R., McLeod K. & Paape M. J. Mammary cell number, proliferation, and apoptosis during a bovine lactation: relation to milk production and effect of bST. J Dairy Sci 84, 2177–2187 (2001). [DOI] [PubMed] [Google Scholar]
- Huston G. E. & Patton S. Factors related to the formation of cytoplasmic crescents on milk fat globules. J Dairy Sci 73, 2061–2066 (1990). [DOI] [PubMed] [Google Scholar]
- Boutinaud M. & Jammes H. Potential uses of milk epithelial cells: a review. Reprod Nutr Dev 42, 133–147 (2002). [DOI] [PubMed] [Google Scholar]
- Medrano J. F., Rincón G. & Islas A. D. Comparative analysis of bovine milk and mammary gland transcriptome using RNA-Seq. 9th World Congress of Genetics Applied to Livestock Production (WCGALP), Leipzig-Germany, No. 0852 (2010).
- Boutinaud M., Rulquin H., Keisler D. H., Djiane J. & Jammes H. Use of somatic cells from goat milk for dynamic studies of gene expression in the mammary gland. J Anim Sci 80, 1258–1269 (2002). [DOI] [PubMed] [Google Scholar]
- Feng S., Salter A. M., Parr T. & Garnsworthy P. C. Extraction and quantitative analysis of stearoyl-coenzyme A desaturase mRNA from dairy cow milk somatic cells. J Dairy Sci 90, 4128–4136 (2007). [DOI] [PubMed] [Google Scholar]
- Boutinaud M., Ben Chedly M. H., Delamaire E. & Guinard-Flament J. Milking and feed restriction regulate transcripts of mammary epithelial cells purified from milk. J Dairy Sci 91, 988–998 (2008). [DOI] [PubMed] [Google Scholar]
- Ben Chedly H. et al. Use of milk epithelial cells to study regulation of cell activity and apoptosis during once-daily milking in goats. Animal 5, 572–579 (2011). [DOI] [PubMed] [Google Scholar]
- Boutinaud M., Lollivier V., Finot L., Bruckmaier R. M. & Lacasse P. Mammary cell activity and turnover in dairy cows treated with the prolactin-release inhibitor quinagolide and milked once daily. J Dairy Sci 95, 177–187 (2012). [DOI] [PubMed] [Google Scholar]
- Brenaut P. et al. Validation of RNA isolated from milk fat globules to profile mammary epithelial cell expression during lactation and transcriptional response to a bacterial infection. J Dairy Sci 95, 6130–6144 (2012). [DOI] [PubMed] [Google Scholar]
- Marioni J. C., Mason C. E., Mane S. M., Stephens M. & Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res 18, 1509–1517 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A., Williams B. A., McCue K., Schaeffer L. & Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5, 621–628 (2008). [DOI] [PubMed] [Google Scholar]
- Wickramasinghe S., Rincon G., Islas-Trejo A. & Medrano J. F. Transcriptional profiling of bovine milk using RNA sequencing. BMC Genomics 13, 45 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner S. A., Jones D. E. & Apley M. D. Effect of endotoxic mastitis on epithelial cell numbers in the milk of dairy cows. Am J Vet Res 70, 796–799 (2009). [DOI] [PubMed] [Google Scholar]
- Piantoni P., Wang P., Drackley J. K., Hurley W. L. & Loor J. J. Expression of metabolic, tissue remodeling, oxidative stress, and inflammatory pathways in mammary tissue during involution in lactating dairy cows. Bioinform Biol Insights 4, 85–97 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrosio M. R. et al. The alteration of lipid metabolism in Burkitt lymphoma identifies a novel marker: adipophilin. PLoS One 7, e44315 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munch E. M. et al. Transcriptome profiling of microRNA by Next-Gen deep sequencing reveals known and novel miRNA species in the lipid fraction of human breast milk. PLoS One 8, e50564 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisson G., Payken H. F., Sharpe J. P. & Jimenez-Flores R. Characterization of Lactobacillus reuteri interaction with milk fat globule membrane components in dairy products. J Agric Food Chem 58, 5612–5619 (2010). [DOI] [PubMed] [Google Scholar]
- Chong B. M., Reigan P., Mayle-Combs K. D., Orlicky D. J. & McManaman J. L. Determinants of adipophilin function in milk lipid formation and secretion. Trends Endocrinol Metab 22, 211–217 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heid H. W., Moll R., Schwetlick I., Rackwitz H. R. & Keenan T. W. Adipophilin is a specific marker of lipid accumulation in diverse cell types and diseases. Cell Tissue Res 294, 309–321 (1998). [DOI] [PubMed] [Google Scholar]
- Finucane K. A., McFadden T. B., Bond J. P., Kennelly J. J. & Zhao F. Q. Onset of lactation in the bovine mammary gland: gene expression profiling indicates a strong inhibition of gene expression in cell proliferation. Funct Integr Genomics 8, 251–264 (2008). [DOI] [PubMed] [Google Scholar]
- VanKlompenberg M. K., McMicking H. F. & Hovey R. C. Technical note: A vacuum-assisted approach for biopsying the mammary glands of various species. J Dairy Sci 95, 243–246 (2012). [DOI] [PubMed] [Google Scholar]
- Farr V. C. et al. An improved method for the routine biopsy of bovine mammary tissue. J Dairy Sci 79, 543–549 (1996). [DOI] [PubMed] [Google Scholar]
- Cánovas A., Rincon G., Islas-Trejo A., Wickramasinghe S. & Medrano J. F. SNP discovery in the bovine milk transcriptome using RNA-Seq technology. Mamm Genome 21, 592–598 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary file